Abstract
From a collection of 2,800 Tn5-TC1 transposon mutants of Salmonella typhimurium F98, 18 that showed reduced intestinal colonization of 3-week-old chicks were identified. The sites of transposon insertion were determined for most of the mutants and included insertions in the lipopolysaccharide biosynthesis genes rfaK, rfaY, rfbK, and rfbB and the genes dksA, clpB, hupA, and sipC. In addition, identification was made of an insertion into a novel gene that encodes a protein showing similarity to the IIC component of the mannose class of phosphoenolpyruvate-carbohydrate phosphotransferase systems, which we putatively called ptsC. Transduction of most of the transposon mutations to a fresh S. typhimurium F98 genetic background and construction of defined mutations in the rfbK, dksA, hupA, sipC, and ptsC genes of S. typhimurium F98 supported the role in colonization of all but the pts locus. The virulence of the rfbK, dksA, hupA, sipC, and ptsC defined mutants and clpB and rfaY transductants in 1-day-old chicks was tested. All but the ptsC and rfaY mutants were attenuated for virulence. A number of other phenotypes associated with some of the mutations are described.
Poultry are a major source of Salmonella food poisoning for humans. More than 2,000 serotypes have been identified, mainly as a result of human food poisoning, the vast majority of which produce little or no systemic disease in healthy adult animals. Salmonellae are usually associated with food poisoning by virtue of their ability to colonize the alimentary tracts of livestock, particularly poultry. This results in considerable contamination of carcasses at slaughter, with entry of salmonellae into human food. A small number of serotypes typically produce typhoid-like diseases in adults of a restricted number of host species. These include Salmonella typhi and S. paratyphi in humans, S. cholerae-suis in pigs, S. dublin in cattle, and S. gallinarum in birds. Although transmission of these serotypes is primarily fecal-oral, they do not colonize the alimentary tract extensively in their respective host species and are not excreted in the feces in large numbers, except in the advanced stages of systemic disease. Thus, carcass contamination by most of these serotypes is infrequent, and they rarely enter the human food chain. S. typhimurium and S. enteritidis are exceptional in being able to colonize the alimentary tracts of poultry and cause typhoid in mice.
While infection of adult poultry leads to limited excretion of Salmonella in the feces, infection of newly hatched chicks, which have a relatively simple gut flora, results in rapid multiplication and extensive excretion. This can lead to rapid spread of a Salmonella strain through a flock as the housing and water and feeding systems become contaminated. Present methods for control are either inadequate or create additional problems. Improvements in hygienic measures are costly, and antibiotic therapy or prophylaxis can result in the production and spread of antibiotic-resistant strains (43). Vaccination with live, attenuated Salmonella vaccines is becoming more attractive and acceptable (11). Colonization of young chickens by Salmonella can be reduced by oral inoculation with preparations of gut flora derived from adult chickens. This has been termed competitive exclusion and provides the young chicken with an established gut flora early in life (28). The level of resistance obtained is, nevertheless, less than that of an adult bird that has been vaccinated.
In the chicken alimentary tract, ingested foodstuff reaches the crop, which acts as a storage organ where little, if any, digestion occurs. The crop is lined with squamous epithelium which is normally colonized by lactobacilli, and, as a result of fermentation, this organ has a pH of 4 to 5. Further along, the proventriculus secretes HCl and is immediately followed by the gizzard, which is a muscular organ in which food is ground. Distal to the small intestine, the ceca branch off from the ileum as two closed tubes, just before it opens into the cloaca. Facultative anaerobic bacteria such as lactobacilli, streptococci, and Escherichia coli are found in moderately high densities throughout the gut, whereas obligate anaerobes are found in high numbers only in the cecum. Upon colonization of the alimentary tract, the highest viable counts of Salmonella are obtained from the cecum, cloaca, and ileum and, to a lesser extent, from the crop (3). The higher counts obtained from the more distal regions of the gut may be the result of the lower flow rate (42), and the reabsorption of fluids from the intestinal contents may also contribute to this.
Very few data regarding the mechanism whereby Salmonella colonize the alimentary tracts of chickens are available. Potentially, it could include the ability to multiply in the intestinal contents, growth in or migration through the mucus layer, and attachment to and invasion of the epithelium. Some Salmonella serotypes colonize more efficiently than others (3, 44); thus, S. montevideo persists in the guts of chickens and is consequently excreted in the feces for longer than S. typhimurium. Serotypes that produce typhoid-like disease in poultry, such as S. gallinarum and S. pullorum, or other animals, such as S. cholerae-suis, colonize the alimentary tracts of chickens very poorly in the absence of clinical disease.
Physical attachment has been suggested as a mechanism for colonization (45), and there is some evidence that fimbriae promote colonization of the murine intestine by S. typhimurium (47, 48) and of the chicken intestine by S. infantis (3) and S. enteritidis (49). Barrow et al. (3) also found that a nonmotile mutant of S. infantis did not colonize as well as the wild type, whereas nonmotility made no difference to colonization by S. typhimurium phage type 14. One interpretation of this apparent anomaly was that the nonspecific chemical method used for mutagenesis may have generated additional mutations affecting colonization. However, the possibility that different serotypes use alternative mechanisms for colonization also needs to be considered.
To improve our understanding of colonization of the chicken alimentary tract by Salmonella, we sought to identify some of the microbial determinants required. We report here the isolation of transposon mutants of S. typhimurium F98 which showed reduced excretion in the feces following experimental oral inoculation and have identified the sites of transposon insertion. The role of the identified genes in colonization was confirmed by transducing the transposon mutations to a fresh S. typhimurium F98 genetic background with a derivative of bacteriophage P22 and by the construction of defined mutations. Many of the identified genes were also found to be required for virulence of S. typhimurium F98 in newly hatched chicks.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains used in this study are listed in Table 1. S. typhimurium F98 was isolated from chickens, and its virulence and colonization characteristics have been studied under a variety of conditions (2, 3, 43, 44). Unless otherwise stated, broth cultures consisted of Luria-Bertani (LB) broth (Difco) and were incubated for 24 h at 37°C in a shaking incubator (150 rpm). These contained 1 × 109 to 3 × 109 CFU ml−1.
