Abstract
GNAS-activating somatic mutations give rise to Fibrous Dysplasia/McCune–Albright syndrome (FD/MAS). The low specificity of extra-skeletal signs of MAS and the mosaic status of the mutations generate some difficulties for a proper diagnosis. We studied the clinical and molecular statuses of 40 patients referred with a clinical suspicion of FD/MAS to provide some clues. GNAS was sequenced using both Sanger and Next-Generation Sequencing (NGS). We were able to identify the pathogenic variants in 25% of the patients. Most of them were identified in the affected tissue, but not in blood. Additionally, NGS demonstrated the ability to detect more patients with mosaicism (8/34) than Sanger sequencing (4/39). Even if in some cases, the clinical information was not complete, we confirmed that, as in previous works, when the patients were young children with a single manifestation, such as hyperpigmented skin macules or precocious puberty, the molecular diagnosis was usually negative. In conclusion, as FD/MAS is caused by mosaic variants, it is essential to use sensitive techniques that allow for the detection of low percentages and to choose the right tissue to study. When not possible, and due to the low positive genetic rate, patients with FD/MAS should only be genetically tested when the clinical diagnosis is really uncertain.
Keywords: GNAS, genetic mosaicism, Fibrous Dysplasia/McCune–Albright syndrome, Sanger sequencing, Next-Generation Sequencing
1. Introduction
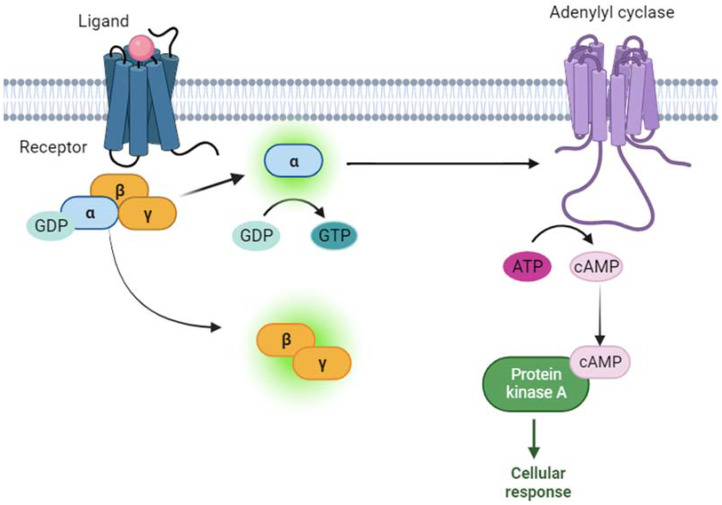
GNAS is a gene that encodes for the α subunit of the stimulatory G protein (Gsα). Gsα is part of a heterotrimeric G protein that binds to G-protein-coupled receptors (GPCRs). When the hormone binds to its GPCR, this causes a conformational change in the GPCR, which allows it to act as a guanine nucleotide exchange factor (GEF). The GPCR can then activate the Gsα by exchanging the guanosine diphosphate (GDP) bound to the G protein for a guanosine triphosphate (GTP). The activated Gsα can then dissociate from the β and γ subunits. Following this, the Gsα activates adenylyl cyclase, which stimulates the production of cyclic AMP (cAMP). cAMP is a second messenger, which finally activates cAMP-dependent protein kinase A (PKA), which phosphorylates different target proteins [1,2] (Figure 1).
Figure 1.
Signaling cascade of Gsα protein is schematically represented. Briefly, when the ligand (pink ball) binds to the receptor (blue), the α subunit of G protein (Gsα) exchanges guanosine diphosphate (GDP) with guanosine triphosphate (GTP). The activated α subunit activates adenylyl cyclase (purple), which produces cyclic adenosine monophosphate (cAMP) from adenosine triphosphate (ATP). Finally, cAMP activates protein kinase A (green), which activates different cellular effectors, generating a cellular response.
Researchers have identified activating and inactivating mutations, either germinal or somatic, in the GNAS gene that impair its function. The vast majority of the variants found in patients are inactivating, with more than 400 unique variants reported [3], and they are associated with pseudohypoparathyroidism type 1A (PHP1A), pseudopseudohypoparathyroidism (PPHP), or progressive osseous heteroplasia (POH) (for review [4]), which is currently renamed as inactivating PTH/PTHrP signaling disorder type 2 (iPPSD2) [2]. A dual effect has been described for a missense variant (p.A366S) affecting both the stability and the activity of the Gsα in a patient with typical PHP1A features combined with testotoxicosis [5,6].
On the other hand, GNAS gain-of-function mutations lead to the constitutive production of cAMP, independent of the ligand binding to the receptor, and, consequently, dysregulated downstream signaling [7,8,9]. In the germline, they have recently been associated with Nephrogenic Syndrome of Inappropriate Antidiuresis (NSIAD), where patients present an impaired capacity to release water in urine, hyponatremia, skeletal developmental problems, and precocious puberty [10,11].
