Abstract
The underlying disease mechanisms for feline degenerative joint disease (DJD) are mostly unidentified. Today, most of what is published on mammalian arthritis is based on human clinical findings or on mammalian models of human arthritis. However, DJD is a common occurrence in the millions of domestic felines worldwide. To get a better understanding of the changes in biological pathways that are associated with feline DJD, this study employed a custom-designed feline GeneChip, and the institution’s unique access to large sample populations to investigate genes and proteins from whole blood and serum that may be up- or down-regulated in DJD cats. The GeneChip results centered around three main pathways that were affected in DJD cats: immune function, apoptosis and oxidative phosphorylation. By identifying these key disease-associated pathways it will then be possible to better understand disease pathogenesis and diagnose it more easily, and to better target it with pharmaceutical and nutritional intervention.
Introduction
There is increasing interest in feline degenerative joint disease (DJD). 1 Retrospective studies suggest, and more recent prospective studies indicate, that radiographic evidence of DJD is common, with up to 90% of cats being affected.2–5 Other studies suggest that this DJD can be associated with pain, making it an important feline disease.6–9 As has been discussed recently, little is known about the etiology of feline DJD. 1
Although several authors have suggested that a large proportion of DJD in the cat is primary, there is currently no evidence of this.2,3 Two primary forms of osteoarthritis (OA) are fairly well recognized in cats: Scottish Fold osteochondrodysplasia and mucopolysaccharidosis.10–20 Currently, the documented secondary causes of DJD in cats are nutritional, hip dysplasia, the non-infectious polyarthropathies and infectious arthropathies.21–31 Obesity has also been suggested as a driving or confounding factor. Despite this information, the etiology of the vast majority of feline DJD seen in practices is unknown.
Research suggests a wide variety of contributors to DJD in humans that range from genetic to environmental.32,33 The first and most commonly cited contributor is a genetic risk factor. 34 This is particularly true for the autoimmune types of arthritis because the body’s immune system seems to undergo a shift in gene expression that elicits an inappropriate intolerance for self- antigens.35,36 Gene expression studies in the human literature illustrate great heterogeneity in gene expression patterns between tested individuals, which complicates the search for possible therapeutic targets.34,37–43 Genes that do appear routinely to be differentially expressed between human rheumatoid arthritis (RA) patients and age-matched controls include immune system and inflammation-associated genes, such as major histocompatibility complex (MHC) genes I, II and III, and tumor necrosis factor (TNF)-α, signaling pathways, such as the interferon (IFN)/signal transducer and activation of transcription factor (STAT), and extracellular proteins, like the matrix metalloproteinases (MMP).34,37–40,44 Genes and proteins that have been found to be differentially expressed in human OA patients include structural proteins collagen I and collagen II alpha I (COL2A1), transcription factors SOX9 and 11, MMP13, the cytokine interleukin-1 beta (IL1B) and the proteoglycan aggrecan.45,46 The majority of arthritis gene expression studies reported in the literature have been carried out in humans with cartilage, bone or synovial tissue. These assorted samples may be a cause of variability in the findings of what is actually occurring in the disease state.
The aim of this study was to investigate feline-specific gene expression differences between DJD cats and age-matched controls in order to obtain a clearer picture of the underlying mechanisms of the feline disease.
Materials and methods
Experimental design
All aspects of this work were conducted in accordance with the Hill’s Pet Nutrition Global Animal Welfare Policy and were approved by Hill’s Institutional Animal Care and Use Committee. All owners provided written informed consent prior to enrollment of their pet in the study. Owing to the provisions of the Global Animal Welfare Policy of Hill’s Pet Nutrition, only whole blood samples were collected from enrolled cats over the course of a period of 3 weeks. Cats diagnosed with clinical DJD (from clinics throughout the USA and Canada) were compared with whole blood samples taken from age-matched non-DJD cats from the Hill’s Pet Nutrition internal colony. The colony cats, although originating from different suppliers, may be more uniform at the genotype level. This could have been a source of bias in the data; however, the risk was mitigated to a certain extent, at least at the technical level, through the use of routine quality control measures, such as GeneChip quantile normalization and overall probe intensity normalization. Furthermore, all cats underwent the same examination process regardless of whether they were DJD or control cats. The eligibility of each DJD candidate was assessed by complete physical examination (including orthopedic evaluation), medical, drug and dietary histories, laboratory evaluation (complete blood count, serum biochemistry panel, total T4, feline immunodeficiency virus and feline leukemia virus) and radiography. Blood samples were collected by percutaneous vene-puncture following an overnight fast. Each tube was labeled with the study number, animal identification and date, and submitted to Antech Laboratories using the sample kit mailed to investigators by Antech Laboratories.
Adult cats with clinical signs and radiographic changes consistent with DJD of the stifles, hips, shoulders, elbows, tarsus or carpus were potential candidates for this study. Exclusion criteria were: (i) cats that had suffered recent traumatic injuries; (ii) cats with severe concurrent systemic disease, for example kidney failure, diabetes, hyperthyroidism or obesity; (iii) cats with technically inadequate radiographic studies; (iv) cats with erosive polyarthropathy (diagnosed radiographically) considered to be due to immune-mediated or infectious causes; (v) cats that had suffered previous fractures, luxations or osteomyelitis involving any part of the skeleton; (vi) cats with non-joint soft tissue mineralization, for example, muscle and tendon; (vii) Manx cats or any other cat with a spinal anomaly; (viii) cats with significant neurologic deficits (significant spinal cord disease other than lumbosacral degeneration); and (ix) cats with severe osteopenia (eg, due to renal disease or senile changes). Supplements or foods with increased levels of n-3 fatty acids were not permitted. Cats eating any of the following wet foods within the previous 3 months were excluded from the study: Friskies Variety Pack Ocean Whitefish, Iams Ocean Fish, Iams Turkey, Iams Chicken, Hill’s Science Diet Kitten, Hill’s Prescription Diet k/d, Hill’s Prescription Diet a/d, and Purina Veterinary Diets DM. Cats eating any of the following dry foods within the previous 3 months were excluded: Hill’s Science Diet Kitten, Iams Kitten Food, Natural Balance Ultra Premium, Iams Cat Original Formula, Hill’s Science Diet Advanced Protection, ProPlan Chicken & Rice, Iams Less Active/Weight Control, Purina Veterinary Diets DM, Purina Veterinary Diets OM, and Iams Restricted Calorie. Concomitant analgesic or anti-inflammatory medications, for example, corticosteroids or non-steroidal anti-inflammatory drugs (NSAIDs) were permitted if started at least 4 weeks prior to the screening visit and continued at the same dose for the entire feeding period.
Gene expression analysis — RNA isolation
Whole blood was collected and processed according to the PAXgene Blood RNA Isolation Kit (Qiagen) manufacturer’s instructions. All extracted RNA samples obtained from the whole blood were quantified by absorbance readings at 260 and 280 nm with a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). The RNA quality was determined with a 2100 Bioanalyzer (Agilent Technologies), according to the manufacturer’s instructions. RNA integrity was determined by 28S:18S ribosomal RNA ratio and RNA integrity number (RIN; Agilent 2100 RIN Beta Version Software). Purified RNA samples were stored at −80°C.
