Abstract
Porphyromonas gingivalis, a putative periodontopathogen, can bind to human saliva through its fimbriae. We previously found that salivary components from the submandibular and sublingual glands bind to P. gingivalis fimbriae and that acidic proline-rich protein (PRP) and statherin function as receptor molecules for fimbriae. In this study, we investigated the fimbria-binding components in parotid saliva. Fractionated human parotid saliva by gel-filtration chromatography was immobilized onto nitrocellulose membranes for the overlay assay following sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The salivary components on the membrane were allowed to interact with fimbriae purified from P. gingivalis ATCC 33277, and the interacted fimbriae were probed with anti-fimbria antibodies. The fimbriae were shown to bind to two forms of proline-rich glycoproteins (PRGs) as well as to acidic PRPs and statherin. Moreover, fimbriae bound to several components of smaller molecular size which appeared to be acidic PRP variants and basic PRPs. Fimbriae bound strongly to the purified PRGs adsorbed onto hydroxyapatite (HAP) beads. In contrast, PRGs in solution failed to inhibit the fimbrial binding to the immobilized PRGs on the HAP beads. These findings suggest that the appearance of binding site(s) of PRGs can be ascribed to their conformational changes. We previously identified the distinct segments within PRP and statherin molecules that are involved in fimbrial binding. The peptides analogous to the binding regions of PRP and statherin (i.e., PRP-C and STN-C) markedly inhibit the binding of fimbriae to PRP and statherin immobilized on the HAP beads, respectively. The PRP-C significantly inhibited the binding of fimbriae to PRG-coated HAP beads as well as to PRP on HAP beads. The peptide did not affect the binding of fimbriae to statherin, whereas the STN-C showed no effect on the fimbrial binding to PRPs or PRGs. In the overlay assay, the PRP-C clearly diminished the interactions between the fimbriae and the various salivary components, including PRPs, the PRGs, and the components with smaller molecular sizes but not statherin. These results strongly suggest that fimbriae bind to salivary components (except statherin) via common peptide segments. It is also suggested that fimbriae bind to saliva through the two distinct binding domains of receptory salivary components: (i) PRGs and PRPs and (ii) statherin.
Porphyromonas gingivalis, a gram-negative anaerobic rod, has been well recognized as a major etiologic agent of periodontal diseases (32). Dental plaque accumulation around the gingival crevice and other oral surfaces is a predisposing factor for the initiation of periodontal diseases. Among plaque bacterial species, P. gingivalis has been shown to prevail in various saliva-coated surfaces of oral matrices such as mucosal membrane (9), healthy crevices (8), and supragingival plaques (35). Therefore, saliva would be a critical factor for the organism to adhere to and colonize the oral cavity. Among the various adhesive factors of the organism, fimbriae are thought to play a major role in its interaction with saliva (19). We previously searched for submandibular and sublingual salivary components that specifically interact with P. gingivalis fimbriae (4). We found that fimbriae strongly bind to acidic proline-rich protein (PRP) and statherin that had been immobilized onto nitrocellulose membranes or hydroxyapatite (HAP) beads. These bindings occur via protein-protein interactions through definitive domains of the fimbriae (3) and salivary proteins, i.e., PRP and statherin (2, 16).
Human salivary PRPs (SPRPs) are composed of heterogeneous molecules that are rich in saliva from the parotid and submandibular and sublingual glands. SPRPs comprise about 75 to 80% of the parotid salivary components (5) and are classified into three groups: acidic (molecular mass, ≤16 kDa), basic (molecular mass, 6 to 9 kDa), and glycosylated (molecular mass, 39 kDa) (31). These SPRPs are coded for by a multigene family of six genes, resulting in more than 20 SPRPs by both differential RNA splicing and posttranslational modification (proteolytic cleavages) after secretion (24). Acidic PRPs have been reported to act as salivary receptors for several plaque-forming bacteria (4, 10, 13).
The proline-rich glycoprotein (PRG) is a basic protein (with an isoelectric point of >8.2) that is especially rich in proline, glycine, and glutamic acid. PRG contains 40% carbohydrate and is found abundantly only in parotid saliva (7, 30). The role of PRG has been demonstrated in lubrication, a property of oral pellicles which may protect surfaces against mechanical disruption or abrasion (7). PRG can bind to Streptococcus oralis, Streptococcus gordonii (7), and Fusobacterium nucleatum (14) in a lectin-like fashion and to Actinomyces viscosus by protein-protein interactions (6). Although PRG appears to be able to bind to various oral bacteria, little information is available concerning the binding mechanisms involved.
