Abstract
Catalase plays a key role as an antioxidant, protecting aerobic organisms from the toxic effects of hydrogen peroxide, and in some cases has been postulated to be a virulence factor. To help elucidate the function of catalase in Candida albicans, a single C. albicans-derived catalase gene, designated CAT1, was isolated and cloned. Degenerate PCR primers based on highly conserved areas of other fungal catalase genes were used to amplify a 411-bp product from genomic DNA of C. albicans ATCC 10261. By using this product as a probe, catalase clones were isolated from genomic libraries of C. albicans. Nucleotide sequence analysis revealed an open reading frame encoding a protein of 487 amino acid residues. Construction of a CAT1-deficient mutant was achieved by using the Ura-blaster technique for sequential disruption of multiple alleles by integrative transformation using URA3 as a selectable marker. Resulting mutants exhibited normal morphology and comparable growth rates of both yeast and mycelial forms. Enzymatic analysis revealed an abundance of catalase in the wild-type strain but decreasing catalase activity in heterozygous mutants and no detectable catalase in a homozygous null mutant. In vitro assays showed the mutant strains to be more sensitive to damage by both neutrophils and concentrations of exogenous peroxide that were sublethal for the parental strain. Compared to the parental strain, the homozygous null mutant strain was far less virulent for mice in an intravenous infection model of disseminated candidiasis. Definitive linkage of CAT1 with virulence would require restoration of activity by reintroduction of the gene into mutants. However, initial results in mice, taken together with the enhanced susceptibility of catalase-deficient hyphae to damage by human neutrophils, suggest that catalase may enhance the pathogenicity of C. albicans.
Candida albicans, an opportunistic pathogenic fungus, is the predominant etiologic agent of mycoses in immunocompromised hosts (2, 29, 41). The fact that the organism is diploid and has no known sexual cycle has hindered the genetic study of potential virulence factors of this increasingly common cause of serious infections. Recent advances in molecular genetic techniques have considerably enhanced the ability to identify and analyze factors that may contribute to the pathogenicity of C. albicans (11, 13). Many cellular properties have been postulated to contribute to the virulence of C. albicans. However, it has been possible to relate only a few factors directly to decreased virulence in vivo by using disruption of specific genes or isolation of variants deficient in particular enzymes. Such putative pathogenicity factors include secreted aspartyl proteinases (20, 24, 34), topoisomerase I (16), 1,3-β-glucosyltransferase (38), and chitin synthase III (3).
Since the host immune response also plays a key role in the ability of C. albicans to cause infection, many studies have focused on the mechanisms involved in the killing of this organism. Neutropenia has been shown to be a major factor contributing to susceptibility to disseminated candidiasis in humans. Our own data (6–8) and those of others (22, 39) emphasize the primary importance of oxidative fungicidal mechanisms by human neutrophils (polymorphonuclear leukocytes [PMNs]) and monocytes. These mechanisms depend largely on the ability of PMNs to synthesize potent oxidants primarily derived from hydrogen peroxide, including H2O2 itself, as well as hydroxyl radical, hypochlorous acid, and chloramines (6–8). This crucial role of oxidant-mediated fungicidal effects dictates a need to define fungal antioxidant defenses. Because exogenous antioxidants, including catalase, impair killing of C. albicans hyphae by PMNs (6, 7, 44) and production of H2O2 by PMNs correlates directly with fungicidal activity (9), catalase is presumed to be an important antioxidant defense in C. albicans. Therefore, we investigated the role of catalase in resistance of the fungus to leukocyte-mediated killing. Since H2O2 is highly diffusible across cell walls, catalases provide essential intracellular antioxidant activity for many organisms, including Saccharomyces cerevisiae and other fungi (4, 15, 17, 42). Catalases are also known to contribute to growth regulation and development in many eukaryotic organisms (17, 26, 28, 43). To study the significance of catalase in the pathogenicity of C. albicans, a single gene was cloned and subsequently disrupted, producing a homozygous mutant that was morphologically identical to the parental wild-type strain and that grew at a comparable rate. Deletion of the catalase gene was associated with increased susceptibility to leukocyte-mediated killing of organisms and to decreased virulence for mice in an experimental model of disseminated candidiasis.
(This work was presented in part at the ASM Conference on Candida and Candidiasis: Biology, Pathogenesis, and Management, San Diego, Calif., 24–27 March 1996.)
MATERIALS AND METHODS
Strains, media, and culture conditions.
The C. albicans strains used in this study were ATCC 10261, CAI4, 1006, 4918, and SC5314. Strains ATCC 10261, 1006, 4918, and SC5314 are clinical isolates. CAI4 is a homozygous ura3 strain isogenic to the clinical isolate SC5314 and was originally obtained from W. Fonzi (11). All strains were routinely propagated in YEPD (2% Bacto Peptone [Difco Laboratories, Detroit, Mich.], 1% yeast extract, 2% glucose), Sabouraud-dextrose, or SD (0.7% yeast nitrogen base, 2% glucose) medium at 30°C. Agar was added (2%) for solid media. Yeast cells were induced to form germ tubes in either RPMI 1640 or synthetic amino acid-rich medium (21) at 37°C. Media were supplemented with 50 μg of uridine ml−1 as required. Selection of auxotrophs was on medium containing 5-fluoroorotic acid (5-FOA) as described previously (11). DNA cloning was carried out in Escherichia coli DH5α.
Gene cloning.
Degenerate primers were used in a PCR to amplify a CAT1 gene fragment from C. albicans genomic DNA, which was subcloned into pBluescript SK(+) (Stratagene, La Jolla, Calif.), creating plasmid pCAT01. This PCR fragment was verified as the CAT1 gene by sequence analysis and used to probe C. albicans genomic DNA after digestion with restriction enzymes. Genomic clones were obtained from libraries constructed from genomic DNA of strains 1006 and 4918. The libraries were originally obtained from D. Miller and X. J. Zhao, respectively. Briefly, a C. albicans genomic library was prepared from strain 1006 on a URA3 2μm vector. Strain 4918 was used to construct a XhoI genomic library in λFIXII (48). Fragments isolated from clones were subcloned into pBluescript SK(+) and subsequently analyzed by sequencing both DNA strands.
DNA sequencing.
Sequencing of both DNA strands was carried out by applying the chain termination method of Sanger et al. (36) with the SequiTherm cycle sequencing kit (Epicentre Technologies, Madison, Wis.) and internal labeling with [α-35S]dATP. Synthetic oligonucleotide primers for sequencing were obtained from either Gibco/BRL (Gaithersburg, Md.) or the Biopolymer Laboratory at the Massachusetts Institute of Technology. DNA and amino acid sequences were analyzed by using DNAStar software.
Catalase gene disruption.
