Abstract
Transport and surface expression of the invasion plasmid antigens (Ipa proteins) is an essential trait in the pathogenicity of Shigella spp. In addition to the type III protein secretion system encoded by the mxi/spa loci on the large virulence plasmid, transport of IpaB and IpaC into the surrounding medium is modulated by IpaD. To characterize the structural topography of IpaD, the Geysen epitope-mapping system was used to identify epitopes recognized by surface-reactive monoclonal and polyclonal antibodies produced against purified recombinant IpaD or synthetic IpaD peptides. Surface-exposed epitopes of IpaD were confined to the first 180 amino acid residues, whereas epitopes in the carboxyl-terminal half were not exposed on the Shigella surface. By using convalescent-phase sera from 10 Shigella flexneri-infected monkeys, numerous epitopes were mapped within a surface-exposed region of IpaD between amino acid residues 14 and 77. Epitopes were also identified in the carboxyl-terminal half of IpaD with a few convalescent-phase sera. Comparison of IpaD epitope sequences with Salmonella SipD sequences indicated that very similar epitopes may exist in the carboxyl-terminal region of each protein whereas the IpaD epitopes in the surface-exposed amino-terminal region were unique for the Shigella protein. Although the IpaD and SipD homologs may play similar roles in transport, the dominant serum antibody response to IpaD is against the unique region of this protein exposed on the surface of the pathogen.
The triad of invasion, intracellular replication, and intercellular spreading within the colonic epithelium are essential steps that lead to the dysentery syndrome caused by virulent shigellae. Expression of several plasmid-encoded proteins (IpaB, IpaC, IpaD, Spa/Mxi proteins, and VirG or IcsA) is required for the complete virulence phenotype of Shigella spp. (1, 13, 29). The Ipa proteins act as the invasins, while the Spa/Mxi proteins make up a type III secretion apparatus which is involved in the presentation and transport of the Ipa proteins to and beyond the Shigella surface (9, 15). A similar invasin/secretion apparatus has been characterized in Salmonella, in which both Ipa and Spa/Mxi homologs are present (6–9). Upon exposure to host cells, surface-localized IpaB, IpaC, and IpaD are released into the surrounding environment (14, 32) by a Spa32-mediated event (31). One potential host cell target of the released Ipa proteins appears to be the α5β1 integrin (30). Among the released proteins is an Ipa invasin complex consisting of IpaB, IpaC, and possibly IpaA and IpaD (12, 15). Another Ipa protein complex (IpaB-IpaD) has been proposed to occur in the Shigella outer membrane and appears to play a role in modulating the transport of IpaC and IpaB (14). In IpaD deletion mutants, the IpaB-IpaD complex is not present, resulting in higher than normal levels of IpaB and IpaC being secreted into the surrounding medium. An almost identical result is found in Salmonella SipD mutants, in that they no longer modulate the secretion of SipC and SipB (8). Although the SipD and IpaD mutants both exhibit altered transport of the invasins, on a structural basis only the carboxyl halves of SipD and IpaD have a significant degree of homology (46%) (6, 8). The sequence and functional similarity between SipD and IpaD suggests that the carboxyl-terminal region may be involved in the transport modulation. Structurally, IpaD and SipD are very hydrophilic, suggesting that they are not integral or membrane-spanning proteins and that their particular role in transport may be a restriction of movement through a channel or pore in the outer membrane (8, 14). Even though IpaD has been identified as an outer membrane protein exposed on the surface of shigellae, its structural topography is not known. By determining the regions of the IpaD protein which are surface exposed and extrapolating this to SipD, it may provide a better understanding of the structural environment in which these proteins reside and how it relates to their similar functions.
IpaD, like IpaB and IpaC, is an essential virulence factor that is also a major antigen recognized by humans and monkeys infected with shigellae (10, 20, 21). The host immune response to the Ipa proteins, VirG, and lipopolysaccharide (LPS), appears to be an attempt to neutralize key virulence factors of the pathogen. Clearly, antibodies to LPS are important in protection against future disease (5, 23), but what is less clear is the role that antibodies to the Ipa proteins play in immunity and disease. One reason that the immune response to the Ipa proteins is not well understood is that even though most infected individuals respond to a particular Ipa protein, as determined by Western blot analysis, it has been determined that in monkeys the antibody response to specific epitopes within each protein varies from individual to individual (26). For example, three epitope domains of IpaC have been identified, each of which contains multiple epitopes recognized by infected monkeys. Interestingly, the animals that responded to all three IpaC epitope regions were less likely to have shigellosis than were animals that responded to either one or two of the IpaC epitope regions. Due to the epitope response variability, it may be impossible to correlate a response to the Ipa proteins with a particular outcome unless epitope analysis is performed. Further epitope analysis of other Ipa proteins may identify a series of epitopes or epitope domains within each protein, the composite of which is correlated with immunity or disease outcome. Such a composite epitope response pattern by the host immune system might indicate that multiple epitopes must be recognized on the Ipa invasin complex to effectively neutralize invasion. If this immune response is possible, it would potentially cross-react with all Shigella spp., since the Ipa proteins are conserved in all Shigella spp. as well as in enteroinvasive Escherichia coli strains.
In addition to identifying specific peptide epitopes recognized by antibodies in immune sera, it is necessary to determine the availability of these epitopes in the native antigen as it exists in the bacterium. By combining epitope mapping with antibodies affinity purified against surface-exposed epitopes, it is possible to identify the epitopes exposed on the surface of the bacterium. In this study, the antigenic structure of IpaD was investigated with monoclonal antibodies (MAbs) and polyclonal sera produced against purified recombinant IpaD. IpaD epitopes exposed on the surface of intact Shigella cells were identified consistently within the amino-terminal half of the protein. In addition, epitopes recognized by antibodies in immune serum from infected monkeys were characterized and found to be located predominantly in the exposed amino-terminal portion of IpaD. Further analysis of the sequence of the IpaD epitopes indicated that the epitopes within the exposed region were in the portion of IpaD that is least homologous to the Salmonella invasins SspD and SipD.
MATERIALS AND METHODS
Peptide synthesis.