We recently determined that the minimum active domain of PRP1 (a major variant of acidic PRP) for the binding to P. gingivalis fimbriae is Pro-Gln-Gly-Pro-Pro-Gln (PQGPPQ) (16). The synthetic PRP1 peptide (i.e., PRP-C) analogous to the carboxyl-terminal 21 amino acid residues containing PQGPPQ and PQGPPPQ showed almost 100% inhibition in the binding of fimbriae to PRP1 (16). We describe here the finding that PRG is specific for the binding to P. gingivalis fimbriae via the common domain found in various SPRP variants as a typical repeating sequence (PQGPPQG or PQGPPPQG). It was also found that the peptide containing the repeating sequence inhibits fimbrial binding to all salivary proteins except statherin.
MATERIALS AND METHODS
Bacterial culture and purification of fimbriae.
Fimbriae were mechanically detached from P. gingivalis ATCC 33277 cells grown anaerobically and purified chromatographically by the method of Yoshimura et al. (34). The iodination of purified fimbriae was performed as described previously (4). The specific activity of iodinated protein was 1.8 mCi/μmol of fimbrillin.
Preparation of salivary components.
Human parotid saliva was obtained from nine 22-year-old male donors by citric acid stimulations with collecting devices as described previously (6). The individual saliva samples were mixed together. Ten percent (vol/vol) of the enzyme inhibitor solution containing KCl buffer (50 mM KCl, 1 mM KH2PO4, 1 mM CaCl2, 0.1 mM MgCl2 [pH 6.0]), 2% Na2EDTA, 10% 2-propanol, and 2 mM phenylmethylsulfonyl fluoride (29) was added to the saliva. After centrifugation at 12,000 × g for 10 min at 4°C, the clarified supernatant was concentrated by 100% solid ammonium sulfate precipitation. The pellet was dissolved in 20 mM Tris-HCl buffer (pH 8.0) containing 0.5 M NaCl and was dialyzed extensively with a 3,000-molecular-mass cutoff membrane (Spectrum Medical Ind., Laguna Hills, Calif.) against the same buffer at 4°C. The material was then applied to a Sephacryl S-200 column (150 by 2 cm; Pharmacia LKB Biotechnology, Piscataway, N.J.) equilibrated with the same buffer by the method of Strömberg et al. (33). The flow rate was 4 ml/h at 4°C, and the eluate was collected as 3-ml fractions. The fractions containing PRGs were further separated by high-performance liquid chromatography with a cation-exchange column (PolyCAT A column; PolyLC, Inc., Columbia, Md.) with 20 mM potassium phosphate buffer (pH 5.5), and the adsorbed proteins were eluted with a linear gradient of the same buffer containing 1 M NaCl. Fractions containing PRGs were dialyzed and lyophilized. The purified preparations were used for the binding experiments after the purities of the proteins were assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Unstimulated human whole saliva was collected by expectoration from a 22-year-old male into a chilled container as described previously (19). PRP1 and statherin were prepared as outlined in our previous study (4).
Overlay assay.
Saliva samples were dissolved in 0.125 M Tris-HCl buffer (pH 6.8) containing 2% SDS, 10% glycerol, and 0.001% bromophenol blue and were incubated for 30 min at room temperature. Preparations separated on SDS-PAGE gels were transferred to nitrocellulose membranes (Trans-Blot, 0.45 μm pore size; Bio-Rad Laboratories, Hercules, Calif.). Unoccupied binding sites were blocked by incubating the membrane for 1 h with KCl buffer containing 1% lipid-free bovine serum albumin (BSA; A-7030; Sigma Chemical Co., St. Louis, Mo.) as a blocking agent. The membranes were incubated with 5 ml of fimbriae (41 μg/ml; 1 nmol of fimbrillin/ml) and, if necessary, inhibitors in KCl buffer overnight at 4°C and were washed three times with KCl buffer containing 0.3 M NaCl and 0.1% Tween 20. The fimbriae bound to salivary proteins on the membranes were probed with rabbit anti-P. gingivalis fimbriae serum diluted 1:2,000 in 5 ml of phosphate-buffered saline (pH 7.4).
Binding of fimbriae to salivary protein-coated HAP.
The binding assays of 125I-labeled fimbriae to salivary protein-coated HAP beads were carried out as follows. HAP beads (3 mg) in a tube were incubated with 100 μl of salivary protein solution (100 μg/ml) including whole saliva, PRP1, statherin, and PRGs overnight at room temperature. Aliquots (100 μl) of 125I-labeled fimbriae (5 nmol/ml) and possible inhibitors such as synthetic peptide or purified salivary proteins were added to tubes containing the salivary protein-coated HAP beads and incubated at room temperature for 1 h. The specific binding was calculated by subtracting the nonspecific binding which was obtained by the preincubation of saliva-coated HAP beads with nonlabeled fimbriae (500 μl of 50 nmol/ml) at room temperature for 1 h. The inhibitory rate was calculated by comparing the specific binding levels with or without inhibitors. All assays were performed in triplicate on three separate occasions.
Preparation of PRP1 and statherin peptides.