A 2,036-bp VspI fragment containing the entire C. albicans CAT1 gene was blunted with Klenow fragment of DNA polymerase I and nucleoside triphosphates and then subcloned into the EcoRV site of pBluescript SK(+) to create plasmid pCAT10. A 1,351-bp BglII-EcoRV fragment was removed from the open reading frame (ORF) of CAT1, followed by addition of BglII linkers and ligation of a BamHI-BglII fragment of plasmid p5921 (1, 14) which contained the C. albicans URA3 gene flanked by direct repeats of the 1.1-kb Salmonella typhimurium hisG gene. The resulting plasmid, designated pCAT20, was fragmented by digestion with restriction endonucleases and used to transform strain CAI4 by a lithium acetate method (12). Selection of Ura3− isolates resulting from cis recombination between the hisG repeats was facilitated by plating transformants onto medium containing 0.1% (wt/vol) 5-FOA and 50 μg of uridine ml−1. The disruption transformation was repeated until a null mutant was generated. Both pre- and post-5-FOA isolates were verified by Southern blot analyses.
Southern and Northern blot analyses.
Standard electrophoretic techniques and formaldehyde RNA gels were employed (35, 37). DNA fragments used as hybridization probes were isolated from agarose gels by using a gel extraction kit (QIAGEN Inc., Santa Clarita, Calif.) and labeled with [α-32P]dCTP (New England Nuclear, Boston, Mass.) by random oligonucleotide priming as described in the manufacturer’s instructions (Boehringer Mannheim, Indianapolis, Ind.). Blotting was carried out with Hybond-N nylon membranes (Amersham, Chicago, Ill.) as described in the manufacturer’s instructions.
Chromosomal location of CAT1.
Chromosomal DNA was resolved by pulsed-field gel electrophoresis with a contour-clamped homogeneous electric field system and transferred to a nylon membrane (46). The chromosomal blot was kindly provided by J. Sturtevant at Georgetown University Medical Center. A 1,156-bp EcoRI fragment of C. albicans CAT1 was labeled with [α-32P]dCTP by the random priming method and used to probe the chromosomal blot by the same methods of Southern hybridization as those described above.
Fungal growth rates.
The growth rates of the parental and mutant strains at 25 and 37°C were compared. To measure yeast-phase growth, both strains were incubated at 25°C with shaking, in YEPD, Lee’s (21), or RPMI 1640 medium. Absorbance at 650 nm was measured after 4 and 8 h and then every 2 h up to 24 h. The ability of yeasts to form hyphae was analyzed by induction of mycelia at 37°C in RPMI 1640 medium. After 2, 3, and 4 h of incubation, organisms were examined microscopically to determine both the relative size of hyphae and the percentage of organisms exhibiting formation of germ tubes.
Hydrogen peroxide sensitivity.
C. albicans hyphae (3 × 106 ml−1) were exposed to H2O2 in concentrations ranging from 0.125 to 4.0 mM. After incubation at 37°C for 1 h, fungal damage was assessed by using the tetrazolium dye (2,3)-bis-(2-methoxy-4-nitro-5-sulfenyl)-(2-H)-tetrazolium-5-carboxanilide (XTT). Hyphae were washed to remove any residual H2O2 and then incubated in 400 μl of 0.5 mg of XTT ml−1 with the addition of 8 μg of coenzyme Q (2,3-dimethoxy-5-methyl-1,4-benzoquinone) ml−1 for 1 h at 37°C. Prior studies established that XTT was reduced only by live organisms, yielding a water-soluble formazan product, which is a sensitive and reproducible indicator of metabolic inactivation (27). Absorbance of supernatants at 450 nm was used to calculate the percentage of damaged hyphae.
Enzymatic assay of catalase activity.
A standard enzymatic assay (42) was used to measure catalase levels of C. albicans parental and mutant strains. Germinated organisms from each strain were disrupted by vortexing with glass beads in 100 mM potassium phosphate buffer followed by centrifugation to obtain cell extracts of the organisms. Catalase activity was measured by the rate of decomposition of H2O2 as measured spectrophotometrically at 240 nm. Enzymatic activity was standardized for protein content.
Assay of fungal damage by PMNs.
PMNs were purified from normal human whole blood by dextran sedimentation followed by Hypaque-Ficoll centrifugation and hypotonic lysis of contaminating erythrocytes (9). C. albicans yeasts were induced to form hyphae by incubation in RPMI 1640 medium, with shaking for 3 h at 37°C. PMNs and serum-opsonized hyphae were incubated together at various ratios for 60 min at 37°C. The reaction of PMNs and hyphae was stopped by the addition of ice-cold H2O followed by centrifugation to ensure lysis of PMNs. Survival of hyphae was measured by reduction of the tetrazolium dye XTT, as described above.
Experimental infections in mice.
Male outbred ICR mice, 4 to 5 weeks old (Harlan-Sprague-Dawley, Indianapolis, Ind.), were housed 7 to 10 per cage. Food and water were given ad libitum, according to National Institutes of Health guidelines. Strains of C. albicans were grown in Sabouraud-dextrose broth at 26°C to a cell density of 107 conidia ml−1. Cells were harvested, washed, and resuspended in sterile H2O. Immunocompetent mice were infected by intravenous inoculation of 0.1 ml of a suspension of 105 to 109 conidia per ml, via the lateral tail veins. Cages were checked daily for dead or moribund mice. Moribund mice were euthanized by CO2 asphyxiation. The lungs, livers, and kidneys were removed from two euthanized mice from each group on days 1, 5, and 32 postinfection. Organs were homogenized, diluted, and plated onto Sabouraud-dextrose agar and incubated for 24 to 48 h at 37°C to determine CFU per organ. Necropsy was performed on lungs, livers, and kidneys of mice from each group to detect the presence of C. albicans. Paraffin sections were prepared from organs preserved in buffered formalin and stained with Grocott’s methenamine silver nitrate and counterstained with hematoxylin and eosin.
Statistical analyses.
The Wilcoxon rank sum test was used to compare survival rates of control and mutant groups of mice. In vivo groups consisted of 7 to 10 animals per inoculum, and virulence studies were performed twice. In vitro experiments were done in triplicate and repeated at least three times. Means ± standard deviations were determined for all data points. Groups were compared by using the Student t test for paired samples.
Nucleotide sequence accession number.
The GenBank accession number for the C. albicans catalase gene CAT1 is U40704.
RESULTS
Cloning of the C. albicans CAT1 gene.