Endocrine or non-endocrine tumors have been identified when the activating variants are somatic, in which the GNAS gene acts as an oncogene [7]. Moreover, they can also cause a spectrum of diseases, known as Fibrous Dysplasia/McCune–Albright syndrome (FD/MAS; OMIM#174800), which is a rare disorder. Although OMIM#174800 refers to McCune–Albright syndrome as the main term, it includes FD among the other entities represented in the entry. Additionally, since 2017 [12], the international consortium for the study of these disease (https://www.icfdmas.com/, accessed on 8 January 2024) has referred to it as FD/MAS and promotes its use. Its exact prevalence is unknown, but it is estimated that FD is responsible for approximately 5% of benign bone lesions [13]. The first visible features are cutaneous affections, specifically café au lait spots on the skin with jagged and irregular borders. Secondly, the bones are also affected, shown as fibrous dysplasia of the bone (FD); in monostotic fibrous dysplasia, only one skeletal site is affected, and when more than one site is involved, FD is classified as polyostotic [8]. The last distinctive sign is peripheral precocious puberty (PP), which is less common in boys than in girls [8,14,15,16,17]. Other manifested signs include hyperthyroidism, growth hormone (GH) excess, and Cushing’s syndrome [16]. Due to its heterogeneity and phenotypic variability, the clinical diagnosis and management of FD/MAS is very challenging. The current guidelines summarize all of the clinical and molecular characteristics of this disorder and indicate that a clinical diagnosis of MAS can be established by the presence of the combination of FD and one or more extra-skeletal features or by the presence of two or more extra-skeletal features [8].
Genetically, FD/MAS is mainly associated with variants occurring in the Arginine in position 201 (R201) of the Gsα protein [18], which can be substituted by two different amino acids: Cysteine (NM_000516:c.601C>T, p.R201C) or Histidine (NM_000516:c.602G>A, p.R201H) [19]. A mutation in the 227 codon replacing Glutamine with Lysine (NM_000516:c.680A>T, p.Q227L) [19] has been identified in approximately 5% of patients [20].
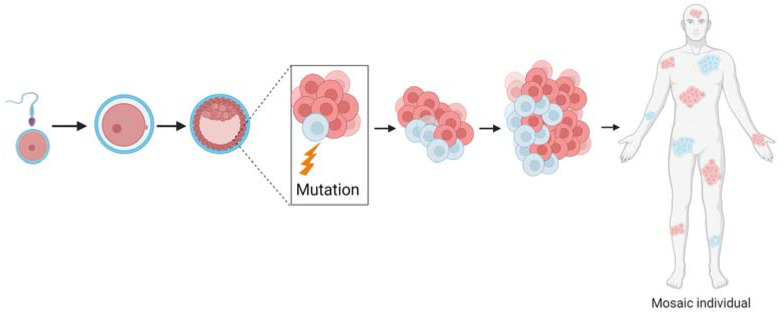
As FD/MAS occurs in a sporadic manner, and due to the pattern of the café au lait pigmentation of the skin, the scientific community suspected that FD/MAS follows a somatic dominant inheritance [21]. In fact, the activating mutation in GNAS occurs postzygotically, and the resulting individual is a genetic mosaic [22] (Figure 2).
Figure 2.
Genetic mosaicism is generated when a mutation occurs in early stages of development. As the genetic variant occurs postzygotically, there will be two or more cell populations. This means that, in the same individual, there can be at least two different genotypes.
Genetic mosaicism is the existence of two or more genetically different cell types within one single tissue or individual [23]. That is, an organism is created from one zygote that presents genotype heterogeneity [24,25]. To form a mosaic, a de novo mutation in the genome occurs during the first stages of development [26]. As the mutation is postzygotic, different cell lineages will develop, and that is why each tissue of the organism will have one different genotype [25] (Figure 2). Because in mosaic organisms, only some tissues are affected, the choice of the adequate tissue to proceed to a correct diagnosis is mandatory [8,23,27]. If an unaffected cell sort is taken, the mutation will remain hidden, and a wrong diagnosis could be given [28,29].
Because of the somatic mosaic nature of the disease, some variants can only be observed in very low levels. Due to this problem, conventional sequencing technologies, such as Sanger sequencing, cannot detect mosaics below 10% [27] or 15–20% according to other authors [30]. Errors in detecting mosaicism have direct consequences in clinical practice [30]. On one side, we can issue a normality diagnosis in a patient, rejecting the clinical suspicion [28,31,32]. On the other side, mosaicism in progenitors can be a cause of disease in the offspring. However, if mosaicism is not detected in the parents, there will be a wrong de novo diagnosis [28,33,34]. Therefore, the genetic counselling will be compromised.
To overcome the problem of the detection limit of Sanger sequencing, other techniques that are more sensitive and can detect mosaicism more accurately have been developed. One of the most used techniques is Next-Generation Sequencing (NGS), thanks to its deep coverage [27,35]. Another more unknown technique is droplet-digital polymerase chain reaction (ddPCR). In this case, specific variants are interrogated and can detect even very-low-percentage mosaics [27]. This has been used to identify mosaicism in several diseases [36,37,38], including in FD/MAS [19,39]. In other rare diseases, quantitative real-time PCR (qRT-PCR) has also been used for the detection of mosaicism [40].
Taking all of the above into account, the main goal of this study is to clinically and genetically characterize all patients with a clinical suspicion of FD/MAS received in the laboratory.
2. Materials and Methods
2.1. Sample Selection
We included all probands with FD/MAS suspicion that have been referred to the Molecular (epi)Genetics Laboratory of the Araba University Hospital—Bioaraba Health Research Institute (Vitoria-Gasteiz, Spain) for GNAS molecular analysis since 2010. Different tissues (blood, bone, skin, etc.) were analyzed according to the availability.
2.2. Nucleic Acid Extraction
Genomic DNA of the patients was extracted from peripheral blood or different tissues using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany), following the corresponding manufacturer’s instructions.
2.3. Molecular Analyses
The presence of point variants in GNAS was investigated using Sanger sequencing as previously described [4]. Next-Generation Sequencing was performed with a custom panel as previously described [41].