IpaD peptides for the generation of immune sera were kindly synthesized by Ettore Appella, National Cancer Institute, National Institutes of Health, Bethesda, Md. These peptides were selected for synthesis based on regions of high antigenic index in the IpaD amino acid sequence as determined by the MacVector sequence analysis program (International Biotechnologies, Inc., New Haven, Conn.). The sequences of the synthesized IpaD peptides are given in Table 1. Where necessary, a carboxyl-terminal cysteine residue was added to the peptide for coupling to carrier proteins.
TABLE 1.
Identification of IpaD epitopes exposed on the surface of shigellae with rabbit antipeptide and anti-rIpaD sera
| Rabbit no. | Immunizing agenta | Surface-exposed epitopeb | Inaccessible epitopeb |
|---|---|---|---|
| 18 | 55-RTTNQALKKELSQKTL-70-C-KLH | 57-TNQALKKE-64 | 63-KELSQKTL-70 |
| 20 | 102-NEYPINKDARELLH-115-C-KLH | 106-INKDAREL-113 | |
| 30 | 195-LKKALEELKEKYKDKPLYPANNTVSQ-220-C-KLH | 202-LKEKYKDK-209 | |
| 23 | 280-NAGFSAEDETMKNNLQ-295-C-KLH | 283-FSAEDETM-290 | |
| 62 | rIpaD | 48-NDTLHNIR-55 | 15-FSPNNTNG-22 |
| 88-DVNKSAQL-95 | 237-VSQKNGGY-244 | ||
| 104-YPINKDAR-111 | 247-SINMTPID-254 | ||
| 117-APKEAELD-124 | 260-LDNLGGNGE-268 | ||
| 322-CTDTDKLF-329 | |||
| 63 | rIpaD | 13-SSFSPNNT-20 | 22-GSSTETVN-29 |
| 40-PVSSLTML-47 | 122-ELDGDQMF-129 | ||
| 47-LNDTLHNI-54 | 211-LYPANNTV-218 | ||
| 73-TSLEEIAL-80 | 213-PANNTVSQ-220 | ||
| 107-NKDARELL-114 | 237-VSQKNGY-244 | ||
| 119-KEAELDGD-126 | 246-VSINMTPIDN-255 | ||
| 256-MLKSLDNL-263 | |||
| 260-LDNLGGNG-267 | |||
| 322-CTDTDKLF-329 |
Single-letter abbreviations are used for amino acid residues of the epitope.
Surface exposed and inaccessible epitopes as determined with antibodies affinity purified with intact, virulent S. flexneri 5.
Epitope mapping was performed with a series of octameric peptides which overlap by 7 amino acid residues and were synthesized in duplicate as previously described (26). These overlapping octamers represented the entire IpaD sequence (28). Control pins consisted of the previously described IpaC peptide epitope H3N-SKLGLNKQ-COOH as the positive control (26) and the peptide H3N-GLAQ-COOH as a negative control. Additional epitope mapping was performed by an enzyme-linked immunosorbent assay (ELISA) with larger synthetic peptides of IpaD consisting of amino acid residues 12 to 36, 30 to 45, 53 to 77, and 71 to 95.
Immune sera.
Immune sera were collected from rhesus monkeys (Macaca mulatta) challenged with 2 × 1010 CFU of S. flexneri 2a 2457T. Blood was drawn before challenge and 9 and 24 days after challenge (5). Sera used in this study were also used in a similar epitope-mapping study involving IpaC (26).
Rabbits were used for the production of immune sera to IpaD synthetic peptides and to purified recombinant IpaD (rIpaD). For immunization, peptides were conjugated to preactivated maleimide-keyhole limpet hemocyanin (KLH) with the Imject immunogen conjugation kit (Pierce Chemical Co., Rockford, Ill.). The purified rIpaD (11) was kindly provided by Bill Picking.
Two rabbits per peptide conjugate were immunized with 200 μg of the KLH-peptide conjugate on four occasions separated by 2-week rests. The first injection was done with the conjugate mixed with complete Freund’s adjuvant (CFA), the second was done with the conjugate mixed with incomplete Freund’s adjuvant (IFA), and the third and fourth boosters were done with no adjuvant. Rabbits immunized with rIpaD were initially immunized with 10 μg of rIpaD in CFA followed by a second immunization with 10 μg of rIpaD in IFA and a third with 100 μg of rIpaD in 0.9% saline. All immunizations in rabbits were given intramuscularly.
Immunization of mice, production of hybridomas, and isotype determination.
BALB/cByJ female mice were immunized subcutaneously on days 0 and 21 with 10 μg of purified rIpaD mixed with Ribi adjuvant (RIBI ImmunoChem Research, Hamilton, Mont.) as specified by the manufacturer. On day 26, all the mice had serum antibody responses as measured in a rIpaD ELISA (20). Subsequently, on day 31, three mice were immunized intravenously with 5 μg of rIpaD (without adjuvant). Three independent fusions were performed on splenocytes (obtained from these three mice on day 34) and SP2/0 myeloma cells with 50% polyethylene glycol 1450. Seven IpaD-positive hybridoma cultures were cloned by limiting dilution and used in further analysis of IpaD. The isotype of the selected IpaD monoclonal cultures was determined with a mouse antibody isotyping kit (Gibco/BRL).
Water extract and rIpaD ELISA.
ELISAs for IpaD and the water extract of S. flexneri 5 were performed as previously described (19, 20). Recombinant IpaD and the water-extractable antigens were used at 0.05 and 1.0 μg/well, respectively. Goat anti-mouse immunoglobulin G, conjugated to alkaline phosphatase (Kirkegaard and Perry, Gaithersburg, Md.), was used to detect mouse antibodies bound to the antigens.
Surface analysis (colony blots).
Virulent shigellae were grown overnight on nitrocellulose disk overlays on tryptic soy agar plates. The nitrocellulose disks with attached shigellae were blocked with 2% casein and subsequently incubated with anti-IpaD hybridoma culture supernatants. After 4 h, the nitrocellulose was washed and then probed with goat anti-mouse IgG conjugated with alkaline phosphatase. The precipitable substrate Fast Red TR salt/ASMX phosphate was used to detect alkaline phosphatase reporter activity.