TABLE 1.
Bacterial strains used
| Name | Comment(s) | Reference |
|---|---|---|
| S. typhimurium F98 (Nalr) | Wild type | 44 |
| S. typhimurium F98 (Spcr) | Wild type | 44 |
| S. typhimurium (Nalr) Tn5-TC1 mutants | ||
| 17/12/90-117 | rfbB::Tn5-TC1; no O antigen | This work |
| 14/12/90-201 | rfbB::Tn5-TC1; no O antigen | This work |
| 30/11/90-85 | rfbK::Tn5-TC1; no O antigen | This work |
| 12/6/91-86 | rfaK::Tn5-TC1; no O antigen | This work |
| 7/12/90-66 | No O antigen | This work |
| 13/2/91-163 | No O antigen | This work |
| 5/6/91-184 | No O antigen | This work |
| 30/11/90-93b | No O antigen | This work |
| 30/11/90-94 | No O antigen | This work |
| 14/12/90-7 | rfaY::Tn5-TC1 | This work |
| 14/12/90-243 | dksA::Tn5-TC1 | This work |
| 14/12/90-43 | dksA::Tn5-TC1 | This work |
| 21/12/90-82 | dksA::Tn5-TC1 | This work |
| 21/12/90-83 | dksA::Tn5-TC1 | This work |
| 14/12/90-19 | hupA::Tn5-TC1 | This work |
| 8/5/91-309 | sipC::Tn5-TC1 | This work |
| 6/2/91-62 | clpB::Tn5-TC1 | This work |
| 10/4/91-86 | ptsC::Tn5-TC1 | This work |
| 14/12/90-14 | ptsC::Tn5-TC1 | This work |
| S. typhimurium F98 (Spcr) transductants | ||
| T14/12/90-7i | Transductant derived from 14/12/90-7 | This work |
| T14/12/90-7ii | Transductant derived from 14/12/90-7 | This work |
| T14/12/90-7iii | Transductant derived from 14/12/90-7 | This work |
| T14/12/90-7iv | Transductant derived from 14/12/90-7 | This work |
| T14/12/90-243 | Transductant derived from 14/12/90-243 | This work |
| T14/12/90-19 | Transductant derived from 14/12/90-19 | This work |
| T8/5/91-309 | Transductant derived from 8/5/91-309 | This work |
| T6/2/91-62 | Transductant derived from 6/2/91-62 | This work |
| T10/4/91-86 | Transductant derived from 10/4/91-86 | This work |
| S. typhimurium F98 (Nalr) defined mutants | ||
| rfbK | Kma insertion in rfbK | This work |
| dksA | Km insertion in dksA | This work |
| hupA | Km insertion in hupA | This work |
| sipC | Km insertion in sipC | This work |
| ptsC | Km insertion in ptsC | This work |
| E. coli strains | ||
| SY327λpir | 29 | |
| SM10λpir | 41 |
Km, kanamycin.
Transposon mutagenesis.
Plasmid pCHR82 is an R388 derivative, is temperature sensitive for replication, encodes resistance to trimethoprim (Tpr), and carries a Tn5 derivative, Tn5-TC1, which encodes resistance to tetracycline (Tcr) (38). The plasmid was transferred from E. coli MC1061 to a nalidixic acid-resistant (Nalr) derivative of S. typhimurium F98 by conjugation in LB broth at 30°C. LB broth was inoculated with a Nalr Tpr Tcr transconjugant and distributed into 96-well microtiter trays, which were then incubated, with three daily passages to fresh trays at 43°C. Growth in the wells was then screened for Tcr and Tps. This identified derivatives from which the plasmid had been lost, but the transposon Tn5-TC1 had been retained presumably by transposition to the chromosome. A collection of 2,800 S. typhimurium F98 Tcr derivatives was produced in this way and stored at −70°C.
Chickens and screening procedure.
For colonization studies, unsexed, specific-pathogen-free, 3-week-old Light Sussex chicks from the Institute for Animal Health flock were used. They were housed in metal cages in groups of 30 and reared under conditions described previously (43). For assessment of virulence, Rhode Island Red chickens, which are more susceptible to salmonellosis than the Light Sussex breed, were used from the same source. Initial studies were carried out with a noncolonizing mutant of S. typhimurium F98 Nalr produced previously (3) and the wild type to determine the optimal conditions under which chickens could be inoculated individually with poorer colonizers and identified by cloacal swabbing. Screening of mutants was, therefore, carried out with individual birds housed in groups of 30, each marked with a numbered wing band. The birds were inoculated orally with 0.2 ml of LB broth culture incubated statically at 37°C for 24 h and containing 3 × 108 to 5 × 108 CFU ml−1 of the Tcr S. typhimurium F98 mutant to be tested. Two days later, a cloacal swab was taken from each bird and mixed in 2 ml of LB broth before plating directly onto a half plate of Brilliant Green agar (Oxoid; CM263, Basingstoke, United Kingdom) containing sodium nalidixate (20 μg ml−1) and novobiocin (1 μg ml−1), which allows a semiquantitative assessment of excretion in the feces (43). Mutants identified as being excreted in lower numbers were checked in groups of 3 chickens and then rechecked in groups of 10 chickens. The excretion pattern of each identified mutant after oral inoculation of 30 birds with 0.3 ml of an undiluted shaken broth culture was then monitored for a 3-week period with cloacal swabbing as described above. Statistical analysis was done by calculating the χ2 distribution.
