Abstract
The importance of the two major extracellular enzymes of Aeromonas salmonicida, glycerophospholipid:cholesterol acyltransferase (GCAT) and a serine protease (AspA), to the pathology and mortality of salmonid fish with furunculosis had been indicated in toxicity studies. In this study, the genes encoding GCAT (satA) and AspA (aspA) have been cloned and mutagenized by marker replacement of internal deletions, and the constructs have been used for the creation of isogenic satA and aspA mutants of A. salmonicida. A pSUP202 derivative (pSUP202sac) carrying the sacRB genes was constructed to facilitate the selection of mutants. The requirement of serine protease for processing of pro-GCAT was demonstrated. Processing involved the removal of a short internal fragment. Surprisingly, pathogenicity trials revealed no major decrease in virulence of the A. salmonicida ΔsatA::kan or A. salmonicida ΔaspA::kan mutants compared to the wild-type parent strains when Atlantic salmon (Salmo salar L.) were challenged by intraperitoneal injection. Moreover, using a cohabitation model, which more closely mimics the natural disease, there was also no significant decrease in the relative cumulative mortality following infection with either of the deletion mutants compared to the parent strain. Thus, although these two toxins may confer some competitive advantage to A. salmonicida, neither toxin is essential for the very high virulence of A. salmonicida in Atlantic salmon. This first report of defined deletion mutations within any proposed extracellular virulence factor of A. salmonicida raises crucial questions about the pathogenesis of this important fish pathogen.
The fish disease furunculosis derived its name from the characteristic lesions or furuncules formed on the surface of fish as a result of chronic infection with Aeromonas salmonicida. Early demonstration that injection of crude preparations of products secreted from the bacterium were lethal and reproduced typical symptoms of the disease (including darkening of the skin, hemorrhaging at the base of the fins, a bleeding vent externally, and necrotic lesions and general liquefaction of tissues internally) led to a search for the toxin(s) responsible for furunculosis. Formation of classic hemorrhagic furuncules has been reproduced following intramuscular injection of a combination of the two major secreted enzymes, the AspA serine protease (9) and glycerophospholipid:cholesterol acyltransferase (7) (GCAT) (14, 18). More importantly, a high-molecular-weight GCAT– lipopolysaccharide (LPS) complex (50% lethal dose [LD50] of 45 ng of protein/g of fish) is considered to be the major lethal toxin secreted by A. salmonicida (4, 14, 24). Alone, the GCAT-LPS complex resulted in only very limited tissue necrosis, and the cause of death appeared to be respiratory failure. Thus, the above two extracellular enzymes have been implicated as the most important secreted products in A. salmonicida pathogenesis. In addition to tissue destruction, proposed roles of serine protease include nutrient acquisition (14, 29) and activation of protoxins, in particular GCAT (13, 21).
GCAT is an unusual bacterial lipase in that it can use cholesterol as an acyl acceptor (7). The protein is synthesized as a preproenzyme of 37.4 kDa (21). Following proteolytic activation of pro-GCAT, the surface activity of the enzyme increases, permitting hydrolysis of membrane phospholipids (19), in turn resulting in lysis of fish erythrocytes, although not directly of mammalian erythrocytes (13, 14). The same enzyme activity has been detected in spent culture supernatants from several members of the closely related genera Aeromonas and Vibrio (25). Recent computer analysis suggests that this enzyme may belong to a new family of lipases (5, 16). Although the biochemical properties and activity of A. salmonicida GCAT (AsGCAT) (7) and later A. hydrophila GCAT (AhGCAT) (5, 19, 21) have been studied in detail, no GCAT-deficient mutant has yet been created to ascertain the importance of this unusual enzyme to the pathogenesis of any bacterium. Serine protease-deficient mutant strains created by chemical mutagenesis have been used to examine the role of this protease in virulence, with conflicting results (12, 29). Indeed, in view of the economic importance of A. salmonicida to the salmonid farming industry, surprisingly little is known about the factors essential to the pathogenesis of this microorganism.
In this study, isogenic satA and aspA deletion mutants of A. salmonicida were created to define the relative roles of these two secreted toxins in furunculosis. Further evidence to support the role of serine protease in pro-GCAT processing was provided. However, the unexpected result that deletion of GCAT had little effect on virulence underscores the need for more extensive studies with defined genetic mutants to identify factors important to A. salmonicida pathogenesis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The properties and source of all bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or relevant characteristic | Source or reference |
|---|---|---|
| Strains | ||
| A. salmonicida | ||
| AS1102 | Type strain, A-layer negative | NCIMB 1102 |
| 644Rb | Virulent isolate, Nalr, A-layer positive | 35 |
| MT1326 | Virulent isolate, A-layer positive | 3 |
| CB3 | Undefined Tn5 mutant, decreased synthesis of extracellular proteins including GCAT and serine protease | 6 |
| 644RbΔG | ΔsatA::kan, serine protease, A-layer positive | This study |
| 644RbΔSP | ΔaspA::kan, pro-GCAT, A-layer positive | This study |
| MT1326ΔG | ΔsatA::kan, serine protease, A-layer positive | This study |
| MT1326ΔSP | ΔaspA::kan, pro-GCAT, A layer positive | This study |
| E. coli | ||
| HB101 | Δ(gpt-proA)62 leuB6 supE44 ara-14 galK2 lacY1 Δ(mcrC-mrr) rpsL20 xyl-5 mtl-1 recA13 | 1 |
| DH5α | recA1gyrA relA1 Δ(lacIZYA-argF)U169 deoR [φ80dlac lacZΔM15] | 1 |
| S17-1 | thi pro hsdR hsdM+ recA [RP4 2-Tc::Mu-Km::Tn7 (Tpr Smr) Tra+] | 32 |
| Plasmids | ||
| pUC18/19 | Cloning and sequencing | 1 |
| pBSK | Bluescript SKII, cloning and sequencing | Stratagene |
| M13mp18/19 | Sequencing and mutagenesis | 1 |
| pSUP202 | Mobilizable suicide vector (Ampr Tetr Camr ColE1 ori Mob+) | 32 |
| pSM20 | Levansucrase confers sucrose sensitivity (sacRB+ Ampr) | 31 |
| pRAT1 | Kanamycin resistance determinant, lacking transcriptional terminator, on 1.436-kb HaeII fragment from Tn903 | D. Evans, University of Reading |
| pMMB66HE | Broad-host-range expression vector (Ampr) | 17 |
| pSUP202sac | Mobilizable suicide vector, confers Sucs (Mob+sacRB+ Tetr) | This study |
| pUC18G | satA on 1.2-kb PstI fragment, cloned in PstI site of pUC18 in reverse orientation to plac | This study |
| pUC19S | satA on 5.5-kb SacI fragment in SacI site of pUC19 | This study |
| pUC19SΔG | pUC19S with Kan cassette replacing an internal fragment of satA | This study |
| pSUP202sacΔG | SacI fragment from pUC19SΔsatA::kan inserted in pSUP202sac | This study |
| pBSKSP | aspA cloned in pBSK | This study |
| pBSKΔSP | pBSKSP with Kan cassette replacing small internal fragment of ΔaspA | This study |
| pSUP202sacΔSP | ΔaspA::kan on PstI fragment from pBSKSPΔaspA::kan in PstI site of pSUP202sac | This study |
| pMMBG | satA subcloned on 1.2-kb PstI fragment in PstI site of pMMB66HE, under tac control | This study |
| M13mp18G+/− | satA on 1.2-kb PstI fragment in forward or reverse orientation in PstI site of M13mp18 | This study |
| pMMBG228-Ala | pMMBG encoding mutation of Gly-228 to Ala-228 | This study |
Media and growth conditions.