Affinity purification of antibodies.
To determine the antibody-accessible epitopes of IpaD on intact shigellae, invasive shigellae were used for affinity purification of the surface-reactive rabbit antibodies. Antisera from rabbits immunized with various peptides of IpaD were first absorbed with S. flexneri 5 (M90T-55), a plasmid-free derivative of S. flexneri 5 (M90T-W) (2). Antisera from rabbits immunized with the rIpaD were affinity purified against both M90T-W and M90T-55. Samples (approximately 250 ml) of late-log-phase cultures of the virulent and avirulent cultures were collected by centrifugation and each resuspended in 10 ml of antiserum (final dilution, 1/10 [diluted in Tris-buffered casein]). The antiserum was incubated with the organism for 2 h at 25°C with constant, slow agitation. Next, the mixture was centrifuged and the resulting pellet was washed twice with Tris-saline buffer (pH 7.4). After being washed, the pellet was treated with 1 ml of 0.2 M glycine in 0.9% saline (pH 2.8) for 30 min to elute the bound antibodies. The bacteria were removed by centrifugation at 17,000 × g. The supernatant containing the eluted antibodies was neutralized with Tris base (1.3 M), diluted 1:3 with casein, and stored at −20°C.
Multipin ELISA.
After peptide synthesis, the pins were initially blocked with a 2% casein–5% bovine serum albumin filler (26). All the pins were probed with each monkey’s prechallenge and 9-day-postchallenge sera (diluted 1:400 in filler) for 2 h at room temperature. For rabbit sera, the pins were incubated for 2 h at room temperature (final dilution, 1:400) and with each affinity-purified serum diluted 1:100 in filler. When mapping MAbs epitopes, ammonium sulfate-concentrated culture supernatants were diluted 1:100 in filler and incubated with the pins overnight at 4°C. After incubation with antibody, the pins were washed with phosphate-buffered saline containing 0.05% Tween-20 (PBS-Tween) and probed with either alkaline phosphatase-conjugated goat anti-human IgG or goat anti-rabbit IgG, diluted 1:500 in filler, for 1 h at room temperature. The pins were washed with PBS-Tween and incubated with p-nitrophenyl phosphate at 37°C for 1 h (26). The pins were also probed separately with each conjugate diluted 1:500 in filler. Readings of optical density at 405 nm (OD405) were averaged for the duplicate pins and plotted against the corresponding IpaD amino acid residue number of the first amino acid of the synthetic octameric peptide.
RESULTS
Characterization of IpaD MAbs.
All mice immunized with rIpaD responded with high-titer antibody levels in serum (data not shown). Fusion of immune splenocytes with SP2/0 myeloma cells resulted in 134 hybridoma cultures producing antibodies reactive with rIpaD. Of these, seven IpaD antibody-producing cell lines were cloned twice by limiting dilution and expanded for subsequent studies on IpaD. Six MAbs reacted with IpaD in a Western blot analysis with whole-cell lysates from virulent shigellae, and most IpaD MAbs reacted with the water extract antigen preparation from virulent shigellae (Table 2). Isotype analysis determined that four of the MAbs were IgG1, two were IgG3, and one was IgG2b.
TABLE 2.
Characteristics of IpaD MAbs
| MAb | Isotype | OD in water extract ELISAa | Reactivity in Western blotb | Titer in IpaD ELISAc | Surface reactivityd |
|---|---|---|---|---|---|
| 16A6 | IgG1 | 0.133 | + | 256 | − |
| 13B2 | IgG3 | 0.776 | + | 32 | + |
| 2A9 | IgG1 | 0.666 | + | 4 | + |
| 1C1 | IgG2b | 0.680 | + | 1,024 | + |
| 4B4 | IgG1 | 0.406 | + | 512 | + |
| 18A8 | IgG3 | 0.084 | − | 128 | − |
| 16F8 | IgG1 | 3.278 | + | 128 | + |
ELISA OD after incubation of undiluted hybridoma culture supernatant with water extract from virulent S. flexneri 5 (19). The OD of the culture medium control was 0.080.
+, reactivity with IpaD from whole-cell lysates of virulent S. flexneri 5; −, no reactivity.
Reciprocal titers of ammonium sulfate-concentrated MAb culture supernatants in a recombinant IpaD ELISA.
Positive (+) or negative (−) reaction indicates MAb activity with colony lifts of virulent S. flexneri 5.
Only five of seven MAbs reacted with intact shigellae bound to nitrocellulose, indicating that in the intact organism only a portion of IpaD is exposed on the surface and accessible to antibodies (Table 2). These results were confirmed by a whole-cell ELISA (data not shown). Attempts to map the epitopes of the IpaD MAbs by the multipin ELISA technique were successful only for MAb 2A9. This particular surface-reactive MAb reacted with peptides 130-SHRELWAK-137 and 175-AGWISPGG-182. The use of larger synthetic peptides in an ELISA allowed the epitope recognized by surface-reactive MAb 16F8 to be localized to peptide 30-SDIKTTTSSHPVSSLT-45.
Identification of IpaD sequences exposed on the surface of shigellae.
The results with the MAbs indicated that portions of IpaD are exposed on the surface of shigellae and other regions are not exposed. One way to localize specific domains of a protein which are exposed on the surface of a microorganism is to use whole, unlysed organisms as a matrix for affinity purification of antibodies reactive with the exposed epitopes. This type of experiment was performed with antisera produced against IpaD synthetic peptides and also with antisera made against purified rIpaD. A late-log-phase culture of virulent S. flexneri 5 (strain M90T-W) was used to capture the surface-reactive antibodies. Once eluted from the intact bacteria, the surface-reactive antibodies were used in the multipin ELISA to identify the peptide epitope sequences of IpaD that were exposed on the surface of the bacterium.