Virulence in groups of approximately 20 Rhode Island Red chicks less than 24 h old was assessed. These were inoculated orally with 0.1 ml of a neat-shaken broth culture of the Salmonella strain containing approximately 108 CFU, and the numbers of birds which died or were killed humanely during a 3-week period because of severe salmonellosis were recorded.
Recombinant DNA procedures.
Chromosomal DNA was extracted from the mutant strains of S. typhimurium by the method described by Silhavy et al. (40). DNA prepared in this way was digested with one of the restriction enzymes PstI, XhoII, and HincII and was then diluted to approximately 10 ng ml−1 with 1× T4 ligase buffer containing ligase (Boehringer Mannheim) at 5 U ml−1, and the dilutions were incubated at 20 to 24°C for 1 to 3 h. The ligation mixture was then purified with the Wizard DNA Cleanup System (Promega), and the DNA was eluted into 50 μl of double-processed water for tissue culture (Sigma). An aliquot of 1 to 5 μl of this DNA was used in PCRs. Inverse PCR (5) was performed with oligonucleotides with the sequences 5′-GCGGATCCCAGGACGCTACTTGTGTATA and 5′-GCGGAATTCAC GGAACCTTTCCCGTTTTC, which prime in the outside edge of the IS50 of Tn5-TC1. Reactions were performed in 50-μl volumes with Taq DNA polymerase (Gibco BRL or Amersham) according to the manufacturer’s instructions. PCR products were purified by electrophoresis through 1 to 1.3% low-gelling-temperature agarose as described in the Wizard PCR Preps DNA Purification System (Promega). The nucleotide sequences of the purified PCR products were determined with a PRISM Ready Reaction DyeDeoxy Terminator Cycle Sequencing kit (Applied Biosystems, Inc.) and the Applied Biosystems 373A DNA sequencer. Nucleotide sequences were analyzed with the Genetics Computer Group Wisconsin package (version 8) (15a).
Rapid genetic mapping with Mud-P22 prophages.
A set of Mud-P22 strains was kindly provided by K. E. Sanderson of the Salmonella Genetic Stock Center (University of Calgary, Calgary, Canada). Lysates were prepared, and DNA was extracted from them, as described by Youderian et al. (53). The concentrations of the DNA samples extracted from lysates were estimated with agarose gels and were equalized at approximately 20 μg ml−1. Dilutions of 1 in 10 to 1 in 100 were used in PCRs with relevant oligonucleotides as primers to identify DNA samples containing the gene of interest.
Transduction of Tn5-TC1 mutations between strains of S. typhimurium.
Transduction was performed with bacteriophage P22 HT105/1int as described previously (1). Transduction from S. typhimurium F98 Nalr derivatives to S. typhimurium F98 spectinomycin-resistant (Spcr) derivatives was performed.
Construction of defined mutants.
Defined mutations were constructed by inserting a short PCR-amplified fragment of the relevant open reading frame into the multiple cloning site of the suicide vector pGP704 (30). A kanamycin resistance (Kmr) GenBlock (Pharmacia) was PCR amplified with primers containing a KpnI site in the 5′ ends and was inserted into the middle of a KpnI site (see below) in the middle of each short fragment by standard procedures (36). SY327λpir was the initial host strain for pGP704 and its derivatives. The mutations were then transferred into E. coli SM10λpir by transformation and from there were transferred to S. typhimurium F98 Nalr by conjugation. A number of Kmr ampicillin-sensitive transconjugants were tested by PCR with relevant primers to confirm that they carried the Kmr determinant in the appropriate gene. In particular, primers were chosen outside the mutated region, and in each case PCR amplification resulted in a DNA fragment which was approximately 1 kb larger (the approximate length of the Kmr GenBlock) than the wild type. In each mutant, the smaller wild-type fragment was absent from these PCRs. For the defined mutations, the Kmr determinant was inserted at the following positions from the start of the relevant open reading frames: hupA, 146 bp; ptsC, 176 bp; rfbK, 268 bp; dksA, 218 bp; and sipC, 757 bp. For the dksA and ptsC mutations, the KpnI site into which the Kmr GenBlock was inserted was generated artificially by first PCR amplifying the fragment in two parts with relevant primers containing a KpnI site.
Analysis of LPS.
Analysis of lipopolysaccharide (LPS) was performed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (27).
Analysis of RNA contents of mutants.
RNA was isolated by minor modifications to the method used for isolation of plasmid DNA by alkaline lysis (36). Cells from 10 ml of 24-h-old cultures were harvested and resuspended in 0.2 ml of 50 mM Tris (pH 8)–10 mM EDTA and were held on ice. The optical density of 20 μl of this suspension in 1 ml of water was determined to give an estimate of the cell mass per milliliter. The cell suspension was then lysed with 0.4 ml of 1% SDS–0.2 M NaOH, and 0.3 ml of chilled 3 M potassium acetate in 2 M acetic acid was added immediately, followed promptly by 0.5 ml of phenol-chloroform. This was mixed thoroughly and then centrifuged for 5 min in a microcentrifuge. The aqueous phase was removed to a clean tube, and nucleic acids were precipitated by the addition of a 0.6 volume of isopropanol. The pellet was resuspended in 10 mM Tris (pH 8)–1 mM EDTA, the volume of which depended on the optical density of the cell suspension. Thus, any variation in the yield of RNA was not due to variation in the number of cells harvested. To reduce experimental variation, RNA from each mutant or the wild type was prepared simultaneously in triplicate, and RNA samples within each triplicate were then pooled.