Degenerate PCR primers were designed based on the highly conserved amino acid sequences encoded by genes for catalases in other fungi (Fig. 1). These primers amplified a 411-bp product from genomic DNA of C. albicans ATCC 10261. The PCR product was used to probe C. albicans genomic DNA of several strains after digestion with a variety of restriction enzymes. A 1.2-kb EcoRI fragment was identified in each strain. No other fragments were observed in these digests. An EcoRI-digested C. albicans genomic library was screened by colony hybridization, which resulted in the identification of several positive clones. After selective purification and subcloning of a 5-kb PstI fragment into pBluescript SK(+), the nucleotide sequence of the catalase isolate was determined. Ultimately, since analyses of several clones revealed identical but partial sequences, another library was screened for catalase-positive clones. Sequence analysis of a portion of a 10.5-kb SpeI fragment isolated from a C. albicans 4918 XhoI genomic library in λFIXII confirmed that the complete gene was present. The SpeI fragment was cloned into pBluescript SK(+) and used for sequence analysis.
FIG. 1.
Alignment of the deduced amino acid sequences of the C. albicans catalase (C. alb.) with the catalases of C. tropicalis (C. trop.) and S. cerevisiae (Yeast A and Yeast T). Identical residues are boxed. Dashes indicate gaps in the amino acid sequence when compared to other sequences. Amino acids in boldface type indicate regions used to design degenerate primers. Arrows indicate directions of degenerate PCR primers. Alignment was generated by using MegAlign (DNAStar).
Nucleotide and deduced amino acid sequences of the CAT1 gene.
The nucleotide sequence of the entire catalase gene and flanking sequences both upstream and downstream of the start codon were determined on both strands of a full-length clone by using a series of synthetic oligonucleotides. The DNA sequence containing the putative C. albicans CAT1 homolog contained a single ORF of 1,461 bp with the potential to encode a protein of 487 amino acids, without introns. The first ATG of the ORF is located 12 bases downstream from a putative TATA box, while one in-frame TAA stop codon was observed 24 bases upstream from the proposed start codon. Analysis of the sequence also revealed the presence of a purine nucleotide (A) located at position −3 and another (G) at position +4 (19). These have been identified as important for recognition by eukaryotic ribosomes (19). A TAA translation termination codon was located 13 bases upstream from a possible yeast transcriptional stop signal (47), followed by a putative polyadenylation signal at the 3′ end of the nucleotide sequence. The protein displayed 92.4, 63.2, and 39.6% identities to the amino acid sequences of the catalase of Candida tropicalis, S. cerevisiae catalase A, and S. cerevisiae catalase T, respectively (Fig. 1).
Southern analysis of disrupted alleles of C. albicans CAT1.
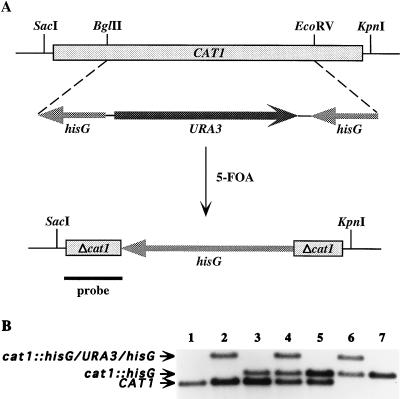
To disrupt the CAT1 alleles, a 1,351-bp BglII/EcoRV fragment of the ORF was replaced by the Ura-blaster cassette (Fig. 2A), a 4-kb BamHI/BglII fragment of p5921 that consists of a functional C. albicans URA3 gene flanked by direct repeats of the Salmonella typhimurium hisG (14). The resulting plasmid was digested with KpnI and SacI and used to transform CAI4, a homozygous ura3 strain isogenic to the clinical isolate SC5314. The repeats of hisG promote homologous cis recombination, resulting in the excision of URA3 and the loss of one copy of hisG, leading to regeneration of Ura3− auxotrophs (14). Several independent Ura3− transformants were selected by resistance to 5-FOA and analyzed by Southern hybridization to verify the loss of one copy of the wild-type CAT1 gene. Analysis revealed that 67% of these primary transformants were heterozygous mutants (designated CADW1). Six of these heterozygous mutants were subjected to a second round of transformation using URA3 as a selectable marker. Southern analysis of 36 secondary transformants after 5-FOA selection resulted in eight mutants that contained only wild-type CAT1, suggesting the possibility that gene conversion with a wild-type allele had led to regeneration of the parental pattern or that there were contaminating wild-type organisms in some of the transformations. The remaining 28 transformants were heterozygous, showing evidence of both wild-type and disrupted CAT1 alleles. Six of these heterozygous mutants (designated CADW2) were subjected to a third round of transformation that yielded 50% homozygous mutants containing no wild-type CAT1 and 50% that still exhibited both wild-type and disrupted alleles after Southern analysis. Homozygous null mutants obtained were designated CADW3. The patterns observed in the autoradiogram of the Southern blot (Fig. 2B) suggested that complete elimination of CAT1 from the wild-type strain required three successive disruptions. After one round of transformation and subsequent 5-FOA selection, lane 3 showed a heterozygous mutant with at least one wild-type allele and one disrupted allele containing a copy of hisG. Lane 5 also showed a heterozygous mutant, obtained after the second round of transformation and 5-FOA selection. Bands on the autoradiogram indicated the presence of a wild-type allele and at least one copy of the disrupted CAT1 allele containing a single copy of hisG. As shown in lanes 6 and 7 of Fig. 2B, no bands were observed at the original location of the wild-type allele. The presence of hisG in each of the disrupted alleles in lanes 3 to 7 (cat1::hisG) was confirmed by hybridization with a hisG probe (data not shown). As expected, the hisG probe also hybridized with cat1::hisG/URA3/hisG bands (lanes 2, 4, and 6) but not with any of the CAT1 bands (lanes 1 to 5). The homozygous null mutant was produced only after three successive rounds of transformation. In addition, four other independent primary transformants that were selected by 5-FOA resistance were found to produce the same patterns on Southern blots with each successive transformation, until formation of null mutants.
FIG. 2.
Sequential disruption of the CAT1 gene of C. albicans by using the Ura-blaster technique. (A) Partial restriction map of the CAT1 allele before and after regeneration of the ura3 genetic marker by 5-FOA selection. The plasmid used for the gene disruption construct encompassed the entire 2,036 bp containing the entire 1,461-kb CAT1 ORF, indicated by the boxed and shaded rectangle at the top. Restriction sites BglII, EcoRV, KpnI, and SacI are indicated. A 1,351-bp BglII-EcoRV fragment of CAT1 was replaced by the his-URA-his construct; this is shown below the CAT1 ORF. The his-URA-his construct consisted of URA3 flanked by a hisG insert at each end. Beneath this construct are shown the final transformants selected by 5-FOA, which contained the disrupted CAT1 gene, missing the 1,351-bp BglII-EcoRV fragment and retaining one copy of hisG (1,149 bp) flanked by small fragments of the disrupted CAT1 gene (Δcat1). The solid black line at the bottom represents the location of a portion of CAT1 used as a 32P-labeled probe for Southern analysis. It is important to note that the lengths of each of the fragments are not drawn to scale. (B) Southern analysis of the parental, heterozygous, and homozygous mutants after digestion with KpnI/SacI, followed by hybridization using a fragment of CAT1 as a probe. Lanes: 1, CAI4 parental DNA; 2 to 7, pre- and post-5-FOA isolates after the first (lanes 2 and 3), second (lanes 4 and 5), and third (lanes 6 and 7) rounds of transformation. Not shown in this figure, Southern analysis after hybridization with a hisG probe confirmed that hisG was present in each of the cat1::hisG bands (lanes 3 to 7) and cat1::hisG/URA3/hisG bands (lanes 2, 4, and 6) but not with any of the CAT1 bands (lanes 1 to 5).
