Abstract
Attenuated strains of Salmonella typhimurium have been widely used as vehicles for delivery and expression of vaccine antigens in murine models of infectious disease. In mice, early bacterial replication following infection with S. typhimurium is controlled by the gene (Nramp1, formerly Ity/Lsh/Bcg) encoding the natural-resistance-associated macrophage protein (Nramp1). Nramp1 regulates macrophage activation and has multiple pleiotropic effects, including regulation of tumor necrosis factor alpha, interleukin 1β (IL-1β), and major histocompatibility complex class II molecules, all of which influence antigen processing and presentation. Nramp1 also has a direct effect on antigen processing, possibly by regulating the activity of proteases in the late endosomal compartment. Hence, there are multiple ways (regulation of bacterial load or recombinant antigen dose, class II molecule expression, costimulatory or adjuvant activity, and antigen processing) that Nramp1 might influence responses to recombinant salmonella vaccines. To test the hypothesis that Nramp1 influences responses to vaccination, congenic mouse strains have been used to analyze immune responses to recombinant antigens (tetanus toxoid antigen and leishmanial gp63) carried by live attenuated S. typhimurium aroA aroD mutants. Results show that congenic mice carrying the wild-type (S. typhimurium resistance) Nramp1 allele mount a predominantly T-helper-1 (IL-2 and gamma interferon) response to vaccination and show enhanced resolution of lesions following challenge infection with Leishmania major. In contrast, mice carrying mutant (S. typhimurium susceptibility) Nramp1 mount a T-helper-2 (immunoglobulin E and IL-4) response and show exacerbated lesion growth upon challenge.
Attenuated strains of Salmonella have been widely used as vehicles for delivery and expression of a heterogeneous range of antigens (Ag) from other pathogens (for a review, see reference 18). In murine models of infectious disease, genetically engineered Salmonella typhimurium has been used to induce immunity to viral (influenza virus A [40]), bacterial (tetanus toxin [10], Streptococcus pyogenes M1 protein [31], and Francisella tularensis [36]), protozoan (Leishmania major [44, 45] and Plasmodium yoelii circumsporozoite protein [33]), and helminth (schistosomiasis [22, 23]) Ag. Since a similar vaccine strategy (e.g., attenuated Salmonella typhi aro mutants in human trials [16, 39]) might ultimately be adopted for use in genetically diverse human populations, it is important to investigate the influence that host genetics might have on the ability to induce protective immune responses to recombinant salmonella vaccines. While major histocompatibility complex (MHC) class I and class II molecule genes with polymorphisms will be obvious candidate genes because of their ability to restrict vaccine responses to certain antigenic epitopes, a role for non-MHC genes acting independently of Ag specificity should also be considered.
In mice, early bacterial replication following infection with S. typhimurium is regulated by the gene (Nramp1, formerly Ity/Lsh/Bcg) encoding the natural-resistance-associated macrophage protein (Nramp1) (for reviews, see references 4, 6, and 37). Nramp1 was identified by positional cloning (42). Gene knockout was then used (41) to formally demonstrate that the gene (renamed Nramp1) mediated resistance to all three intramacrophage pathogens S. typhimurium (Ity), Leishmania donovani (Lsh), and Mycobacterium bovis (Bcg). The protein (Nramp1) encoded by Nramp1 regulates the cascade of gene-inductive events which follow interaction of macrophages with bacterial lipopolysaccharide (LPS) and/or natural killer- or T-cell-derived gamma interferon (IFN-γ). The gene has multiple pleiotropic effects, including regulation of tumor necrosis factor alpha (TNF-α), interleukin 1β (IL-1β), and MHC class II molecules. All of these influence Ag processing and presentation, either directly (class II) or indirectly through their costimulatory or adjuvant (IL-1β and TNF-α) activity. Recent studies also demonstrate that Nramp1 has a more direct effect on Ag processing, possibly by regulating the activity of proteases in the late endosomal compartment (26). Hence, there are multiple ways (regulation of bacterial load or recombinant Ag dose, class II molecule expression, costimulatory or adjuvant activity, and Ag processing) that Nramp1 might influence responses to recombinant salmonella vaccines.
To test the hypothesis that Nramp1 influences responses to vaccination, congenic mouse strains have been used to analyze immune responses to recombinant Ag (tetanus toxoid Ag and leishmanial gp63) carried by live attenuated S. typhimurium aroA aroD mutants. Results show that congenic mice carrying the wild-type (S. typhimurium resistance) Nramp1 allele mount a predominantly T-helper-1 (IL-2 and IFN-γ) response to vaccination and show enhanced resolution of lesions following challenge infection with L. major. In contrast, mice carrying mutant (S. typhimurium susceptibility) Nramp1 mount a T-helper-2 (immunoglobulin E [IgE] and IL-4) response and show exacerbated lesion growth upon challenge.
MATERIALS AND METHODS
Construction of salmonella vaccines.
The attenuated S. typhimurium BRD847 aroA aroD double mutant vaccine strain carrying the expression plasmid pTETnir15 containing DNA sequence encoding fragment C of the tetanus toxoid Ag (TetC) was kindly provided by S. N. Chatfield (Medeva Group Research, Imperial College of Science and Technology, London, England). For preparation of the leishmanial gp63-salmonella vaccine, primers (GAGGATCCGTGCGCGACGTGAACTGG and AAACTAGTACCACGACGACCAGCCGC, engineered to provide BamHI restriction enzyme sites at either end of the PCR product) were designed to PCR (Pfu polymerase; New England Biolabs, Beverly, Mass.) amplify the gp63 gene (excluding the region containing the signal sequence for addition of the glycosylphosphatidylinositol anchor) from L. major expression clone pBS10Rb.1, in which codon usage had been corrected for bacterial expression (kindly provided by Robert McMaster, Department of Medical Genetics, University of British Columbia, Vancouver, British Columbia, Canada). One or two copies of the gp63 gene were cloned in tandem with the TetC gene into the salmonella expression vector pTECH2 (22, 23), a derivative of pTETnir15. Plasmids (pTECH2 alone or carrying genes encoding gp631TetC or gp632TetC; see below) were transfected into the single aroA mutant vaccine strain S. typhimurium SL3261 (kindly provided by B. A. D. Stocker, Department of Microbiology and Immunology, Stanford University School of Medicine, Stanford, Calif.). Expression of gp63 was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting. Briefly, cells growing at mid-log phase under ampicillin selection were harvested by centrifugation and the proteins were fractionated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis. Proteins were transferred to nitrocellulose membranes by electroblotting, and the presence of gp63TetC was detected with the mouse anti-gp63 monoclonal antibody (MAb) CP3.235 also kindly provided by Robert McMaster or with rabbit polyclonal anti-TetC prepared in-house. Appropriate-size bands were seen for clones bearing one (gp631TetC = 100 kDa) or two (gp632TetC = 150 kDa) copies of the gp63 gene. S. typhimurium SL3261 was used as the salmonella-only control where appropriate.