RESULTS
Isolation of Tn5-TC1 mutants of S. typhimurium F98 that showed reduced colonization in the chicken alimentary tract.
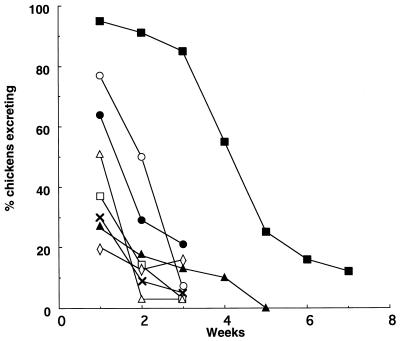
From a collection of 2,800 S. typhimurium F98 derivatives carrying independent Tn5-TC1 insertions, 18 were identified as being excreted in the feces over a time period shorter than that for the parent strain (Fig. 1). Of these, 9 were rough, as determined by agglutination with acriflavine, and examination of the LPS profiles by SDS-PAGE (27) showed an absence of O-antigen repeats in the 9 mutants. All of the mutants grew at a rate similar to that of the wild-type parent strain in LB broth at 37°C (data not shown).
FIG. 1.
Numbers of chicks from groups of approximately 30 animals (expressed as percentages) excreting mutants of Nalr S. typhimurium F98 in the feces after oral inoculation with approximately 108 CFU. ▪, mutant 14/12/90-14 (positive control); ○, mutant 14/12/90-7; •, mutant 8/5/91-309; ▵, mutant 14/12/90-43; □, mutant 30/11/90-85; ×, mutant 21/12/90-82; ▴, mutant 10/4/91-86; ◊, mutant 17/12/90-117.
Identification of Tn5-TC1 insertion sites.
The nucleotide sequence adjacent to the site of Tn5-TC1 insertion was determined for four of the nine mutants which lacked LPS O antigen. For two of these mutants, the transposon had inserted into the rfbB gene (immediately after bp 551 and 559, respectively, from the start of the rfbB open reading frame). Another insertion was in the rfbK gene (immediately after bp 204 of the open reading frame). The rfb genes are required for O-antigen synthesis of the LPS. For the fourth mutant, the site of insertion was the rfaK gene (immediately after bp 304 of the open reading frame), which is involved in completion of outer core synthesis required for the subsequent attachment of the O antigen. Therefore, mutations in rfaK give rise to LPS in which no O antigen is detectable by SDS-PAGE, and our mutant also showed this phenotype. In light of these data, it was deemed unnecessary to determine the sites of insertion for the other five mutants defective for LPS.
The remaining nine mutants had LPS profiles that showed amounts of O antigen similar to that of the wild type by SDS-PAGE (not shown). For one of these mutants, the site of insertion was the rfaY gene (somewhere between bp 272 and 276 of the open reading frame). The nucleotide sequence (GenBank accession no. AF001397) indicated that this gene was at the end of the rfaIJ operon (8). The 3′ end of the rfaY gene was identified previously (25) and, together with our sequence, confirms that rfaY is in the middle of an operon, i.e., rfaIJYZK.
For four mutants, the nucleotide sequence at the site of insertion was similar to the dksA gene from E. coli (17). The nucleotide sequence of this locus was determined for S. typhimurium (GenBank accession no. AF010249) and showed 86% similarity to the dksA gene from E. coli. An open reading frame of 151 codons was identified, the amino acid sequence of which showed 97% identity to and was the same length as that of DksA from E. coli. Upstream of the open reading frame was a putative ς70 promoter sequence and ribosome binding site, while downstream the sequence showed a short inverted repeat sequence that may function as a transcriptional terminator.
For one mutant, the nucleotide sequence adjacent to the site of the transposon insertion showed similarity to the E. coli clpB gene. This gene is 2,573 bp long (20) (GenBank accession no. ecd887). Across the 1 kb of S. typhimurium nucleotide sequence obtained (GenBank accession no. AF010250), there was 85% identity compared to that of E. coli, and the amino acid sequence of the partial open reading frame showed 99% identity to that of E. coli. Therefore, it is likely that this S. typhimurium nucleotide sequence is part of the clpB gene. If the E. coli and S. typhimurium nucleotide sequences align exactly, the Tn5-TC1 insertion is immediately after bp 928 of the open reading frame.
The nucleotide sequence at the site of one transposon insertion showed no significant similarity to any nucleotide sequences in any of the data bases searched (GenBank accession no. L47170). However, 1.2 kbp of nucleotide sequence was obtained and found to contain an open reading frame, the amino acid sequence of which showed 30% similarity to the IIC components of the mannose class of the phosphoenolpyruvate-sugar phosphotransferases of bacteria (reviewed by Postma et al. [34]). We have tentatively called this gene ptsC. Parts of open reading frames upstream and downstream were also found, and the amino acid sequences of these showed 30% similarity to the IIB and IID components of the same class of transferase, respectively. It is likely, therefore, that the Tn5-TC1 transposon had inserted into a gene from a novel phosphoenolpyruvate-carbohydrate phosphotransferase system of S. typhimurium.
For two of the smooth transposon mutants, the sites of insertion were Salmonella genes that have been identified previously (immediately after nucleotide 154 from the start of the open reading frame in each case). One was into the hupA gene (16), whose product is involved in maintaining the nucleoid structure, and the other was into sipC of the sicAsipBCDA operon, which encodes Salmonella invasins (18, 19).
Chromosomal map locations of genes associated with colonization of the chicken alimentary tract.