Rabbits immunized with synthetic IpaD peptides conjugated to KLH recognized at least one IpaD peptide epitope (Table 1) within the immunizing peptide. Of five peptide epitopes identified in animals immunized with IpaD peptide conjugates, only two (57-TNQALKKE-64 and 106-INKDAREL-113) were surface exposed (Table 1). Interestingly, immunizing peptide 55–70 elicited antibodies in rabbit 18 to two distinct epitopes, one (57-TNQALKKE-64) exposed on the surface of shigellae and one (63-KELSQKTL-70) not accessible to antibody in intact shigellae.
Antisera from two rabbits immunized with purified rIpaD recognized numerous epitopes throughout the IpaD protein (Table 1; Fig. 1). By using affinity-purified antibodies from the same sera in the multipin ELISA, it was possible to identify a subset of the IpaD peptide epitopes which were located on the surface of shigellae (Fig. 1). Antibodies to several peptide epitopes were totally absent in the affinity-purified product compared to the whole, non-affinity-purified serum. This was especially true for any epitope located in the carboxyl-terminal half of the IpaD protein. Peptide epitopes located on the surface and reactive with antibody were located predominantly in the amino-terminal half of the protein. Table 1 lists the epitopes located on the surface and those inaccessible to antibodies. The identification of surface-exposed regions and antibody-inaccessible regions was consistent regardless of the source of antisera (i.e., made against IpaD peptides or rIpaD). Similar peptide epitopes were recognized by different rabbit sera as surface accessible, i.e., 48-NDTLHNIR-55 (rabbit 62) and 47-LNDTLHNI-54 (rabbit 63); 106-INKDAREL-113 (rabbit 20), 104-YPINKDAR-111 (rabbit 62), and 107-NKDARELL-114 (rabbit 63); and 117-APKEAELD-124 (rabbit 62) and 119-KEAELDGD-126 (rabbit 63) (Table 1).
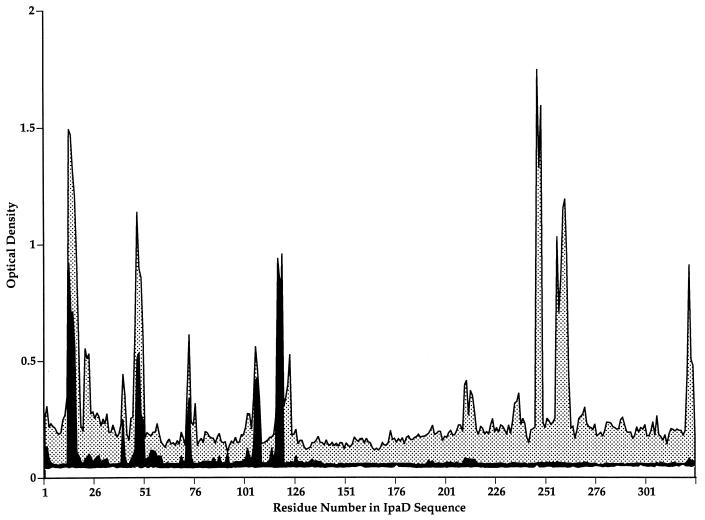
FIG. 1.
Identification of IpaD epitopes exposed on the surface of S. flexneri. Antibodies produced in rabbits immunized with purified recombinant IpaD were affinity purified against intact, virulent S. flexneri 5 strain M90T-W. Untreated serum (before affinity purification) (stippled peaks) and affinity-purified antibodies (solid peaks) were used to probe overlapping synthetic peptides representing the entire IpaD sequence. A subset of peptide epitopes mapped in the untreated serum samples were also identified with the affinity-purified antibodies, indicating that these IpaD peptide epitopes were exposed on the surface of virulent shigellae.
In no instance was the same peptide identified as both surface exposed and also inaccessible, although in one case, two similar (sharing 6 of 8 amino acids) peptide epitopes were recognized as surface accessible by one rabbit and as surface inaccessible by another, i.e., 13-SSFSPNNT-20 (rabbit 63) and 15-FSPNNTNG-22 (rabbit 62) (Table 2). Although the difference is slight, this may indicate an area of IpaD in the outer membrane of intact shigellae that is at the interface between a surface-exposed epitope and an inaccessible epitope.
Epitope mapping of IpaD with convalescent-phase monkey sera.
The multipin ELISA was also used to identify epitopes recognized by convalescent monkeys infected with virulent shigellae. Of 13 S. flexneri-infected animals, 10 (77%) had antibodies in their 9-day-postinfection sera which recognized peptide epitopes on IpaD (Fig. 2), with each responder recognizing a unique set of IpaD peptide epitopes. The epitope response pattern for asymptomatic monkeys (monkeys 10, 11, and 12) appeared less complex, with an average of three unique epitopes per animal, than the pattern in animals with symptoms (diarrhea, blood, and mucus), which had an average of seven epitopes per animal. Similar to what was previously reported for IpaC (26), it was possible to identify a region of IpaD, between amino acids 14 and 77, in which all 10 IpaD responders recognized at least one peptide epitope. The majority of epitopes within this highly antigenic 63-amino-acid region were clustered within two stretches, from amino acid 32-IKTTTSSHPVSSLTMLN-48 (recognized by monkeys 5, 10, 15, 20, 37, and 41) and from amino acid 55-RTTNQALKKELSQKTLTKTSLEE-77 (recognized by monkeys 3, 5, 8, 10, 15, and 41).
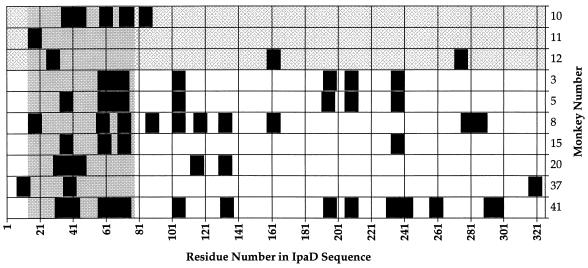
FIG. 2.