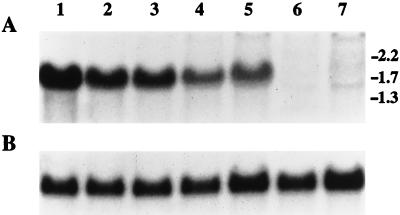
Northern analysis of C. albicans CAT1 expression.
Expression levels of CAT1 were compared for the heterozygous and homozygous mutants by preparing total RNA from wild-type and each mutant strain followed by Northern hybridization and autoradiography. There was a progressive decrease in expression of CAT1 after each transformation (Fig. 3). The size of the transcript was approximately 1.8 kb, based on ethidium bromide-stained standards. The expression of CAT1 in the yeast phase of the wild-type strain was also compared to expression in the mycelial or hyphal phase. Preliminary data from this experiment indicated that both yeast and hyphae expressed the message and suggested a slight relative increase in the expression of CAT1 in the yeast phase of growth (data not shown).
FIG. 3.
Northern blot analysis of CAT1 expression in strain CAI4 and in CAT1 mutants. All isolates were grown as yeast cells in YEPD medium at 30°C. Total RNA was extracted from cultures, and 10 μg of each sample was analyzed by Northern blot hybridization. (A) CAT1 expression was detected by using a 1,156-bp EcoRI fragment of CAT1 as a hybridization probe. Lanes: 1, CAI4 parental DNA; 2 and 3, pre- and post-5-FOA isolates of the heterozygous mutant after first transformation; 4 and 5, pre- and post-5-FOA isolates of the heterozygous mutant after second transformation; 6 and 7, pre- and post-5-FOA isolates of the homozygous mutant after third transformation. The sizes (in kilobases) and positions of electrophoretic RNA standards are indicated on the right. (B) Control hybridization using a 1.4-kb ACT1 fragment from plasmid pLV4 as a probe to demonstrate equal loading in each lane.
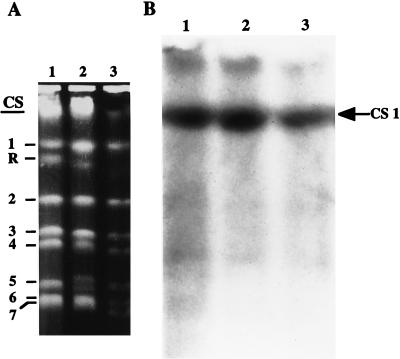
Chromosomal location of C. albicans CAT1.
Separations of C. albicans 4918, ATCC 10261, and CAI4 by a contour-clamped homogeneous electric field system were probed under conditions of high stringency with a 1.2-kb EcoRI fragment of C. albicans CAT1, which hybridized to chromosome 1 in all three strains (Fig. 4).
FIG. 4.
Electrophoretic karyotypes of C. albicans CAI4, ATCC 10261, and 4918 were probed with the 1,156-bp EcoRI fragment of CAT1. Numbers to the left of the photograph of the ethidium bromide-stained gel (A) indicate the positions of the C. albicans chromosomes. The arrow to the right of the Southern blot (B) indicates the location of chromosome 1.
Phenotypic analysis of the CAT1 disruptant.
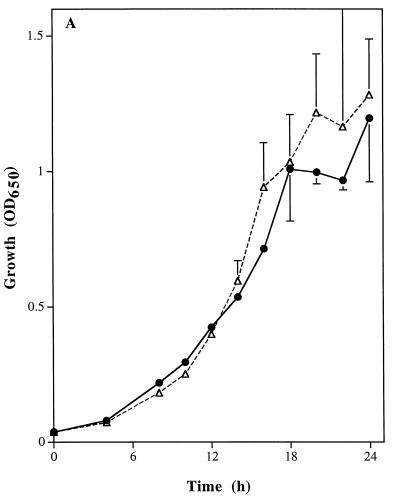
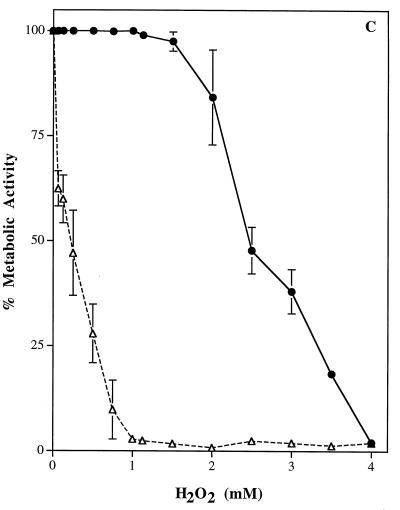
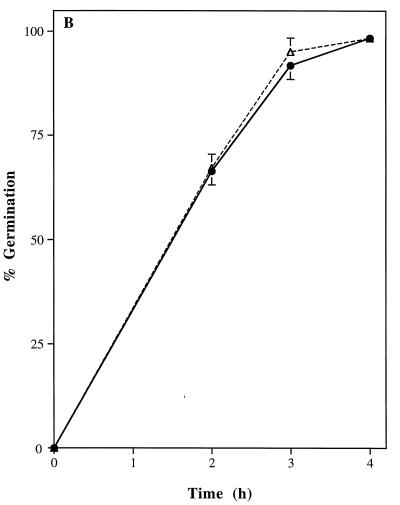
There were no significant differences in growth rates of the wild-type parental strain, SC5314, and the homozygous CAT1 mutant, strain CADW3. Rates of yeast-phase growth were compared by incubating both strains at 25°C and measuring absorbance at 650 nm (Fig. 5A). Hyphal germination and rates of extension formation were also compared. Both strains were induced to form germ tubes in RPMI 1640 medium at 37°C. Comparisons of percentages germinated (Fig. 5B) and lengths of hyphae (data not shown) revealed no differences between the strains after 2, 3, and 4 h of incubation. Microscopic examination of the strains revealed no evident morphological differences between SC5314 and CADW3. Conversely, compared to the parental SC5314 strain, the catalase-deficient mutant strain, CADW3, had a decreased survival rate upon exposure to various concentrations of hydrogen peroxide (Fig. 5C).