2.4. Bioinformatic Approach
As a first-tier approach, the MiniSeq integrated DNA Enrichment Analysis Module was used for secondary analysis (alignment with BWA 0.7.7 on GRCh37/hg19 and Variant Call Format (VCF), and bam/bai files were obtained with Somatic Variant Caller v3.5.2.3 [42] and SAMtools v0.1.19, respectively). Then, output bam and VCF files were visualized with Integrative Genomics Viewer (IGV) [43] to check for the presence of variants associated with FD/MAS.
3. Results
A total of 40 patients were referred to our center, and their GNAS gene was sequenced to look for variants associated with FD/MAS. The main results of the available clinical data and the genetic studies are summarized in Table 1.
Table 1.
Summary of the clinical and genetic results of patients with FD/MAS suspicion. -: no sample available; F: female; M: male; GH: growth hormone; ND: no data; *: tissue is paraffin-embedded. Samples with a positive genetic result are highlighted in grey (dark grey are samples detected with the integrated software, and light grey are samples detected with IGV).
| Patient Code | Sex | Age at Study | Skeletal Abnormalities |
Skin Abnormalities |
Precocious Puberty | Other Manifestations | Sanger (Blood) | Sanger (Tissue) | NGS (Blood) | NGS (Tissue) |
Mutation |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GS0113 | M | 12 | ND | Yes | ND | ND | Negative | Negative (skin) | - | Negative (skin) | - |
| GS1029 | M | 17 | Yes | No | No | ND | Negative | Positive (bone) |
Negative | Positive (34%) C:225× T:114× (bone) |
p.R201C |
| GS1030 | F | 8 | Yes | Yes | ND | ND | Negative | Negative (bone) |
Negative | Negative (bone) |
- |
| GS1043 | F | 14 | Yes (polyostotic) | Yes | ND | ND | Negative | Negative (skin) |
Negative | Negative (skin) |
- |
| GS1045 | F | ND | ND | ND | ND | ND | Negative | Negative (buccal swab) |
Negative | - | - |
| GS1048 | F | 8 | No | No | Yes | Ovarian cyst | Negative | Negative (ovarian cyst *) | Negative | Sequence of bad quality | - |
| GS1050 | F | ND | ND | Yes | ND | ND | Negative | - | Negative | - | - |
| GS1066 | F | ND | ND | ND | ND | ND | Negative | - | - | - | - |
| GS1072 | F | 10 | ND | Yes | Yes | Ovarian cyst | Negative | Negative (ovarian cyst) |
Negative | Negative (ovarian cyst) |
- |
| GS1077 | F | ND | ND | Yes | ND | ND | Negative | Negative (buccal swab) |
Negative | - | - |
| GS1079 | M | ND | ND | ND | ND | ND | Negative | - | Negative | - | - |
| GS1081 | M | 29 | Yes | Yes | Yes | Hepatic affection | Sample of bad quality | Negative (liver) |
Sample of bad quality | Negative (liver) |
- |
| GS1082 | F | ND | ND | ND | ND | ND | Negative | - | Negative | - | - |
| GS1087 | F | ND | ND | Yes | ND | ND | Negative | Negative (skin) | Negative | Negative (skin) | - |
| GS1091 | F | ND | ND | ND | ND | ND | Negative | - | Negative | - | - |
| GS1098 | F | ND | ND | ND | Yes | ND | Negative | - | Sequence of bad quality | - | - |
| GS1102 | M | 0 | Yes | Yes | ND | Hyperthyroidism | Negative | Negative (skin) |
Positive (5%) G:345× A:18× |
Positive (4%) G:388× A:15× (skin) |
p.R201H |
| GS1105 | M | 8 | Yes (polyostotic) | ND | ND | ND | Negative | Negative (bone *) |
Positive (3%) G:230× A:8× |
Negative (bone *) |
p.R201H |
| GS1109 | F | 1 | Yes | Yes | ND | Hyperthyroidism | Negative | Positive (thyroid) |
Positive (1%) G:256× A:2× |
Positive (25%) G:207× A:71× (thyroid) |
p.R201H |
| GS1128 | F | 29 | Yes | No | No | ND | - | Positive (bone) |
- | - | p.R201C |
| GS1144 | F | 9 | ND | Yes | Yes | ND | Negative | - | Negative | - | - |
| GS1172 | M | 9 | Yes (polyostotic) | Yes | No | ND | Negative | - | Negative | - | - |
| GS1176 | F | 3 | ND | Yes | No | ND | Negative | - | - | - | - |
| GS1178 | F | 11 | Yes | Yes | Yes | ND | Negative | - | - | - | - |
| GS1189 | M | 21 | Yes | No | Yes | Hydrocephaly | Negative | Positive pituitary *; Negative bone * | Negative | Sequence of bad quality (G:13× A:14×) * |
p.R201H |
| GS1224 | M | 29 | Yes | Yes | No | Nephrotic syndrome | - | Sample of bad quality (kidney *) |
- | Sequence of bad quality (kidney *) |
- |
| GS1228 | F | 3 | Yes (polyostotic) | Yes | ND | ND | Negative | Negative (skin) |
Negative |
Positive (1%) C:215× T:2× (skin) |
p.R201C |
| GS1240 | F | ND | ND | ND | ND | ND | - | Negative | - | Negative | - |
| GS1241 | F | 1 | Yes | ND | ND | ND | - | Negative (bone) |
- | Negative (bone) |
- |
| GS1255 | F | 10 | ND | Yes | Yes | Ovarian cyst | Negative | - | Negative | - | - |
| GS1258 | F | 0 | ND | ND | ND | Neonatal Cushing’s syndrome | Negative | Negative (skin) |
Negative | Negative (skin) |
- |
| GS1281 | M | ND | ND | ND | ND | ND | - | Negative (skin) | - | Positive (1%) C:177× T:2× (skin) |
p.R201C |
| GS1297 | M | 14 | Yes (polyostotic) | No | No | GH and prolactin hypersecretion | - | Negative (bone) |
- | Positive (27%) C:208× T:78× (bone) |
p.R201C |
| DX1180 | F | 6 | No | Yes | Yes | ND | Negative | - | Negative | - | - |