Salmonella Ag preparation.
To prepare salmonella Ag, a stationary overnight culture of S. typhimurium C5 was sedimented by centrifugation and cells were washed and resuspended in sterile phosphate-buffered saline (PBS). Bacteria were lysed by sonication (Soniprep 150; Fisher Scientific UK, Loughborough, England) and cell debris was removed by centrifugation. The lysate was filter sterilized through 0.22-μm-pore-size filters, and the protein content was estimated with the bicinochoninic acid kit (Pierce Biochemicals, Rockford, Ill.).
Mice.
Female CBA/Ca (Nramp1 wild-type) and BALB/c (Nramp1 mutant) mice and male and female C57BL/10ScSn (B10; Nramp1 mutant) mice were purchased from Harlan Olac (Blackthorn, Bicester, England) or were bred in-house from original breeding stock obtained from Harlan Olac. Male and female B10.L-Lshr (N20; Nramp1 wild-type) congenic mice (7) were bred in-house in a conventional facility. All mice were cohoused in this facility prior to experimental use.
Vaccination protocol.
Salmonella vaccine strains were grown in Luria-Bertani broth supplemented with 50 μg of ampicillin per ml and incubated overnight aerobically at 37°C. Cultures were aliquoted (0.5 ml) and cryopreserved in liquid nitrogen. Cryopreserved aliquots were thawed in a water bath at 45°C and 200 μl was diluted in sterile PBS to 1 in 10−6. A 200-μl portion was further diluted in 20 ml of molten Luria-Bertani agar (45°C) and plated with or without 50 μg of ampicillin per ml. Plates were incubated at 37°C overnight, and the bacterial concentration was established by colony counting (CFU). The original cryopreserved bacterial culture aliquots could then be diluted in sterile PBS as required for consistent provision of the appropriate concentration of bacteria for vaccination in different experiments. Mice were vaccinated at the age of 8 to 10 or 10 to 12 weeks (age matched within experiments). Mice were inoculated intravenously with 1 × 106 to 2 × 106 CFU in sterile PBS. Viable counts in the livers and spleens of mice sacrificed at intervals from weeks 1 to 6 postvaccination were determined as described previously (17).
Spleen cell assays.
Spleens from three mice per vaccine group sacrificed at weeks 1 to 6 were pooled, and cell suspensions were prepared in RPMI 1640 (Sigma, Poole, Dorset, England) by sieving. Erythrocytes were lysed by incubation in Gey’s solution (28) at 4°C for 4 min. Cells were washed by centrifugation at 260 × g for 7 min and finally resuspended in CRPMI (RPMI 1640 supplemented with 10% fetal calf serum [FCS] [Sigma], 0.02 M HEPES buffer, 0.5 mM 2-mercaptoethanol, 100 U each of penicillin and streptomycin per ml, and 0.2 mM glutamine. Cells were dispensed in triplicate to round-bottomed 96-well plates at 4 × 105 cells per well in 200 μl of CRPMI. Salmonella Ag or TetC (Boehringer Mannheim, Lewes, Sussex, England) was diluted in CRPMI and added in 10- to 20-μl volumes to obtain final concentrations as indicated below. Cell culture supernatants were collected at 48 h for IL-2 detection and 72 h for IFN-γ detection and stored at −80°C. For IL-4 detection in culture supernatants, cells were incubated with TetC for 120 h prior to addition of 50 ng of phorbol myristate acetate (PMA) per ml and 1 μM ionophore (Calbiochem, La Jolla, Calif.). Supernatants were collected 15 h after stimulation with PMA-ionophore.
Cytokine detection.
Cytokines were detected by enzyme-linked immunosorbent assay (ELISA). Flat-bottomed 96-well plates (Maxisorb, Nunclon; GIBCO BRL, Life Technologies Ltd., Paisley, Scotland) were coated with one of the following capture MAbs in 0.1 M NaHCO3: anti-IL-2 JES6-1A12, anti-IFN-γ XMG1.2, and anti-IL-4 11B11 (Pharmingen, San Diego, Calif.). Nonspecific binding sites were blocked with CRPMI. Plates were washed with PBS–0.05% Tween. Supernatants and standards (Pharmingen) were diluted with CRPMI. Capture Abs (biotinylated anti-IL-2 JES6-5H4, anti-IFN-γ 18112D, or anti-IL-4 18191D) were diluted according to the manufacturer’s instructions in PBS–10% FCS. Second-layer detection was done with avidin peroxidase (0.0025 mg/ml in PBS–10% FCS), followed by orthophenylenediamine-H2O2 as a substrate. Plates were read at 490 nm on a microtiter plate reader (Biotek Instruments Inc., ANACHEM, Luton, England).
Anti-TetC Ab detection.
Anti-TetC Abs were measured by ELISA with sera prepared from weekly tail bleeds of immunized mice and stored at −20°C. Plates were coated with 0.1 μg of recombinant TetC (Boehringer Mannheim) per well and washed with PBS–0.05% Tween. For total anti-TetC Ig, sera were diluted 1:25 in 2% casein in PBS and Ig levels were determined by direct second-layer incubation with a 1:1,000 dilution of goat anti-mouse total Ig horseradish peroxidase (HRP) conjugate (Dako Ltd., High Wycombe, Buckinghamshire, England). Subclass Ab levels were determined by using rat anti-mouse IgE or rabbit anti-mouse IgG1, IgG2a, or IgM (Zymed Laboratories Inc., Cambridge BioScience, Cambridge, England) at a dilution of 1:5,000. Third-layer detection was done with a 1:500 dilution of rabbit anti-rat HRP conjugate or a 1:500 dilution of swine anti-rabbit HRP conjugate (Dako), as appropriate. Sera and second- and third-layer Abs were diluted in 1% bovine serum albumin in PBS. Plates were developed with orthophenylenediamine-H2O2. The reaction was stopped with 3 M sulfuric acid. Plates were read at 490 nm.
Antisalmonella Ab detection.
Non-O-Ag-specific antisalmonella Abs were detected by ELISA. Plates were coated with lysate of a rough derivative of S. typhimurium C5 in Reggiardo’s buffer (0.01 M glycine, 0.02 M NaCl, 0.2 mM EDTA [pH 8.0], 0.01 M NaF, 0.1% sodium deoxycholate). Sera were diluted 1:400 in 1% bovine serum albumin (Sigma) in PBS. Total Ig and subclass Ab levels were detected as before.
Challenge infection.