FIG. 5.
Phenotypic comparisons between the parental strain, SC5314 (•), and the homozygous mutant strain, CADW3 (▵). (A) Both strains were grown in RPMI 1640 medium at 25°C. Growth rates were compared by measuring the optical density at 650 nm (OD650) after 4 and 8 h and then every 2 h up to 24 h. (B) Both strains were grown in RPMI 1640 medium at 37°C. The percentage of organisms exhibiting phase change from yeast to mycelia was determined by microscopic examination after 2, 3, and 4 h of incubation. (C) Sensitivity to H2O2 was measured by incubating yeast-phase organisms from both strains with 0.125 to 4.0 mM H2O2 for 60 min followed by washing of the organisms. The percentage of metabolically active organisms remaining was determined by measuring the absorbance at 450 nm after reduction of XTT, as described in Materials and Methods.
Quantitation of catalase by enzymatic assay.
Catalase activity from each strain was determined by H2O2 decomposition. Each successive disruption of a CAT1 allele was associated with a progressive decrease in catalase enzymatic activity per milligram of protein in hyphal cell extracts from each strain. Relative activities of catalase in hyphae from the heterozygous mutants lacking one or two alleles of wild-type catalase (strains CADW1 and CADW2, respectively) were reduced significantly to 62.6% ± 13.7% and 37.2% ± 5.0%, respectively, of catalase activity in the parental strain (P < 0.001 by the Student t test for paired samples). No catalase activity was detected in the homozygous null mutant (CADW3).
In vitro assay of neutrophil damage.
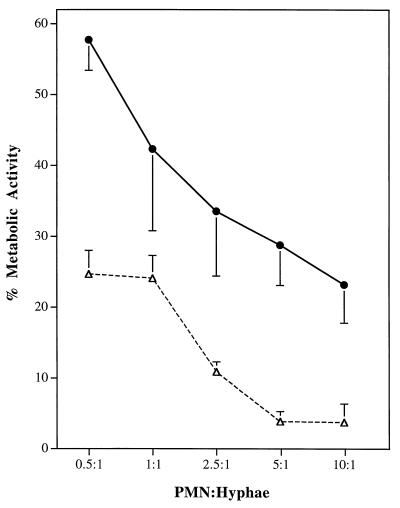
C. albicans hyphae from both the parental strain and the catalase-deficient homozygous mutant strain were damaged during incubations with neutrophils, as indicated by a reduced ability to metabolize the tetrazolium dye XTT. When hyphae were mixed for 1 h with neutrophils at PMN:hypha ratios ranging from 0.5:1 to 10:1, the metabolic activity of catalase-deficient organisms was significantly lower than that of the parental strain (Fig. 6). In experiments done at a PMN:hypha ratio of 25:1, the metabolic activities of both the heterozygous and homozygous mutants also were significantly less than that of the parental strain (data not shown), although the differences were not as striking as those seen at the lower PMN:hypha ratios.
FIG. 6.
Effect of C. albicans CAT1 deletion on susceptibility to damage by PMNs. Hyphae of both the parent strain, SC5314 (•), and the homozygous mutant strain, CADW3 (▵), were incubated at 37°C for 60 min with PMNs. Percentages of metabolically active hyphae were determined by measuring the absorbance of supernatants at 450 nm after reduction of XTT, as described in Materials and Methods.
Experimental infections in mice.
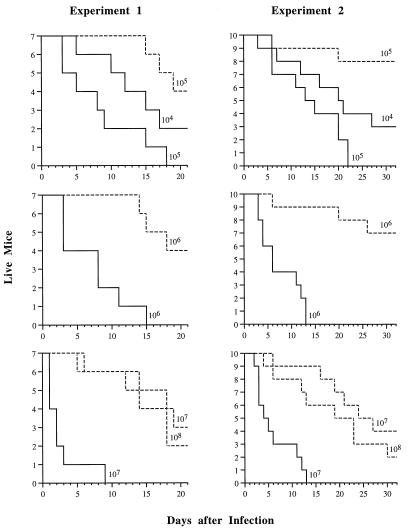
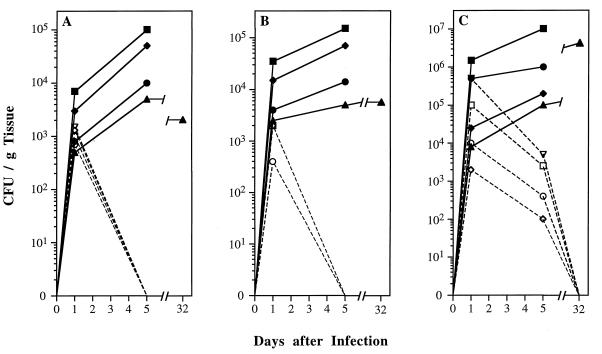
To test the effect of the CAT1 deletion on virulence in an animal model, strains SC5314 and CADW3 (Δcat1) were used to infect immunocompetent mice. Since it has been shown that virulence is decreased by the ura3 mutation (18, 25, 32, 40), only the Ura+ strain of the homozygous null mutant, isolated prior to treatment with 5-FOA, was used to infect mice. For each inoculum of organisms tested, mice infected with the catalase-deficient CADW3 strain survived significantly longer than did mice infected with the wild-type SC5314 parental strain (P < 0.001 by Wilcoxon rank sum test for all inocula in each of two separate experiments). In vivo experiments were performed twice with similar results. Data from both experiments are shown in Fig. 7. In experiment 2, an inoculum of 107 CFU of the wild-type strain produced lethal infections in 100% of the mice within 13 days, while 40% of mice infected with an equal inoculum of the CAT1 mutant remained alive at day 30. With a smaller inoculum of 105 CFU, all mice infected with the wild-type strain died within 22 days, compared with an 80% survival rate (at day 30) of mice inoculated with the mutant strain (experiment 2). Quantitative cultures of lungs, livers, and kidneys from two euthanized mice in each group at each time point (Fig. 8) revealed that clearance of C. albicans from the lungs and the liver was achieved by day 5 postinoculation in mice infected with the catalase-deficient CADW3 strain. In contrast, live organisms persisted for the entire 32-day duration of the experiment in mice infected with sublethal doses of SC5314 (Fig. 8A).
FIG. 7.
Virulence of SC5314 and the CAT1 disruptant in an immunocompetent mouse model. Mice (7 to 10 per group) were inoculated with 104, 105, 106, 107, or 108 CFU of the parental strain, SC5314 (solid lines), or the homozygous mutant strain, CADW3 (dashed lines), via the lateral tail vein. The size of the inoculum is indicated at the end of each plotted line. The time of death (days after infection) was recorded. The results of two separate experiments are shown.