Probe preparation from RNA
Labeling and amplification reagents were obtained from NuGEN Technologies and biotinylated cDNA targets were prepared according to the manufacturer’s instructions. Double-stranded cDNA was synthesized from 30 ng total RNA, followed by a linear isothermal amplification (SPIA Amplification) step to produce single-stranded cDNA. Fragmentation was followed by a direct labeling process that attached biotin to the amplified probe. Probe purifications were performed using DNA Clean and Concentrator– 25 (Zymo Research) and the DyeEx 2.0 Spin Kit (Qiagen).
Gene array hybridization and processing
After pre-hybridization for 20 mins at 45°C, 1.3 µg of each target cDNA was mixed with Affymetrix hybridization controls (Affymetrix) in hybridization buffer and hybridized with the Hills-Feline-2 GeneChip for 16 h at 45°C. After the hybridization cocktails were removed, the chips were washed in a fluidics station with low-stringency buffer (6X standard saline phosphate with ethylenediaminetetraacetic acid and 0.01% Tween20) and high stringency buffer (100 mM N-morpholino-ethanesulfonic acid, 0.1 M NaCl and 0.01% Tween20) and stained with streptavidin phycoerythrin (SAPE). This process was followed by incubation with normal goat IgG and biotinylated mouse anti-streptavidin antibody (Vector Laboratories) followed by re-staining with SAPE. The chips were scanned in a GeneChip Scanner 3000 7G (Affymetrix) to detect hybridization signals. Image inspection was performed manually immediately after each scan.
Data analysis
The Genomic Suite for Gene Expression Data software (Partek) was used for data analysis. The robust multichip average algorithm was used for background adjustment, normalization and probe-level summarization of the GeneChip samples. 47 Analysis of variance was performed to find significantly differentially expressed genes between any two groups with a minimal false discovery rate control at 0.1 and a fold change of 1.3 in each direction. Empirical studies have revealed that the Hills-Feline-2 GeneChip has an associated background noise level of 1.3-fold. Therefore, all analyses presented here employ a ± 1.3-fold cut-off. Additionally, the false discovery rate threshold of 0.1 (meaning that 10% of observations are due to chance alone) was chosen as the minimum level of acceptable statistical significance. Principal component analysis (PCA) was performed on the data from the GeneChip. Genes that were identified as differentially expressed were then put into the MetaCore Pathway Analysis and Data Mining package (Thompson Reuters Bioinformatics Software) in order to ascertain pertinent biological pathways that varied between the experimental groups.
Proteomic analysis
The proteomic study was performed with serum as opposed to the whole blood used in the genomic study. The pooling of samples allows for the enhancement of differentiating signals without the exaggeration of noise within the experiment and is commonly employed in proteomic studies to improve signal:noise ratios and reduce the time for analysis.
Sample preparation
One milliliter of serum from each sample pool was treated with the ProteoMiner Protein Enrichment and Sequential Elution Kit, according to the manufacturer’s instructions (BioRad Laboratories). This was performed to normalize proteins within the samples. Protein concentration was then determined using the 2-D Quant Kit (GE Healthcare).
Two-dimensional electrophoresis
For two-dimensional (2D) electrophoresis, samples were labeled with Cydye 3 or Cydye 5 (GE Healthcare). An internal global standard was generated by pooling equivalent amounts of all samples into the standard pool. This standard pool was labeled with Cydye 2 (GE Healthcare). Equal amounts of protein from the control group, arthritic group and global standard were loaded on the same immobilized pH gradient (IPG) strip (24 cm, pH 4–7; GE Healthcare) and rehydrated at 30V for 12 h. Isoelectric focusing conditions were as follows: step 1 at 500 V for 1 h, step 2 at 1000 V for 1 h and step 3 at 8000 V for 80,000 VHr. Focused IPG strips were equilibrated and then loaded onto 9–14% gradient sodium dodecyl sulfate polyacrylamide gels for second dimension separation. Gels were then scanned with a Typhoon 9400 fluorescent imager (GE Healthcare) for quantification. Differentially-expressed protein spots were cut using ProtPic (Genomic Solutions), enzymatically digested and analyzed using an Orbitrap Discovery mass spectrometer (Thermo Fisher Scientific).
Data analysis
The 2D experimental data were analyzed using Non-Linear’s Progenesis SameSpots software (Nonlinear Dynamics). Differentially-expressed proteins (P <0.05 and fold change ≥1.3) were cut, and tryptic digests were performed in gel and identified using the liquid chromatography tandem mass spectrometry method. Protein identification was ascertained by using PEAKS Studio software (Bioinformatics Solution) combined with X! Tandem (Global Proteome Machine Organization Proteomics Database and Open Source Software, see www.thegpm.org) and SEQUEST (Thermo Fisher Scientific) search engines. A confidence score of 0.99 was set for positive identification. The MetaCore Pathway Analysis and Data Mining package (Thompson Reuters Bioinformatics Software) was again used, this time for differentially regulated proteins, in order to ascertain pertinent biological pathways that varied between the experimental groups.
Results
Twenty-nine cats diagnosed with clinical DJD were recruited from over 300 cats screened and data were collected from 35 age-matched non-DJD cats from the Hill’s Pet Nutrition internal colony (Table 1).
Table 1.
Demographics of the study cats
| Age in years (median; maximum, minimum) | Weight in kg (median; maximum, minimum) | Sex | |
|---|---|---|---|
| DJD cats | 10; 14, 5 | 5.5; 8.4, 3.7 | F: 18; M: 11 |
| Control cats | 8.9; 14, 4 | 4.6; 6.8, 3.0 | F: 24; M: 11 |
DJD = degenerative joint disease
Genomic analysis was performed on samples from all 64 cats; however, only 18 viable serum samples were received from the DJD cats. Therefore, for the proteomic analysis, only these samples and 18 age-matched controls were used. To streamline the processing of all 18 samples per group for proteomic analysis, the samples were used to prepare nine pools of two samples each within each experimental group (the samples were randomly selected for pooling).