Escherichia coli strains were routinely grown at 37°C in Luria-Bertani broth or agar (1). A. salmonicida strains were grown at 15 to 18°C in tryptic soy agar (TSA) or broth (tryptic soy broth [TSB; Oxoid]) supplemented with Davis minimal medium (6). Media used for screening or selection included egg yolk emulsion (EYE) (20% [vol/vol] egg yolk in 10% [wt/vol] NaCl), egg yolk agar (EYA) (10% [vol/vol] EYE in salt-free nutrient agar), skim milk (50% [vol/vol]) in salt-free nutrient agar (SMA), sucrose (15% [wt/vol]) in TSA (TSAS), and Congo Red (50 μg/ml) in TSA (CRA). Antibiotics were added at the following final concentrations: for E. coli, ampicillin (50 μg/ml), tetracycline (10 μg/ml), and kanamycin (25 μg/ml); for A. salmonicida, ampicillin (100 μg/ml), tetracycline (2 μg/ml), kanamycin (40 μg/ml), and nalidixic acid (40 μg/ml).
DNA isolation, manipulation, and sequencing.
Chromosomal DNA was isolated as previously described (11). Plasmid and cosmid DNA was routinely prepared from E. coli DH5α by alkali lysis, and pMMB66 derivatives were isolated by the boiling method (1). Fragments were isolated with Prep-a-gene (Bio-Rad). Restriction endonucleases, DNA-modifying enzymes, and polymerases were used as specified by the manufacturers (Gibco BRL and Boehringer Mannheim) and by routine procedures (1). Southern blotting was carried out under standard conditions with probes incorporating digoxigenin-dUTP (DIG-dUTP) created by random hexanucleotide priming or by PCR with specific primers as described (DIG labelling kit; Boehringer Mannheim). Oligonucleotides were supplied by Genosys Ltd. DNA sequencing was performed with a Sequenase version 2.0 kit (Amersham) or by cycle sequencing with an fmol DNA sequencing kit (Promega Corp.).
Cloning and sequencing of the GCAT gene (satA). (i) 1.2-kb PstI fragment.
A. salmonicida AS1102 chromosomal DNA was digested to completion with PstI. Fragments in the 0.8- to 1.4-kb size range were purified from a TAE agarose (1) (0.75%) gel and ligated into PstI-digested, dephosphorylated pUC18. Transformants in E. coli HB101 were screened for clearing on EYA plates. Two positive transformants were confirmed by PCR with primers based on the sequence of the gene encoding A. hydrophila GCAT (EMBL accession no. X07279). The cloned gene was sequenced in part in pUC18G, and the sequencing was completed in M13mp18G. Although the gene was cloned in the reverse orientation with respect to the lac promoter in pUC18G, weak activity was observed on EYA plates and by acyltransferase activity in cell lysates.
(ii) pLAFR3 cosmid bank.
Clones from an A. salmonicida AS1102/pLAFR3 cosmid bank (22) were lifted, denatured on a positively charged nylon membrane, and screened with a 724-bp satA probe, created by PCR incorporation of DIG-dUTP (Boehringer Mannheim) with pUC18G as the template and primers SMR10 (5′ GCGACAACCTCTCCGATACCGG) and SMR13 (5′ GGCGTTCCTGCGGACTGAAG) as described previously (22) (DIG kit). Restriction and Southern analysis of one positive cosmid (p7G7) revealed that the satA gene was located on the 10-kb EcoRV, 6-kb NotI, 5.5-kb NcoI, and 5.5-kb SacI fragments and that it was centrally located on the SacI fragment.
Cloning of aspA.
aspA was cloned, in a pMMB66HE derivative, after PCR amplification of the gene from an AS1102 genomic DNA template (100 ng) with tailed primers (200 ng each) SP1 (5′ GCGAGTCGACATGGAGTTATAAATGAAAAACATCG and SP2 (5′ CCGGATCCATCAGGAACGGGCTGCGTCGTGACC), based on the published sequence (EMBL accession no. X67043). Cycle conditions were 95°C for 1 min followed by 30 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 2 min and a single step of 72°C for 10 min with Pfu DNA polymerase (Stratagene). For subsequent manipulation, the aspA gene was transferred on a 2.0-kb EcoRI-HindIII fragment from pMMBSP to EcoRI-HindIII-digested pBluescript SKII to form pBSKSP.
Marker replacement mutagenesis. (i) pSUP202sac suicide vector.
The suicide vector pWS233 (31), which carries the sucrose sensitivity sacRB genes, appeared to replicate in A. salmonicida. Therefore, to facilitate the selection of double-crossover mutants following marker exchange mutagenesis, the sacRB cassette was excised from pSM20 on a PstI-EcoRI fragment and ligated into the large PstI-EcoRI fragment of pSUP202 to create pSUP202sac. pSUP202sac had lost the functional bla and cam genes but retained tetracycline resistance and the PstI and EcoRI cloning sites (see Fig. 1). Although strains of A. salmonicida expressing sacRB were resistant to 5% (wt/vol) sucrose, sucrose sensitivity was achieved with 15% (wt/vol) sucrose.
FIG. 1.