The locations on the S. typhimurium chromosome of the rfa, rfb, hupA, and sip loci have already been determined (18, 19, 37). The positions on the S. typhimurium chromosome of the dksA, clpB, and ptsC genes were approximately and rapidly mapped by PCR with primers derived from the known nucleotide sequences of the genes, with bacteriophage DNA from a series of locked-in Mud-P22 S. typhimurium lysogens (6) as the template. The results indicated that the dksA, clpB, and ptsC genes were located between 3 and 7 min, 54 and 63 min, and 93 and 97 min, respectively, on the S. typhimurium chromosome. This is similar to the positions of the dksA and clpB genes on the E. coli chromosome (3.4 and 58.7 min) (7). As a positive control in this experiment, the map position of the rfbK gene was checked, and a value of 40.5 to 50 min was obtained, which compared favorably with the known value of 45 min (37).
Transduction of Tn5-TC1 mutations to S. typhimurium F98.
To confirm that the respective Tn5-TC1 insertions were responsible for the reduced ability to colonize, the hupA, rfaY, dksA, clpB, and ptsC mutations were transduced to a wild-type S. typhimurium F98 Spcr isolate to provide a fresh genetic background. When tested, all except the ptsC transductant showed a reduced ability to colonize the alimentary tracts of 3-week-old chicks.
Construction of defined mutants of S. typhimurium F98.
Based on the nucleotide sequences from previous work, or those derived as part of this work, defined mutations in the rfbK, dksA, hupA, sipC, and ptsC genes were constructed by insertion of a kanamycin resistance GenBlock (Pharmacia) into the respective open reading frames. These constructs were then transferred into Nalr S. typhimurium F98. Attempts to introduce a clpB mutation based on the nucleotide sequence of the E. coli gene failed.
Phenotypes of the rfbK, dksA, hupA, sipC, and ptsC defined mutants and the rfaY and clpB transductants.
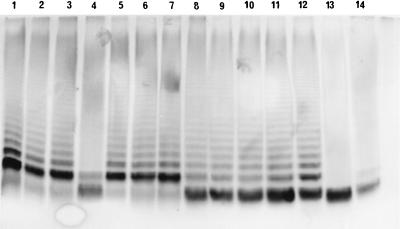
The LPS profiles of the defined mutants and the rfaY and clpB transductants were then checked by SDS-PAGE. All except the rfbK mutant and the rfaY transductants showed a wild-type LPS profile by SDS-PAGE. The LPS profiles are shown in Fig. 2, which includes LPS from the rfaK::Tn5-TC1 mutant and the wild type for comparison. For the rfbK mutant (lane 4), the ladder of bands which represents the O-antigen repeats was absent and there was more staining in the lowest band of the gel. This lowest band in the gel was the only band obtained with the rfaK mutant (Fig. 2, lane 13), representing the lipid A with attached core component. For the rfaY mutants (lanes 8 to 12), the ladder of bands representing the O-antigen repeats was present at an intensity similar to those of the wild type and other mutants showing a wild-type LPS. However, the lowest band in the gel showed an intensity similar to that of the rfaK::Tn5-TC1 mutant (lane 13), while the second lowest band (present in the rfbK mutant but absent from the rfaK mutant) was less intense than those obtained for the wild type and the other mutants showing a wild-type LPS.
FIG. 2.
LPS profiles of S. typhimurium F98, both wild type and mutants carrying mutations in genes required for colonization of the chicken alimentary tract. LPS was extracted from the strains and visualized by SDS-PAGE. Lanes: 1, wild type; 2, dksA; 3, hupA; 4, rfbK; 5, ptsC; 6, sipC; 7, clpB; 8 to 11, T14/12/90-7i, -ii, -iii, and -iv, respectively (rfaY::Tn5-TC1 transductants); 12, 14/12/90-7 (rfaY::Tn5-TC1 transposon mutant); 13, 12/6/91-86 (rfaK::Tn5-TC1 transposon mutant); and 14, 30/11/90-85 (rfbK::Tn5-TC1 transposon mutant).
All of the defined mutants and the clpB and rfaY transductants grew at similar rates in LB broth at 41°C (the body temperature of the chicken) and then remained viable in numbers similar to that for the wild type for 1 week at this temperature (data not presented). Under the light microscope, all of the mutants and transductants except the dksA and hupA mutants resembled the wild-type cell in shape and size and were motile. The dksA bacteria were elongated with respect to the wild type but were still motile, while the hupA mutant bacteria grew mainly in clumps, which caused them to settle out of culture and severely impeded the motility of the majority of the cells.
The S. typhimurium dksA mutant grew much more poorly than the parent strain on M9 minimal agar (36) with glucose as the sole carbon source but grew as well as the wild type when Casamino Acids were added at 0.4%. The dksA mutant also grew well, although not as well as the wild type, when M9 minimal salts were supplemented with glutamine or glutamate (at 0.04%) as the sole carbon source but not when they were supplemented with histidine. The ptsC defined mutant was tested for its ability to ferment 48 different carbohydrates with API 50 CH fermentation strips (API, Marcy-l’Etoile, France). The mutant showed no difference from the wild type in this respect.
The initial observation that the dksA mutant required amino acids for growth suggested to us that this gene may have some involvement in the stringent response, when a lack of amino acids results in some repression of genes encoding components of the translation machinery (rRNA, ribosomal proteins, and tRNA) and induction of some amino acid biosynthetic enzymes (9). Therefore, we decided to check the levels of RNA that were extractable from 24-h-old cultures of the wild type and the mutants. The dksA mutant yielded more RNA than the wild type or the hupA mutant (Fig. 3) or any of the other mutants, as determined from 2% agarose gels. Apart from the overall intensity, the RNA profiles for the wild type and mutants gave the same number of bands with similar mobilities. The greater yield of RNA from the dksA mutant was confirmed by measuring the optical density at 260 nm, which gave average values of 1.7 for the mutant and 0.8 for the wild type. When an overnight culture was used to inoculate fresh broth and RNA from these was prepared after 5 h of incubation at 37°C, there was no significant difference in the yield between all of the mutants and the wild type.