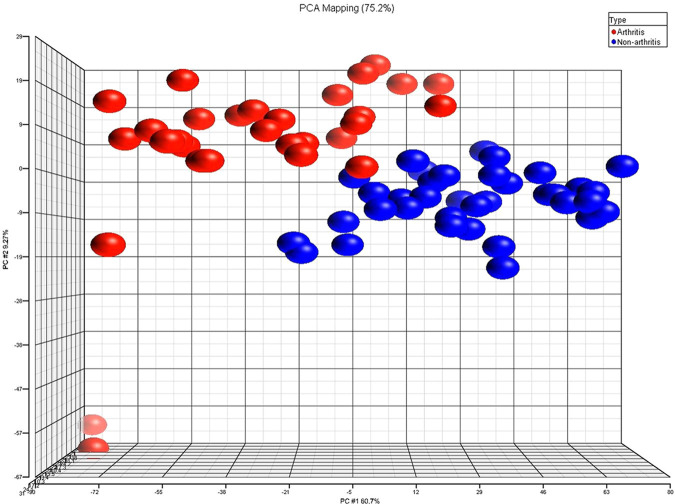
PCA was performed on the data from the GeneChip expression results for 29 DJD and 35 non-DJD cats to get a global picture of differences between the two experimental groups. The PCA was performed on 2842 genes that passed the quality and statistical criteria out of more than 20,000 genes total on the custom feline GeneChip. The results of the Partek Genomics Suite software are shown in Figure 1. In PCA analysis, researchers strive to get a high level view of patterns within complex data to more easily ascertain differences between experimental samples. In other words, PCA attempts to summarize large sets of data. For genomics work it allows one to examine general themes in gene expression between groups. Therefore, in this application of PCA, data of animals with more similar gene expression patterns will appear more compactly clustered on a graph, whilst those with less similar gene expression patterns will be located further apart on the graph. In Figure 1, non-DJD cats (in blue) were closely grouped together, indicating similar occurrences of gene expression between those animals. DJD cats (in red) were also clustered together, yet much more loosely. Subjectively, there appeared to be more variation in gene expression amongst the DJD animals than in non-DJD animals, which supports findings already reported in the literature.34,37–40,48 This could also reflect some variability due to the aforementioned potential differences in genetic uniformity between the two groups that could not be eliminated technically.
Figure 1.
Principal component analysis (PCA) of 2842 genes found to be associated with degenerative joint disease (DJD) cats as compared with control populations. DJD cat data (n = 29) are shown in red, whilst non-DJD cats (n = 35) are represented by blue; each spot represents one animal
Out of the 2842 total genes entered into the MetaCore Pathway Analysis software, there were 400 genes that the software could not assign to a specific pathway mainly due to database discrepancies. These genes were not examined further in this study. The output from the software for the other 2442 genes from the feline GeneChip demonstrates the likelihood that the up- or down-regulated genes from the GeneChip are involved in a particular pathway (Table 2). The most likely affected pathways in DJD cats, according to the GeneChip analysis and MetaCore software, include antigen presentation by MHC class I and II molecules, as well as T-cell receptor signaling components of the immune system and oxidative phosphorylation [energy production by the cell in the form of adenosine triphosphate (ATP)].
Table 2.
Top pathways represented by the differentially expressed genes in arthritic versus control populations. MetaCore by Thompson Reuters identified the likelihood that 2442 of the 2842 differentially expressed genes identified in the expression profile are involved in these pathways. The other 400 genes could not be assigned to a pathway by the software. The smaller the P-value, the greater the likelihood that the genes are involved in that pathway and the less likely the genes were differentially expressed by chance alone
| Pathway name | GeneGo p-value |
|---|---|
| Antigen presentation by MHC class I and II | 1.2e-08 |
| T-cell receptor signaling | 2.5e-08 |
| Oxidative phosphorylation | 2.2e-08 |
MHC = major histocompatibility complex
To closely investigate expression levels of key genes involved in antigen presentation by the immune system, the genes were grouped into three subcategories (Figure 2a–c). The subcategories within antigen presentation by the immune system were MHC class I and II molecules (a), proteasome molecules (b) and cathepsins (c). In general, all the genes involved in immune response that were analyzed in this study displayed a decrease in expression in the DJD animals when compared with non-DJD cats. The specific molecules outlined here in the figure all work together to ensure that the body’s immune system properly identifies foreign antigens. Proteasome and cathepsin molecules are thought to be primarily responsible for first digesting and then presenting foreign peptides to the immune system via MHC class I and II molecules.49–54 Figure 2 (a–c) illustrates the gene expression fold differences between the two experimental groups for these specific antigen presentation genes.
Figure 2.
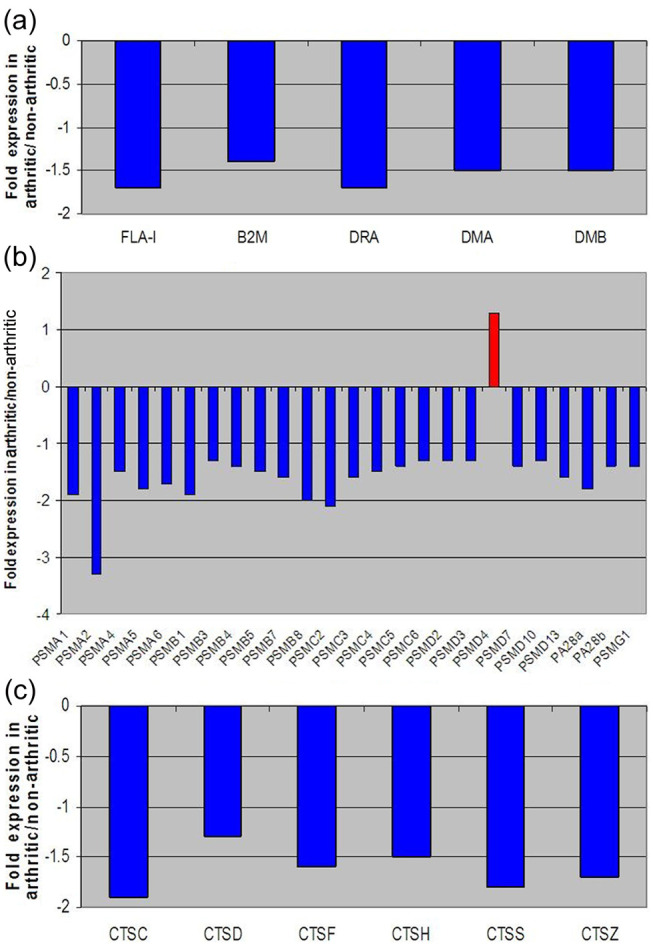
Fold change expression levels of key genes involved in antigen presentation. (a) Major histocompatibility complex (MHC) molecules; (b) proteasome and (c) cathepsin in degenerative joint disease (DJD) versus control populations. These graphs list fold changes in specific MHC class I and II (a), proteasome (b) and cathepsin (c) genes between DJD and non-DJD populations. All negative fold changes reflect down-regulation of the gene in DJD animals compared with non-DJD animals, and are represented in blue. All positive fold changes reflect up-regulation of the gene in DJD animals compared with non-DJD animals, and are represented in red
Figure 3 displays the fold change of some T-cell receptor genes found within DJD cats compared with controls. All the T-cell receptor and associated genes examined here were down-regulated in DJD animals compared with non-DJD animals. T-cell specific receptors, for example CD3, 4 and 8, to name a few, were all notably decreased in expression within DJD cats.
Figure 3.
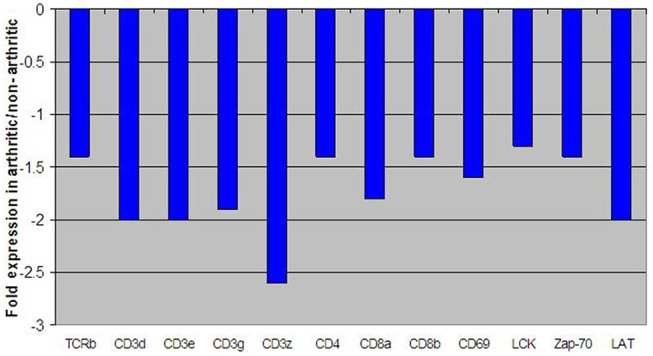
Fold change expression levels of key genes involved in T-cell receptor signaling. All negative fold changes reflect down-regulation of the gene in degenerative joint disease (DJD) animals compared with non-DJD animals and are represented in blue
Apoptosis-associated genes were also found to be differentially expressed between the two groups. Figure 4 shows the fold change expression levels of key genes involved in apoptosis; anti-apoptotic genes were up-regulated in DJD animals. 55
Figure 4.