The peptides corresponding to the carboxyl-terminal segment composed of 21 amino acid residues of PRP1 (16) and the carboxyl-terminal segment composed of 15 amino acid residues of statherin (2) were synthesized and purified. The amino acid sequences of these peptides are as follows: the PRP1-carboxyl-terminal peptide (PRP-C [PQGPPPQGGRPQGPPQGQSPQ]) and the statherin-carboxyl-terminal peptide (STN-C [LYPQPYQPQYQQYTF]). The amino acid sequence and mass value of the product were confirmed with a 477A/120 gas-phase automatic sequencer and with fast atom bombardment mass spectrometry and the JMS-HX100/JMA-3100 data system (Jeol Ltd., Tokyo, Japan), respectively.
Analytical methods.
SDS-PAGE was performed with a precast 10-20 gel gradient (Daiichikayaku, Kyoto, Japan) according to the manufacturer’s recommendations. The saliva samples were dissolved in PAGE sample buffer (10 mM Tris-HCl buffer [pH 8.0] with 2.5% SDS, 1 mM EDTA, and 0.01% bromophenol blue) and, in some experiments, the samples were immersed in a boiling water bath for 5 min. The proteins on SDS-PAGE were stained with Coomassie brilliant blue (CBB) R-250. The protein content of samples was determined with a protein assay kit (Bio-Rad) with BSA as a standard according to the manufacturer’s manual.
RESULTS
Binding of parotid saliva components to P. gingivalis fimbriae.
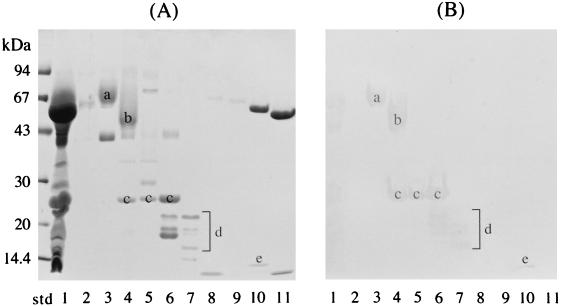
The overlay assay was performed to detect salivary components capable of binding to fimbriae. The parotid saliva was fractionated by gel filtration chromatography, and 10 peak fractions were obtained. These 10 fractions and the parotid saliva were separated by SDS-PAGE (Fig. 1A), and a replica was prepared by the transference of salivary components to a nitrocellulose membrane (Fig. 1B). The parotid saliva components in lanes 3 to 7 and 10 clearly bound to fimbriae (Fig. 1B); the protein bands c and e shown in Fig. 1 were PRP1 and statherin, whereas the broad bands a and b were novel fimbria-binding proteins. The latter two bands belonged to PRGs as judged by the relative mobility and pink-violet coloration with CBB R-250. Other reacting bands d with molecular weights lower than that of PRP1 were also observed in lanes 6 and 7. The relative mobilities of these bands and the pink-violet coloration with CBB R-250 indicate that these components are acidic and basic PRP variants.
FIG. 1.
Binding of parotid saliva components to P. gingivalis fimbriae. (A) SDS-PAGE profiles of parotid saliva and fractionated components. Parotid saliva and the 10 peak fractions were dissolved in SDS sample buffer without heating and were then separated by SDS-PAGE. (B) The salivary components on the nitrocellulose replica were incubated with 5 ml of fimbriae (41 μg/ml; 1 nmol of fimbrillin/ml of KCl buffer). The salivary proteins interacting with fimbriae were probed with anti-fimbria antibodies. It should be noted that PRG and PRPs migrated anomalously with respect to molecular mass in the SDS-PAGE gels. Lanes: std, molecular mass standard; 1, whole parotid saliva; 2 to 11, fractionated parotid salivary components. a and b, novel fimbria-binding proteins found to be PRGs; c, PRP1; d, small-molecular-size components; e, statherin.
Purification of PRG variants and binding to fimbriae.
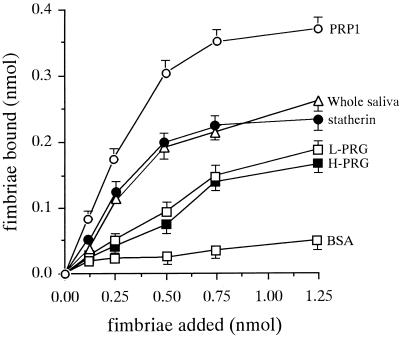
The PRGs were newly found to bind to P. gingivalis fimbriae and were purified to homogeneity. The purities of the final preparations of PRGs with a higher molecular size (H-PRG) and a lower molecular size (L-PRG) were confirmed by SDS-PAGE results stained by CBB (data not shown). The abilities of the H- and L-PRGs to bind to fimbriae were then compared to those of PRP1 and statherin by using HAP beads (Fig. 2). The binding levels of 125I-labeled fimbriae to H- and L-PRG-coated HAP beads were very similar. In addition, the binding abilities of fimbriae to PRP1, statherin, and whole saliva were higher than those to PRGs, whereas the fimbrial bindings to the H- and L-PRGs were significantly greater than the binding to BSA. The inhibition assay was carried out with various reagents; however, no significant inhibitors were found among amino sugars (0.1 M N-acetylgalactosamine, N-acetylglucosamine, galactosamin, and glucosamine), neutral sugars (0.1 M glucose, mannose, galactose, fucose, fructose, lactose, and maltose), and amino acids (0.1 M l-arginine and l-lysine).