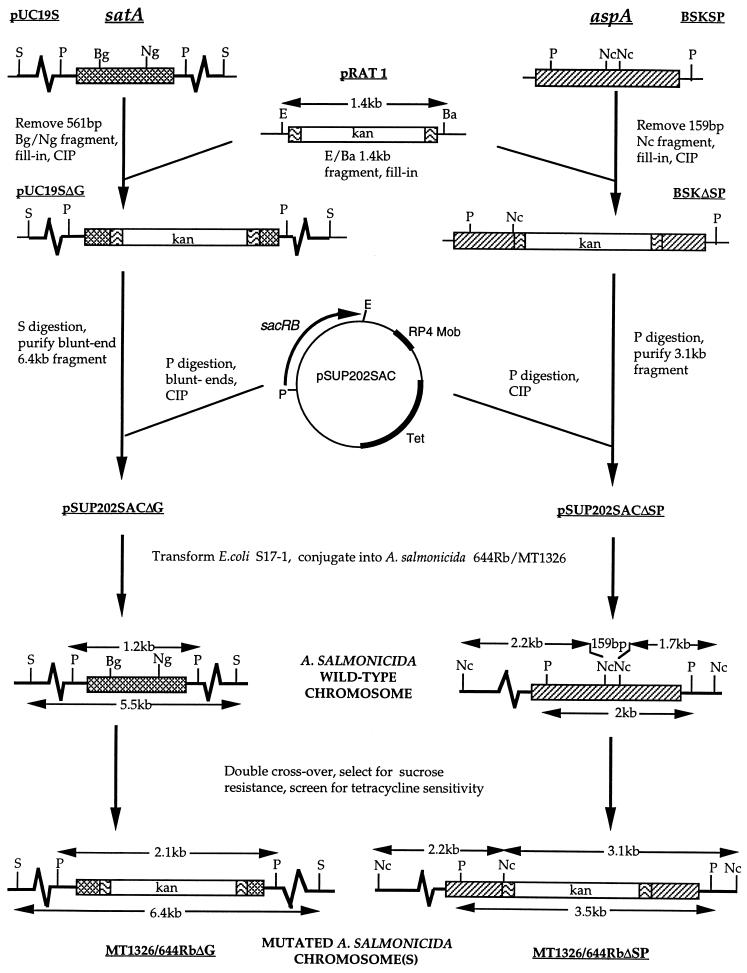
Schematic diagram of the procedure used to create the satA and aspA deletion mutant strains of A. salmonicida. Thick line, A. salmonicida chromosomal DNA; thin line, vector DNA; cross-hatched box, satA gene encoding GCAT; hatched box, aspA gene encoding serine protease; wavy lines, 1,436-bp HaeII fragment originating from Tn903, encompassing the kanamycin resistance gene, kan (blank); sacRB, levansucrase gene conferring sucrose sensitivity. Ba, BamHI; Bg, BglII; E, EcoRI; Nc, NcoI; Ng, NgoIV; P, PstI; S, SacI; CIP, calf intestinal phosphatase.
(ii) Conjugation and mutant selection.
Mutated genes (Fig. 1) were conjugated into A. salmonicida 644Rb or strain MT1326 from E. coli S-17 on the suicide vector pSUP202sac by filter mating (32) at 15°C for 2 days. A. salmonicida primary integrants were then selected in the presence of tetracycline and kanamycin. Nalidixic acid was also added for strain 644Rb; for MT1326, selection against E. coli was based solely on growth on TSA at 15°C for 3 days. Selected primary integrants were grown in TSB plus kanamycin prior to growth for 3 to 4 days on TSA containing 15% (wt/vol) sucrose (TSAS) and kanamycin to select for double-crossover mutants. Alternatively, double-crossover mutants were selected directly by growth on TSAS plus kanamycin at 15°C for 4 to 5 days.
Site-directed mutagenesis of Gly-228 of GCAT.
Site-directed mutagenesis was carried out on M13mp18G with the mismatch (mutation in boldface) oligonucleotide 5′-CTACGACGCCGGCTATGTGTG by the method of Kunkel (23). The mutation was confirmed by the presence of an additional NgoAIV restriction site (underlined) and sequencing, prior to transfer of the mutation on a 0.86-kb BamHI-BglII fragment to similarly restricted pMMB66G to form pMMB66G228-Ala.
In vitro processing of proGCAT and SDS-polyacrylamide gel electrophoresis (PAGE). (i) Immunoblots.
The source of proGCAT was an overnight (18-h) culture supernatant of A. salmonicida CB3/pMMBG following a 30-min induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The source of protease was 60-h culture supernatant from MT1326ΔG. Following incubation of 0.9 ml of a pro-GCAT sample at 25°C with 100 μl of protease, proteins were precipitated with 10% trichloroacetic acid (TCA), electrophoresed on Laemmli sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gels, and analyzed by immunoblotting with an ECL detection kit (Amersham) and 1:10,000 dilution of rabbit anti-pro-GCAT serum.
(ii) Identification of the C-terminal fragment of processed GCAT.
Induction of A. salmonicida CB3/pMMBG was extended to 3 h. Proteins in the recovered culture supernatant were concentrated by precipitation with ammonium sulfate (65% saturation) at 4°C, and pro-GCAT was isolated by high-pressure liquid chromatography on a Superose 12 column (bed volume, 24 ml) equilibrated with 20 mM Tris-HCl (pH 7.5)–0.15 M NaCl at a flow rate of 0.5 ml/min. pro-GCAT was recovered, >98% pure, between the 33- and 35-min fractions. For processing, 3 μg of purified pro-GCAT was incubated with 5 μl of overnight culture supernatant from AS1102ΔG, as a source of serine protease, in a final volume of 100 μl at 25°C for 1 h, at which time serine protease was inhibited by the addition of phenylmethylsulfonyl fluoride (PMSF; final concentration, 6.25 mM). The protein was then concentrated by TCA precipitation and analyzed on an SDS–16% polyacrylamide gel.
Enzyme assays.
The AspA serine protease was assayed with azocasein as the substrate, as previously described (2). Typically, 0.1 to 0.5 ml of culture supernatant was used in a final reaction volume of 2 ml and activity was measured after 10, 20, 60, and 120 min at 37°C. Protease units were calculated as the change in absorbance at 436 nm per minute per milliliter of supernatant. General protease activity against Azocoll was determined from the mean of duplicate samples assayed as described previously (8). Acyltransferase activity of GCAT was monitored qualitatively by thin-layer chromatography of the neutral lipids following incubation of 1 ml of spent bacterial culture supernatant with 3.3 ml of EYE at 37°C for 16 h (25).
Pathogenicity trials.
Fish trials were performed at the FRS Experimental Fish Production Unit, Aultbea, Scotland, where Atlantic salmon (Salmo salar L.) were raised in 1-m3 aquaria containing approximately 450 liters of salt water for smolts and fresh water for parr, maintained at 14°C and exchanging at 4 liters/min. The fish were fed with BP Mainstream pellets at 2% of body weight per day. Both the fish stock and water source were certified disease free. Salmon parr, all siblings, were potential S2 fish, with an average weight of 100 g, and they were 12 to 17 months old at challenge. Smolts (17 months) were potential S1 fish, full siblings of the members of the first group, and had been in seawater (9 to 15°C) for 3 months prior to challenge. During challenge, the temperature was maintained at 14°C.
(i) i.p. injection.
The fish were anesthetised by immersion in tricaine methane sulfonate solution (MS-222; Sigma), injected intraperitoneally (i.p.) with 0.1 ml of a given dilution of bacterial suspension or with phosphate-buffered saline (control), dye marked with Alcian blue (Sigma), and returned to the parent tank. A separate tank was used for each bacterial strain. Samples of head kidney from fish that died were plated onto TSA or SMA, and the kanamycin resistance of the recovered A. salmonicida was subsequently confirmed. The genotype of randomly selected samples was confirmed by PCR and Southern blotting analysis of isolated DNA.
(ii) Cohabitation challenge.