FIG. 3.
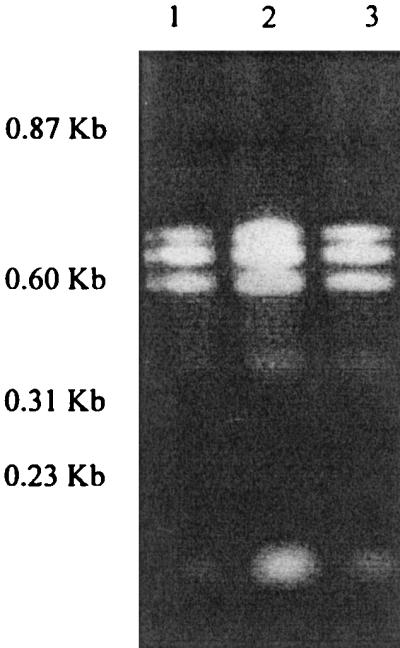
RNA contents of the S. typhimurium F98 parental strain (lane 1) and defined dksA::km (lane 2) and hupA::km (lane 3) mutants. The bands are stable low-molecular-weight RNA, which is probably rRNA (upper three bands) and tRNA (lower band).
Like the transductants (see above), all of the defined mutants tested showed a reduced ability to colonize the alimentary tracts of 3-week-old chicks, confirming that LPS, dksA, hupA, and at least one of the invasion proteins are required for S. typhimurium colonization of the chicken alimentary tract. In the case of the sipC mutant with the experimental system used, this reduction was of statistical significance only when the heavier excretion was considered and only for the cloacal swabs at 2 weeks postinoculation (χ2 = 4.29, P = 0.02) and for the ceca (χ2 = 4.32, P = 0.02). The ptsC defined mutant was not tested. The reduced colonization ability in these defined mutants was much more marked for dksA, in which hardly any reisolations were made, than for the others (Table 2).
TABLE 2.
Fecal excretion of defined mutants of S. typhimurium F98 (Nalr) by groups of 30 3-week-old chickens
| Wk post- infection | % of chickens excreting the following mutants at the indicated times after inoculationa
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parent strain
|
dks::km
|
sipC::km
|
hupA::km
|
rfbK::km
|
|||||||||||
| ≥50 | D | T | ≥50 | D | T | ≥50 | D | T | ≥50 | D | T | ≥50 | D | T | |
| 1 | 5 | 52 | 90 | 0 | 0 | 5 | 7 | 33 | 73 | 0 | 0 | 70 | 10 | 25 | 65 |
| 2 | 5 | 14 | 81 | 0 | 0 | 0 | 0 | 0 | 67 | 0 | 0 | 35 | 0 | 0 | 25 |
| 3 | 0 | 10 | 52 | 0 | 0 | 5 | 0 | 13 | 40 | 0 | 0 | 10 | 0 | 0 | 0 |
| 3 (ceca)b | 38 | 67 | 90 | 0 | 5 | 5 | 47 | 80 | 80 | 10 | 25 | 35 | 0 | 5 | 5 |
≥50, 50 or more colonies isolated per direct plate; D, one or more colonies isolated per direct plate; T, Salmonella organisms isolated by direct plating or by enrichment culture.
Cecal contents sampled immediately after killing.
Attenuation of S. typhimurium virulence for newly hatched chicks.
The rfbK, dksA, hupA, sipC, and ptsC defined mutants and the clpB and rfaY Tn5-TC1 transductants were tested for virulence in 1-day-old chicks. Since S. typhimurium is highly virulent for very young chicks, this is a sensitive assay for attenuation. Although S. typhimurium F98 can colonize the alimentary tracts of older chickens, mortality in such birds resulting from systemic disease caused by this strain is generally low. Like the wild type, all of these mutants colonized the alimentary tracts of 1-day-old chicks rapidly and in high numbers (data not shown), so that attenuation of virulence was not a direct result of their reduced colonization under these conditions. All of the chicks inoculated with the wild type became very ill, showing anorexia and lethargy with diarrhea, necessitating humane killing. Chicks inoculated with the defined ptsC mutant and the rfaY::Tn5-TC1 transductant behaved similarly. Each of the other mutants showed a considerable degree of attenuation, with small proportions of the birds inoculated showing signs of disease (Table 3).
TABLE 3.
Mortality in 1-day-old chicks infected with wild-type S. typhimurium F98 or mutants carrying mutations in genes associated with colonization of the chicken alimentary tract
| Strain | Mortality |
|---|---|
| Wild type | 21/21 |
| S. typhimurium (hupA) | 0/19 |
| S. typhimurium (dksA) | 0/18 |
| S. typhimurium (ptsC) | 21/21 |
| S. typhimurium (sipC) | 0/21 |
| S. typhimurium (rfbK) | 0/21 |
| T6/2/91-62 (clpB::Tn5-TC1) | 2/22 |
| T14/12/90-7i (rfaY::Tn5-TC1) | 19/22 |
| 14/12/90-201 (rfbB::Tn5-TC1) | 3/23 |
| 12/6/92-86 (rfaK::Tn5-TC1) | 0/23 |
DISCUSSION
To improve our understanding of the mechanism of colonization of the chicken alimentary tract by S. typhimurium, we identified 18 Tn5-TC1 transposon mutants from a collection of 2,800 that showed reduced ability to colonize. Half of these mutants had a defective LPS (rough) phenotype, and nucleotide sequence data from 4 of these mutants confirmed that the transposon had inserted into the rfa or rfb LPS biosynthesis gene. In the remaining 9 smooth mutants, the sites of transposon insertion were the rfaY, dksA, clpB, hupA, and sipC genes and a gene that we putatively called ptsC. The locations on the S. typhimurium F98 chromosome of the dksA and clpB genes were similar to the locations of the homologous genes on the E. coli chromosome, while the ptsC gene was determined to be at 93 to 97 min.