Mice were challenged by subcutaneous inoculation of 5 × 106 metacyclic L. major LV39 promastigotes in the right hind footpad. Infection was monitored at weekly intervals by measurement of footpad depth with a vernier dial caliper (Manostat; CP Instrument Co., Bishop Stortford, Hertfordshire, England).
Statistics.
Unpaired two-tailed Student’s t tests were used to compare groups of mice at each time point.
RESULTS
Bacterial loads postvaccination.
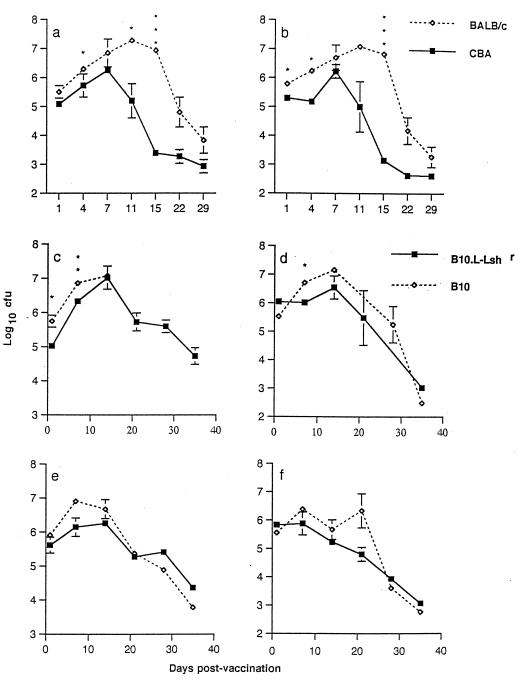
To test the hypothesis that Nramp1 wild-type (S. typhimurium-resistant) and mutant (S. typhimurium-susceptible) mice would respond differently to foreign Ag presented to the immune system in the context of a salmonella vaccine vehicle, experiments were performed initially using the live attenuated aroA aroD mutant S. typhimurium BRD847 strain expressing TetC alone. As expected, Nramp1 wild-type CBA/Ca mice showed lower bacterial loads in liver and spleen at day 4 after infection than did Nramp1 mutant BALB/c mice and an enhanced rate of clearance of the vaccine strain thereafter (Fig. 1a and b). The early (days 1 to 7) Nramp1-regulated differences in bacterial growth and clearance were not as dramatic as those observed with the more virulent S. typhimurium C5 strain (17, 30) and were barely detectable in experiments using the pair of B10 congenic mouse strains (B10 and N20 [Fig. 1c to f]). We did, however, observe an unexpected 33% mortality in B10 mice, in contrast to 14% in N20 male mice, over the first 14 days postvaccination. Bacterial loads in dead mice were not determined and hence do not contribute to the bacterial loads provided in Fig. 1. Only 1 of 36 female mice of each strain died during this time period. In subsequent experiments, female mice were used in conjunction with a lower bacterial inoculum. In the initial experiment, there was no subsequent mortality, and the B10 congenic mouse strains showed equivalent rates of bacterial clearance after day 10 postvaccination (Fig. 1c to f), suggesting that other genes might influence the later course of infection in BALB/c and CBA/Ca mice (Fig. 1a and b). For our purposes, it was preferable that there were not large differences in bacterial loads between Nramp1 wild-type and Nramp1 mutant mouse strains, since the resulting difference in vaccine Ag load could complicate the interpretation of results. Hence, using the B10 congenic mice, we could be more confident that any differences in immune response to TetC observed between strains would be attributable to Nramp1-regulated differences in Ag handling and presentation to the immune system, rather than simply to Ag dose.
FIG. 1.
Recovery of S. typhimurium BRD847 carrying pTETnir15 (mean log10CFU ± standard deviation/organ; three mice per time point) from the spleens (a, c, and e) and livers (b, d, and f) of vaccinated mice. Results of two separate experiments are shown; one compared BALB/c Nramp1 mutant and CBA/Ca wild-type mice (a and b), and one compared male (c and d) or female (e and f) B10 and N20 congenic mice. Closed symbols, Nramp1 wild-type strains; open symbols, Nramp1 mutant strains. Where error bars are not visible, the standard error was within the area occupied by the symbol. Significance levels determined by t tests are indicated as follows: ∗, P < 0.05, and ∗∗∗, P < 0.001. Similar results were obtained in two subsequent experiments in which B10 mice were injected with S. typhimurium SL3261 carrying pTECH2.
Ab responses to TetC.
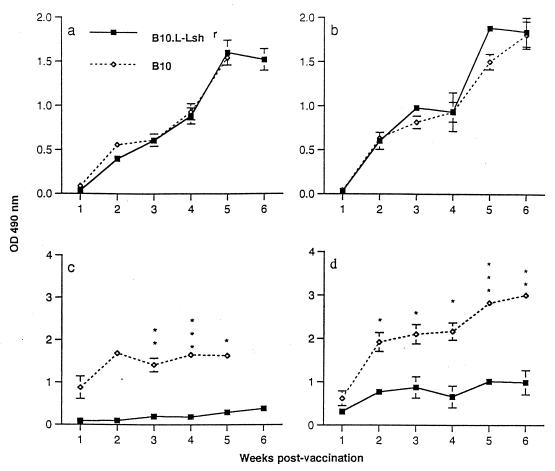
No significant differences in total anti-TetC Ig levels (or total antisalmonella Ig levels [data not shown]) were observed between BALB/c and CBA/Ca mice (data not shown) or between B10 and N20 male (Fig. 2a) or female (Fig. 2b) mice. In striking contrast, the B10 congenic mouse strains showed dramatic differences in anti-TetC IgE levels from weeks 1 through 5 postvaccination for male mice (Fig. 2c) and weeks 2 through 6 for female mice (Fig. 2d), indicative of a strong T-helper-2-driven response in the B10 Nramp1 mutant strain. Because of the unexpected mortality in male B10 mice, immune response measurements were taken only up to 5 weeks postvaccination. As indicated above, female mice were used in all subsequent experiments to avoid this problem. BALB/c mice also showed a significantly (P < 0.05) higher anti-TetC IgE response at 20 days postvaccination than did CBA/Ca mice (data not shown). There were, however, no consistent differences in IgM responses, or in T-helper-1-driven IgG2a versus T-helper-2-driven IgG1 responses, to TetC between mouse strains (data not shown). Nevertheless, given the very strong evidence for linkage between IgE responses and the IL-4–IL-9 T-helper-2 cytokine gene cluster (11, 27), this very dramatic difference in IgE responses is strongly indicative of a bias in the T-helper-2 arm of the immune response in Nramp1 mutant mice.
FIG. 2.