FIG. 2.
Binding of P. gingivalis fimbriae to purified PRG-coated HAP beads. HAP beads (3 mg) in a tube were incubated overnight at room temperature with 100 μl of salivary protein solution (100 μg/ml) containing purified PRGs, whole saliva, PRP1, and statherin. 125I-labeled fimbriae were added to a tube containing salivary protein-coated HAP beads and incubated at room temperature for 1 h. The specific binding level was calculated by subtracting the nonspecific binding level, which was obtained by the preincubation of HAP beads with nonlabeled fimbriae (500 μl of 50 nmol/ml) at room temperature for 1 h. All assays were performed in triplicate on three separate occasions. Data are expressed as means ± standard deviations.
We examined whether H- and L-PRGs functioned as inhibitors on the binding of fimbriae to the HAP beads pretreated with H- and L-PRGs, respectively (Fig. 3). Although increasing amounts of PRGs were added (up to 1,000 μg/ml), no significant inhibition of the fimbrial binding to the PRG-immobilized HAP surfaces was observed, a finding that is probably due to cryptic receptors.
FIG. 3.
Effects of H- and L-PRG solutions on the binding of P. gingivalis fimbriae to HAP beads coated with H- and L-PRGs, respectively. Increasing concentrations of H- and L-PRGs in KCl buffer were used as inhibitors for 125I-labeled fimbriae (0.5 nmol). Inhibition studies were performed by the addition of H-PRG in solution to H-PRG-coated HAP beads or of L-PRG to L-PRG-coated HAP beads. All assays were performed in triplicate on three separate occasions. Data are expressed as means ± standard deviations.
Inhibition studies by synthetic peptides (PRP-C and STN-C).
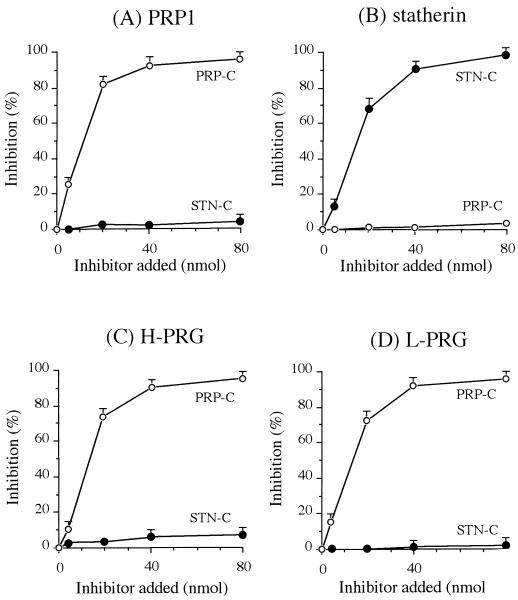
The bindings of fimbriae to the immobilized PRP1 and statherin have been shown to be strongly inhibited by synthetic peptides containing the binding sites of PRP1 and statherin for fimbriae. Synthetic peptides, PRP-C (the carboxyl-terminal 21-amino-acid segment of PRP1 [PQGPPPQGGRPQGPPQGQSPQ]), and STN-C (the carboxyl-terminal 15-amino-acid segment of statherin [LYPQPYQPQYQQYTF]), were used to determine whether the fimbria-PRG interactions would be mediated by the same mechanism as those of PRP1 or statherin. As shown in Fig. 4, PRP-C strongly inhibited the binding of fimbriae to the immobilized PRP1, whereas it was ineffective for the fimbrial binding to statherin (Fig. 4A). On the other hand, STN-C, but not PRP-C, inhibited the fimbrial binding to the statherin-coated HAP beads (Fig. 4B). In the interaction between fimbriae and immobilized PRGs, PRP-C was markedly inhibitory, whereas STN-C showed no inhibitory effect (Fig. 4C and D). These results suggest that P. gingivalis fimbriae recognize the domain common to H- and L-PRGs and to PRP1 and that the binding site(s) of statherin for fimbriae is distinct from that of PRP or PRG.
FIG. 4.
Effects of the peptides analogous to the carboxyl-terminal segments of PRP1 (PRP-C [PQGPPPQGGRPQGPPQGQSPQ]) and statherin (STN-C [LYPQPYQPQYQQYTF]) on the binding of fimbriae to H- and L-PRG immobilized to HAP beads. Increasing concentrations of the peptides in KCl buffer were used as inhibitors for the binding of 125I-labeled fimbriae (0.5 nmol) to HAP beads coated with salivary proteins: PRP1 (A), statherin (B), H-PRG (C), and L-PRG (D). All assays were performed in triplicate on three separate occasions. Data are expressed as means ± standard deviations.