| GS1304 | F | 9 | Yes (monostotic) | No | Yes | ND | Negative | - | Negative | - | - |
| GS1311 | F | 9 | Yes | Yes | Yes | Ovarian cyst | Negative | - | Negative | - | - |
| GS1312 | F | 9 | ND | Yes | Yes | ND | Negative | - | Positive (1%) G:225× A:4× |
- | p.R201H |
| GS1313 | F | ND | ND | ND | ND | ND | - | Negative | - | Negative | - |
| GS1329 | F | 5 | ND | ND | Yes | ND | Negative | - | Negative | - | - |
| GS1333 | M | 68 | Yes (polyostotic) | ND | ND | ND | Negative | - | Negative | - | - |
According to the received clinical information, 6 patients presented isolated fibrous dysplasia, whereas 17 satisfied the clinical criteria for MAS. The rest of the patients had some of the characteristics of MAS in isolation and, probably, a screening test was requested for the genetic study. It must be outlined that four females had ovarian cysts, two patients had hyperthyroidism, one had GH excess, and one patient had neonatal Cushing’s syndrome.
Regarding the samples, only blood (or DNA extracted from blood) was received from 17 patients, only the affected tissue (either fresh (6) or paraffin-embedded (1)) was received from 7 patients, and from 16 patients, both blood and the affected tissue (either fresh (13) or paraffin-embedded (3)) samples were received.
Sanger sequencing did not detect genetic variants in the DNA extracted from peripheral blood, but in four affected tissues (two bones, one pituitary, and one thyroid), the causative variant was identified. Of these, two carried the p.R201C mutation, and the other two carried the p.R201H mutation.
Regarding the NGS results, in seven samples, the variant was called (i.e., present in the variant calling file, shown in dark grey in Table 1) using the integrated software. During the visualization of these variants by IGV, we noticed that the other samples (shown in light grey in Table 1) also showed a possible presence of genetic alteration with a frequency of ≤4%, which is below Sanger’s detection limit [27].
So, finally, NGS was able to identify the presence of the mutation in four of the blood samples analyzed from all cases in a very low mosaicism percentage (<5%). Regarding the tissues, in four of them, the p.R201C mutation was detected, and in the other three, the p.R201H mutation was detected. Of all of the positive tissues examined using NGS, three samples were also detected by Sanger sequencing, probably due to the high percentage in which the variant was present. In one of the samples identified as positive using Sanger sequencing, the NGS sequence quality was not good enough (<30× GS1189 in Table 1) to consider the identification of the variant as positive. This low depth was probably caused by the tissue being paraffin-embedded.
When correlating the identification or lack of identification of the variant using any of the sequencing techniques with the patient’s clinical data, we observed that it was identified in four of the six patients with FD, in five of the seventeen patients with MAS, and in a skin sample from a patient for whom no clinical data were sent when the genetic study was requested.
4. Discussion
Fibrous Dysplasia/McCune–Albright syndrome (FD/MAS) comprises a spectrum of disorders caused by activating somatic mutations in the GNAS gene [16]. As the genetic variants occur postzygotically at any stage of development, the patient may have the variant in some of its cells and not in others [23,26]. In general, in somatic mosaicism, different mosaic ratios may be found in a patient’s distinct body tissues. This means that, depending on the tissue analyzed and the sensitivity of the technique used, alterations that are actually present may not be identified (false negatives).
Currently, there are several techniques that allow for the detection of mosaicism [19,35,44,45]. In this research work, we tried to identify somatic activating pathogenic variants that cause FD/MAS in a cohort of patients received in the laboratory. Using conventional Sanger sequencing, the variant was identified in four tissue samples but in none of the blood samples. On the contrary, when the same samples were studied using NGS, the variants were detected in six affected tissues (including three of those identified through Sanger sequencing) and five blood samples.
This difference in the results obtained using the two techniques has been acutely reported in the literature [27,30]. Even though Sanger sequencing has been considered the gold-standard, it has been deeply demonstrated that its detection limit is compromised; mosaics below 10–20% are hardly detected and, in most cases, they may not be seen [27,30]. To overcome this outcome, in recent years, the tools and the sensitivity needed to detect mosaicism have increased substantially. To detect variants at the genetic level (short deletions/insertions and single-nucleotide variants (SNVs)), droplet-digital polymerase chain reaction (ddPCR) and real-time PCR using TaqMan probes [19,46] have been used.