Atlantic salmon smolts were maintained as above, and 10% of the population were injected with a potentially lethal dose (105 in 100 μl of sterile phosphate-buffered saline [pH 7.4]) of the appropriate strain and returned to the parent tank, with two separate tanks being used for the wild-type strain and two for each mutant strain. The injected fish died within 2 to 3 days and were immediately removed. The uninjected cohabitants began to die after a further 4 to 5 days and were removed and screened as above. The fish were monitored for 21 days.
Statistics.
The final LD50 and effective doses giving 50% cumulative mortality (ED50) on a given day were calculated by logistic regression analysis, with 95% confidence intervals calculated from to Fieller’s theorem with Genstat 5 release 3.2 (Lawes Agricultural Trust, Rothamstead Experimental Station). Independence of tanks and strains (cohabitation trial) was determined by the chi-square test, and differences between groups were determined by Student’s t test (36).
RESULTS
Cloning and properties of the GCAT gene (satA).
The gene encoding GCAT was initially cloned and sequenced from A. salmonicida AS1102 on a 1,176-bp PstI fragment. Subsequently, to facilitate allelic replacement, it was cloned from a pLAFR3 cosmid (p7G7) on a 5.5-kb SacI fragment (see Materials and Methods for details). The same 35.4-kDa pro-GCAT was produced and secreted in reasonable levels from both the PstI and SacI fragments following transfer of the fragments individually to the broad-host-range plasmid pMMB66HE and conjugation into A. salmonicida CB3. The DNA sequence of the cloned gene, which encoded a 335-residue protein including an 18-amino-acid signal sequence, was identical to that recently published (27) for satA from A. salmonicida FT449 (GenEmbl Accession no X70686). It had 93.7% amino acid identity to the AhGCAT precursor (SwissProt accession no. P10480), including the proposed active-site residues, Ser-16, Asp-116, and His-291 (5). The only notable difference between the two sequences was the conservation of tandem Gly residues but with a single residue shift: 225-CYGGSYV for AhGCAT and 225-CYDGGYV for AsGCAT.
Marker replacement mutagenesis. (i) A. salmonicida ΔsatA::kan mutants.
ΔsatA::kan mutants (referred to as ΔG) of two virulent isolates of A. salmonicida (strains MT1326 and 644Rb) were created by marker replacement mutagenesis as outlined in Fig. 1. Deletion of the 561-bp BglII-NgoAIV fragment removed approximately half of the satA open reading frame including DNA encoding Asp-116, which is proposed to form part of the active site (5). Southern analysis revealed that the genotypes of three tetracycline-sensitive recombinants for each strain were identical with respect to satA (examples are shown in Fig. 2). For all six isolates, the single wild-type satA gene had been replaced with the mutated ΔsatA::kan gene. Southern analysis also confirmed that the kan cassette was inserted only in the target satA gene. Phenotypic analysis confirmed loss of GCAT (data not shown). No GCAT could be detected by immunoblotting, and no acyltransferase activity was observed in the spent-culture supernatants taken at several different stages of growth of A. salmonicida 644ΔG and A. salmonicida MT1326ΔG. Immunoblotting also excluded the accumulation of any truncated product of GCAT inside the cells. Serine protease production in the satA mutant strains was unaffected, as was the growth pattern, outer membrane profile, and presence of the A-layer (defined by Congo red binding together with the outer membrane profiles) (data not shown).
FIG. 2.
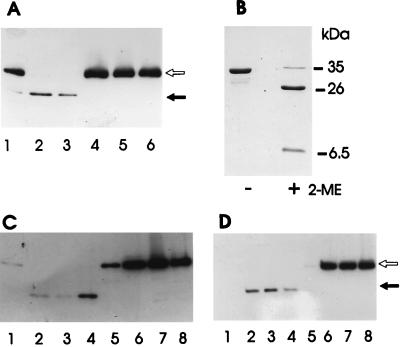
Southern blot analysis of chromosomal deletion mutants. (A) Chromosomal DNA from wild-type A. salmonicida 644Rb (lanes 5 and 6), A. salmonicida 644RbΔG (lanes 1 and 2) and A. salmonicida MT1326ΔG (lanes 3 and 4) digested with PstI (lanes 1, 3, and 5) and SacI (lanes 2, 4, and 6) and hybridized with a satA or kanamycin cassette probe (kan) as indicated. Wild-type satA fragments were 1.2 kb (PstI) and 5.5 kb (SacI); satA::kan-derived fragments were 2.1 kb (PstI) and 6.4 kb (SacI). (B) Wild-type MT1326 (lanes 1 and 2) and MT1326ΔSP (lanes 3 and 4) digested with NcoI (lanes 1 and 3) or PstI (lanes 2 and 4) and detected with an aspA or kanamycin cassette probe (kan) as indicated. Relevant wild-type aspA fragments were 1.7 kb (NcoI) and 2 kb (PstI); aspA::kan-derived fragments were 3.1 kb (NcoI) and 3.5 kb (PstI). Three independent ΔsatA::kan isolates for each strain were identical, as were three independent ΔaspA::kan isolates.
(ii) A. salmonicida ΔaspA::kan mutants.
The aspA gene was cloned as a 2-kb PCR product from A. salmonicida AS1102 (see Materials and Methods). Kanamycin replacement mutagenesis resulted in removal of the central 159-bp NcoI fragment encoding the active-site Ser-330 (9) (Fig. 1). Mutants of A. salmonicida MT1326 and 644Rb were created by allelic replacement. The genotypes of three isolates of each strain were confirmed by Southern blotting (examples are given in Fig. 2).
The AspA serine protease-deficient phenotype of each mutant was confirmed by using azocasein as substrate. No activity could be detected in 60-h culture supernatants (Table 2), and no product was detected by immunoblotting of 24-, 40-, or 60-h culture supernatants (data not shown) from any of the ΔaspA::kan mutants (referred to as ΔSP). It was noted from both the enzyme assays and the immunoblots that the parent A. salmonicida MT1326 produced considerably more of this enzyme (5.33 U) than did the wild-type A. salmonicida 644Rb (0.18 U). While the azocasein substrate is apparently specific to serine protease, Azocoll is degraded by a wider range of proteases. The use of Azocoll as the substrate also reflected loss of AspA in the mutants (compare the activities without inhibitor and in the presence of EDTA [Table 2]). For MT1326ΔSP, the remaining activity could be attributed largely to the secreted metalloprotease. However, low levels of a second serine protease (PMSF sensitive) were detectable in samples from all mutant strains. With Azocoll substrate and EDTA and PMSF inhibitors, synergistic activity between the metalloenzyme(s) and serine protease(s) was also evident in all samples. A large increase in metalloprotease activity was evident in the MT1326ΔSP strains. Since the mutant and parent strains grew at similar rates (all in absence of kanamycin), the most likely explanation for this is the absence of serine protease-mediated degradation of metalloprotease in the mutant strains.
TABLE 2.