Distribution of IpaD peptide epitopes recognized by rhesus monkeys challenged with virulent S. flexneri 2a. The figure shows the peptide epitopes recognized by the convalescent-phase serum samples from monkeys infected with virulent S. flexneri 2a (black rectangles). Peptide epitopes were defined as the peaks with the highest OD405 in the multipin ELISA and were plotted by the residue number of the first amino acid of the peptide as related to the sequence of IpaD. Animals were grouped by symptoms: asymptomatic animals (monkeys 10, 11, and 12) are at the top (grey checkerboard shading), and animals with shigellosis are towards the bottom (no shading). Each of the 10 animals which recognized peptide epitopes on IpaD recognized at least one peptide epitope within the antigenic region between amino acid residues 14 and 77 (solid gray region).
Secondary structure of IpaD.
By using the PROTEAN analysis software (DNASTAR, Inc., Madison, Wis.), the amino acid sequence of IpaD was analyzed with algorithms to predict the antigenic index, hydrophilicity, and surface probability. These predictions were compared to the experimental data for all monkey peptide epitopes recognized after infection with shigellae, the surface-exposed epitopes determined with affinity-purified rabbit antibodies, and the MAbs (Fig. 3). Computer analysis of the 63-amino-acid immunogenic region (between amino acids 14 and 77) of IpaD indicated that this region contains amino acid sequences with high antigenic indices, hydrophilic character, and surface exposure. These predictions were validated by surface-labeling experiments which identified several surface-exposed peptide epitopes which lie within this region (Fig. 3), including two surface epitopes (residues 40 to 47 and residues 57 to 64) which are contained within the epitope clusters between residues 32 and 48 and residues 55 and 77, respectively (Fig. 3). The IpaD protein contains many predicted hydrophilic domains and sequences with potential antigenicity throughout the protein. Epitopes identified with convalescent-phase sera or rabbit antibodies confirmed the high level of antigenicity from one end to the other of the protein. In contrast, there is a clear distinction between the surface exposure of the amino-terminal half of membrane-bound IpaD and the inaccessibility of the carboxyl-terminal half of IpaD to antibodies. Even though the carboxyl-terminal half of the protein contains hydrophilic domains and defined epitopes, these epitopes were not accessible to antibody on the surface of intact shigellae.
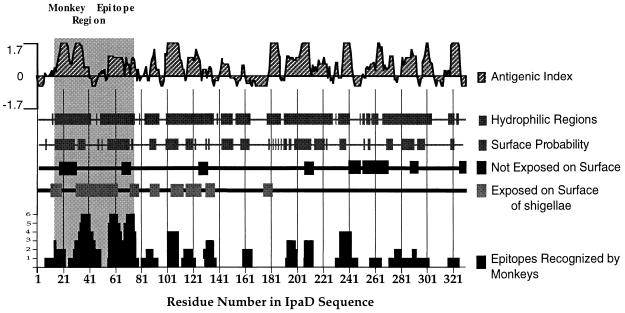
FIG. 3.
Relationship of the predicted secondary structure of IpaD with experimentally identified epitopes. By using the deduced amino acid sequence of IpaD (28) and the PROTEAN analysis software, the antigenic index, hydrophilic regions, and surface probability were calculated. Peptide epitopes exposed on the surface or not accessible to antibodies are indicated. Surface epitopes were identified with rabbit sera or MAbs 2A9 and 16F8 made against IpaD peptides or rIpaD. Peptide epitopes recognized by 10 different monkeys challenged with S. flexneri 2a are also plotted. The height of the column corresponds to the number of monkeys (vertical axis, 1 to 6) recognizing a particular residue within a peptide epitope. All secondary structure and epitope data are plotted against the amino acid residue number in the IpaD sequence; the highly immunogenic region between amino acid residues 14 and 77 is also indicated (grey shaded area).
Relationship of IpaD to Salmonella SspD and SipD proteins.
Homologs to the Ipa proteins called Ssp or Sip proteins have been recently identified in Salmonella spp. (6, 7). Similar functional properties such as critical roles in invasion and transport to the exterior of the bacterial cell by a type III secretion system, along with regions of high sequence identity, suggest that the Shigella and Salmonella invasins have similar ancestral genes. Homology between IpaD and Ssp/SipD is highest in the carboxyl-terminal end of the protein (6, 7). Interestingly, the region of IpaD that is exposed on the surface and is most likely to react with convalescent-phase sera is the amino-terminal region, which is least similar to the Salmonella SspD or SipD. Alignment of IpaD epitope sequences recognized by S. flexneri-infected monkeys with Salmonella SspD/SipD sequences verifies that the antibody response is directed predominantly at the region of IpaD that is unique to Shigella (Table 3). However, it is noteworthy that several epitopes in the carboxyl-terminal region of IpaD, in particular those after residue 270, have sequences that are almost identical to sequences in both the SspD and SipD proteins. It is possible that such peptide stretches are a source of immunological cross-reactivity between the Shigella and Salmonella invasins. Similar analysis of IpaC epitopes (26) and SspC/SipC did not identify any Shigella epitopes that had greater than 3 of 8 identical amino acid residues in the Salmonella sequence (data not shown).
TABLE 3.