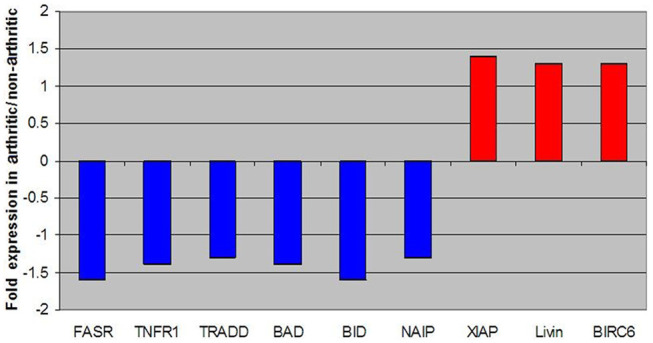
Fold change expression levels of key genes involved in apoptosis. All negative fold changes reflect down-regulation of the gene in degenerative joint disease (DJD) animals compared with non-DJD and are represented in blue. All positive fold changes reflect up-regulation of the gene in DJD animals compared with non-DJD, and are represented in red
Expression levels of genes that participate in oxidative phosphorylation were also distinct between DJD and non-DJD cats. The differentially expressed genes play important roles within the oxidative phosphorylation pathway that exists inside the mitochondria of cells (Figure 5). Of the five main protein complexes that comprise the oxidative phosphorylation pathway, all five were significantly down-regulated within DJD cats. Figure 5 shows down-regulation of genes within complexes I, II, III, IV and ATP synthase (also known as complex V).
Figure 5.
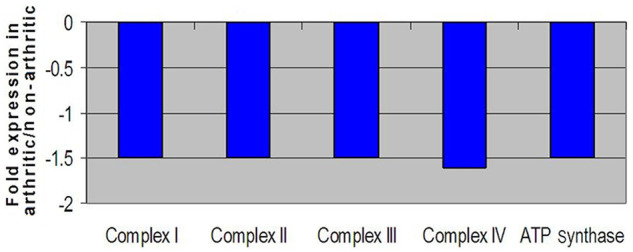
Fold change expression levels of key genes involved in oxidative phosphorylation. All negative fold changes reflect down-regulation of the gene in degenerative joint disease (DJD) animals compared with non-DJD and are represented in blue
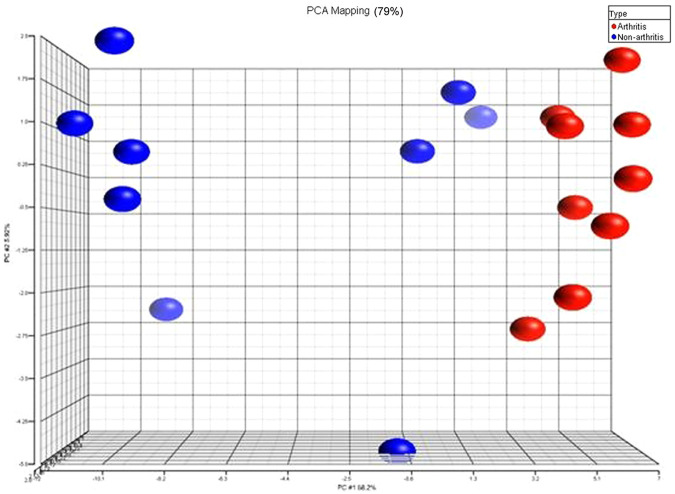
For the protein expression levels, Figure 6 exhibits the PCA plot for the 87 differentially expressed proteins between the two experimental groups. Once more, discrete differences in protein expression between the two groups were evident by the data point clustering on the graph. This analysis confirms that the experimental groups could be distinguished by differentially expressed serum proteins, as well as genes from whole blood.
Figure 6.
Principal component analysis (PCA) of the differentially expressed proteins found in differentially joint disease (DJD) cats compared with control populations. DJD cat data (n = 9) are shown in red, whilst non-DJD cats (n = 9) are represented by blue, and each spot represents one pooled sample
Figure 7 outlines differences in protein expression for the classical complement component of the immune system and clusterin, the latter representing a regulator of the complement system.56,57 Major proteins within the classical complement component of the immune system, such as C1s, C1r and C3, were notably up-regulated in DJD animals, whilst clusterin protein showed a decrease in expression compared with controls.
Figure 7.
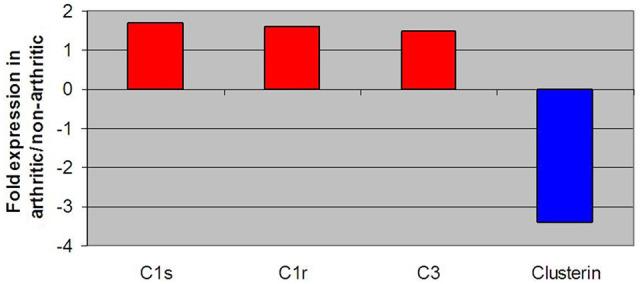
Fold change expression levels of key proteins involved in the complement component of immune system response and clusterin. All negative fold changes reflect down-regulation of the gene in degenerative joint disease (DJD) animals compared with non-DJD, and are represented in blue. All positive fold changes reflect up-regulation of the gene in DJD animals compared with non-DJD, and are represented in red
To control for age-specific differences between the DJD and non-DJD cats used in this study, gene expression differences were examined on the basis of age alone (Table 3). This was performed to test whether gene differences between the experimental groups could be attributed to the disease state and not to the difference in age between the two groups. Table 3 displays the results of two separate evaluations. The subjects were separated into >12 years of age versus 8–11 years of age and then separated into >11 years of age versus 7–10 years of age with the number of differentially expressed genes listed. When age alone was a factor, gene differences between the experimental groups were either non-existent or significantly smaller than when the subjects were grouped based on disease state alone. These findings indicate that the results reported here between the DJD and non-DJD populations were not greatly affected by age differences between the subjects.
Table 3.
Comparison of differentially-expressed genes based on age as opposed to disease state. To control for age-specific differences between the experimental groups, differentially-expressed genes were examined on the basis of age alone to compare between the mean ages of the two populations. The number and magnitude of differentially-expressed genes between groups when separated by age alone were significantly less than when the animals were grouped by disease state alone
| Age group comparison (years) | Number of samples | Number of differentially-expressed genes |
|---|---|---|
| >12 vs 8–11 | 27/25 | 159 |
| >11 vs 7–10 | 32/29 | 1014 |
Discussion
The proprietary Hills-Feline-2 GeneChip with more than 20,000 feline genes is a novel tool to evaluate the differences between cats with and without DJD. These investigations allow further insight into the disease, which is important given the prevalence of the disease.1,5 Such data may also uncover possible treatment opportunities.