On the other side, NGS produces single sequence reads, which enhances sensitivity and allows for a quantification of mosaicism. As the single reads can be counted, a precise percentage of mosaicism can be calculated, even in very low percentages [27,35]. In the present work, the sensitivity of the technique allowed us to find the mutation in seven samples (six different patients) in which the mosaicism was observed in very low levels (≤5%), which means that there are very few reads of the altered allele. As the number of reads in these mosaics is low, it is usually difficult to differentiate between a sequencing artifact and a real mosaic. The somatic variant caller uses a Poisson model to establish the Q score of a variant, and it excludes the variant if it presents a Q score below Q20 (which means there is a 1/100 probability of obtaining a false positive) [42]. Based on this model, it is not possible to call a 1% variant allele frequency (VAF) variant with confidence if the coverage is 200×, as the error rate is 1% (2/200 expected miscalls), i.e., there is an important overlap between noise and 1% VAF true variants. Probably, a coverage of 500–600×would enable variants below 5% VAF to be called. Of course, in a case where there is access to different tissues from the same person and there is more than one tissue confirming the presence of the variant, the confidence regarding the presence of the variant is high. But, if not, one way to solve this issue is to analyze the number of reads in each sense; if the reads are approximately the same, this provides an idea that it is a real variant, and not an artifact [47]. It is also important to confirm that the identified variant has a quality score over Q20 and that the quality of the reads containing it is good [48]. Moreover, the fact that the mutation identified is the one specific to that pathology also provides confidence, as seen in all of our samples.
Various biological materials may be tested in mosaicism investigation. Primarily, low invasive procedures are recommended in the sample collection process. Blood is one of the most easily obtained samples, and the collection procedure is not very invasive. However, genetic alterations in a mosaic are not usually present (or found) in blood. In this study, the variants were found only in 4 out of the 33 blood samples, which means there was a 12.12% rate of success (if all of the patients really present FD/MAS). On the other hand, 23 affected tissues were studied and, in 8, the variant was identified, which translates into a 34.78% success rate. As described previously, blood is not a good tissue to find mosaicism, as it is very rare to find it in blood [27,30]. So, in order to obtain more accurate results, it is important to study the affected tissues, even if obtaining samples is not so easy. Some studies also point to the preference of using biopsies more than blood in FD/MAS [27,30]. Nevertheless, we have to take into account that determining the total extent of the affected tissue is not an easy task [8]. Furthermore, different tissues may show different degrees of mosaicism levels, and if the biopsy includes normal tissue, false negatives may occur [8,35], i.e., if the sample is not adequately selected, the results that are obtained from the genetic study may not correspond to reality. Furthermore, we must not forget that it is not only important to choose the right tissue to study, but also the quality in which it has been preserved. In this study, several of the tissues were embedded in paraffin, which generated massive sequences of poor quality (with a low reading depth), which prevented us from considering the findings of the variants as true positives [8,49].
However, we must keep in mind that maybe some of the negative results can be true negatives (due to misdiagnosis) instead of false negatives (due to technical problems). For example, the success rate in identifying the genetic variant was higher in the samples received from patients with suspected FD than those received with extra-skeletal features. In fact, in some of the samples we received, the GNAS test was requested as a screening test, as the patients were still very young. However, just in this type of patients, it is important to bear in mind that the fewer symptoms present, the more difficult it seems to be to identify the presence of the mutation [19], even if essential for a clinical diagnosis that is really difficult in children who only present a single manifestation, such as hyperpigmented skin macules or precocious puberty. In these patients, both the clinical and the molecular diagnoses are difficult to confirm.
In brief, despite the improvements in techniques or in the selection of tissues to be analyzed, our data and those of other teams confirm that the success rate in detecting mosaicism in patients with MAS depends on (i) the severity of the disease, (ii) the tissue(s) studied, and (iii) the technique used. So, as already proposed in the international guidelines, a genetic study is only recommended when the diagnosis is uncertain, especially in patients with monostotic fibrous dysplasia without other skeletal or extra-skeletal abnormalities [8].
With the arrival of new genetic and genomic technologies, it may be worthwhile to continue researching and validating their usefulness for the diagnosis of this disease in different tissues, as suggested in the novel proposal presented by Roszko et al. [19]. It may also be advisable to update the current clinical diagnostic guidelines (mainly in the early stages of the disease) for better diagnosis and follow-up for these patients.
5. Conclusions
Genetic mosaicism (especially in low frequency) is difficult to detect using conventional techniques, such as Sanger sequencing. Due to its capacity to detect mosaicism, NGS is a reliable tool that facilitates the detection of mosaicism, even in very low levels, and it is better when analyzing the affected tissue. Nevertheless, it is important to establish a good bioinformatic pipeline to detect these variants and to differentiate real variants from artefacts.
Acknowledgments
The authors would like to thank the Spanish Fibrous Dysplasia Association (ADF) for inspiring this work. We would also like to thank all of the professionals who have sent us samples from their patients, giving us the opportunity to consider how to improve our work. And, of course, we thank the anonymous patients whose samples we have analyzed.