Anti-TetC total Ig (a and b) or IgE (c and d) antibody levels (means ± standard errors; three mice per time point) in postvaccination sera from male (a and c) or female (b and d) B10 (open symbols) and N20 (closed symbols) mice. Where error bars are not visible, the standard error was within the area occupied by the symbol. Significance levels determined by t tests are indicated as follows: ∗, P < 0.05; ∗∗, P < 0.01; and ∗∗∗, P < 0.001. Results are from the same B10 experiment as presented in Fig. 1. Similar results were obtained in a repeat experiment in which S. typhimurium SL3261 carrying pTECH2 was injected into female B10 mice. OD, optical density.
Cytokine responses to TetC.
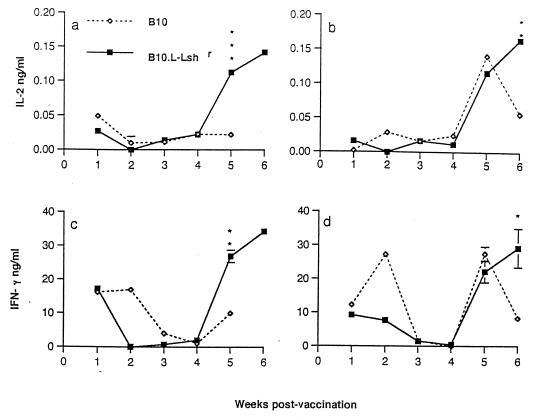
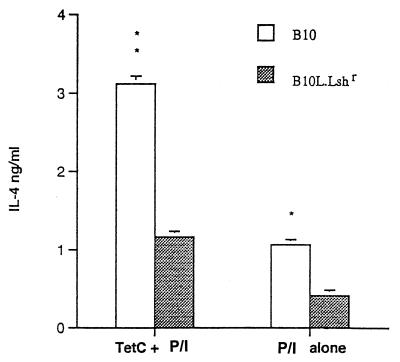
To determine whether a divergence in T-helper-1 and T-helper-2 responses between Nramp1 wild-type and mutant congenic mouse strains could also be detected at the cytokine level, IL-2 and IFN-γ responses to TetC were measured following restimulation of spleen cells in vitro. Figure 3 demonstrates that following clearance of the bacterial load in liver and spleen (Fig. 1), a strong recall memory T-cell response was established for both IL-2 (Fig. 3a and b) and IFN-γ (Fig. 3c and d) production. For both of these T-helper-1-associated cytokine responses, N20 congenic Nramp1 wild-type mice showed an enhanced response compared to B10 Nramp1 mutant mice at weeks 5 (male mice) and/or 6 (female mice) postvaccination. A very similar pattern of responses between the strains was observed in experiments examining the response to restimulation with whole crude salmonella lysates (data not shown), suggesting that there is a generally enhanced ability to show a T-helper-1 CD4 response to all salmonella-derived Ag in congenic Nramp1 wild-type mice. Attempts to measure IL-4 cytokine responses in supernatants from this experiment were not successful. In follow-up experiments, IL-4 was measured in spleen cell populations expanded by TetC stimulation for 120 h prior to being triggered with PMA plus ionophore. Supernatants were collected 15 h after the PMA-ionophore trigger. The rationale for this experiment is that the PMA and ionophore act together as a potent trigger for rapid IL-4 production by this expanded cell population. This was used as a measure of whether a bias towards a committed T-helper-2 population has occurred. Figure 4 demonstrates that, as would be predicted from the anti-TetC IgE data, B10 mice showed an enhanced PMA-ionophore-elicited IL-4 response in this TetC-expanded population compared to that of N20 mice. In these experiments, stimulation with TetC alone did not lead to significant IL-4 release, presumably because IL-4 produced is consumed within the culture system. Interestingly, the spleen cell population from vaccinated B10 mice which had not been expanded in vitro on TetC also showed a higher PMA-ionophore-elicited IL-4 response than the parallel cell population from N20 mice, indicating that the cells ex vivo also showed a bias towards a T-helper-2 response at 6 weeks after salmonella TetC vaccination. Spleen cells from naive B10 and N20 mice did not show this difference in baseline response to PMA-ionophore (data not shown).
FIG. 3.
TetC-stimulated IL-2 (a and b) or IFN-γ (c and d) production (means ± standard errors; three mice per time point) in spleen cells isolated from male (a and c) or female (b and d) B10 (open symbols) and N20 congenic (closed symbols) mice after vaccination with S. typhimurium BRD847. Where error bars are not visible, the standard error was within the area occupied by the symbol. Significance levels determined by t tests are indicated as follows: ∗, P < 0.05; ∗∗, P < 0.01; and ∗∗∗, P < 0.001. Background IL-2 and IFN-γ production in wells without Ag were subtracted from the TetC-stimulated response. The background IFN-γ levels were highest (12 to 24 ng/ml) at weeks 1 and 2 postvaccination, suggesting that endogenous salmonellae were stimulating a natural killer cell response. Backgrounds for IL-2 were <0.02 ng/ml throughout. Results are from the same B10 experiment as presented in Fig. 1. Similar results were obtained in three repeat experiments in which S. typhimurium SL3261 carrying pTECH2 was injected in female B10 congenic mice.
FIG. 4.
PMA-ionophore (P/I)-triggered IL-4 production (means ± standard errors; three mice per group) in TetC-expanded spleen cells isolated from B10 (open bars) and N20 congenic (filled bars) female mice 6 weeks after vaccination with S. typhimurium SL3261 carrying pTECH2. The differences between mouse strains for P/I-elicited IL-4 production were significant with or without restimulation with TetC in vitro, indicating that the splenocyte population ex vivo was also biased towards a T-helper-2-committed population in the B10 strain. No differences were observed for P/I-elicited IL-4 production in splenocytes from naive B10 and N20 mice (data not shown). Similar results were obtained in a repeat experiment for S. typhimurium SL3261 carrying pTECH2 in female B10 congenic mice. Significance levels determined by t tests are indicated as follows: ∗, P < 0.05, and ∗∗, P < 0.01.
Response to challenge infection.