IpaD peptide epitopes and related sequences in SspD and SipD
| IpaD epitope sequencea | No. of identical residues in SspD/SipDb | IpaD epitope sequencea | No. of identical residues in SspD/SipDb |
|---|---|---|---|
| 7-TNSISTSS-14 | 1/2 | 112-ELLHSAPK-119 | 3/0 |
| 14-SFSPNNTN-21* | 0/0 | 114-LHSAPKEA-121 | 4/0 |
| 25-TETVNSDI-32* | 2/0 | 122-ELDGDQMI-129 | 1/2 |
| 29-NSDIKTTT-36* | 1/0 | 130-SHRELWAK-137 | 3/3 |
| 32-IKTTTSSH-39* | 2/1 | 158-SSYTQMYQ-165 | 4/4 |
| 33-KTTTSSHP-40* | 2/1 | 191-QVNSLKKA-198 | 4/4 |
| 34-TTTSSHPV-41* | 2/1 | 192-VNSLKKAL-199 | 5/4 |
| 35-TTSSHPVS-42* | 2/1 | 205-KYKDKPLY-212 | 3/1 |
| 37-SSHPVSSL-44* | 1/1 | 230-LGGTIGKY-237 | 1/1 |
| 41-VSSLTMLN-48* | 1/1 | 233-TIGKVSQK-240 | 0/0 |
| 55-RTTNQALK-62* | 2/2 | 234-IGKVSQKN-241 | 0/0 |
| 56-TTNQALKK-63* | 2/2 | 238-SQKNGGYV-245 | 3/3 |
| 57-TNQALKKE-64* | 2/2 | 256-MLKSLDNL-263 | 3/2 |
| 62-KKELSQKT-69* | 1/1 | 271-LDNAKYQA-278 | 7/7 |
| 67-QKTLTKTS-74* | 0/0 | 275-KYQAWNAG-282 | 6/6 |
| 68-KTLTKTSL-75* | 0/0 | 283-FSAEDETM-290 | 4/3 |
| 69-TLTKTSLEE-77* | 1/1 | 289-TMKNNLQT-296 | 5/4 |
| 81-HSSQISMD-88 | 1/1 | 293-NLQTLVQK-300 | 6/6 |
| 85-ISMDVNKS-92 | 1/1 | 316-SSTISSCT-323 | 6/6 |
| 101-RNEYPINK-108 | 1/0 |
The single-letter abbreviations are used for the amino acid residues of the epitopes. Epitopes marked with asterisks are located in the 63-amino-acid immunogenic region of IpaD.
DISCUSSION
Upon infection with virulent shigellae, the host responds vigorously with both nonspecific immune mechanisms (17, 24) and specific immune responses to antigen targets including the Ipa proteins, VirG, and LPS (4, 10, 19–21, 27). Characterization of the pathogenic mechanisms of shigellae, the components involved in virulence, and the subsequent host immune response has led to the development of several different vaccine strategies for shigellosis. Although LPS will be a major antigenic component of any Shigella vaccine because of its documented role in species-specific protection (5), it is still conceivable that the immune system can neutralize virulent shigellae with antibodies to proteins involved in the invasiveness of the organism. In fact, studies have indicated that antibodies against the Ipa proteins alter the invasiveness of shigellae (16, 25). Clearly, the infected host responds to IpaA, IpaB, IpaC, IpaD, and VirG (19–21), but, as was recently shown with IpaC, the response to individual epitopes within a specific protein antigen can be highly variable (26). For example, in a group of monkeys infected with S. flexneri, all of which responded to IpaC, it was possible to demonstrate that a different set of IpaC epitopes was recognized by each animal. Although the individuality of the IpaC immune response was striking, it was clear that three regions of IpaC were the dominant targets of the serum antibody response (26).
In this study, the antigenic structure and surface-exposed domains of IpaD have been characterized. Previous studies have shown that most, but not all, monkeys infected with Shigella produce serum antibodies against IpaD (20). Epitope analysis of serum antibodies from S. flexneri-infected monkeys indicated that a 63-amino-acid sequence from residues 14 to 77 was recognized by 10 of 13 animals. Some animals produced antibodies to many other epitopes throughout the hydrophilic IpaD protein. Unlike the previously described IpaC response (26), it was not possible to associate a particular disease pattern with the IpaD epitope patterns, although it did appear that several of the animals with more severe conditions (diarrhea, blood, and mucus) presented a more complex, multiple-epitope pattern throughout the IpaD protein (Fig. 2).
The role that IpaD plays in the invasion of host cells is not clear, although it is known that IpaD mutants are noninvasive, nonhemolytic, and presumably unable to escape from the phagosome (13, 33). IpaD does not appear to be involved in the attachment phase of invasion, since IpaD mutants are still able to attach to host cells (13) and purified recombinant IpaD does not bind to eukaryotic host cells (11). More recently, it has been suggested that IpaD, along with IpaB, interacts with the secretion apparatus (encoded by the spa/mxi genes) to modulate the secretion of IpaC, IpaB and several other proteins (14, 22). Based on cell fractionation studies, IpaD is present in the cytoplasm and is also associated with the outer membrane (14). In addition, IpaD, along with IpaB and IpaC, is released from the Shigella surface after contact with host cells (32). The ability of some IpaD MAbs to react with intact shigellae indicates that portions of IpaD are accessible to antibody in intact virulent shigellae. By mapping surface-exposed epitopes of IpaD with antibodies affinity purified against whole shigellae, we have demonstrated that the amino-terminal half of IpaD is exposed on the surface of shigellae. The antibody-inaccessible carboxyl-terminal half of IpaD, which is mostly hydrophilic (much like the amino-terminal end), may be associated with other outer membrane surface structures such as IpaB (14), Spa32 (31), LPS hydrophilic O side chains, or even IpaD itself (11), thereby preventing antibodies from binding to IpaD in the intact organism. It is unlikely that IpaD is a membrane-spanning protein, since it is a hydrophilic protein which does not have a hydrophobic region long enough to span the outer membrane. Furthermore, IpaD is easily released from the shigella surface by procedures used to detach proteins that are weakly associated with the surface of bacteria (14, 18). Our proposed model (Fig. 4) of the antigenic topography of membrane-bound IpaD is consistent with these observations.
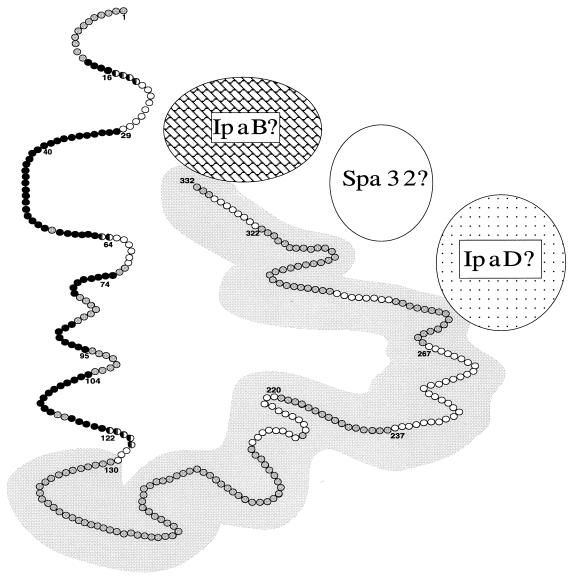
FIG. 4.