Inhibitory effect by PRP-C in the binding of fimbriae to salivary components.
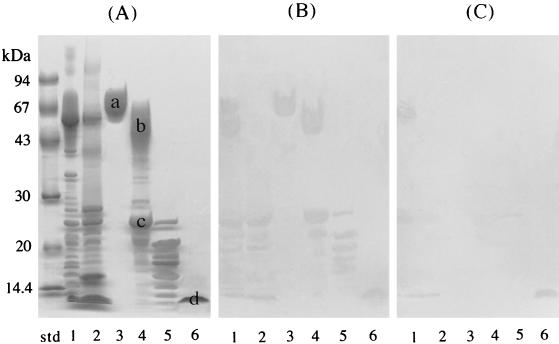
Fimbriae bound to several unknown salivary components which appeared to be acidic and basic PRP variants (protein bands d in Fig. 1). These variants have been reported to possess the typical repeating amino acid sequences, PQGPPQ and PQGPPPQ, as well as SPRP (5). Therefore, the effects of PRP-C on the binding of fimbriae to these components with smaller molecular sizes were examined. As shown in Fig. 5, the overlay assays were performed with or without the addition of PRP-C. The salivary proteins that had reacted with the fimbriae were clearly probed with anti-fimbria antibodies (Fig. 5B). The addition of PRP-C (100 nmol/ml) diminished the binding of fimbriae to most of the saliva proteins, including the smaller molecular size components, the PRGs and PRP1, whereas the fimbrial binding to statherin was not affected (Fig. 5C). These results suggest that P. gingivalis fimbriae recognize a common peptide segment shared by various salivary components (except statherin).
FIG. 5.
Inhibitory effect of PRP-C on the binding of fimbriae to salivary components. Since the peptide PRP-C (PQGPPPQGGRPQGPPQGQSPQ) was found to be significantly inhibitory for the fimbria-PRG interactions, the overlay assays as shown in Fig. 1 were performed with the addition of PRP-C. (A) SDS-PAGE profiles of salivary components which bound to fimbriae. (B) The overlay assay performed as shown in Fig. 1 without the addition of PRP-C. The replica membrane was incubated with 5 ml of fimbriae (41 μg/ml, 1 nmol/ml of KCl buffer). (C) The overlay assay performed with the simultaneous additions of PRP-C (100 nmol/ml) and fimbriae (1 nmol/ml) in 5 ml of KCl buffer. Lanes: std, molecular mass standard; 1, parotid saliva; 2, submandibular and sublingual saliva; 3, H-PRG; 4, L-PRG–PRP1 fraction; 5, small-molecular-size components that bind to fimbriae; 6, statherin.
DISCUSSION
The flow of saliva in the oral cavity may contribute to the promotion and/or retardation of the adherence of oral bacteria to oral and tooth surfaces. Many bacterial species are known to be agglutinated in the presence of saliva, which results in the detachment of oral bacteria and their eventual clearance from the oral cavity (30). It was reported that several saliva components, including the SPRPs, mucin, amylase, and statherin can bind to various oral bacteria (30). For the clearance of bacterial cells from the oral cavity, flowing saliva components must interact with the organisms. However, PRG, PRP, and statherin in flowing saliva are unlikely to bind to P. gingivalis cells. PRP1, a major variant of acidic PRP, has been thought to possess unique hidden receptors termed “cryptitopes” that promote bacterial adherence (11, 12). The PRP1 molecule in solution shows little ability to bind to bacteria. However, once adsorbed to HAP surfaces, the protein undergoes a conformational change. As a result, the cryptitopes are exposed, resulting in the promotion of bacterial adherence to the surfaces. Similarly, it was found that PRGs and statherin also bound to fimbriae only when immobilized to the solid surface of HAP beads or a nitrocellulose membrane. All of these phenomena may be ascribed to the appearance of hidden receptors caused by a conformational change of the saliva proteins. Although the nature of these cryptitopes remains to be elucidated, it is suggested that salivary cryptitopes enhance the adherence of the pathogenic bacteria but not the host defense.