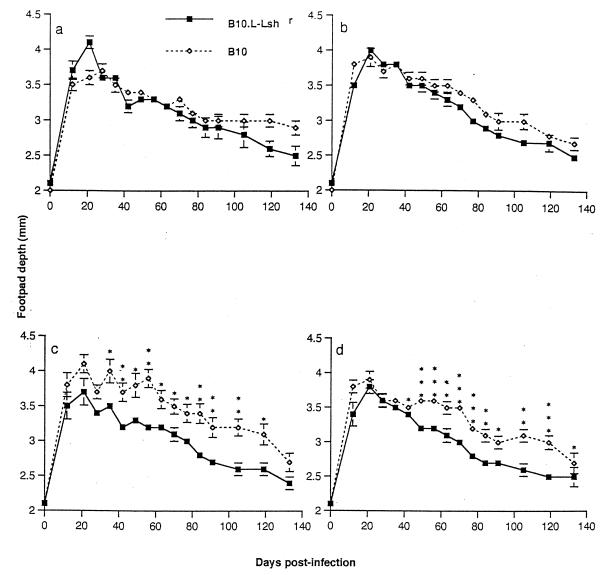
To determine whether the strong bias in T-helper-1 versus T-helper-2 responses in Nramp1 congenic mouse strains would influence their responses to challenge infection in a model disease system, an attenuated salmonella vaccine was engineered to incorporate leishmanial gp63 in tandem with TetC. Previous studies (22, 23) had demonstrated that TetC enhances the response to vaccine Ag in this cloning strategy. Two types of construct were prepared in which one or two copies of the gp63 gene were cloned in tandem with one copy of the TetC gene. To ensure complete bacterial clearance before challenge infection, mice were challenged with L. major 12 weeks after vaccination. Monitoring infection in control mice, either in naive infected mice (Fig. 5a) or in mice receiving the S. typhimurium SL3261 pTECH2 control vaccination (Fig. 5b), demonstrated that as shown previously (5, 19), Nramp1 does not have a dramatic influence over the course of L. major infection in the footpad. Among mice vaccinated with S. typhimurium SL3261 transfected with expression plasmids containing one (Fig. 5c) or two (Fig. 5d) copies of the gp63 gene in tandem with the TetC gene, resolution of lesion size was moderately enhanced in N20 mice and moderately exacerbated in B10 mice. When footpad measurements for B10 pTECH2 control-vaccinated mice were compared to those for the B10 gp63-vaccinated mice, it was found that the mean footpad depth for the gp63-vaccinated mice was higher than for mice vaccinated with pTECH2 alone at every time point. Similarly, when the footpad measurements for N20 pTECH2 control-vaccinated mice were compared to those for the N20 gp63-vaccinated mice, it was found that the mean footpad depth for the gp63-vaccinated mice was lower than that for mice vaccinated with pTECH2 alone at every time point. Hence, although the between-group differences within mouse strains do not achieve statistical significance, the sum of these two effects produces highly statistically significant differences in lesion size in B10 gp63-vaccinated and N20 gp63-vaccinated mice over 17 weeks of infection (Fig. 5c and d). Two copies of the gp63 gene did not significantly alter this response to vaccination, in terms of either protection in the Nramp1 wild-type mice or disease exacerbation in mutant mice. The results are consistent with the hypothesis that an enhanced T-helper-1 response in congenic Nramp1 wild-type mice is protective, while the T-helper-2 response of Nramp1 mutant mice leads to exacerbated disease. The latter did not, however, overcome the overall genetic resistance of the B10 background to L. major infection (19), because all groups of mice ultimately resolved their footpad lesions.
FIG. 5.
Course of L. major infection (means ± standard errors) in B10 (open symbols) and N20 congenic (closed symbols) mice. (a) Naive control mice; (b) mice vaccinated with S. typhimurium SL3261 carrying control vector (pTECH2) only; (c) mice vaccinated with S. typhimurium SL3261 carrying pTECH2 containing one copy of the leishmanial gp63 gene; (d) mice vaccinated with S. typhimurium SL3261 carrying pTECH2 containing two copies of the leishmanial gp63 gene. Where error bars are not visible, the standard error was within the area occupied by the symbol. Significance levels determined by t tests are indicated as follows: ∗, P < 0.05; ∗∗, P < 0.01; and ∗∗∗, P < 0.001. Similar results were obtained in two repeats of this experiment.
DISCUSSION
Few studies have examined the influence of host genetics on immune responses to vaccination. We knew that one of the consequences of the influence of Nramp1 on macrophage activation is enhanced induction and long-term maintenance of a T-helper-1 response in congenic Nramp1 wild-type versus mutant mice following both L. donovani (20) and Mycobacterium bovis (24) infections. Hence, we hypothesized that recombinant Ag delivered to the immune system by using salmonella or M. bovis BCG as a vaccine vehicle might also elicit enhanced T-helper-1 responses in Nramp1 wild-type mice. Such differences in vivo may reflect the influence of Nramp1 on upregulation of MHC class II molecule expression (21, 46), as well as the enhanced ability of Nramp1 wild-type macrophages to process and present protein epitopes to CD4 T cells (26). Alternatively, when virulent organisms like L. donovani and M. bovis are used, there are also large Nramp1-regulated differences in parasite load which might influence subsequent induction of T-cell responses in vivo. It is well known, for example, that high Ag load is associated with a suppressed T-helper-1 response (8). In a recent study Pie and colleagues (29) specifically examined the role of IL-10 in inhibiting IFN-γ responses during the first 5 days after S. typhimurium C5 infection. IL-10 mRNA expression and serum IL-10 levels were high in Nramp1 mutant compared to congenic wild-type mice. However, the use of neutralizing anti-IL-10 and attenuated S. typhimurium strains demonstrated that the level of IL-10 expression correlated with bacterial load and was secondary to, rather than functionally related to, the action of the Nramp1 gene. In contrast, our studies using attenuated S. typhimurium aroA aroD mutant strains have demonstrated a dramatic polarization of T-cell responses in Nramp1 congenic mouse strains in the presence of equivalent bacterial loads. Congenic mice carrying the wild-type Nramp1 allele mounted a predominantly T-helper-1 (IL-2 and IFN-γ) response to recombinant TetC synthesized by the salmonellae, as well as more generally to salmonella Ag. On the other hand, Nramp1 mutant mice mounted a T-helper-2 (IgE and IL-4) response. Hence, in contrast to the results of the study by Pie et al. (29), it seems likely that differences in the abilities of Nramp1 wild-type and mutant macrophages to process and present Ag are directly and functionally responsible for polarization of the T-cell response in this vaccine model.
Many previous studies (21, 26, 46) have demonstrated that IFN-γ induces differences in MHC class II molecule expression in macrophages carrying wild-type and mutant Nramp1 alleles. In the early phases of salmonella infection, bacterial LPS provides a potent trigger for macrophage IL-12 production, which in turn triggers IFN-γ release from natural killer cells. Hence, even in the early postvaccination phase there is likely to be a source of endogenously produced IFN-γ inducing MHC class II molecule expression. Indeed, we observed a small burst of IFN-γ at weeks 1 and 2 postvaccination in unstimulated spleen cells, which may have resulted from natural killer cell activity driven by endogenous salmonellae. However, there were no differences in TetC-induced IFN-γ production by spleen cells from Nramp1 wild-type and mutant mice at this time. The recall memory T-cell response to TetC and salmonella Ag was not evident until week 5 postvaccination, and it was then that differences between Nramp1 congenic mouse strains became apparent.