The gene expression studies reported here were performed on whole blood. Previous gene expression studies of mammalian arthritis in the literature have reported variations in gene expression between human RA and OA patients, and non-arthritic subjects.34,37–43,45,46 However, these studies have been performed on joint tissue. The use of whole blood, as opposed to certain blood components, bone, cartilage or synovial samples, is a unique approach. Using whole blood and blood derivatives, such as serum and plasma, might afford a more global view of what is happening in the disease state because blood touches all tissue types in the body and many disease-associated proteins are shed and can be found in the plasma or serum. Furthermore, establishing a blood-associated molecular signature for DJD in cats can potentially provide practitioners with easier means to diagnose the disease and follow its progression and the effects of treatment.
Data reported here support the literature’s findings that gene expression differences within arthritic populations tend to be more variable overall than gene expression patterns in non-DJD individuals.34,37–40,48 This result serves to further emphasize the complexity of the illness and the difficulties in finding effective treatment options.
Based on our gene expression data, two of the most likely differentially expressed pathways involve immune system response: antigen presentation by MHC molecules and T-cell receptor signaling, both of which are necessary for normal immune response to foreign antigens. The remaining pathway that is most likely affected contains genes with a role in oxidative phosphorylation. These immune system-related genes were uniformly down-regulated in DJD cats compared with non-DJD cats. The literature supplies considerable evidence that immune system function generally declines and becomes less regulated with age in all mammals.58–60 Additionally, the risks for developing arthritis typically increase with age,44,61 although the data presented here suggest that arthritis is more than just normal ageing. Taken as a whole, this suggests that feline DJD arises, at least in part, owing to a dysfunctional immune system, which can become pathologically unregulated as the animal ages.
The act of antigen presentation by the immune system involves the proteasome complex, cathepsins, and MHC class I and II molecules. Proteasome and cathepsin molecules work in concert to break down foreign antigens that the body encounters; the digested fragments are then presented to the immune system via MHC class I and II molecules.49–52 Part of the adaptive immune response, MHC class I molecules are located on the surface of all nucleated cells,53,54 whilst MHC class II molecules reside mainly on antigen presentation cells of the immune system.62,63 Collectively, MHC class I and II molecules are necessary to present antigens to T-cells, which then elicit the body’s immune response by releasing cytokines to recruit macrophages and/or promote antibody release by B cells.53,54,62,63 It is noteworthy that all the antigen processing and presentation players examined here were down-regulated in cats with clinical DJD. Taken together, the data suggest a reduced ability to process and present antigens in DJD cats, thereby decreasing the immune system’s ability to identify foreign, and potentially harmful, cells and proteins within those animals.
In addition to a reduced ability to identify foreign antigens, the next step in the immune cascade, which is mediated by T-cells, was also diminished within DJD animals. Specifically, CDs 3, 4 and 8 were down- regulated in DJD animals compared with controls. The CDs 3, 4 and 8, transmembrane glycoproteins that bind to MHC molecules and act as co-receptors, are essential for mediating T-cell based immunity.53,54,63,64 CD3 is a part of the T-cell receptor complex which initiates a T-cell-mediated response against antigens, CD4 binds with MHC class II molecules, and CD8 is a glycoprotein that specifically binds to MHC class I molecules to recognize targets and mount an attack.62–64 Because all of these key T-cell players were found to be down-regulated in DJD cats, it is probable that cats with DJD have dampened T-cell activity and an overall impaired immune response. Whether these altered processes result in, or occur concurrently with, DJD remains to be uncovered.
Genes representing apoptotic cascades also differed between DJD and non-DJD cats. Specifically, pro-apoptotic genes BAD, BID and TRADD were down-regulated and anti-apoptotic genes, such as XIAP and BIRC6, were up-regulated in DJD animals. This finding has implications for many pathways throughout the body. For the immune system in particular, this could mean an abnormal build-up of non-functional cells that the body is unable to clear. Equilibrium between pro-apoptotic and anti-apoptotic mechanisms is vital to the proper maintenance and function of the immune system.65–67 Functionality of immune system cells is largely determined by the specificity of interactions between antigen presentation components, namely MHC and T-cell-associated molecules. 66 As DJD cats demonstrated a decrease in both MHC and T-cell associated genes, this suggests that dysfunctional immune cells exist in the DJD population. A deficiency in apoptotic cascades to remove these dysfunctional cells might compound the existing problem within these diseased animals. Evidence in the literature suggests that disequilibrium between apoptotic forces can lead to many ailments, such as immunodeficiency; autoimmunity, including systemic lupus erythematosus; and various degenerative disorders.68–71 Additionally, these findings appear to explain why corticosteroids are sometimes effective during the treatment of certain arthritidies. The literature suggests that glucocorticoids, such as dexamethasone, have a pro-apoptotic effect with lymphocytes; 66 therefore, the use of corticosteroidal compounds in DJD may help not only to reduce inflammation, but also to eliminate non-functional immune system cells from the body.
Another major area of differential gene expression between DJD and control animals was in the area of oxidative phosphorylation. Oxidative phosphorylation is the means by which the mitochondrial electron transport chain produces cellular energy in cells throughout the body.72,73 In this feline study, all of the protein complexes (I–V) that make up the electron transport chain revealed down-regulated genes compared with controls; these results, combined with the data generated here that control for age, strongly suggest that the differences in oxidative phosphorylation in the DJD population were above and beyond the changes that occur in the electron transport chain during the normal ageing process. 74 Mitochondrial dysfunction could be an important factor in the development of DJD. The literature solidly supports the theory of mitochondrial dysfunction in the realm of mammalian arthritis. One such study used human articular chondrocytes to investigate gene changes between osteoarthritic and control populations. That report demonstrated that electron transport chain complexes II and III were down-regulated and mitochondrial mass increased in the osteoarthritic, but not in the normal, human articular chondrocytes. 75 The authors of that study also concluded that mitochondrial dysfunction could be contributing to the pathology of arthritis. In cats, it seems evident from the current study’s findings that DJD cats demonstrated reduced activity within the electron transport chain and therefore produced less cellular energy. Taken together, mitochondrial involvement in the disease process of DJD seems credible and further research into mitochondrial imbalances within arthritic subjects is clearly necessary.
PCA on the 87 differentially-expressed proteins (Figure 6) confirmed that the two experimental groups could be separated based on protein expression in the serum. Furthermore, proteomic analysis provided authentication for many of the genomic differences seen between the DJD and non-DJD animals. The clustering amongst the subjects within each group was not as tight on the proteomics graph of the data as it was in the genomics version. There are a few explanations for this. It is important to remember that the genomics PCA was performed using thousands of differentially expressed genes, while the proteomics PCA was carried out using just 87 differentially-expressed proteins. When comparing gene and protein changes, a direct, one-to-one relationship between the two molecule types is never expected. This is because the dynamic range and half-lives of messenger RNA molecules and proteins are quite different, and the sensitivity of equipment used to measure each molecule type also differs. Consequently, more variation within a group with respect to one molecule type over another is not considered unusual. Also, the proteomics work used samples from pooled as opposed to individual subjects, so more variation within the proteomics graph is to be expected. Lastly, if any of the control animals (in blue) were in early, pre-clinical stages of DJD disease, they would demonstrate higher variability in serum protein levels than the control animals. This last point may explain why some of the control data points are grouped closer to DJD rather than fellow non-DJD cats on the PCA graph. Although this possibility cannot be discounted, none of these cats had developed any outward signs of clinical disease when this article was accepted for publication.