Author Contributions
Conceptualization, G.P.d.N.; methodology, G.P.d.N., Y.V. and A.P.; validation, A.M.-A. and A.P.; formal analysis, A.M.-A. and A.P.; investigation, Y.V., A.M.-A. and A.P.; data curation, A.P.; writing—original draft preparation, Y.V.; writing—review and editing, G.P.d.N. and A.P.; supervision, G.P.d.N.; funding acquisition, G.P.d.N. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of Euskadi- Basque Country (CEIC-E) (protocol code PI2017018).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in the study are deposited in the European Nucleotide Archive (ENA) repository (https:/www.ebi.ac.uk/ena, accessed on 18 December 2023), accession number PRJEB71103.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Instituto de Salud Carlos III (ISCIIII) of the Ministry of Economy and Competitiveness (Spain), which was co-financed by the European Regional Development Fund, grant number PI20/00950; the Department of Health of the Basque Government, grant number GV2021/111056; and the 2019 Research Unit Grant from the European Society of Paediatric Endocrinology (ESPE).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Weinstein L.S., Chen M., Liu J. Gs(Alpha) Mutations and Imprinting Defects in Human Disease. Ann. N. Y. Acad. Sci. 2002;968:173–197. doi: 10.1111/j.1749-6632.2002.tb04335.x. [DOI] [PubMed] [Google Scholar]
- 2.Thiele S., Mantovani G., Barlier A., Boldrin V., Bordogna P., De Sanctis L., Elli F.M., Freson K., Garin I., Grybek V., et al. From Pseudohypoparathyroidism to Inactivating PTH/PTHrP Signalling Disorder (IPPSD), a Novel Classification Proposed by the EuroPHP Network. Eur. J. Endocrinol. 2016;175:P1–P17. doi: 10.1530/EJE-16-0107. [DOI] [PubMed] [Google Scholar]
- 3.Fokkema I.F.A.C., Taschner P.E.M., Schaafsma G.C.P., Celli J., Laros J.F.J., den Dunnen J.T. LOVD v.2.0: The next Generation in Gene Variant Databases. Hum. Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 4.Elli F.M., Linglart A., Garin I., de Sanctis L., Bordogna P., Grybek V., Pereda A., Giachero F., Verrua E., Hanna P., et al. The Prevalence of GNAS Deficiency-Related Diseases in a Large Cohort of Patients Characterized by the EuroPHP Network. J. Clin. Endocrinol. Metab. 2016;101:3657–3668. doi: 10.1210/jc.2015-4310. [DOI] [PubMed] [Google Scholar]
- 5.Nakamoto J.M., Zimmerman D., Jones E.A., Loke K.Y., Siddiq K., Donlan M.A., Brickman A.S., Van Dop C. Concurrent Hormone Resistance (Pseudohypoparathyroidism Type Ia) and Hormone Independence (Testotoxicosis) Caused by a Unique Mutation in the Gαs Gene. Biochem. Mol. Med. 1996;58:18–24. doi: 10.1006/bmme.1996.0027. [DOI] [PubMed] [Google Scholar]
- 6.Iiri T., Herzmark P., Nakamoto J.M., van Dop C., Bourne H.R. Rapid GDP Release from Gsα in Patients with Gain and Loss of Endocrine Function. Nature. 1994;371:164–168. doi: 10.1038/371164a0. [DOI] [PubMed] [Google Scholar]
- 7.Landis C.A., Masters S.B., Spada A., Pace A.M., Bourne H.R., Vallar L. GTPase Inhibiting Mutations Activate the α Chain of Gs and Stimulate Adenylyl Cyclase in Human Pituitary Tumours. Nature. 1989;340:692–696. doi: 10.1038/340692a0. [DOI] [PubMed] [Google Scholar]
- 8.Javaid M.K., Boyce A., Appelman-Dijkstra N., Ong J., Defabianis P., Offiah A., Arunde P., Shaw N., Pos V.D., Underhil A., et al. Best Practice Management Guidelines for Fibrous Dysplasia/McCune-Albright Syndrome: A Consensus Statement from the FD/MAS International Consortium. Orphanet J. Rare Dis. 2019;14:139. doi: 10.1186/s13023-019-1102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ringel M.D., Schwindinger W.F., Levine M.A. Clinical Implications of Genetic Defects in G Proteins. The Molecular Basis of McCune-Albright Syndrome and Albright Hereditary Osteodystrophy. Medicine. 1996;75:171–184. doi: 10.1097/00005792-199607000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Miyado M., Fukami M., Takada S., Terao M., Nakabayashi K., Hata K., Matsubara Y., Tanaka Y., Sasaki G., Nagasaki K., et al. Germline-Derived Gain-of-Function Variants of Gs α-Coding GNAS Gene Identified in Nephrogenic Syndrome of Inappropriate Antidiuresis. J. Am. Soc. Nephrol. 2019;30:877–889. doi: 10.1681/ASN.2018121268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biebermann H., Kleinau G., Schnabel D., Bockenhauer D., Wilson L.C., Tully I., Kiff S., Scheerer P., Reyes M., Paisdzior S., et al. A New Multisystem Disorder Caused by the Gαs Mutation p.F376V. J. Clin. Endocrinol. Metab. 2019;104:1079–1089. doi: 10.1210/jc.2018-01250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyce A.M., Turner A., Watts L., Forestier-Zhang L., Underhill A., Pinedo-Villanueva R., Monsell F., Tessaris D., Burren C., Masi L., et al. Improving Patient Outcomes in Fibrous Dysplasia/McCune-Albright Syndrome: An International Multidisciplinary Workshop to Inform an International Partnership. Arch. Osteoporos. 2017;12:21. doi: 10.1007/s11657-016-0271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leet A.I., Collins M.T. Current Approach to Fibrous Dysplasia of Bone and McCune–Albright Syndrome. J. Child. Orthop. 2007;1:3–17. doi: 10.1007/s11832-007-0006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albright F., Butler A.M., Hampton A.O., Smith P. Syndrome Characterized by Osteitis Fibrosa Disseminata, Areas of Pigmentation and Endocrine Dysfunction, with Precocious Puberty in Females. N. Engl. J. Med. 1937;216:727–746. doi: 10.1056/NEJM193704292161701. [DOI] [Google Scholar]