In a recent study (26) using Nramp1-transfected macrophage clones, it was observed that LPS can also induce enhanced processing of recombinant protein Ag in transfectants bearing wild-type Nramp1 compared to that in transfected and parental macrophages bearing the Nramp1 mutant allele. In the vaccine model described here, where recombinant Ag is synthesized by live attenuated salmonella, this LPS-dependent enhanced Ag processing may be a significant component of the enhanced T-helper-1 response observed in Nramp1 wild-type mice. The environment in which Ag presentation occurs might also be important in determining whether the CD4 T-cell response biases towards T-helper-1 or T-helper-2 responses. Nramp1 wild-type macrophages stimulated with LPS also show enhanced TNF-α release (13, 32) and IL-1β expression (34), factors which bias towards stimulation of a T-helper-1 response. Hence, the vaccine vehicle itself might be important in determining the bias of the CD4 T-cell response.
The influence of Nramp1 on Ag processing and presentation may be directly related to its localization and function. The Nramp1 gene was identified by positional cloning (42), and the full-length cDNA sequence was obtained (3). Computer-assisted analysis of the deduced amino acid sequence shows that Nramp1 encodes a 53-kDa protein with homology to bacterial and lower-eukaryotic membrane-bound transporter proteins (42). Further structural analysis (9) suggests that it functions as an ion channel/transporter. Confocal and electron microscope studies demonstrate that Nramp1 localizes to the late endosomal/lysosomal compartment and can be seen around the leishmanial parasitophorous vacuole (2, 35a). This vacuole also contains MHC class II molecules without the invariant chain Ii (25), i.e., ready for loading with processed foreign Ag epitopes. Class II molecules generally accumulate in a specialized endosomal compartment, referred to as the MHC class II compartment or the compartment for peptide loading, during their transport from the trans-Golgi network to the cell surface. In this compartment, foreign peptides acquired through the endocytic or phagocytic pathways displace self-peptides on class II molecules and are thus transported to the cell surface for presentation to CD4+ T cells. Salmonella also occupies a similar acidified receptor-mediated phagocytic compartment (1). Processing of salmonella-derived Ag may thus be directly affected by Nramp1 function. One hypothesis (26) is that Nramp1 regulates ion uptake to the late endosome, where Ag processing occurs, perhaps influencing ion-dependent peptidase activity. In yeast, the homologous SMF1 protein has been shown to play a role in manganese uptake to the cell (38). The SMF1 gene was first identified for its ability to complement a defect in protein import into the mitochondria (43). Its ability to complement this function was due to the mangenese-dependent peptidase activity required to cleave proteins being imported into the mitochondria (38). The mammalian homolog Nramp2 has now also been shown to be a metal ion channel (14), transporting a wide variety of metal ions, including Fe2+, Zn2+, Mn2+, Cu2+, Co2+, and Cd2+, with equivalent efficiencies and by a pH-dependent electrogenic process. Nramp1 could play a parallel role in providing an essential ion for peptidase activity required for Ag processing or in regulating peptidase activity by controlling pH in the late endosomal compartment. Whatever its role, pinpointing the processing defect in Nramp1 mutant macrophages has important implications for further analysis of genetic regulation of response to vaccination, particularly for recombinant vaccines delivered by using viable salmonella or BCG.
As a corollary of the polarization of the CD4 T-cell response, we hypothesised that mice bearing Nramp1 wild-type allele and mutant mice would behave differently following vaccination and challenge infection with an organism for which protective immunity is T helper 1 dependent. We decided to test this hypothesis using the L. major infection model (15, 35), in which T-helper-1 responses are vital to development of a protective immune response while T-helper-2 responses are associated with exacerbated disease. We demonstrated that Nramp1 wild-type mice showed enhanced protection against challenge infection following immunization with a gp63 salmonella vaccine, while Nramp1 mutant mice show exacerbated disease. Although these experiments were carried out with congenic mice with a genetic background (B10) where self-healing L. major infection occurs, we were able to demonstrate significant alterations in the rates of healing in vaccinated Nramp1 wild-type and mutant mice. A more rigorous test for recombinant leishmanial Ag vaccines would be to test their efficacies in a BALB/c genetic background where a polarized T-helper-2 response to L. major infection (under separate genetic control from Nramp1) leads to extreme susceptibility (15, 35). Previous studies have shown (12) that Nramp1 mutant BALB/c mice produce significantly higher titers of total serum Ab against salmonella-carried recombinant E. coli MalE protein than do their congenic wild-type C.D2 [Idh1bPep3b-(N7)F12] and C.CB [Ityr(N9)F5] counterparts, but subclass differences and T-cell responses were not examined. Whether the enhanced protective T-helper-1 response we have observed here in Nramp1 wild-type mice is sufficient to mediate strong protection in an L. major-susceptible BALB genetic background is the subject of our continuing research. We are also examining whether recombinant Ag delivered to Nramp1 congenic mice via DNA vaccines engineered for endogenous versus secretory protein trafficking pathways also elicit polarized CD4 T-cell responses. Such studies should allow us to determine the importance of Nramp1 in regulating the Ag-presenting-cell environment in response to the adjuvant activity of the salmonella vehicle, as opposed to a more direct role in regulating Ag processing, in determining T-cell subset expansion. In either case, it is clear that the mutation in murine Nramp1 has a major impact on response to vaccination, supporting the concept that host genetic variation may be an important factor in determining vaccine efficacy.
ACKNOWLEDGMENTS
This work was funded by The Wellcome Trust.
We thank Sandra Toole for her technical assistance during this project and Raquel Demarco De Hormaeche for helpful advice on Ab and cytokine assays.