The antigen presentation aspect of the immune response was not the only aspect of the immune system to be differentially regulated in DJD cats. Through proteomic analysis of serum, the classical complement component of the immune system and clusterin protein were also found to be altered in cats with DJD compared with non-DJD cats. The classical complement component, also known as the complement system, is a bridge between the adaptive system, represented by the antigen presentation and processing components (described above) and the innate system of a body’s immune function. 76 It is comprised of a regulated set of blood proteins that work to lyse non-specific foreign matter. 76 In this study, DJD cats were found to have a marked increase in complement components C1s, C1r and C3, while exhibiting down-regulation of the complement system regulator clusterin. An over-activated, improperly activated or unregulated complement system has been identified as problematic because these situations lead to a perpetuation of inflammation and disease in mammals. 77 Research in the literature reports up-regulated complement system gene components and down-regulated complement system regulators within juvenile idiopathic arthritis, RA and OA.43,78,79
As a whole, the altered expression levels of these genes and proteins involved in both the adaptive immune response and classical complement component of the immune system point to a defective immune response within cats with DJD that could cause and exacerbate an arthritic condition. Specifically targeting the immune system for intervention in the areas outlined above is an obvious step for future pharmaceutical and nutritional therapies.
Conclusions
The chances of developing DJD are increased with increasing age; furthermore, increasing weight could also be a risk factor for the disease. However, the potential contribution of weight differences was not assessed directly in this study and will be within the scope of future studies. The differentially-expressed genes detailed here are important because they represent changes within DJD animals that far exceed the changes observed with normal ageing. As noted previously, the chances of developing DJD greatly increase with age; therefore, it is important that the study addresses this issue so that only those genes and proteins specifically related to DJD are examined and that the results are not clouded by factors unrelated to disease state. This study controlled for subject age to ensure that the differentially-expressed genes discussed here represented true differences between disease states and were not complicated by this confounding factor. There were certainly gene expression differences in common between the groups when subjects were classified by disease or age, indicating commonalities between natural ageing and DJD pathways. However, significantly more up- or down-regulated genes were identified between disease groups than when the animals were grouped by age alone, and those genes that were similarly altered when the animals were grouped based on other factors were found to be changed to a lesser extent than when the animals were classified by disease alone. The data suggest that feline DJD could be a form of ‘accelerated ageing’ and support the assertion that the differentially expressed genes shown here are, in fact, related to disease and not due to the confounding ageing process alone. Furthermore, the molecular processes associated with feline DJD in the affected joint tissues may be different than those reported here for whole blood and its derivatives, and further work is needed to evaluate the gene expression and proteomic changes in peripheral tissue.
Acknowledgments
The authors would like to thank Rebecca J Palmer PhD for her assistance in the preparation of this article.
Footnotes
Funding: Full financial support for this work was provided by Hill’s Pet Nutrition.
All authors were employees of Hill’s Pet Nutrition, Inc. except B. Duncan X. Lascelles.
Accepted: 15 November 2012
References
- 1. Lascelles BD, Henry JB, Brown J, Robertson I, Sumrell AT, Simpson W, et al. Cross-sectional study of the prevalence of radiographic degenerative joint disease in domesticated cats. Vet Surg 2010; 39: 535–544. [DOI] [PubMed] [Google Scholar]
- 2. Clarke SP, Mellor D, Clements DN, Gemmill T, Farrell M, Carmichael S, et al. Prevalence of radiographic signs of degenerative joint disease in a hospital population of cats. Vet Rec 2005; 157: 793–799. [DOI] [PubMed] [Google Scholar]
- 3. Godfrey DR. Osteoarthritis in cats: a retrospective radiological study. J Small Anim Pract 2005; 46: 425–429. [DOI] [PubMed] [Google Scholar]
- 4. Hardie EM, Roe SC, Martin FR. Radiographic evidence of degenerative joint disease in geriatric cats: 100 cases (1994–1997). J Am Vet Med Assoc 2002; 220: 628–632. [DOI] [PubMed] [Google Scholar]
- 5. Slingerland LI, Hazewinkel HA, Meij BP, Picavet P, Voorhout G. Cross-sectional study of the prevalence and clinical features of osteoarthritis in 100 cats. Vet J 2011; 187: 304–309. [DOI] [PubMed] [Google Scholar]
- 6. Clarke SP, Bennett D. Feline osteoarthritis: a prospective study of 28 cases. J Small Anim Pract 2006; 47: 439–445. [DOI] [PubMed] [Google Scholar]
- 7. Gunew MN, Menrath VH, Marshall RD. Long-term safety, efficacy and palatability of oral meloxicam at 0.01-0.03 mg/kg for treatment of osteoarthritic pain in cats. J Feline Med Surg 2008; 10: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lascelles BD, Hansen BD, Roe S, DePuy V, Thomson A, Pierce CC, et al. Evaluation of client-specific outcome measures and activity monitoring to measure pain relief in cats with osteoarthritis. J Vet Intern Med 2007; 21: 410–416. [DOI] [PubMed] [Google Scholar]
- 9. Lascelles BD, Henderson AJ, Hackett IJ. Evaluation of the clinical efficacy of meloxicam in cats with painful locomotor disorders. J Small Anim Pract 2001; 42: 587–593. [DOI] [PubMed] [Google Scholar]
- 10. Malik R, Allan GS, Howlett CR, Thompson DE, James G, McWhirter C, et al. Osteochondrodysplasia in Scottish Fold cats. Aust Vet J 1999; 77: 85–92. [DOI] [PubMed] [Google Scholar]
- 11. Mathews KG, Koblik PD, Knoeckel MJ, Pool RR, Fyfe JC. Resolution of lameness associated with Scottish Fold osteodystrophy following bilateral ostectomies and pantarsal arthrodeses: a case report. J Am Anim Hosp Assoc 1995; 31: 280–288. [DOI] [PubMed] [Google Scholar]
- 12. Partington BP, Williams JF, Pechman RD, Beach RT. What is your diagnosis? Scottish Fold osteodystrophy. J Am Vet Med Assoc 1996; 209: 1235–1236. [PubMed] [Google Scholar]
- 13. Cowell KR, Jezyk PF, Haskins ME, Patterson DF. Mucopolysaccharidosis in a cat. J Am Vet Med Assoc 1976; 169: 334–339. [PubMed] [Google Scholar]