FIG. 8.
Clearance of C. albicans from tissue after infection with strain SC5314 (solid lines) or the catalase-deficient mutant, CADW3 (dashed lines). Colony counts were done to determine average numbers of CFU per gram of tissue from the lungs (A), the liver (B), and the kidney (C). Inoculation of mice is described in the legend to Fig. 7. Closed symbols indicate the parental strain, SC5314, while open symbols represent the homozygous mutant strain, CADW3. Inocula consisted of 104 (▴ and ▵), 105 (⧫ and ◊), 106 (• and ○), 107 (▪ and □), and 108 (▾ and ▿) conidia.
The fact that organisms were recovered from organs 1 day after infection indicated that the catalase-deficient mutant strain, CADW3, was capable of in vivo survival. While clearance of organisms from the kidneys took much longer to achieve, by day 32 postinoculation, no C. albicans organisms were detected in the kidneys of mice infected with CADW3, while organism counts in the kidneys of mice infected with SC5314 had increased substantially from the original inoculum (Fig. 8C). Organisms cultured from the kidneys of mice infected 5 days previously with the catalase-deficient mutant strain evidenced no catalase activity. In contrast, the parental strain retained catalase activity after culture from infected murine kidneys. Histopathological specimens examined microscopically revealed far fewer organisms in organs of mice that had been infected with the catalase-deficient strain. In addition, there was no evidence of clumped organisms, suggesting that perceived differences in colony counts were real and not attributable to selective aggregation of the mutant Candida strain.
It has not been possible, so far, to restore active CAT1 to mutants despite the use of multiple techniques. Therefore, an additional experiment using heterozygote strains was performed to further investigate whether certain other factors besides CAT1 inactivation might have reduced the virulence of homozygous null mutants. It also was conceivable that spontaneous in vivo excision of URA3 with reversion to nonpathogenic Ura− auxotrophic mutants might have occurred. Groups of mice were challenged with 105 C. albicans organisms from either the parental strain (SC5314), the CADW1 strain (with a single deletion of CAT1), the CADW2 strain (with a double deletion of CAT1), or the CADW3 strain (with a triple CAT1 deletion; Δcat1). Mice infected with strain CADW1, the likeliest spontaneous revertant, had a median survival after infection of 8.5 days, not significantly different from the survival after infection with parental strain SC5314 (6 days, P = 0.156 by Wilcoxon rank sum test). By comparison, median survival times were 12 days with the double deletion CAT1 (CADW2) strain and 13 days with the triple deletion mutant strain (CADW3; Δcat1) in this experiment. Mice infected with strain CADW1 also had significantly more organisms in kidneys cultured 5 days after infection than did mice infected with either strain CADW2 or strain CADW3. These data further supported the relationship between reduced virulence and the loss of CAT1. Differences in virulence between the homozygous null mutant and a heterozygous mutant strain with URA3 at the same site also made it unlikely that instability of the URA3 marker caused attenuation of virulence.
DISCUSSION
These experiments describe the first isolation and derived sequence of the C. albicans CAT1 gene by use of a PCR probe constructed on the basis of homologous sequences of other fungal catalases. The predicted amino acid sequence encoded by the gene showed a high level of identity to the sequences of catalases from other fungi, suggesting that catalases are highly conserved.
Sequential disruptions of three apparent copies of the gene were required for total elimination of expression and enzymatic activity of catalase. Since C. albicans has been shown to be a diploid organism by others (31, 33), it seems possible that an additional copy of the gene may have been introduced during early transformation. Alternatively, the parental strain may have had a duplication of part or all of chromosome 1, on which the catalase gene was localized. Others have also reported requiring three successive disruptions to eliminate activity of a chitin synthase in C. albicans SGY243 (14). Our data did indicate that each successive disruption of a catalase allele resulted in an incremental decrease in catalase enzymatic activity. Both enzymatic assays of catalase activity and Northern blot analyses of the two heterozygous and the homozygous mutants revealed a progressive decrease in expression of CAT1.
Our data showed that deletion of CAT1 did not alter the phenotype or morphology of the strain but did dramatically attenuate the ability of the organism to resist damage by human leukocytes in vitro. In addition, organisms that totally lacked catalase activity had a significantly reduced capacity to cause progressive infection in vivo. These differences could not be explained by altered growth or germination of catalase-deficient mutants. In multiple in vitro assays measuring both yeast-phase growth and extent and time required for germination, there were no discernible differences in growth rate or hyphal development between the parental strain with normal catalase activity and the completely catalase-deficient mutant strain. Thus, CAT1 did not appear to be an essential gene for growth or germination.
Catalase-deficient mutant organisms also retained the ability to survive in vivo after infection of mice, although they were ultimately cleared from tissues of surviving animals. Combined with the increased susceptibility of catalase-deficient organisms to injury by neutrophils, the reduced ability of CAT1 disruptants to cause progressive murine disseminated candidiasis suggests that catalase may contribute to the virulence of C. albicans. Our data cannot eliminate the possibility that integration of transforming DNA occurred at an ectopic site, potentially altering the activity of an unrelated virulence factor. However, it seems unlikely that this would have fortuitously yielded an identical band of expected size on Southern blots. Moreover, although many C. albicans mutants have been constructed by other investigators, these mutants have behaved as expected and there are no reports, so far, suggesting that transformation of Candida induced mutations at other sites. While this does not eliminate the concern, it at least suggests that secondary genotypic alterations do not occur frequently. There also remains a possibility that inadvertent introduction of duplicated genes during the transformation process, if it occurred, might conceivably have changed dosages of other genes controlling potential virulence factors. Although direct experimental evidence is necessary to address these and other issues, current published data suggest that secondary genetic alterations do not occur at high frequency. Even so, additional studies will be required to definitively determine the role of catalase as a virulence factor.
Since auxotrophic strains of C. albicans have exhibited greatly attenuated virulence (18, 25, 32, 40), our studies were done with the prototrophic Ura+ isolate of the homozygous null mutant, recovered prior to 5-FOA treatment. The reduced virulence of the homozygous null mutant suggests that the CAT1 gene is expressed during the course of infection and that this mutant strain has a reduced capacity to cause infection. Histological examination of lung, liver, and kidney tissues revealed high levels of wild-type hyphae and yeast but few mutant organisms. Although the CAT1 deletion strain (CADW3) initially survived in mice and was maintained in murine kidneys for at least 5 days, complete clearance of the organism was eventually achieved. Invasion of the kidneys is often associated with late fatalities in immunocompetent mice since C. albicans has been known to persist and grow preferentially in hyperosmolar areas of the kidney during disseminated infections (29). Clearance of the null mutant strain from lung and liver tissue was much faster, with no organisms being detected in these tissues as early as day 5 postinoculation. One of the vagaries of colony counts from infected organs is that organisms may be clumped within infected tissues, leading to falsely low colony counts. This was not the case in these studies of the CAT1-deficient null mutant, as evidenced by the histological examination of organs containing very few organisms, which gave no indication of any clumping.