The size variants of the PRG molecule are expressed due to different lengths of the tandemly repetitive exon 3 portions of the allele (PRB2) and the differences of glycosylation (20, 21, 24). It has been reported that the electrophoretic patterns of PRG variants are expressed to give two size-variant PRG proteins (15, 33). Various variants of the SPRP family have been found, the SPRP-coding genes have been identified, and the whole amino acid sequences of the core proteins have been deduced by analyzing the genes, including PRH1 and PRH2 and PRB1 to PRB4 (22–24). Many of the characteristics of the repeating amino acid sequences are observed in the SPRPs (5). For example, a typical repeating sequence in acidic PRPs is Pro-Gln-Gly-Pro-Pro-Gln-Gly (PQGPPQG), while the sequence PQGPPPQG dominates in basic and glycosylated PRPs. The segment PQGPPQ has been shown to be an active domain of PRP1 to bind to fimbriae, whereas the peptide PQGPPPQ is also inhibitory for the fimbrial interaction with PRP1 (16). It is of interest that the majority of SPRPs possess the characteristic repeating sequences which would act as the binding domains for P. gingivalis fimbriae. Although it is not yet known whether all of the purified SPRPs bind to fimbriae, the findings in this study suggest that smaller-molecular-size components, presumably acidic and basic PRP variants, also bind to fimbriae via the repeating sequences. It is likely that SPRPs act as anchoring proteins for P. gingivalis to oral surfaces, including dental pellicle and plaque. It is unknown whether the binding sites of the smaller-molecular-size components are masked. These protein components might agglutinate P. gingivalis and interfere with the adherence to the oral cavity by the organism. Further study is necessary to understand the biological function of the smaller acidic and basic PRP variants in the interaction with fimbriae.
The findings obtained so far strongly suggest that fimbriae bind to salivary components (except statherin) via a common peptide segment. The active peptide regions of statherin for the binding to fimbriae have been reported to be LY and TYF in the amino- and carboxyl-terminal ends of STN-C (2). It is suggested that fimbriae bind to saliva through the two distinct binding domains of receptory salivary components: (i) PRGs and PRPs and (ii) statherin. Although many plaque-forming bacteria have been shown to bind to PRGs and PRPs and to statherin (13, 30), few studies on the mechanisms of these interactions are available. It has been demonstrated that S. gordonii recognizes the PQ residues located at the carboxyl-terminal end of PRP1 and that the PQ sequence of the internal residues of PRP1 does not promote the adhesion (11). Various interactions between bacteria and salivary components are known (30). Further analyses of these interactions will increase our understanding of the diversity of adherence of many species of oral organisms to different oral surfaces.
Fimbriated P. gingivalis strains exhibit higher binding abilities for collagen type I and IV (28). Since human collagen alpha (types I and IV) contains multiple repeating regions of PQGPP, it is reasonable to speculate that the binding of P. gingivalis to collagen might be mediated by the interaction between fimbriae and the PQGPP regions found in collagen. It would be of interest to determine whether P. gingivalis fimbriae can recognize these characteristic PRP regions ubiquitously present in various host tissues, including those of the periodontal area.
The fimbrial binding to salivary proteins is not inhibited by the addition of l-arginine (4). However, Kontani et al. (17, 18) recently reported the presence of cryptic receptors in fibronectin and the surface components of fibroblasts for P. gingivalis fimbriae, in which the arginine residue plays a critical role. They showed that the adherence of fimbriae to the fibroblasts and fibronectin immobilized on the culture plates was significantly enhanced by the treatment of proteinase of the organism that split the arginine residue. The proteolysis likely exposes a cryptitope in the host matrix proteins, i.e., the carboxyl-terminal Arg residue, so that the fimbriae can bind to the cryptitope through a fimbria-Arg interaction. In this regard, the aggregation of S. oralis cells induced by fimbriae was inhibited by l-arginine (1). Fibrinogen is a potent inhibitor of P. gingivalis coaggregation with S. oralis, and chemical modification of the Arg residues of fibrinogen markedly reduced the inhibitory ability (27). Histatin likely binds to the fimbriae and hemagglutinin of P. gingivalis, and arginine residues of histatin seem to be critically important for the inhibition of coaggregation (25). Although little is known regarding cryptic receptors in the coaggregation, fibrinogen seems to possess cryptic receptors for fimbriae (26). These cryptic receptors might mediate the selective colonization of P. gingivalis in the oral cavity.
In conclusion, it has been shown that PRGs, which are found only in parotid saliva, can bind to P. gingivalis fimbriae via the common cryptic peptide segment shared with SPRPs, the major salivary components. It is also suggested that fimbriae can bind to saliva through the two distinct binding domains of receptory salivary components, SPRPs, and statherin.
ACKNOWLEDGMENT
This work was supported in part by Grants-in-Aid (09557139 and 09557175) from the Ministry of Education, Science, and Sports of Japan.