- 14. Crawley AC, Yogalingam G, Muller VJ, Hopwood JJ. Two mutations within a feline mucopolysaccharidosis type VI colony cause three different clinical phenotypes. J Clin Invest 1998; 101: 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haskins ME, Jezyk PF, Desnick RJ, McDonough SK, Patterson DF. Mucopolysaccharidosis in a domestic short-haired cat — a disease distinct from that seen in the Siamese cat. J Am Vet Med Assoc 1979; 175: 384–387. [PubMed] [Google Scholar]
- 16. Haskins ME, Jezyk PF, Patterson DF. Mucopolysaccharide storage disease in three families of cats with arylsulfatase B deficiency: leukocyte studies and carrier identification. Pediatr Res 1979; 13: 1203–1210. [DOI] [PubMed] [Google Scholar]
- 17. Jezyk PF, Haskins ME, Patterson DF, Mellman WJ, Greenstein M. Mucopolysaccharidosis in a cat with arylsulfatase B deficiency: a model of Maroteaux-Lamy syndrome. Science 1977; 198: 834–836. [DOI] [PubMed] [Google Scholar]
- 18. Konde LJ, Thrall MA, Gasper PW, Dial S, McBiles K, Colgan S, et al. Radiographically visualized skeletal changes associated with mucopolysaccharidosis VI in cats. Vet Radiol 1987; 28: 223–228. [Google Scholar]
- 19. Macri B, Marino F, Mazzullo G, Trusso A, De Maria R, Amedeo S, et al. Mucopolysaccharidosis VI in a Siamese/short-haired European cat. J Vet Med A Physiol Pathol Clin Med 2002; 49: 438–142. [DOI] [PubMed] [Google Scholar]
- 20. Vinayak A, Cross AR, Newell S. What is your diagnosis? Mucopolysaccharidosis (MPS) type VI. J Am Vet Med Assoc 2005; 226: 351–352. [DOI] [PubMed] [Google Scholar]
- 21. Polizopoulou ZS, Kazakos G, Patsikas MN, Roubies N. Hypervitaminosis A in the cat: a case report and review of the literature. J Feline Med Surg 2005; 7: 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seawright AA, English PB. Hypervitaminosis A and deforming cervical spondylosis of the cat. J Comp Pathol 1967; 77: 29–39. [DOI] [PubMed] [Google Scholar]
- 23. Seawright AA, English PB, Gartner RJ. Hypervitaminosis A and hyperostosis of the cat. Nature 1965; 206: 1171–1172. [DOI] [PubMed] [Google Scholar]
- 24. Langenbach A, Green P, Giger U, Rhodes H, Gregor TP, LaFond E, et al. Relationship between degenerative joint disease and hip joint laxity by use of distraction index and Norberg angle measurement in a group of cats. J Am Vet Med Assoc 1998; 213: 1439–1443. [PubMed] [Google Scholar]
- 25. Bennett JA, Goodchild CS, Kidd C, McWilliam PN. Inhibition of brain stem neuronal activity by cardiac and pulmonary vagal afferent fibres in the cat. Q J Exp Physiol 1988; 73: 959–972. [DOI] [PubMed] [Google Scholar]
- 26. Pedersen KB. The serology of Bordetella bronchiseptica isolated from pigs compared with strains from other animal species. Acta Pathol Microbiol Scand Suppl 1975; 83: 590–594. [DOI] [PubMed] [Google Scholar]
- 27. Pedersen NC, Pool RR, O’Brien T. Feline chronic progressive polyarthritis. Am J Vet Res 1980; 41: 522–535. [PubMed] [Google Scholar]
- 28. Moise NS, Crissman JW, Fairbrother JF, Baldwin C. Mycoplasma gateae arthritis and tenosynovitis in cats: case report and experimental reproduction of the disease. Am J Vet Res 1983; 44: 16–21. [PubMed] [Google Scholar]
- 29. Zeugswetter F, Hittmair KM, de Arespacochaga AG, Shibly S, Spergser J. Erosive polyarthritis associated with Mycoplasma gateae in a cat. J Feline Med Surg 2007; 9: 226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liehmann L, Degasperi B, Spergser J, Niebauer GW. Mycoplasma felis arthritis in two cats. J Small Anim Pract 2006; 47: 476–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tisdall PL, Martin P, Malik R. Cryptic disease in a cat with painful and swollen hocks: an exercise in diagnostic reasoning and clinical decision-making. J Feline Med Surg 2007; 9: 418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pedersen M, Jacobsen S, Klarlund M, Pedersen BV, Wiik A, Wohlfahrt J, et al. Environmental risk factors differ between rheumatoid arthritis with and without auto-antibodies against cyclic citrullinated peptides. Arthritis Res Ther 2006; 8: R133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reckner Olsson A, Skogh T, Wingren G. Comorbidity and lifestyle, reproductive factors, and environmental exposures associated with rheumatoid arthritis. Ann Rheum Dis 2001; 60: 934–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clarke A, Vyse TJ. Genetics of rheumatic disease. Arthritis Res Ther 2009; 11: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hasler P, Zouali M. Immune receptor signaling, ageing, and autoimmunity. Cell Immunol 2005; 233: 102–108. [DOI] [PubMed] [Google Scholar]
- 36. van den Berg WB. Lessons from animal models of arthritis over the past decade. Arthritis Res Ther 2009; 11: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averageing of multiple internal control genes. Gen Biol 2002; 3: research0034.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Teixeira VH, Olaso R, Martin-Magniette ML, Lasbleiz S, Jacq L, Oliveira CR, et al. Transcriptome analysis describing new immunity and defense genes in peripheral blood mononuclear cells of rheumatoid arthritis patients. PLoS One 2009; 4: e6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Baarsen LG, Bos CL, van der Pouw Kraan TC, Verweij CL. Transcription profiling of rheumatic diseases. Arthritis Res Ther 2009; 11: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Verweij CL. Transcript profiling towards personalised medicine in rheumatoid arthritis. Neth J Med 2009; 67: 364–371. [PubMed] [Google Scholar]
- 41. Katsuragawa Y, Saitoh K, Tanaka N, Wake M, Ikeda Y, Furukawa H, et al. Changes of human menisci in osteoarthritic knee joints. Osteoarthritis Cartilage 2010; 18: 1133–1143. [DOI] [PubMed] [Google Scholar]
- 42. Kumarasinghe DD, Perilli E, Tsangari H, Truong L, Kuliwaba JS, Hopwood B, et al. Critical molecular regulators, histomorphometric indices and their correlations in the trabecular bone in primary hip osteoarthritis. Osteoarthritis Cartilage 2010; 18: 1337–1344. [DOI] [PubMed] [Google Scholar]
- 43. Geyer M, Grassel S, Straub RH, Schett G, Dinser R, Grifka J, et al. Differential transcriptome analysis of intraarticular lesional vs intact cartilage reveals new candidate genes in osteoarthritis pathophysiology. Osteoarthritis Cartilage 2009; 17: 328–335. [DOI] [PubMed] [Google Scholar]
- 44. Boileau C, Martel-Pelletier J, Caron J, Msika P, Guillou GB, Baudouin C, et al. Protective effects of total fraction of avocado/soybean unsaponifiables on the structural changes in experimental dog osteoarthritis: inhibition of nitric oxide synthase and matrix metalloproteinase-13. Arthritis Res Ther 2009; 11: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brew CJ, Clegg PD, Boot-Handford RP, Andrew JG, Hardingham T. Gene expression in human chondrocytes in late osteoarthritis is changed in both fibrillated and intact cartilage without evidence of generalised chondrocyte hypertrophy. Ann Rheum Dis 2010; 69: 234–240. [DOI] [PubMed] [Google Scholar]