Proteolytic activity of 60-h culture supernatants from wild-type and aspA mutant A. salmonicida
| Straina | Activity (unitsb) in supernatant with substratec:
|
|||
|---|---|---|---|---|
| Azocasein (no inhibitor) | Azocoll
|
|||
| No inhibitor | 10 mM EDTA | 6.25 mM PMSF | ||
| MT1326 (wt) | 5.33 | 1.981 | 0.84 | 0.21d |
| MT1326ΔSP-1 | <0.015 | 1.461 | 0.21 | 0.86 |
| MT1326ΔSP-2 | <0.015 | 1.371 | 0.24 | 0.83 |
| 644Rb (wt) | 0.18 | 0.492 | 0.273 | 0.07 |
| 644RbΔSP-1 | <0.015 | 0.222 | 0.123 | 0.05 |
| 644RbΔSP-2 | <0.015 | 0.252 | 0.173 | 0.1 |
wt, wild type. 1 and 2 indicate isolate 1 and isolate 2, respectively.
One unit of protease activity = 1 OD unit change/ml of culture supernatant/h/OD650 culture density.
<0.015, below the limit of detection, which was 0.015 unit. Superscript numbers indicate that the mutants were significantly different from the respective parental wild type as calculated by Student’s t test with 4 degrees of freedom: 1, P = 0.0026; 2, P = 0.0013; 3, P = 0.0067.
In the presence of EDTA plus PMSF, the activity of the MT1326 wild-type supernatant against Azocoll was reduced to 0.05 unit.
Serine protease-mediated processing of pro-GCAT.
In vitro studies had previously indicated that the AspA serine protease was responsible for pro-GCAT processing in A. salmonicida (13). Culture supernatants from A. salmonicida MT1326 also mediated PMSF-sensitive processing of pro-GCAT (Fig. 3A). Confirmation of the role of the aspA-encoded serine protease in pro-GCAT processing was obtained by analysis of GCAT in culture supernatants from the A. salmonicida ΔaspA::kan mutants. No processed GCAT could be detected in samples from either A. salmonicida 644ΔSP or A. salmonicida MT1326ΔSP, while only the mature form of GCAT was seen in samples taken at the same time points from the parent strains (Fig. 3C and D). Lower levels of processed GCAT than of pro-GCAT were consistently observed in immunoblots. This could be attributed to the lower reactivity of the N-terminal fragment of GCAT with the antibody and possibly also to further proteolytic degradation of processed GCAT.
FIG. 3.
Importance of the 64-kDa serine protease in pro-GCAT processing. (A) Processing of pro-GCAT by MT1326ΔG culture supernatant in the absence (lanes 1 to 3) or presence (lanes 4 to 6) of 6.25 mM PMSF. Samples were incubated for 10, 20, or 60 min (lanes 1 and 4, 2 and 5, and 3 and 6, respectively) before being analyzed by SDS-PAGE and immunoblotting. (B) Coomassie blue-stained SDS-polyacrylamide gel of purified and processed GCAT solubilized in the absence (−) or presence (+) of 5% β-mercaptoethanol (2-ME). The approximate sizes of intact GCAT and processed fragments are indicated. (C and D) Absence of in vivo processing of pro-GCAT in A. salmonicida 644RbΔSP (C) and A. salmonicida MT1326ΔSP (D) mutants. Culture supernatant samples were taken after growth for 24, 40, 50, or 65 h (lanes 1 and 5, 2 and 6, 3 and 7, and 4 and 8, respectively). Lanes: 1 to 4, parent strains; 5 to 8, ΔSP mutants. An immunoblot of an SDS-polyacrylamide gel of 10-fold-concentrated samples is shown. In all cases, the open arrow indicates pro-GCAT (35.1 kDa) and the solid arrow indicates the N-terminal fragment of processed GCAT (approximately 26 kDa).
Previous estimates of the size of processed AsGCAT vary from 24 to 26 kDa (7, 13, 24). However, in this study, the use of nonreducing conditions in SDS-PAGE and overproduced pro-GCAT demonstrated that serine protease-processed GCAT was approximately 33 kDa. The C-terminal fragment (approximately 6.5 kDa) was not degraded but remained associated with the N-terminal fragment via a disulfide bond between Cys-225 (N-terminal fragment) and Cys-281 (C-terminal fragment) (Fig. 3B). Clearly, on processing, only a small fragment of pro-GCAT was lost. Maintenance of the C-terminal fragment is consistent with the similar structure of trypsin-processed AhGCAT (19) and the proposal that Ser-16, Asp-116, and His-291 constitute the catalytic triad of the active site of this lipase (5).
The possibility that conservation of the double Gly sequence (see above) is important to proteolytic processing of GCAT was considered. However, replacement of Gly-228 with Ala-228 in AsGCAT did not inhibit the processing of proGCAT (data not shown).
Importance of GCAT and serine protease to pathogenesis. (i) i.p. injection.
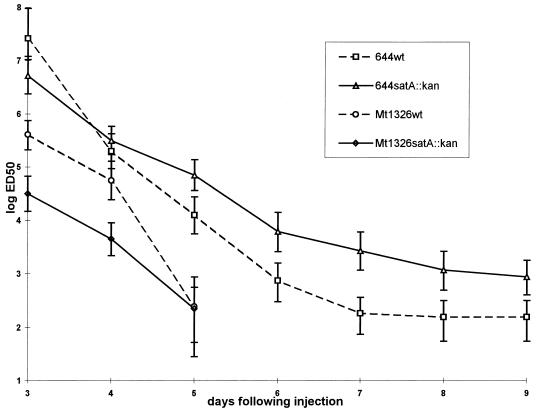
The virulence of the satA deletion mutants was compared to that of their respective parent strains, A. salmonicida MT1326 and A. salmonicida 644Rb, by i.p. challenge of Atlantic salmon (parr, trial 1 [Fig. 4], smolts, trial 2 [Table 3]). In this model system, GCAT was clearly not a major virulence factor, as might be predicted from the earlier toxicity studies (24). Following injection of Atlantic salmon parr with the highly virulent strain A. salmonicida MT1326, one could only conclude that the LD50 of both the parent and satA mutant was <102. Even at the lowest injected dose of 102, the mortality was higher than 80% for both strains. At the end of trial 1 (9 days), the LD50 of the A. salmonicida 644Rb parent strain was estimated to be 1.55 × 102 (95% confidence intervals = 3.16 × 102 and 5.48 × 101) compared to an LD50 of A. salmonicida 644RbΔG of 8.7 × 102 (95% confidence intervals = 1.77 × 103 and 4 × 102) (Fig. 4). The fish injected with the parent or the respective isogenic satA mutant died at very similar rates. The estimated ED50 was already below the smallest number of bacteria injected (100 CFU) on day 6 for both parent and mutant strains of A. salmonicida MT1326. Also, very similar patterns of time to death were followed for A. salmonicida 644Rb parent and A. salmonicida 644Rb satA. In this analysis, a slightly higher but statistically different ED50 was maintained from days 5 to 9 for the 644Rb satA mutant compared to its parent (Fig. 4).
FIG. 4.