REFERENCES
- 1.Alpuche-Aranda C M, Racoosin E L, Swanson J A, Miller S I. Salmonella stimulate macrophage macropinocytosis and persist within spacious phagosomes. J Exp Med. 1994;179:601–608. doi: 10.1084/jem.179.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson P G P, Blackwell J M, Barton C H. The Nramp1 locus encodes 65kD IFNγ-inducible protein in murine macrophages. Biochem J. 1997;325:779–786. doi: 10.1042/bj3250779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton C H, White J K, Roach T I A, Blackwell J M. NH2-terminal sequence of macrophage-expressed natural resistance-associated macrophage protein (Nramp) encodes a proline/serine-rich putative Src homology 3-binding domain. J Exp Med. 1994;179:1683–1687. doi: 10.1084/jem.179.5.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwell J M. Structure and function of the natural-resistance-associated macrophage protein (Nramp1), a candidate protein for infectious and autoimmune disease susceptibility. Mol Med Today. 1996;2:205–211. doi: 10.1016/1357-4310(96)88773-9. [DOI] [PubMed] [Google Scholar]
- 5.Blackwell J M, Alexander J. Different host genes recognise and control infection with taxonomically distinct Leishmania species. In: Rioux J A, Peters W, editors. Leishmania. Taxonomie et phylogenèse. Applications éco-épidémiologiques. Montpellier, France: IMEEE; 1986. pp. 211–219. [Google Scholar]
- 6.Blackwell J M, Barton C H, White J K, Roach T I A, Shaw M-A, Whitehead S H, Mock B A, Searle S, Williams H, Baker A-M. Genetic regulation of leishmanial and mycobacterial infections: the Lsh/Ity/Bcg gene story continues. Immunol Lett. 1994;43:99–107. doi: 10.1016/0165-2478(94)00161-8. [DOI] [PubMed] [Google Scholar]
- 7.Blackwell J M, Toole S, King M, Dawda P, Roach T I, Cooper A. Analysis of Lsh gene expression in congenic B10.L-Lshr mice. Curr Top Microbiol Immunol. 1988;137:301–309. doi: 10.1007/978-3-642-50059-6_45. [DOI] [PubMed] [Google Scholar]
- 8.Bretscher P A, Wei G, Menon J N, Bielefeldt-Ohmann H. Establishment of stable, cell-mediated immunity that makes “susceptible” mice resistant to Leishmania major. Science. 1992;257:539–542. doi: 10.1126/science.1636090. [DOI] [PubMed] [Google Scholar]
- 9.Cellier M, Belouchi A, Gros P. Resistance to intracellular infections: comparative genomic analysis of Nramp. Trends Genet. 1996;12:201–204. doi: 10.1016/0168-9525(96)30042-5. [DOI] [PubMed] [Google Scholar]
- 10.Chatfield S N, Charles I G, Makoff A J, Oxer M D, Dougan G, Pickard D, Slater D, Fairweather N F. Use of the nirB promoter to direct the stable expression of heterologous antigens in Salmonella oral vaccine strains: development of a single-dose oral tetanus vaccine. Biotechnol N Y. 1992;10:888–892. doi: 10.1038/nbt0892-888. [DOI] [PubMed] [Google Scholar]
- 11.Doull I J M, Lawrence S, Watson M, Begishvili T, Beasley R W, Lampe F, Holgate S T, Morton N E. Allelic association of gene markers on chromosomes 5q and 11q with atopy and bronchial hyperresponsiveness. Am J Respir Crit Care Med. 1996;153:1280–1284. doi: 10.1164/ajrccm.153.4.8616554. [DOI] [PubMed] [Google Scholar]
- 12.Fayolle C, O’Callaghan D, Martineau P, Charbit A, Clément J M, Hofnung M, Leclerc C. Genetic control of antibody responses induced against an antigen delivered by recombinant attenuated Salmonella typhimurium. Infect Immun. 1994;62:4310–4319. doi: 10.1128/iai.62.10.4310-4319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Formica S, Roach T I A, Blackwell J M. Interaction with extracellular matrix proteins influences Lsh/Ity/Bcg (candidate Nramp) gene regulation of macrophage priming/activation for tumour necrosis factor α and nitrite release. Immunology. 1994;82:42–50. [PMC free article] [PubMed] [Google Scholar]
- 14.Gunshin H, Mackenzie B, Berger U V, Gunshin Y, Romero M F, Boron W F, Nussberger S, Gollan J L, Hediger M A. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 15.Heinzel F P, Sadick M D, Holaday B J, Coffman R L, Locksley R M. Reciprocal expression of interferon gamma or interleukin 4 during the resolution or progression of murine leishmaniasis. Evidence for expansion of distinct helper T cell subsets. J Exp Med. 1989;169:59–72. doi: 10.1084/jem.169.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hone D M, Harris A M, Chatfield S, Dougan G, Levine M M. Construction of genetically defined double aro mutants of Salmonella typhi. Vaccine. 1991;9:810–816. doi: 10.1016/0264-410x(91)90218-u. [DOI] [PubMed] [Google Scholar]
- 17.Hormaeche C E. Natural resistance to Salmonella typhimurium in different inbred mouse strains. Immunology. 1979;37:311–318. [PMC free article] [PubMed] [Google Scholar]
- 18.Hormaeche C E. Live attenuated Salmonella vaccines and their potential as oral combined vaccines carrying heterologous antigens. J Immunol Methods. 1991;142:113–120. doi: 10.1016/0022-1759(91)90298-t. [DOI] [PubMed] [Google Scholar]
- 19.Howard J G, Hale C, Chan-Liew W L. Immunological regulation of experimental cutaneous leishmaniasis. I. Immunogenetic aspects of susceptibility to Leishmania tropica in mice. Parasite Immunol. 1980;2:303–314. doi: 10.1111/j.1365-3024.1980.tb00061.x. [DOI] [PubMed] [Google Scholar]
- 20.Kaye P M, Blackwell J M. Lsh, antigen presentation and the development of CMI. Res Immunol. 1989;140:810–815. doi: 10.1016/0923-2494(89)90038-2. [DOI] [PubMed] [Google Scholar]
- 21.Kaye P M, Patel N K, Blackwell J M. Acquisition of cell-mediated immunity to Leishmania. II. Lsh gene regulation of accessory cell function. Immunology. 1988;65:17–22. [PMC free article] [PubMed] [Google Scholar]
- 22.Khan C M A, Villarreal Ramos B, Pierce R J, Demarco de Hormaeche R, McNeill H, Ali T, Chatfield S, Capron A, Dougan G, Hormaeche C E. Construction, expression and immunogenicity of multiple tandem copies of the Schistosoma mansoni peptide 115–131 of the P28 glutathione S-transferase expressed as C-terminal fusions to tetanus toxin fragment C in a live aro-attenuated vaccine of Salmonella. J Immunol. 1994;153:5634–5642. [PubMed] [Google Scholar]