REFERENCES
- 1.Amano A, Fujiwara T, Nagata H, Kuboniwa M, Sharma A, Sojar H T, Genco R J, Hamada S, Shizukuishi S. Porphyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J Dent Res. 1997;76:852–857. doi: 10.1177/00220345970760040601. [DOI] [PubMed] [Google Scholar]
- 2.Amano A, Kataoka K, Raj P A, Genco R J, Shizukuishi S. Binding sites of salivary statherin for Porphyromonas gingivalis recombinant fimbrillin. Infect Immun. 1996;64:4249–4254. doi: 10.1128/iai.64.10.4249-4254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano A, Sharma A, Lee J-Y, Sojar H T, Raj P A, Genco R J. Structural domains of Porphyromonas gingivalis recombinant fimbrillin that mediate binding to salivary proline-rich protein and statherin. Infect Immun. 1996;64:1631–1637. doi: 10.1128/iai.64.5.1631-1637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amano A, Sojar H T, Lee J-Y, Sharma A, Levine M J, Genco R J. Salivary receptors for recombinant fimbrillin of Porphyromonas gingivalis. Infect Immun. 1994;62:3372–3380. doi: 10.1128/iai.62.8.3372-3380.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennick A. Structural and genetic aspects of proline-rich proteins. J Dent Res. 1987;66:457–461. doi: 10.1177/00220345870660021201. [DOI] [PubMed] [Google Scholar]
- 6.Clark W B, Beem J E, Nesbitt W E, Cisar J O, Tseng C C, Levine M J. Pellicle receptors for Actinomyces viscosus type 1 fimbriae in vitro. Infect Immun. 1989;57:3003–3008. doi: 10.1128/iai.57.10.3003-3008.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen R E, Levine M J. Salivary glycoproteins. In: Tenovuo J O, editor. Human saliva: clinical chemistry and microbiology. Vol. 1. Boca Raton, Fla: CRC Press, Inc.; 1989. pp. 101–130. [Google Scholar]
- 8.Conrads G, Mutters R, Fischer J, Brauner A, Lutticken R, Lampert F. PCR reaction and dot-blot hybridization to monitor the distribution of oral pathogens within plaque samples of periodontally healthy individuals. J Periodontol. 1996;67:994–1003. doi: 10.1902/jop.1996.67.10.994. [DOI] [PubMed] [Google Scholar]
- 9.Danser M M, Timmerman M F, van-Winkelhoff A J, van-der-Velden U. The effect of periodontal treatment on periodontal bacteria on the oral mucous membranes. J Periodontol. 1996;67:478–485. doi: 10.1902/jop.1996.67.5.478. [DOI] [PubMed] [Google Scholar]
- 10.Gibbons R J. Bacterial adhesion to oral tissues: a model for infectious diseases. J Dent Res. 1989;68:750–760. doi: 10.1177/00220345890680050101. [DOI] [PubMed] [Google Scholar]
- 11.Gibbons R J, Hay D I, Schlesinger D H. Delineation of a segment of adsorbed salivary acidic proline-rich proteins which promotes adhesion of Streptococcus gordonii to apatitic surfaces. Infect Immun. 1991;59:2948–2954. doi: 10.1128/iai.59.9.2948-2954.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbons R J, Hay D I, Ghilds W C I, Davis G. Role of cryptic receptors (cryptitopes) in bacterial adhesion to oral surfaces. Arch Oral Biol. 1990;35:107–114. doi: 10.1016/0003-9969(90)90139-2. [DOI] [PubMed] [Google Scholar]
- 13.Gibbons R J, Hay D I. Adsorbed salivary proline-rich proteins as bacterial receptors on apatitic surfaces. In: Switalski L M, Hook M, Beachey E, editors. Molecular mechanisms of microbial adhesion. New York, N.Y: Springer-Verlag; 1988. pp. 143–169. [Google Scholar]
- 14.Gillece-Castro B L, Prakobphol A, Burlingame A L, Leffler H, Fisher S J. Structure and bacterial receptor activity of a human salivary proline-rich glycoproteins. J Biol Chem. 1991;266:17358–17368. [PubMed] [Google Scholar]
- 15.Henkin R I, Lippoldt R E, Bilstad J, Wolf R O, Lum C K, Edelhoch H. Fractionation of human parotid saliva proteins. J Biol Chem. 1978;253:7556–7565. [PubMed] [Google Scholar]
- 16.Kataoka K, Amano A, Kuboniwa M, Horie H, Nagata N, Shizukuishi S. Active sites of salivary proline-rich protein for binding to Porphyromonas gingivalis fimbriae. Infect Immun. 1997;65:3159–3164. doi: 10.1128/iai.65.8.3159-3164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kontani M, Kimura S, Nakagawa I, Hamada S. Adherence of Porphyromonas gingivalis to matrix proteins via a fimbrial cryptic receptor exposed by its own arginine-specific protease. Mol Microbiol. 1997;24:1179–1187. doi: 10.1046/j.1365-2958.1997.4321788.x. [DOI] [PubMed] [Google Scholar]