Mortality rate in Atlantic salmon parr following i.p. injection with wild-type or satA mutant strains. Groups of 20 fish were injected with one dose, ranging in a 10-fold series from 109 to 102 CFU of the respective parent or satA mutant of A. salmonicida or PBS, as described in Materials and Methods, and monitored for 9 days. From day 6 onward, the estimated ED50 of both the parent and satA mutant of strain MT1326 was lower than the lowest dose tested (100 CFU). Values for the PBS control (number of dead fish out of 20 fish injected/tank): MT1326, 0, MT1326ΔG, 1 (day 5); 644Rb, 1 (day 7); 644ΔG, 0.
TABLE 3.
Challenge of Atlantic salmon smolts by i.p. injection (trial 2)a
| A. salmonicida strain | Day 8
|
LD50 (CFU) on day 15b | ||
|---|---|---|---|---|
| ED50 (CFU) | +95% CI (CFU) | −95% CI (CFU) | ||
| MT1326 | 2.63 × 103 | 2.01 × 104 | 1.42 × 102 | <102 |
| MT1326ΔG | 1.08 × 103 | 8.18 × 103 | 2.64 × 101 | <102 |
| MT1326ΔSP | 4.55 × 103 | 7.13 × 104 | 3.49 × 101 | 6.25 × 102 |
Groups of six fish were injected with each dose (ranging from 108 to 102 CFU) of the parent or respective mutants of A. salmonicida MT1326 or PBS, as described in Materials and Methods, and monitored for 15 days. There were no deaths among PBS controls.
<102, LD50 lower than the lowest dose tested (102 CFU). The lower estimated 95% confidence interval (CI) for MT1326ΔSP was below zero; therefore, no valid statistical comparison to the parent strain can be made.
Examination of mortalities revealed no discernible difference in the disease pathology caused by wild-type and sat mutant strains. The characteristic pathologic findings of the acute form of furunculosis were observed in both cases, namely, darkened skin; hemorrhages at the base of the fins, along the belly, and internally; and a swollen and hemorrhaging vent. Moribund fish displayed lethargy, increased respiration rate, and loss of equilibrium.
Atlantic salmon smolts were also challenged with A. salmonicida MT1326 and a comparison was made with the condition of the fish infected with the isogenic satA and aspA mutants. This also reflected the retention of very high virulence of both mutants tested. No statistically significant difference in virulence was detected at either 8 or 15 days (in the latter case, most fish had died) (Table 3). As above, no obvious difference in disease pathology between the parent and mutant strains was evident.
Analysis of bacteria isolated from dead fish in both trials confirmed that only kanamycin-resistant A. salmonicida strains were recovered from fish injected with either of the satA or aspA deletion mutants. Southern analysis with satA and kan or aspA and kan probes resulted in blots identical to those shown in Fig. 2A and B for MT1326ΔG and MT1326ΔSP, respectively. This confirmed that the deletion in the satA and aspA open reading frames was stably maintained during the course of the trial and excluded any possibility of cross-contamination.
(ii) Cohabitation challenge.
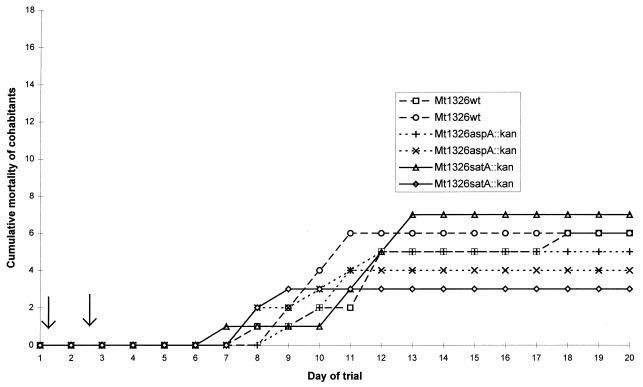
The evidence from the i.p. challenge trials contradicted the widely accepted role of GCAT in virulence. There was some concern that the i.p. challenge model of infection used in the trial was bypassing a critical stage in the natural infection process during which GCAT was required. For this reason, the virulence of both deletion mutants was further assessed by using a cohabitation model, which more closely replicates a natural outbreak of the disease (3, 26). Figure 5 shows the results of the cohabitation challenge for wild-type MT1326 and the isogenic satA and aspA deletion mutants. The average cumulative mortalities of fish infected with MT1326ΔG (27%) and MT1326ΔSP (25%) were only marginally lower than that (33%) of fish infected with the wild-type strain. The difference between these strains was not statistically significant (for independence of tank and bacterial strain, χ2 = 0.95 with 2 degrees of freedom). This is in comparison to results of previous studies, where less virulent strains (A. salmonicida MT046 and MT004, with LD50s of 106 by i.p. injection) failed to induce any mortality in a cohabitation trial (2a). Thus, the cohabitation trial also indicated that both mutant strains retained virulence. It is noteworthy that the mortality rates due to both mutant strains were again indistinguishable from that due to the wild type. Fish began to die between days 6 and 8, and, except for one much later death, all the fish had died by days 11 to 13 (Fig. 5). Screening of the isolates recovered, as above, confirmed the stability of the mutations and absence of cross-contaminants.
FIG. 5.
Mortality rate in Atlantic salmon following cohabitation with fish infected with parent or mutant A. salmonicida MT1326 (trial 3). The cohabitation challenge was performed with duplicate tanks for each strain, as indicated, and 18 test fish plus 2 injected fish per tank (see Materials and Methods for details). Arrows indicate the death of injected fish within 2 or 3 days of injection. A consequence of the nature of this trial is the significant variation between duplicate tanks. Mean (and relative) mortalities were 33% (100%), 27.7% (83%), and 25% (75%), for MT1326 parent, MT1326 satA, and MT1326 aspA.
DISCUSSION
The absence of any significant effect of satA deletion on the virulence of two different isolates of A. salmonicida demonstrates that the lipase, GCAT, is not, as might be predicted from the toxicity studies (24), a major virulence factor of this bacterium. (For comparison, loss of the A-layer is generally associated with an increase in the LD50 from approximately 102 or less to >106 [34].) The possibility that the nonessential role of GCAT is a reflection of the challenge model is highly unlikely. A cohabitation model (3, 26), which closely approximates the natural route of infection and which would reflect requirements for the early establishment of infection, was tested in addition to the more invasive i.p. injection model.
The earlier toxicity studies (24) identified a high-molecular-weight complex of GCAT and LPS as the major lethal toxic product released from plate cultures of this bacterium. More recent studies suggest that much of the polysaccharide in this complex may be derived from the capsule rather than from LPS (4). The purified GCAT-LPS-polysaccharide complex had an LD50 of 45 ng of protein/g Atlantic salmon body weight, whereas the soluble form of GCAT was less toxic, with an LD50 of 338 ng of protein/g of Atlantic salmon body weight. It was concluded that GCAT itself was the major toxin, since up to 0.714 mg of A. salmonicida LPS/g of salmonid fish lacked toxicity (24). Also, toxicity was lost on heating the GCAT complex at 60°C for 30 min, providing additional evidence that the enzyme GCAT, and not any form of LPS or carbohydrate associated with the high-molecular-weight complex, mediated toxicity (24). The LPS or carbohydrate may simply increase the stability of the protein toxin in vivo and possibly enhance the targeting of GCAT to the eucaryotic cell membrane. The toxicity studies and pathogenicity trials are, of course, totally different models, with differences in the levels of GCAT, sites of delivery, and presentation of the toxin. For example, the extent to which GCAT is complexed with LPS following in vivo production in fish is unknown. Certainly, in vitro the ratio of soluble GCAT to GCAT-LPS complex is evidently dependent on the culture method (7, 24). Regardless of the factors that determine the degree of toxicity of GCAT, it is clear from the results presented here that this unusual lipase is not essential to the establishment of acute furunculosis and death in Atlantic salmon.