To check that the reduced colonization shown by these smooth mutants was not a result of an additional mutation elsewhere on the chromosome, the transposon mutations were transduced, with bacteriophage P22 HT105/1int, to an S. typhimurium F98 Spcr derivative to provide a fresh genetic background. Only the ptsC::Tn5-TC1 transductant colonized as well as the wild type, indicating that the ptsC gene is unlikely to be required for colonization. All of the other transductants colonized more poorly than the wild type, supporting the view that the rfaY, dksA, clpB, hupA, and sipC genes are required for colonization. To provide further support and to confirm the results obtained with the transductants, defined mutations were constructed in the hupA, dksA, and sipC genes by insertion of a kanamycin resistance determinant. In addition, since bacteriophage P22 transduction is not possible with rough mutants, a defined mutation was constructed in the rfbK gene to confirm the role of LPS in colonization. These defined mutations were then transferred to Nalr S. typhimurium F98 and tested for colonization. Although greater reductions in colonization ability were seen with the rfbK, hupA, and dksA mutants than with the sipC mutant, it seems likely that all of these genes are likely to be required for colonization, albeit to different degrees. The mutations are either in the gene into which the transposon inserted or in a gene downstream within the operon. The available evidence also strongly supports the view that the rfaY and clpB genes are required for colonization, but this has not yet been confirmed with defined mutations.
The rfbK, hupA, dksA, sipC, and ptsC defined mutants and the rfaY and clpB transductants were then tested for virulence in 1-day-old chickens. S. typhimurium F98 was originally isolated from chickens and is highly virulent for young chicks (2, 44). Young chicks that lack a complex gut flora are rapidly colonized with high numbers of Salmonella following infection (3, 33) and therefore provide a sensitive assay. The defined mutants and transductants that were tested also rapidly colonized the alimentary tracts of the young chickens. Thus, any attenuation of virulence is not a result of reduced colonization in young chickens. The ptsC defined mutant and the rfaY transductant were as virulent as the wild type in 1-day-old chicks, while the rfbK, hupA, dksA, and sipC defined mutants and the clpB transductant showed significant attenuation.
Several earlier studies have suggested that LPS plays a role in virulence and colonization of the alimentary tract. The most recent studies regarding colonization indicate that rough mutants of S. typhimurium are less able to attach to or traverse the mucosal layer of mice (23) or chickens (12) than their smooth counterparts. Thus, it is not surprising that our study also identified LPS-deficient mutants. Other work has also indicated the ability of S. typhimurium to associate with intestinal mucus and to be agglutinated by it. In that case, a 66-kDa heat shock protein was thought to be responsible (14, 15).
Our nucleotide sequence data confirmed that the S. typhimurium rfaY gene was part of an operon, rfaIJYZK. It is known that rfaK mutants are rough, and yet our rfaY transposon mutant was smooth, suggesting that this Tn5-TC1 insertion was nonpolar, as has been observed for a number of other Tn5 insertions (10). The function of RfaY in LPS biosynthesis is unknown (39). While the LPS profile of our rfaY::Tn5-TC1 mutant and those derived by transduction showed quantities of O-antigen repeats similar to that of the wild type, the rfaY mutants showed significantly more LPS without attached O antigen, while the wild-type profile showed more LPS with one O-antigen repeat attached (represented by the band which was absent from the rfaK mutant but present for the rfbK mutant). This aberration in the LPS of the rfaY mutant was sufficient to significantly affect intestinal colonization but did not affect virulence in newly hatched chicks.
The dksA gene was originally identified in E. coli as a suppressor, at high copy number, of the dnaK, dnaJ, and grpE mutations (17). Later, it was shown at high copy number also to be a suppressor of the mukB and prc mutations (4, 52). More recently, it was found that a single-base pair change in dksA suppressed point mutations in the pSC101 plasmid replication origin that were deleterious for replication, suggesting a role for dksA in replication at least for this plasmid (32). The S. typhimurium dksA defined mutant grew quite well with glutamine or glutamate as the sole carbon source, but not with glucose or histidine as the sole carbon source. This suggests that the dksA mutant was defective in its biosynthesis of glutamate, which is required for the biosynthesis of glutamine. Glutamate and glutamine serve as the primary nitrogen donors for all cellular metabolites in S. typhimurium (35), and it has been shown that simultaneous prevention of glutamine biosynthesis and uptake attenuates Salmonella virulence (21).
Our initial observation that the dksA mutant required amino acids for growth suggested to us that this gene may have some involvement in the stringent response, when a lack of amino acids results in repression of genes encoding components of the translation machinery (rRNA, ribosomal proteins, and tRNA) and induction of some amino acid biosynthetic enzymes (9). The yields of RNA from older cultures were significantly higher for the dksA mutant than for any other of the mutants or of the wild type, while yields from younger cultures showed no significant differences. This suggested that DksA plays a role in the regulation of rRNA expression during stationary phase. However, survival of the dksA mutant in stationary-phase LB broth cultures for as much as 1 week was similar to that of the wild type, and there was no obvious ςs promoter in front of the dksA open reading frame. In addition, cells of the dksA mutant were longer than those of the wild type both in younger cultures (data not shown) and in 24-h-old cultures, suggesting that DksA is required for wild-type growth during exponential-phase as well as stationary-phase growth. The pleiotropic nature of the dksA mutation suggests that DksA is a regulator of gene expression and as such at artificially high copy number suppresses the mutations mentioned above but normally plays a role which may include regulation of glutamine and glutamate biosynthesis and the stringent response.