- 46. Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One 2008; 3: e3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003; 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gillis J, Pavlidis P. A methodology for the analysis of differential coexpression across the human lifespan. BMC Bioinformatics 2009; 10: 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gaczynska M, Rock KL, Goldberg AL. Role of proteasomes in antigen presentation. Enzyme Protein 1993; 47: 354–369. [DOI] [PubMed] [Google Scholar]
- 50. Lopez D, Del Val M. Selective involvement of proteasomes and cysteine proteases in MHC class I antigen presentation. J Immunol 1997; 159: 5769–5772. [PubMed] [Google Scholar]
- 51. Katunuma N, Matsunaga Y, Himeno K, Hayashi Y. Insights into the roles of cathepsins in antigen processing and presentation revealed by specific inhibitors. Biol Chem 2003; 384: 883–890. [DOI] [PubMed] [Google Scholar]
- 52. Riese RJ, Mitchell RN, Villadangos JA, Shi GP, Palmer JT, Karp ER, et al. Cathepsin S activity regulates antigen presentation and immunity. J Clin Invest 1998; 101: 2351–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Blanchard N, Shastri N. Cross-presentation of peptides from intracellular pathogens by MHC class I molecules. Ann N Y Acad Sci 2010; 1183: 237–250. [DOI] [PubMed] [Google Scholar]
- 54. Rock KL, Farfan-Arribas DJ, Shen L. Proteases in MHC class I presentation and cross-presentation. J Immunol 2010; 184: 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kufe DW, Holland JF, Frei E. American Cancer Society. Cancer medicine 6. 6th ed. Hamilton, ON, Lewiston, NY: BC Decker, 2003. [Google Scholar]
- 56. Chiang KC, Goto S, Chen CL, Lin CL, Lin YC, Pan TL, et al. Clusterin may be involved in rat liver allograft tolerance. Transpl Immunol 2000; 8: 95–99. [DOI] [PubMed] [Google Scholar]
- 57. Jenne DE, Lowin B, Peitsch MC, Bottcher A, Schmitz G, Tschopp J. Clusterin (complement lysis inhibitor) forms a high density lipoprotein complex with apolipoprotein A-I in human plasma. J Biol Chem 1991; 266: 11030–11036. [PubMed] [Google Scholar]
- 58. Fulop T, Larbi A, Dupuis G, Pawelec G. Ageing, autoimmunity and arthritis: Perturbations of TCR signal transduction pathways with ageing — a biochemical paradigm for the ageing immune system. Arthritis Res Ther 2003; 5: 290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gress RE, Deeks SG. Reduced thymus activity and infection prematurely age the immune system. J Clin Invest 2009; 119: 2884–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Desai A, Grolleau-Julius A, Yung R. Leukocyte function in the ageing immune system. J Leukoc Biol 2010; 87: 1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Murphy L, Schwartz TA, Helmick CG, Renner JB, Tudor G, Koch G, et al. Lifetime risk of symptomatic knee osteoarthritis. Arthritis Rheum 2008; 59: 1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Robinson JH, Delvig AA. Diversity in MHC class II antigen presentation. Immunology 2002; 105: 252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Klein L, Munz C, Lunemann JD. Autophagy-mediated antigen processing in CD4(+) T cell tolerance and immunity. FEBS Lett 2010; 584: 1405–1410. [DOI] [PubMed] [Google Scholar]
- 64. Doucey MA, Goffin L, Naeher D, Michielin O, Baumgartner P, Guillaume P, et al. CD3 delta establishes a functional link between the T cell receptor and CD8. J Biol Chem 2003; 278: 3257–3264. [DOI] [PubMed] [Google Scholar]
- 65. Marsden VS, Strasser A. Control of apoptosis in the immune system: Bcl-2, BH3-only proteins and more. Annu Rev Immunol 2003; 21: 71–105. [DOI] [PubMed] [Google Scholar]
- 66. Ekert PG, Vaux DL. Apoptosis and the immune system. Br Med Bull 1997; 53: 591–603. [DOI] [PubMed] [Google Scholar]
- 67. Opferman JT, Korsmeyer SJ. Apoptosis in the development and maintenance of the immune system. Nat Immunol 2003; 4: 410–415. [DOI] [PubMed] [Google Scholar]
- 68. Strasser A. The role of BH3-only proteins in the immune system. Nat Rev Immunol 2005; 5: 189–200. [DOI] [PubMed] [Google Scholar]
- 69. Ogden CA, Elkon KB. Role of complement and other innate immune mechanisms in the removal of apoptotic cells. Curr Dir Autoimmun 2006; 9: 120–142. [DOI] [PubMed] [Google Scholar]
- 70. Hughes P, Bouillet P, Strasser A. Role of Bim and other Bcl-2 family members in autoimmune and degenerative diseases. Curr Dir Autoimmun 2006; 9: 74–94. [DOI] [PubMed] [Google Scholar]
- 71. Botto M. Links between complement deficiency and apoptosis. Arthritis Res 2001; 3: 207–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Filippin LI, Vercelino R, Marroni NP, Xavier RM. Redox signalling and the inflammatory response in rheumatoid arthritis. Clin Exp Immunol 2008; 152: 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shi Q, Vaillancourt F, Cote V, Fahmi H, Lavigne P, Afif H, et al. Alterations of metabolic activity in human osteoarthritic osteoblasts by lipid peroxidation end product 4-hydroxynonenal. Arthritis Res Ther 2006; 8: R159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hutter E, Renner K, Pfister G, Stockl P, Jansen-Durr P, Gnaiger E. Senescence-associated changes in respiration and oxidative phosphorylation in primary human fibroblasts. Biochem J 2004; 380: 919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Maneiro E, Martin MA, de Andres MC, Lopez-Armada MJ, Fernandez-Sueiro JL, del Hoyo P, et al. Mitochondrial respiratory activity is altered in osteoarthritic human articular chondrocytes. Arthritis Rheum 2003; 48: 700–708. [DOI] [PubMed] [Google Scholar]
- 76. Yazdanbakhsh K. Review: complement receptor 1 therapeutics for prevention of immune hemolysis. Immunohematology 2005; 21: 109–118. [PubMed] [Google Scholar]
- 77. Schlosser RJ, Mulligan RM, Casey SE, Varela JC, Harvey RJ, Atkinson C. Alterations in gene expression of complement components in chronic rhinosinusitis. Am J Rhinol Allergy 2010; 24: 21–25. [DOI] [PubMed] [Google Scholar]
- 78. Barnes MG, Grom AA, Thompson SD, Griffin TA, Pavlidis P, Itert L, et al. Subtype-specific peripheral blood gene expression profiles in recent-onset juvenile idiopathic arthritis. Arthritis Rheum 2009; 60: 2102–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Corallini F, Bossi F, Gonelli A, Tripodo C, Castellino G, Mollnes TE, et al. The soluble terminal complement complex (SC5b-9) up-regulates osteoprotegerin expression and release by endothelial cells: implications in rheumatoid arthritis. Rheumatology (Oxford) 2009; 48: 293–298. [DOI] [PubMed] [Google Scholar]