The results with the defined A. salmonicida MT1326ΔaspA::kan mutant confirm the earlier findings of Drinan and Smith (12) that a mutant obtained by random chemical mutagenesis and deficient in serine protease had a maximum 10-fold increase in LD50. The sequence downstream of aspA has not been determined. However, if there had been any polar effect in the aspA::kan mutation, the results would not have been affected, since this mutation had no dramatic in vivo effect. Like GCAT, AspA serine protease is evidently not essential for acute A. salmonicida subsp. salmonicida-induced furunculosis in salmon, even though one of the primary roles of AspA serine protease might be in activating not only pro-GCAT but also precursors of other secreted enzymes or toxins. In agreement with studies with purified serine protease (13), AspA was shown here to be essential for pro-GCAT processing in broth cultures. Despite the existence of metalloprotease(s) and a second serine protease in spent growth medium from strain MT1326ΔaspA::kan, none of the pro-GCAT was processed. The extent to which host proteases might replace serine protease in precursor processing remains to be seen. Although the larger decrease in virulence reported by Sakai (29) for a protease-deficient mutant could reflect real strain variation, the possibility that this chemically mutagenized mutant possessed multiple mutations cannot be excluded.
A marginal increase was observed in the LD50 of the A. salmonicida 644Rb satA mutant, compared to its respective parent strain (Fig. 4). Whether this reflects a real marginal decrease in virulence of the satA mutant may be tested most effectively in competition trials. Should this be the case, it may reflect a role of GCAT, like serine protease or any of the other secreted enzymes and toxins, in the maximization of bacterial growth during infection. GCAT and serine protease may represent only two of a battery of extracellular aggressins that individually are not essential but together aid the progression of disease, e.g., by enhancing nutrient acquisition, tissue destruction, and attack on components of the host defense system. Loss of one may be compensated by another. For example, preparations of both GCAT and serine protease lead to the development of thrombosis and circulatory failure in Atlantic salmon (30). It is possible that no single potent toxin is produced by A. salmonicida and that maximization of bacterial growth is of prime importance to its pathogenesis. Proliferation and dissemination of the organism might then lead to mortality through acute bacteremia. The requirement for possession of a surface A-layer for the virulence of most strains of A. salmonicida (34) is certainly consistent with this notion.
However, although GCAT in combination with LPS is by far the most toxic product thus far identified in cultures of A. salmonicida, the possibility that this bacterium produces a more potent unidentified toxin(s) should not be excluded. Candidates for this might be the potent cytolysin aerolysin (21) or acetylcholinesterase (28), both of which have been identified in A. hydrophila. The aerA gene has been identified in A. salmonicida, but no active hemolysin is produced, possibly due to lack of correct in vitro processing (20, 21). Confirmation of acetylcholinesterase production still awaits the identification of a gene encoding this enzyme in any Aeromonas spp. In addition, it would not be surprising if A. salmonicida targeted toxins directly into eucaryotic cells via a “contact” (type III) secretion pathway (10). The widespread nature and the importance of this mechanism of toxin delivery are now evident from the recent identification of genes for this system in several different pathogens of the closely related enterobacterial family.
Finally, while secreted products in concert may enhance growth during infection, the importance of the extracellular enzymes to the bacterium may actually be in the final stages of infection, i.e., dissemination, rather than in the disease process itself. We have recently shown that production of AspA protease is affected by the quorum-sensing asaI/R regulatory system and occurs only at high cell density in vitro (33). Serine protease is detectable at high levels in vivo in furuncules (15), but production at earlier stages of infection has not been analyzed. Nutrient limitation is likely to be most severe at high cell density. Thus, should AspA protease also be produced only at high cell density in vivo, its importance would be in maintaining bacterial viability and multiplication prior to release from the fish into the water, where nutrients are limiting and the viability of A. salmonicida is greatly decreased. This would be of particular importance to bacterial infection in the presence of very low densities of fish, as is found in the natural environment. Clearly, understanding of the relative importance of the secreted products of A. salmonicida to the pathogenesis of furunculosis is still in its infancy.
ACKNOWLEDGMENTS
This research was funded by BBSRC grant A00442. R.V. and E.D. were recipients of MRC and University of Reading studentships, respectively.
We thank Tom Buckley for A. salmonicida CB3 and anti-GCAT antiserum; Larry Vaughan for A. salmonicida 644Rb; Ulrich Wagner for anti-AspA antiserum; the Department of Applied Statistics, University of Reading, for advice on statistical analyses; and Andrey Karlyshev for helpful discussions.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Chichester, United Kingdom: John Wiley & Sons; 1992. [Google Scholar]
- 2.Braun V, Schmitz G. Excretion of a protease by Serratia marcescens. Arch Microbiol. 1980;124:55–61. doi: 10.1007/BF00407028. [DOI] [PubMed] [Google Scholar]
- 2a.Bricknell, I. Unpublished data.