- 23.Khan C M A, Villarreal Ramos B, Pierce R J, Riveau G, Demarco de Hormaeche R, McNeill H, Ali T, Fairweather N, Chatfield S, Capron A, et al. Construction, expression and immunogenicity of the Schistosoma mansoni P28 glutathione S-transferase as a genetic fusion to tetanus toxin fragment C in a live Aro attenuated vaccine strain of Salmonella. Proc Natl Acad Sci USA. 1994;91:11261–11265. doi: 10.1073/pnas.91.23.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramnik I, Radzioch D, Skamene E. T-helper 1-like subset selection in Mycobacterium bovis bacillus Calmette-Guérin-infected resistant and susceptible mice. Immunology. 1994;81:618–625. [PMC free article] [PubMed] [Google Scholar]
- 25.Lang T, Hellio R, Kaye P M, Antoine J. Leishmania donovani-infected macrophages: characterization of the parasitophorous vacuole and potential role of this organelle in antigen presentation. J Cell Sci. 1994;107:2137–2150. doi: 10.1242/jcs.107.8.2137. [DOI] [PubMed] [Google Scholar]
- 26.Lang T, Prina E, Sibthorpe D, Blackwell J M. Nramp1 transfection transfers Ity/Lsh/Bcg-related pleiotropic effects on macrophage activation: influence on antigen processing and presentation. Infect Immun. 1997;65:380–386. doi: 10.1128/iai.65.2.380-386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marsh D G, Neely J D, Breazeale D R, Ghosh B, Friedhoff L R, Ehrlich-Kautzky E, Schou C, Krishnaswamy G, Beaty T H. Linkage analysis of IL4 and other chromosome 5q31.1 markers and total serum immunoglobulin E concentrations. Science. 1994;264:1152–1156. doi: 10.1126/science.8178175. [DOI] [PubMed] [Google Scholar]
- 28.Mishell B B, Shiigi S M. Selected methods in cellular immunology. San Francisco, Calif: W. H. Freeman and Company; 1980. pp. 23–24. [Google Scholar]
- 29.Pie S, Matsiota-Bernard P, Truffa-Bachi P, Nauciel C. Gamma interferon and interleukin-10 gene expression in innately susceptible and resistant mice during the early phase of Salmonella typhimurium infection. Infect Immun. 1996;64:849–854. doi: 10.1128/iai.64.3.849-854.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plant J, Glynn A A. Genetics of resistance to infection with Salmonella typhimurium in mice. J Infect Dis. 1976;133:72–78. doi: 10.1093/infdis/133.1.72. [DOI] [PubMed] [Google Scholar]
- 31.Poirier T P, Kehoe M A, Beachey E H. Protective immunity evoked by oral administration of attenuated aroA Salmonella typhimurium expressing cloned streptococcal M protein. J Exp Med. 1988;168:25–32. doi: 10.1084/jem.168.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roach T I A, Kiderlen A F, Blackwell J M. Role of inorganic nitrogen oxides and tumor necrosis factor alpha in killing Leishmania donovani amastigotes in gamma interferon-lipopolysaccharide-activated macrophages from Lshs and Lshr congenic mouse strains. Infect Immun. 1991;59:3935–3944. doi: 10.1128/iai.59.11.3935-3944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadoff J C, Ballou W R, Baron L S, Majarian W R, Brey R N, Hockmeyer W T, Young J F, Cryz S J, Ou J, Lowell G H, et al. Oral Salmonella typhimurium vaccine expressing circumsporozoite protein protects against malaria. Science. 1988;240:336–338. doi: 10.1126/science.3281260. [DOI] [PubMed] [Google Scholar]
- 34.Schurr E, Radzioch D, Malo D, Gros P, Skamene E. Molecular genetics of inherited susceptibility to intracellular parasites. Behring Inst Mitt. 1991;88:1–12. [PubMed] [Google Scholar]
- 35.Scott P, Natovitz P, Coffman R L, Pearce E, Sher A. Immunoregulation of cutaneous leishmaniasis. T cell lines that transfer protective immunity or exacerbation belong to different T helper subsets and respond to distinct parasite antigens. J Exp Med. 1988;168:1675–1684. doi: 10.1084/jem.168.5.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Searle, S., N. Bright, P. Atkinson, C. H. Barton, and J. M. Blackwell. Submitted for publication.
- 36.Sjostedt A, Sandstrom G, Tarnvik A. Immunization of mice with an attenuated Salmonella typhimurium strain expressing a membrane protein of Francisella tularensis. A model for identification of bacterial determinants relevant to the host defence against tularemia. Res Microbiol. 1990;141:887–891. doi: 10.1016/0923-2508(90)90126-b. [DOI] [PubMed] [Google Scholar]
- 37.Skamene E. The Bcg gene story. Immunobiology. 1994;191:451–460. doi: 10.1016/S0171-2985(11)80451-1. [DOI] [PubMed] [Google Scholar]
- 38.Supeck F, Supekova L, Nelson H, Nelson N. A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc Natl Acad Sci USA. 1996;93:5105–5110. doi: 10.1073/pnas.93.10.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tacket C O, Hone D M, Losonsky G A, Guers L, Edelman R, Levine M M. Clinical acceptability and immunogenicity of CVD 908 Salmonella typhi vaccine strain. Vaccine. 1992;10:443–446. doi: 10.1016/0264-410x(92)90392-w. [DOI] [PubMed] [Google Scholar]
- 40.Tite J P, Gao X M, Hughes Jenkins C M, Lipscombe M, O’Callaghan D, Dougan G, Liew F Y. Anti-viral immunity induced by recombinant nucleoprotein of influenza A virus. III. Delivery of recombinant nucleoprotein to the immune system using attenuated Salmonella typhimurium as a live carrier. Immunology. 1990;70:540–546. [PMC free article] [PubMed] [Google Scholar]
- 41.Vidal S, Tremblay M L, Govoni G, Gauthier S, Sebastiani G, Malo D, Skamene E, Olivier M, Jothy S, Gros P. The Ity/Lsh/Bcg locus: natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1 gene. J Exp Med. 1995;182:655–666. doi: 10.1084/jem.182.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidal S M, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 43.West A H, Clark D J, Martin J, Neupert W, Hartl F U, Horwich A L. Two related genes encoding extremely hydrophobic proteins suppress a lethal mutation in the yeast mitochondrial processing enhancing protein. J Biol Chem. 1992;267:24625–24633. [PubMed] [Google Scholar]
- 44.Xu D, McSorley S J, Chatfield S N, Dougan G, Liew F Y. Protection against Leishmania major infection in genetically susceptible BALB/c mice by gp63 delivered orally in attenuated Salmonella typhimurium (AroA− AroD−) Immunology. 1995;85:1–7. [PMC free article] [PubMed] [Google Scholar]
- 45.Yang D M, Fairweather N, Button L L, McMaster W R, Kahl L P, Liew F Y. Oral Salmonella typhimurium (AroA−) vaccine expressing a major leishmanial surface protein (gp63) preferentially induces T helper 1 cells and protective immunity against leishmaniasis. J Immunol. 1990;145:2281–2285. [PubMed] [Google Scholar]
- 46.Zwilling B S, Vespa L, Massie M. Regulation of I-A expression by murine peritoneal macrophages: differences linked to the Bcg gene. J Immunol. 1987;138:1372–1376. [PubMed] [Google Scholar]