- 15.McCune D. Osteitis Fibrosa Cystica: The Case of a Nine-Year-Old Girl Who Also Exhibits Precocious Puberty, Multiple Pigmen-Tation of the Skin and Hyperthyroidism. Am. J. Dis. Child. 1936;52:743–744. [Google Scholar]
- 16.Dumitrescu C.E., Collins M.T. McCune-Albright Syndrome. Orphanet J. Rare Dis. 2008;3:12. doi: 10.1186/1750-1172-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mieszczak J., Eugster E.A. Treatment of Precocious Puberty in McCune-Albright Syndrome. Pediatr. Endocrinol. Rev. 2007;4((Suppl. S4)):419–422. [PMC free article] [PubMed] [Google Scholar]
- 18.Bianco P., Riminucci M., Majolagbe A., Kuznetsov S.A., Collins M.T., Mankani M.H., Corsi A., Bone H.G., Wientroub S., Spiegel A.M., et al. Mutations of the GNAS1 Gene, Stromal Cell Dysfunction, and Osteomalacic Changes in Non-McCune-Albright Fibrous Dysplasia of Bone. J. Bone Miner. Res. 2000;15:120–128. doi: 10.1359/jbmr.2000.15.1.120. [DOI] [PubMed] [Google Scholar]
- 19.Roszko K.L., Guthrie L., Li X., Collins M.T., de Castro L.F., Boyce A.M. Identification of GNAS Variants in Circulating Cell-Free DNA from Patients with Fibrous Dysplasia/McCune Albright Syndrome. J. Bone Miner. Res. 2023;38:443–450. doi: 10.1002/jbmr.4766. [DOI] [PubMed] [Google Scholar]
- 20.Idowu B.D., Al-Adnani M., O’Donnell P., Yu L., Odell E., Diss T., Gale R.E., Flanagan A.M. A Sensitive Mutation-Specific Screening Technique for GNAS1 Mutations in Cases of Fibrous Dysplasia: The First Report of a Codon 227 Mutation in Bone. Histopathology. 2007;50:691–704. doi: 10.1111/j.1365-2559.2007.02676.x. [DOI] [PubMed] [Google Scholar]
- 21.Weinstein L.S., Shenker A., Gejman P.V., Merino M.J., Friedman E., Spiegel A.M. Activating Mutations of the Stimulatory G Protein in the McCune-Albright Syndrome. N. Engl. J. Med. 1991;325:1688–1695. doi: 10.1056/NEJM199112123252403. [DOI] [PubMed] [Google Scholar]
- 22.Riminucci M., Saggio I., Robey P.G., Bianco P. Fibrous Dysplasia as a Stem Cell Disease. J. Bone Miner. Res. 2006;21((Suppl. S2)):P125–P131. doi: 10.1359/jbmr.06s224. [DOI] [PubMed] [Google Scholar]
- 23.Hall J.G. Review and Hypotheses: Somatic Mosaicism: Observations Related to Clinical Genetics. Am. J. Hum. Genet. 1988;43:355–363. [PMC free article] [PubMed] [Google Scholar]
- 24.Youssoufian H., Pyeritz R.E. Mechanisms and Consequences of Somatic Mosaicism in Humans. Nat. Rev. Genet. 2002;3:748–758. doi: 10.1038/nrg906. [DOI] [PubMed] [Google Scholar]
- 25.Mohiuddin M., Kooy R.F., Pearson C.E. De Novo Mutations, Genetic Mosaicism and Human Disease. Front. Genet. 2022;13:983668. doi: 10.3389/fgene.2022.983668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veltman J.A., Brunner H.G. De Novo Mutations in Human Genetic Disease. Nat. Rev. Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 27.Mefford H.C. Mosaicism in Clinical Genetics. Cold Spring Harb. Mol. Case Stud. 2021;7:a006125. doi: 10.1101/mcs.a006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vado Y., Pereda A., Manero-Azua A., Perez de Nanclares G. Frequency of de Novo Variants and Parental Mosaicism in Families with Inactivating PTH/PTHrP Signaling Disorder Type 2. Front. Endocrinol. 2023;13:1055431. doi: 10.3389/fendo.2022.1055431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee M., Lui A.C.Y., Chan J.C.K., Doong P.H.L., Kwong A.K.Y., Mak C.C.Y., Li R.H.W., Kan A.S.Y., Chung B.H.Y. Revealing Parental Mosaicism: The Hidden Answer to the Recurrence of Apparent de Novo Variants. Hum. Genom. 2023;17:91. doi: 10.1186/s40246-023-00535-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohlin A., Wernersson J., Engwall Y., Wiklund L., Björk J., Nordling M. Parallel Sequencing Used in Detection of Mosaic Mutations: Comparison with Four Diagnostic DNA Screening Techniques. Hum. Mutat. 2009;30:1012–1020. doi: 10.1002/humu.20980. [DOI] [PubMed] [Google Scholar]
- 31.Morandi A., Bonnefond A., Lobbens S., Carotenuto M., del Giudice E.M., Froguel P., Maffeis C. A Girl with Incomplete Prader-Willi Syndrome and Negative MS-PCR, Found to Have Mosaic Maternal UPD-15 at SNP Array. Am. J. Med. Genet. Part A. 2015;167:2720–2726. doi: 10.1002/ajmg.a.37222. [DOI] [PubMed] [Google Scholar]