- 3.Bricknell I R. A reliable method for the induction of experimental furunculosis. J Fish Dis. 1995;18:127–133. [Google Scholar]
- 4.Bricknell I R, Bowden T J, Lomax J, Ellis A E. Antibody response and protection of Atlantic salmon (Salmo salar) immunised with an extracellular polysaccharide of Aeromonas salmonicida. Fish Shellfish Immunol. 1997;7:1–16. [Google Scholar]
- 5.Brumlick M J, Buckley J T. Identification of the catalytic triad of the lipase/acyltransferase from Aeromonas hydrophila. J Bacteriol. 1996;178:2060–2064. doi: 10.1128/jb.178.7.2060-2064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckley J T. Purification of cloned proaerolysin released by a low protease mutant of Aeromonas salmonicida. Biochem Cell Biol. 1990;68:221–224. doi: 10.1139/o90-029. [DOI] [PubMed] [Google Scholar]
- 7.Buckley J T, Halasa L N, MacIntyre S. Purification and partial characterisation of a bacterial phospholipid: cholesterol acyltransferase. J Biol Chem. 1982;255:3320–3325. [PubMed] [Google Scholar]
- 8.Chavira R, Burnett T J, Hageman J H. Assaying proteinases with Azocoll. Anal Biochem. 1984;136:446–450. doi: 10.1016/0003-2697(84)90242-2. [DOI] [PubMed] [Google Scholar]
- 9.Coleman G, Whitby P W. A comparison of the amino acid sequence of the serine protease of the fish pathogen Aeromonas salmonicida subsp. salmonicida with those of other subtilisin-type enzymes relative to their substrate-binding sites. J Gen Microbiol. 1993;139:245–249. doi: 10.1099/00221287-139-2-245. [DOI] [PubMed] [Google Scholar]
- 10.Cornelis G R, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 11.Costello G, Vipond R, MacIntyre S. Aeromonas salmonicida possesses two genes encoding homologues of the major outer membrane protein, OmpA. J Bacteriol. 1996;178:1623–1630. doi: 10.1128/jb.178.6.1623-1630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drinan E M, Smith P R. Histopathology of a mutant Aeromonas salmonicida infection in Atlantic salmon (Salmo salar) In: Ellis A E, editor. Fish and shellfish pathology. London, United Kingdom: Academic Press Ltd.; 1985. pp. 79–83. [Google Scholar]
- 13.Eggset G, Bjornsdottir R, McQueen Leifson R, Arnesen J A, Coucheron D H, Jorgensen T O. Extracellular glycerophospholipid:cholesterol acyltransferase from Aeromonas salmonicida: activation by serine protease. J Fish Dis. 1994;17:17–29. [Google Scholar]
- 14.Ellis A E. The extracellular toxins of Aeromonas salmonicida ssp. salmonicida. In: Bernoth E-M, Ellis A E, Midtlyng P J, Olivier G, Smith P, editors. Furunculosis. Multidisciplinary fish disease research. London, United Kingdom: Academic Press Ltd.; 1997. pp. 248–268. [Google Scholar]
- 15.Ellis A E, do Vale A, Bowden T J, Thompson K, Hastings T S. In vivo production of A-protein, lipopolysaccharide, iron-regulated outer membrane proteins and 70 kDa serine protease by Aeromonas salmonicida subsp. salmonicida. FEMS Microbiol Lett. 1997;149:157–163. doi: 10.1111/j.1574-6968.1997.tb10323.x. [DOI] [PubMed] [Google Scholar]
- 16.Fiore A E, Michalski J M, Russell R G, Sears C L, Kaper J B. Cloning, characterisation and chromosomal mapping of a phospholipase (lecithinase) produced by Vibrio cholerae. Infect Immun. 1997;65:3112–3117. doi: 10.1128/iai.65.8.3112-3117.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furste J P, Pansegrau W, Frank R, Blocker H, Scolz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multihost-range expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 18.Fyfe L, Coleman G, Munro A L S. The combined effect of isolated Aeromonas salmonicida protease and haemolysin on Atlantic salmon, Salmo salar L., compared with that of a total extracellular products preparation. J Fish Dis. 1988;11:101–104. [Google Scholar]
- 19.Hilton S, McCubbin W D, Kay C M, Buckley J T. Purification and spectral study of a microbial fatty acyltransferase: activation by limited proteolysis. Biochemistry. 1990;29:9072–9078. doi: 10.1021/bi00490a026. [DOI] [PubMed] [Google Scholar]
- 20.Hirono I, Aoki T. Cloning and characterisation of three hemolysin genes from Aeromonas salmonicida. Microb Pathog. 1993;15:269–282. doi: 10.1006/mpat.1993.1077. [DOI] [PubMed] [Google Scholar]
- 21.Howard S P, MacIntyre S, Buckley J T. Toxins. In: Austin B, Altwegg M, Gosling P J, Joseph S, editors. The genus Aeromonas. Chichester, United Kingdom: John Wiley & Sons; 1996. pp. 267–281. [Google Scholar]
- 22.Karlyshev A V, MacIntyre S. Cloning and study of the genetic organisation of the exe gene cluster of Aeromonas salmonicida. Gene. 1995;158:77–82. doi: 10.1016/0378-1119(95)00139-w. [DOI] [PubMed] [Google Scholar]
- 23.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee K-K, Ellis A E. Glycerophospholipid:cholesterol acyltransferase complexed with lipopolysaccharide (LPS) is a major lethal exotoxin and cytolysin of Aeromonas salmonicida: LPS stabilizes and enhances the toxicity of the enzyme. J Bacteriol. 1990;172:5382–5393. doi: 10.1128/jb.172.9.5382-5393.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacIntyre S, Trust T J, Buckley J T. Distribution of glycerophospholipid:cholesterol acyltransferase in selected bacterial species. J Bacteriol. 1979;139:132–136. doi: 10.1128/jb.139.1.132-136.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy D H. An experimental model for fish furunculosis caused by Aeromonas salmonicida. J Fish Dis. 1983;6:231–237. [Google Scholar]
- 27.Nerland A H. The nucleotide sequence of the gene encoding GCAT from Aeromonas salmonicida ssp. salmonicida. J Fish Dis. 1996;19:145–150. [Google Scholar]
- 28.Nieto T P, Santos Y, Rodriguez L A, Ellis A E. An extracellular acetylcholinesterase produced by Aeromonas hydrophila is a major lethal toxin for fish. Microb Pathog. 1991;11:101–110. doi: 10.1016/0882-4010(91)90003-s. [DOI] [PubMed] [Google Scholar]
- 29.Sakai D K. Loss of virulence in a protease-deficient mutant of Aeromonas salmonicida. Infect Immun. 1985;48:146–152. doi: 10.1128/iai.48.1.146-152.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salte R, Norberg K, Arnesen J A, Odegaard O R, Eggset G. Serine protease and glycerophospholipid:cholesterol acyltransferase of Aeromonas salmonicida work in concert in thrombus formation: in vitro the process is counteracted by plasma antithrombin and α2-macroglobulin. J Fish Dis. 1992;15:215–227. [Google Scholar]
- 31.Selbitschka W, Stefan N, Puhler A. Construction of gene replacement vectors for Gram −ve bacteria using a genetically modified sacRB gene as a positive selection marker. Appl Microbiol Biotechnol. 1993;38:615–618. [Google Scholar]
- 32.Simon R, Priefer U, Puhler A. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 33.Swift S, Karlyshev A V, Fish L, Durant E L, Winson M K, Chhabra S R, Williams P, MacIntyre S, Stewart G A A B. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologues AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J Bacteriol. 1997;179:5271–5281. doi: 10.1128/jb.179.17.5271-5281.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trust T J, Noonan B, Chu S, Lutwyche P, Umelo E. Molecular approaches to understanding the pathogenesis of Aeromonas salmonicida—relevance to vaccine development. Fish Shellfish Immunol. 1996;6:269–276. [Google Scholar]
- 35.Vaughan L M, Smith P R, Foster T J. An aromatic-dependent mutant of the fish pathogen Aeromonas salmonicida is attenuated in fish and is effective as a live vaccine against the salmonid disease furunculosis. Infect Immun. 1993;61:2172–2181. doi: 10.1128/iai.61.5.2172-2181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wardlaw A C. Practical statistics for experimental biologists. Chichester, United Kingdom: John Wiley & Sons; 1985. [Google Scholar]