It has also been suggested that excision of the URA3 gene might produce spontaneous reversion to auxotrophic Ura− mutants. Were that to occur, any auxotrophs would likely be cleared quickly since Ura− strains cannot scavenge enough uracil from surrounding tissues to ensure survival. The fact that catalase-negative organisms caused progressive, ultimately fatal disseminated candidiasis in some mice indicated that at least some of these organisms remained Ura+. In addition, survival of mice challenged with the most likely revertant mutant strain in our studies, CADW1, with only a single deletion of a CAT1 allele, was not significantly different from survival after infection with the parental strain, SC5314.
The relative importance of the catalase gene in the pathogenicity of C. albicans may also depend upon the immune status of the host and the site of infection. The presence of catalase alone does not necessarily have any relevance to the virulence of a fungus since most fungi, including S. cerevisiae, are nonpathogens, causing significant infection in humans only extremely rarely. As with many postulated virulence factors, the CAT1 gene may play a role in the infectious process of C. albicans but seems highly unlikely to be sufficient by itself to account for the ability of the organism to cause disease.
Thus, our data raise the possibility that catalase may be one of several factors that significantly contribute to the virulence of C. albicans. However, definitive evidence will require restoration of the gene and consequent enzymatic activation together with a return of wild-type virulence. The ability to reintroduce such functional, stably active genes has been elusive for many other investigators besides ourselves, but experiments to restore the catalase gene to the mutant strain are in progress. Successful restoration of virulence would address potentially broader effects of catalase gene disruption on gene regulation or additional factors unrelated to deletion of the specific gene itself. However, results of any of these approaches will require extremely cautious interpretations.
Meanwhile, the importance of catalase has already been established in the protection of a variety of organisms against oxidants released by host cells (4, 15, 17) and in its potential ability to regulate growth and development, including phase transition, in some fungi (17, 26, 28, 43). Nevertheless, the present data are not sufficient to establish a definitive role for catalase in the pathogenesis of candidiasis in vivo. However, the clear-cut effects of catalase gene inactivation on sensitivity to neutrophils provide important new evidence supporting the need for further study.
Disseminated candidiasis in nonneutropenic patients continues to be an increasing problem in hospitalized patients (10). Even in neutropenic patients, monocytes and macrophages have been presumed to provide significant, if imperfect, defenses against Candida (23). If future studies show that restoration of catalase activity restores virulence, this may open a potential avenue for combating systemic candidiasis, since even lifelong genetic acatalasemia in humans has not caused severe systemic deleterious effects (30, 45).
ACKNOWLEDGMENTS
This work was supported by grant AI-15338 (to R.D.D.) from the National Institutes of Health and, in part, by the Collaborative Medical Mycology Research Program (Pfizer, Inc., Roerig Division; Phytera; Scriptgen Pharmaceuticals; and the Section of Infectious Diseases, Boston Medical Center).
We thank Xiuping Liu for invaluable assistance with the animal studies.
Footnotes
Publication 011 from the Collaborative Medical Mycology Research Program.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck-Sague C M, Jarvis W R. National nosocomial infections surveillance system. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990. J Infect Dis. 1993;167:1247–1251. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- 3.Bulawa C E, Miller D W, Henry L K, Becker J M. Attenuated virulence of chitin-deficient mutants of Candida albicans. Proc Natl Acad Sci USA. 1995;92:10570–10574. doi: 10.1073/pnas.92.23.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen G, Rapatz W, Ruiz H. Sequence of the Saccharomyces cerevisiae CTA1 gene and amino acid sequence of catalase A derived from it. Eur J Biochem. 1988;176:159–163. doi: 10.1111/j.1432-1033.1988.tb14263.x. [DOI] [PubMed] [Google Scholar]
- 5.Diamond R D, Clark R A. Damage to Aspergillus fumigatus and Rhizopus oryzae hyphae by oxidative and nonoxidative microbicidal products of human neutrophils in vitro. Infect Immun. 1982;38:487–495. doi: 10.1128/iai.38.2.487-495.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diamond R D, Clark R A, Haudenschild C C. Damage to Candida albicans hyphae and pseudohyphae by the myeloperoxidase system and oxidative products of neutrophil metabolism in vitro. J Clin Invest. 1980;66:908–917. doi: 10.1172/JCI109958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diamond R D, Krzesicki R. Mechanisms of attachment of neutrophils to Candida albicans pseudohyphae in the absence of serum, and of subsequent damage to pseudohyphae by microbicidal processes of neutrophils in vitro. J Clin Invest. 1978;61:360–369. doi: 10.1172/JCI108946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamond R D, Krzesicki R, Jao W. Damage to pseudohyphal forms of Candida albicans by neutrophils in the absence of serum in vitro. J Clin Invest. 1978;61:349–359. doi: 10.1172/JCI108945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diamond R D, Lyman C A, Wysong D R. Disparate effects of interferon-γ and tumor necrosis factor-α on early neutrophil respiratory burst and fungicidal responses to Candida albicans hyphae in vitro. J Clin Invest. 1991;87:711–720. doi: 10.1172/JCI115050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards J E. Invasive Candida infections. Evolution of a fungal pathogen. N Engl J Med. 1991;324:1060–1062. doi: 10.1056/NEJM199104113241511. . (Editorial.) [DOI] [PubMed] [Google Scholar]
- 11.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorman J A, Chen W, Gorman J W. Repeated use of GAL1 for gene disruption in Candida albicans. Genetics. 1991;129:19–24. doi: 10.1093/genetics/129.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gow N A R, Robbins P W, Lester J W, Brown A P J, Fonzi W A, Chapman T, Kinsman O S. A hyphal-specific chitin synthase gene (CHS2) is not essential for growth, dimorphism or virulence of Candida albicans. Proc Natl Acad Sci USA. 1994;91:6216–6220. doi: 10.1073/pnas.91.13.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartig A, Ruis H. Nucleotide sequence of the Saccharomyces cerevisiae CTT1 gene and the deduced amino-acid sequence of yeast catalase T. Eur J Biochem. 1986;160:487–490. doi: 10.1111/j.1432-1033.1986.tb10065.x. [DOI] [PubMed] [Google Scholar]