- 32.Sánchez J., Fernández R., Madruga M., Bernabeu-Wittel J., Antiñolo G., Borrego S. Somatic and Germ-Line Mosaicism of Deletion 15q11.2-Q13 in a Mother of Dyzigotic Twins with Angelman Syndrome. Am. J. Med. Genet. A. 2014;164A:370–376. doi: 10.1002/ajmg.a.36281. [DOI] [PubMed] [Google Scholar]
- 33.Schwab A.L., Tuohy T.M.F., Condie M., Neklason D.W., Burt R.W. Gonadal Mosaicism and Familial Adenomatous Polyposis. Fam. Cancer. 2008;7:173–177. doi: 10.1007/s10689-007-9169-1. [DOI] [PubMed] [Google Scholar]
- 34.Campbell I.M., Yuan B., Robberecht C., Pfundt R., Szafranski P., McEntagart M.E., Nagamani S.C.S., Erez A., Bartnik M., Wiśniowiecka-Kowalnik B., et al. Parental Somatic Mosaicism Is Underrecognized and Influences Recurrence Risk of Genomic Disorders. Am. J. Hum. Genet. 2014;95:173–182. doi: 10.1016/j.ajhg.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin L., Wang J., Tian X., Yu H., Truong C., Mitchell J.J., Wierenga K.J., Craigen W.J., Zhang V.W., Wong L.-J.C. Detection and Quantification of Mosaic Mutations in Disease Genes by Next-Generation Sequencing. J. Mol. Diagn. 2016;18:446–453. doi: 10.1016/j.jmoldx.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 36.White T.B., McCoy A.M., Streva V.A., Fenrich J., Deininger P.L. A Droplet Digital PCR Detection Method for Rare L1 Insertions in Tumors. Mob. DNA. 2014;5:30. doi: 10.1186/s13100-014-0030-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daly A.F., Yuan B., Fina F., Caberg J.-H., Trivellin G., Rostomyan L., de Herder W.W., Naves L.A., Metzger D., Cuny T., et al. Somatic Mosaicism Underlies X-Linked Acrogigantism Syndrome in Sporadic Male Subjects. Endocr. Relat. Cancer. 2016;23:221–233. doi: 10.1530/ERC-16-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilbe M., Gudmundsson S., Johansson J., Ameur A., Stattin E.-L., Annerén G., Malmgren H., Frykholm C., Bondeson M.-L. A Novel Approach Using Long-Read Sequencing and DdPCR to Investigate Gonadal Mosaicism and Estimate Recurrence Risk in Two Families with Developmental Disorders. Prenat. Diagn. 2017;37:1146–1154. doi: 10.1002/pd.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vasilev V., Daly A.F., Thiry A., Petrossians P., Fina F., Rostomyan L., Silvy M., Enjalbert A., Barlier A., Beckers A. McCune-Albright Syndrome: A Detailed Pathological and Genetic Analysis of Disease Effects in an Adult Patient. J. Clin. Endocrinol. Metab. 2014;99:E2029–E2038. doi: 10.1210/jc.2014-1291. [DOI] [PubMed] [Google Scholar]
- 40.Baker S.W., Duffy K.A., Richards-Yutz J., Deardorff M.A., Kalish J.M., Ganguly A. Improved Molecular Detection of Mosaicism in Beckwith-Wiedemann Syndrome. J. Med. Genet. 2021;58:178–184. doi: 10.1136/jmedgenet-2019-106498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manero-Azua Á., Pereda A., González Cabrera N., Martínez de Salinas Santamaría M.Á., Cámara Balda A., Pérez de Nanclares G. Vitamin D Deficiency in Adulthood: Presentation of 2familial Cases Simulating Pseudohypoparathyroidism. Med. Clin. 2023;161:493–497. doi: 10.1016/j.medcli.2023.06.009. [DOI] [PubMed] [Google Scholar]
- 42.Illumina . Somatic Variant Caller. Volume 1 Illumina; San Diego, CA, USA: 2014. [Google Scholar]
- 43.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang S., Zhou Y., Xiao G., Qiu X. Application of Various Genetic Analysis Techniques for Detecting Two Rare Cases of 9p Duplication Mosaicism during Prenatal Diagnosis. Mol. Genet. Genomic Med. 2023;11:e2229. doi: 10.1002/mgg3.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alfirevic Z., Navaratnam K., Mujezinovic F. Amniocentesis and Chorionic Villus Sampling for Prenatal Diagnosis. Cochrane Database Syst. Rev. 2017;9:CD003252. doi: 10.1002/14651858.CD003252.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Sanctis L., Galliano I., Montanari P., Matarazzo P., Tessaris D., Bergallo M. Combining Real-Time COLD- and MAMA-PCR TaqMan Techniques to Detect and Quantify R201 GNAS Mutations in the McCune-Albright Syndrome. Horm. Res. Paediatr. 2017;87:342–349. doi: 10.1159/000463384. [DOI] [PubMed] [Google Scholar]
- 47.Li H. Toward Better Understanding of Artifacts in Variant Calling from High-Coverage Samples. Bioinformatics. 2014;30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strom S.P. Current Practices and Guidelines for Clinical Next-Generation Sequencing Oncology Testing. Cancer Biol. Med. 2016;13:3–11. doi: 10.20892/j.issn.2095-3941.2016.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cazzato G., Caporusso C., Arezzo F., Cimmino A., Colagrande A., Loizzi V., Cormio G., Lettini T., Maiorano E., Scarcella V.S., et al. Formalin-Fixed and Paraffin-Embedded Samples for Next Generation Sequencing: Problems and Solutions. Genes. 2021;12:1472. doi: 10.3390/genes12101472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in the study are deposited in the European Nucleotide Archive (ENA) repository (https:/www.ebi.ac.uk/ena, accessed on 18 December 2023), accession number PRJEB71103.