- 18.Kontani M, Ono H, Shibata H, Okamura Y, Tanaka T, Fujiwara T, Kimura S, Hamada S. Cysteine protease of Porphyromonas gingivalis 381 enhances binding of fimbriae to cultured human fibroblasts and matrix proteins. Infect Immun. 1996;64:756–762. doi: 10.1128/iai.64.3.756-762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J-Y, Sojar H T, Bedi G S, Genco R J. Synthetic peptides analogous to the fimbrillin sequence inhibit adherence of Porphyromonas gingivalis. Infect Immun. 1992;60:1662–1670. doi: 10.1128/iai.60.4.1662-1670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levine M J, Ellison S A, Bahl O P. The isolation from human parotid saliva and partial characterization of the protein core of a major parotid glycoprotein. Arch Oral Biol. 1973;18:827–837. doi: 10.1016/0003-9969(73)90053-8. [DOI] [PubMed] [Google Scholar]
- 21.Levine M J, Weill J C, Ellison S A. The isolation and analysis of a glycoprotein from parotid saliva. Biochem Biophys Acta. 1969;188:165–167. doi: 10.1016/0005-2795(69)90060-9. [DOI] [PubMed] [Google Scholar]
- 22.Lyons K M, Azen E A, Goodman P A, Smithies O. Many protein products from a few loci: assignment of human salivary proline-rich proteins to specific loci. Genetics. 1988;120:255–265. doi: 10.1093/genetics/120.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyons K M, Stein J H, Smithies O. Length polymorphisms in human proline-rich proteins genes generated by intragenic unequal crossing over. Genetics. 1988;120:267–278. doi: 10.1093/genetics/120.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeda N, Kim H S, Azen E A, Smithies O. Differential RNA splicing and post-translational cleavages in the human salivary proline-rich protein gene system. J Biol Chem. 1985;260:11123–11130. [PubMed] [Google Scholar]
- 25.Murakami Y, Nagata H, Shizukuishi S, Tsunemitsu A. Role of an arginine residue present in histatin 8 which inhibits coaggregation between Porphyromonas gingivalis and Streptococcus mitis. J Dent Health. 1993;43:221–223. [Google Scholar]
- 26.Nagata H, Sojar H T, Hamada N, Sharma A, Genco R J. Binding of Porphyromonas gingivalis fimbriae to fibrinogen. J Dent Res. 1997;76:222. doi: 10.1177/00220345970760040601. . (Abstract 1668.) [DOI] [PubMed] [Google Scholar]
- 27.Nagata H, Amano A, Hanioka T, Tamagawa H, Shizukuishi S. Inhibition of coaggregation between Porphyromonas gingivalis and Streptococcus oralis by fibrinogen fragments. FEMS Microbiol Lett. 1993;114:31–36. doi: 10.1111/j.1574-6968.1993.tb06546.x. [DOI] [PubMed] [Google Scholar]
- 28.Naito Y, Gibbons R J. Attachment of Bacteroides gingivalis to collagenous substrata. J Dent Res. 1988;67:1075–1080. doi: 10.1177/00220345880670080301. [DOI] [PubMed] [Google Scholar]
- 29.Ramasubbu N, Reddy M S, Bergey E J, Haraszthy G G, Soni S-D, Levine M J. Large-scale purification and characterization of the major phosphoproteins and mucins of human submandibular-sublingual saliva. Biochem J. 1991;280:341–352. doi: 10.1042/bj2800341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scannapieco F A. Saliva-bacterium interactions in oral microbial ecology. Crit Rev Oral Biol Med. 1994;5:203–248. doi: 10.1177/10454411940050030201. [DOI] [PubMed] [Google Scholar]
- 31.Schenkels L C P M, Veerman E C I, Amerongen A V N. Biological composition of human saliva in relation to other mucosal fluids. Crit Rev Oral Biol Med. 1995;6:161–175. doi: 10.1177/10454411950060020501. [DOI] [PubMed] [Google Scholar]
- 32.van Steenbergen T J M, van Winkelhoff A J, de Graaff J. Black-pigmented oral anaerobic rods: classification and role in periodontal disease. In: Hamada S, Holt S C, McGhee J R, editors. Periodontal disease: pathogens and host immune responses. Tokyo, Japan: Quintessence Publishing Co., Ltd.; 1991. pp. 41–52. [Google Scholar]
- 33.Strömberg N, Borén T, Carlén A, Olsson J. Salivary receptors for GalNAcβ-sensitive adherence of Actinomyces spp.: evidence for heterogeneous GalNAcβ and proline-rich protein receptor properties. Infect Immun. 1992;60:3278–3286. doi: 10.1128/iai.60.8.3278-3286.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshimura F, Takahashi K, Nodasaka Y, Suzuki T. Purification and characterization of a novel type of fimbriae from the oral anaerobe Bacteroides gingivalis. J Bacteriol. 1984;160:949–957. doi: 10.1128/jb.160.3.949-957.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zambon J J, Reynolds H S, Dunford R G, DeVizio W, Volpe A R, Berta R, Tempro J P, Bonta Y. Microbial alterations in supragingival dental plaque in response to a triclosan-containing dentifrice. Oral Microbiol Immunol. 1995;10:247–255. doi: 10.1111/j.1399-302x.1995.tb00150.x. [DOI] [PubMed] [Google Scholar]