- 16.Jiang W, Gerhold D, Kmiec E B, Hauser M, Becker J M, Koltin Y. The topoisomerase I gene from Candida albicans. Microbiology. 1997;143:377–386. doi: 10.1099/00221287-143-2-377. [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki L, Wysong D, Diamond R, Aguirre J. Two divergent catalase genes are differentially regulated during Aspergillus nidulans development and oxidative stress. J Bacteriol. 1997;179:3284–3292. doi: 10.1128/jb.179.10.3284-3292.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirsch D R, Whitney R R. Pathogenicity of Candida albicans auxotrophic mutants in experimental infections. Infect Immun. 1991;59:3297–3300. doi: 10.1128/iai.59.9.3297-3300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 20.Kwon-Chung K J, Lehman D, Good C, Magee P T. Genetic evidence for role of extracellular proteinase in virulence of Candida albicans. Infect Immun. 1985;49:571–575. doi: 10.1128/iai.49.3.571-575.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee K L, Buckley H R, Campbell C C. An amino acid liquid synthetic medium for the development of the mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 22.Lehrer R I, Jan R G. Interaction of Aspergillus spores with human leukocytes and serum. Infect Immun. 1970;1:345–350. doi: 10.1128/iai.1.4.345-350.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levitz S M. Overview of host defenses in fungal infections. Clin Infect Dis. 1992;14:S37–S42. doi: 10.1093/clinids/14.supplement_1.s37. [DOI] [PubMed] [Google Scholar]
- 24.Macdonald F, Odds F C. Virulence for mice of a proteinase-secreting strain of Candida albicans and a proteinase-deficient mutant. J Gen Microbiol. 1983;129:431–438. doi: 10.1099/00221287-129-2-431. [DOI] [PubMed] [Google Scholar]
- 25.Manning M, Snoddy C B, Fromtling R A. Comparative pathogenicity of auxotrophic mutants of Candida albicans. Can J Microbiol. 1984;30:31–35. doi: 10.1139/m84-005. [DOI] [PubMed] [Google Scholar]
- 26.Marchler G, Schüller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by a protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meshulam T, Levitz S M, Christin L, Diamond R D. A simplified new assay for assessment of fungal cell damage with the tetrazolium dye, (2,3)-bis-(2-methoxy-4-nitro-5-sulphenyl)-(2-H)-tetrazolium-5-carboxanilide (XTT) J Infect Dis. 1995;172:1153–1156. doi: 10.1093/infdis/172.4.1153. [DOI] [PubMed] [Google Scholar]
- 28.Navarro R E, Stringer M A, Hansberg W, Timberlake W E, Aguirre J. CatA, a new Aspergillus nidulans gene encoding a developmentally regulated catalase. Curr Genet. 1996;29:352–359. [PubMed] [Google Scholar]
- 29.Odds F C. Candida and candidosis: a review and bibliography. 2nd ed. London, United Kingdom: Bailliere Tindall; 1988. [Google Scholar]
- 30.Ogata M. Acatalasemia. Hum Genet. 1991;86:331–340. doi: 10.1007/BF00201829. [DOI] [PubMed] [Google Scholar]
- 31.Olaiya A F, Sogin S J. Ploidy determination of Candida albicans. J Bacteriol. 1979;140:1043–1049. doi: 10.1128/jb.140.3.1043-1049.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polak A. Virulence of Candida albicans mutants. Mycoses. 1992;35:9–16. doi: 10.1111/j.1439-0507.1992.tb00813.x. [DOI] [PubMed] [Google Scholar]
- 33.Riggsby W S, Torres-Bauza L J, Wills J W, Townes T M. DNA content, kinetic complexity, and the ploidy question in Candida albicans. Mol Cell Biol. 1982;2:853–862. doi: 10.1128/mcb.2.7.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross I K, DeBernardis F, Emerson G W, Cassone A, Sullivan P A. The secreted aspartate proteinase of Candida albicans: physiology of secretion and virulence of a proteinase-deficient mutant. J Gen Microbiol. 1990;136:687–694. doi: 10.1099/00221287-136-4-687. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saporito S M, Sypherd P S. The isolation and characterization of a calmodulin gene (CMD1) from the dimorphic fungus Candida albicans. Gene. 1991;106:43–49. doi: 10.1016/0378-1119(91)90564-r. [DOI] [PubMed] [Google Scholar]
- 38.Sarthy A V, McGonigal T, Coen M, Frost D J, Meulbroek J A, Goldman R C. Phenotype in Candida albicans of a disruption of the BGL2 gene encoding a 1,3-β-glucosyltransferase. Microbiology. 1997;143:367–376. doi: 10.1099/00221287-143-2-367. [DOI] [PubMed] [Google Scholar]
- 39.Schaffner A, Douglas H, Braude A. Selective protection against conidia by mononuclear and against mycelia by polymorphonuclear phagocytes in resistance to Aspergillus: observations on these two lines of defense in vivo and in vitro with human and mouse phagocytes. J Clin Invest. 1982;69:617–631. doi: 10.1172/JCI110489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shepherd M G. Pathogenicity of morphological and auxotrophic mutants of Candida albicans in experimental infections. Infect Immun. 1985;50:541–544. doi: 10.1128/iai.50.2.541-544.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sobel J D, Vazquez J. Candidemia and systemic candidiasis. Semin Respir Infect. 1990;5:123–137. [PubMed] [Google Scholar]
- 42.Ueda M, Mozaffar S, Tanaka A. Catalase from Candida boidinii 2201. Methods Enzymol. 1990;188:463–467. doi: 10.1016/0076-6879(90)88074-k. [DOI] [PubMed] [Google Scholar]
- 43.Veenhuis M, Mateblowski M, Kunau W H, Harder W. Proliferation of microbodies in Saccharomyces cerevisiae. Yeast. 1987;3:77–84. doi: 10.1002/yea.320030204. [DOI] [PubMed] [Google Scholar]
- 44.Wagner D K, Collins-Lech C, Sohnle P G. Inhibition of neutrophil killing of Candida albicans pseudohyphae by substances which quench hypochlorous acid and chloramines. Infect Immun. 1986;51:731–735. doi: 10.1128/iai.51.3.731-735.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wen J K, Osumi T, Hashimoto T, Ogata M. Molecular analysis of human acatalasemia. Identification of a splicing mutation. J Mol Biol. 1990;211:383–393. doi: 10.1016/0022-2836(90)90359-T. [DOI] [PubMed] [Google Scholar]
- 46.Wickes B L, Golin J E, Kwon-Chung K J. Chromosomal rearrangement in Candida stellatoidea results in a positive effect on phenotype. Infect Immun. 1991;59:2480–2484. doi: 10.1128/iai.59.5.1762-1771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zaret K S, Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982;28:563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- 48.Zhao X-J, Cihlar R L. Isolation and sequence of the Candida albicans FAS1 gene. Gene. 1994;147:119–124. doi: 10.1016/0378-1119(94)90050-7. [DOI] [PubMed] [Google Scholar]