A working model of the antigenic topography of membrane-bound IpaD. This model shows epitopes of IpaD that are exposed on the surface of shigellae (black circles) and epitopes of IpaD that are not accessible to antibody (white circles). Amino acid residues found in both exposed and inaccessible epitopes are represented by a half-black, half-white circle. The grey circles represent amino acid residues not identified as part of a surface-exposed or inaccessible epitope. Amino acid residues overlying the grey background are within the region of IpaD that is homologous to the Salmonella SipD protein. Amino acid residue numbers at the beginning or end of epitopes are indicated. Based on previously published data, hypothetical associations of IpaD with membrane-associated IpaB, Spa32, and IpaD are also indicated (11, 14, 31).
The significance of the exposed and antibody-inaccessible regions of IpaD may be related to the similarities and differences of IpaD with the Salmonella invasins SspD and SipD (6, 7). A high degree of identity exists, but only in the carboxyl-terminal end of the Shigella and Salmonella proteins. The amino-terminal region (residues 1 to 129) of IpaD, which is exposed on the surface, has very little homology to the Salmonella IpaD homologs (6). Because the Salmonella and Shigella invasins appear to modulate the secretion of SipC and IpaC, respectively (3, 14), it is possible that the antibody-inaccessible, conserved carboxyl-terminal half of the IpaD molecule is involved in this common function. The exposed region, which may have evolved into its present unique sequence because of selective environmental and immunological pressures, may be involved in functions unique to the Shigella Ipa proteins such as lysis and escape from host cell phagosomes.
In addition to the potential biochemical similarities between the Salmonella and Shigella invasins, epitopes have been identified in the sequestered or inaccessible regions of IpaD, which suggests that the carboxyl-terminal half of the protein is presented to the immune system at some point during infection, even though it is inaccessible to antibodies in the intact organism. It is likely that IpaD originating from degraded shigellae or released during invasion of eukaryotic host cells is processed by antigen-presenting cells of the host, thereby exposing epitopes normally sequestered in the bacterium. The high degree of sequence identity between several IpaD epitopes and the SspD/SipD sequences suggests that immunological cross-reactivity between SspD/SipD and IpaD may occur. The significance of any immunological cross-reactivity between Salmonella and Shigella invasins is unknown, but it could be a source of unexplained background in serological assays with the Ipa proteins.
By determining the antigenic fine structure and surface-exposed regions of IpaD, we have established the groundwork for designing future experiments by site-directed mutagenesis of the IpaD protein. Similar progress on each of the Ipa proteins and other proteins involved in the transport and presentation of the Ipa proteins will eventually lead to a structural model of the Ipa invasin complex.
ACKNOWLEDGMENTS
We thank Ettore Apella for synthesis of the IpaD synthetic peptides used for immunization and Fred Puente for superb technical assistance. We also thank Fred Cassels and Bill Picking for comments and suggestions concerning the manuscript and Bill Picking for providing purified IpaD used to generate polyclonal and monoclonal sera.
REFERENCES
- 1.Andrews G P, Hromockyj A E, Coker C, Maurelli A T. Two novel virulence loci, mxiA and mxiB, in Shigella flexneri 2a facilitate excretion of invasion plasmid antigens. Infect Immun. 1991;59:1997–2005. doi: 10.1128/iai.59.6.1997-2005.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buysse J M, Stover C K, Oaks E V, Venkatesan M, Kopecko D J. Molecular cloning of invasion plasmid antigen (ipa) genes from Shigella flexneri: analysis of ipa gene products and genetic mapping. J Bacteriol. 1987;169:2561–2569. doi: 10.1128/jb.169.6.2561-2569.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collazo C M, Galan J E. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect Immun. 1996;64:3524–3531. doi: 10.1128/iai.64.9.3524-3531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinari G, Hale T L, Austin S W, Formal S B. Local and systemic antibody responses to Shigella infection in Rhesus monkeys. J Infect Dis. 1987;155:1065–1069. doi: 10.1093/infdis/155.5.1065. [DOI] [PubMed] [Google Scholar]
- 5.Formal S B, Oaks E V, Olsen R E, Wingfield-Eggleston M, Snoy P J, Cogan J P. Effect of prior infection with virulent Shigella flexneri 2a on the resistance of monkeys to subsequent infection with Shigella sonnei. J Infect Dis. 1991;164:533–537. doi: 10.1093/infdis/164.3.533. [DOI] [PubMed] [Google Scholar]
- 6.Hermant D, Menard R, Arricau N, Parsot C, Popoff M Y. Functional conservation of the Salmonella and Shigella effectors of entry into epithelial cells. Mol Microbiol. 1995;17:781–789. doi: 10.1111/j.1365-2958.1995.mmi_17040781.x. [DOI] [PubMed] [Google Scholar]
- 7.Hueck C J, Hantman M, Bajaj V, Johnston C, Lee C A, Miller S I. Salmonella typhimurium secreted invasion determinants are homologous to Shigella Ipa proteins. Mol Microbiol. 1995;18:479–490. doi: 10.1111/j.1365-2958.1995.mmi_18030479.x. [DOI] [PubMed] [Google Scholar]
- 8.Kaniga K, Trollinger D, Galan J E. Identification of two targets of the type III secretion system encoded in the inv and spa loci of Salmonella typhimurium that share homology to IpaD and IpaA proteins. J Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaniga K, Tucker S, Trollinger D, Galan J E. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J Bacteriol. 1995;177:3965–3971. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li A, Rong Z C, Ekwall E, Forsum U, Lindberg A A. Serum antibody responses against Shigella lipopolysaccharides and invasion plasmid-coded antigens in Shigella infected Swedish patients. Scand J Infect Dis. 1993;25:569–577. doi: 10.3109/00365549309008545. [DOI] [PubMed] [Google Scholar]
- 11.Marquart M E, Picking W L, Picking W D. Structural analysis of invasion plasmid antigen D (IpaD) from Shigella flexneri. Biochem Biophys Res Commun. 1995;214:963–970. doi: 10.1006/bbrc.1995.2380. [DOI] [PubMed] [Google Scholar]
- 12.Menard R, Prevost M-C, Gounon P, Sansonetti P, Dehio C. The secreted Ipa complex of Shigella flexneri promotes entry into mammalian cells. Proc Natl Acad Sci USA. 1996;93:1254–1258. doi: 10.1073/pnas.93.3.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menard R, Sansonetti P, Parsot C. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 1994;13:5293–5302. doi: 10.1002/j.1460-2075.1994.tb06863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menard R, Sansonetti P, Parsot C, Vasselon T. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of Shigella flexneri. Cell. 1994;79:515–525. doi: 10.1016/0092-8674(94)90260-7. [DOI] [PubMed] [Google Scholar]
- 16.Mills J A, Buysse J M, Oaks E V. Shigella flexneri invasion plasmid antigens B and C: epitope location and characterization with monoclonal antibodies. Infect Immun. 1988;56:2933–2941. doi: 10.1128/iai.56.11.2933-2941.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munoz C, Baqar S, Van De Verg L, Thupari J, Goldblum S, Olson J G, Taylor D N, Heresi G P, Murphy J R. Characteristics of Shigella sonnei infection of volunteers: signs, symptoms, immune responses, changes in selected cytokines and acute-phase substances. Am J Trop Med Hyg. 1995;53:47–54. [PubMed] [Google Scholar]
- 18.Oaks, E. V. Unpublished data.
- 19.Oaks E V, Hale T L, Formal S B. Serum immune response to Shigella protein antigens in Rhesus monkeys and humans infected with Shigella spp. Infect Immun. 1986;53:57–63. doi: 10.1128/iai.53.1.57-63.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oaks E V, Picking W D, Picking W L. Antibody response of monkeys to invasion plasmid antigen D after infection with Shigella spp. Clin Diagn Lab Immunol. 1996;3:242–245. doi: 10.1128/cdli.3.2.242-245.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberhelman R A, Kopecko D J, Salazar-Lindo E, Gotuzzo E, Buysse J M, Venkatesan M M, Yi A, Fernandez-Prada C, Guzman M, Leon-Barua R, Sack R B. Prospective study of systemic and mucosal immune responses in dysenteric patients to specific Shigella invasion plasmid antigens and lipopolysaccharides. Infect Immun. 1991;59:2341–2350. doi: 10.1128/iai.59.7.2341-2350.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsot C, Menard R, Gounon P, Sansonetti P J. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol Microbiol. 1995;16:291–300. doi: 10.1111/j.1365-2958.1995.tb02301.x. [DOI] [PubMed] [Google Scholar]
- 23.Phalipon A, Kaufmann M, Michetti P, Cavaillon J-M, Huerre M, Sansonetti P J, Kraehenbuhl J P. Monoclonal IgA antibody directed against serotype-specific epitope of Shigella flexneri lipopolysaccharide protects against murine experimental shigellosis. J Exp Med. 1995;182:769–778. doi: 10.1084/jem.182.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raqib R, Wretlind B, Anderson J, Lindberg A A. Cytokine secretion in acute shigellosis is correlated to disease activity and directed more to stool than to plasma. J Infect Dis. 1995;171:376–384. doi: 10.1093/infdis/171.2.376. [DOI] [PubMed] [Google Scholar]
- 25.Shaikh N M, Balakrish Nair G, Kumar R. Significance of the secreted form of IpaC, a 45 kDa protein of Shigella dysenteriae 1, in the invasive process as determined by monoclonal antibodies. FEMS Microbiol Lett. 1995;125:247–254. doi: 10.1111/j.1574-6968.1995.tb07365.x. [DOI] [PubMed] [Google Scholar]
- 26.Turbyfill K R, Joseph S W, Oaks E V. Recognition of three epitopic regions on invasion plasmid antigen C by immune sera of Rhesus monkeys infected with Shigella flexneri 2a. Infect Immun. 1995;63:3927–3935. doi: 10.1128/iai.63.10.3927-3935.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van De Verg L L, Herrington D A, Boslego J, Lindberg A A, Levine M M. Age-specific prevalence of serum antibodies to the invasion plasmid and lipopolysaccharide antigens of Shigella species in Chilean and North American populations. J Infect Dis. 1992;166:158–161. doi: 10.1093/infdis/166.1.158. [DOI] [PubMed] [Google Scholar]
- 28.Venkatesan M M, Buysse J M, Kopecko D J. Characterization of invasion plasmid antigen genes (ipaBCD) from Shigella flexneri. Proc Natl Acad Sci USA. 1988;85:9317–9321. doi: 10.1073/pnas.85.23.9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkatesan M M, Buysse J M, Oaks E V. Surface presentation of Shigella flexneri invasion plasmid antigens requires the products of the spa locus. J Bacteriol. 1992;174:1990–2001. doi: 10.1128/jb.174.6.1990-2001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watarai M, Funato S, Sasakawa C. Interaction of Ipa proteins of Shigella flexneri with α5β1 integrin promotes entry of the bacteria into mammalian cells. J Exp Med. 1996;183:991–999. doi: 10.1084/jem.183.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watarai M, Tobe T, Yoshikawa M, Sasakawa C. Disulfide oxidoreductase activity of Shigella flexneri is required for release of Ipa proteins and invasion of epithelial cells. Proc Natl Acad Sci USA. 1995;92:4927–4931. doi: 10.1073/pnas.92.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watarai M, Tobe T, Yoshikawa M, Sasakawa C. Contact of Shigella with host cells triggers release of Ipa invasins and is an essential function of invasiveness. EMBO J. 1995;14:2461–2470. doi: 10.1002/j.1460-2075.1995.tb07243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zychlinsky A, Kenny B, Menard R, Prevost M, Holland B, Sansonetti P J. IpaB mediates macrophage apoptosis induced by Shigella flexneri. Mol Microbiol. 1994;11:619–627. doi: 10.1111/j.1365-2958.1994.tb00341.x. [DOI] [PubMed] [Google Scholar]