The ClpB protein is a heat shock protein involved in removal of denatured proteins from the cell, although at present it is unclear whether it acts as a renaturing chaperone or a protease (22, 51). At 44°C, an E. coli clpB mutant grew at half of the rate of the wild type and was killed more rapidly at 50°C, but growth rate was normal at 42°C (46). Our S. typhimurium clpB mutant also grew and survived as well as the wild type in LB broth for 1 week at 41°C, the body temperature of the chicken. The heat shock response is induced by many factors other than heat shock (24, 31), and simultaneous treatment with certain chemicals was shown to induce synergistically the heat shock response (50). This suggests that different stress conditions may act synergistically to cause cellular damage. In the alimentary tract, ClpB probably protects the bacteria from a combination of stress factors, one of which may be the relatively high temperature, that acting alone are not harmful to the clpB mutant.
The hupA gene encodes a subunit of the HU protein, which is one of the most abundant DNA binding proteins in E. coli, and contributes to the architectural structure of the nucleoid. The architectural proteins H-NS and integration host factor have been implicated in DNA reactions (recombination, transposition, and replication) and the regulation of many genes and operons (for a review, see Dorman [13]). Of more direct relevance is the evidence that protein HU is involved in transcription of the osmoresponsive proU operon (26). This suggests that expression of any number of genes required for colonization and for virulence may be affected by mutation in the hupA gene. How a hupA mutation might induce clumping is unclear. Notwithstanding this, the clumping phenotype observed when the hupA mutant bacteria were grown in vitro probably contributed to its reduced virulence in newly hatched chicks and its reduced ability to colonize the alimentary tracts of 3-week-old chicks.
Our data have shown that at least one of the Sip proteins is also required for full colonization ability. Nonpolar mutations in sicA, sipB, sipC, or sipD render S. typhimurium incapable of entering cultured epithelial cells (18, 19) and attenuate Salmonella virulence. This finding suggests that some invasion of the epithelium may be required for colonization by S. typhimurium. Several of the inv genes have been found to contribute to colonization of the ileal wall in very young chicks (33), but this may largely reflect an invasive process. It has been observed that strains of S. typhimurium are more invasive for chickens than is S. montevideo, yet S. montevideo is able to persist longer in the alimentary tract. A suggested explanation for this was that invasion induced a greater immune response which could lead to elimination of the S. typhimurium serotype (3). If this were the case, then it would be expected that a noninvasive S. typhimurium, such as the sipC mutant, would persist longer than the wild type. Our present data indicate that this is not the case and suggest that S. typhimurium colonization involves some invasion of the epithelium, while for S. montevideo, invasion is of less importance for colonization. Therefore, it may be that colonization mechanisms vary between different serotypes and consequently that certain genes are required for various degrees for colonization.
In all likelihood, this study will have identified only a proportion of genes required for intestinal colonization. Screening of a larger number of mutants would have been even more time-consuming and expensive. In addition, Tn5 insertion is not completely random, as illustrated by the high proportion of insertions into dksA, so that some genes involved in colonization could remain undetected even if significantly more numbers of mutants had been screened. The data presented will provide a rational basis upon which to identify other genes required for colonization, which are likely to include other stress response proteins, regulators of gene expression (particularly those involved in stress responses), and invasins. Investigating the behavior of such mutants in the experimental animal with respect to their growth and ability to remain viable in intestinal contents, in the mucosa, and on or in the epithelium could provide some valuable insights. Our investigation identified no new genes that were required for colonization, most likely because very few genes remain to be identified from the E. coli chromosome, which probably contains most of the genes that are required for survival and multiplication in the alimentary tract. It is likely that many other genes already identified will turn out to be required for colonization of the alimentary tracts of animals.
ACKNOWLEDGMENTS
We are grateful to K. Murphy, K. Page, and A. Gray for technical assistance.
This work was funded by the Ministry of Agriculture Fisheries and Food, London, United Kingdom.
REFERENCES
- 1.Barrow P A, Hassan J O, Berchieri A., Jr Reduction in faecal excretion of Salmonella typhimurium strain F98 in chickens vaccinated with live and killed S. typhimurium organisms. Epidemiol Infect. 1990;104:413–426. doi: 10.1017/s0950268800047439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrow P A, Huggins M B, Simpson J M, Lovell M A. Observations on the pathogenesis of Salmonella typhimurium infection in chickens. Res Vet Sci. 1987;42:194–199. [PubMed] [Google Scholar]
- 3.Barrow P A, Simpson J M, Lovell M A. Intestinal colonisation in the chicken by food-poisoning Salmonella serotypes; microbial characteristics associated with faecal excretion. Avian Pathol. 1988;17:571–588. doi: 10.1080/03079458808436478. [DOI] [PubMed] [Google Scholar]
- 4.Bass S, Gu Q, Christen A. Multicopy suppressors of Prc mutant Escherichia coli include two HtrA (DegP) protease homologs (HhoAB), DksA, and a truncated RlpA. J Bacteriol. 1996;178:1154–1161. doi: 10.1128/jb.178.4.1154-1161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäumler A J, Kusters J G, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson N R, Goldman B S. Rapid mapping in Salmonella typhimurium with Mud-P22 prophages. J Bacteriol. 1992;174:1673–1681. doi: 10.1128/jb.174.5.1673-1681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berlyn M K B, Low K B, Rudd K E. , p. 1715–1902. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C. 1996. Linkage map of Escherichia coli K-12, ed. 9 [Google Scholar]
- 8.Cartensius P, Flock J I, Lindberg A. Nucleotide sequence of rfaI and rfaJ genes encoding lipopolysaccharide glycosyltransferases from Salmonella typhimurium. Nucleic Acids Res. 1990;18:6128. doi: 10.1093/nar/18.20.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cashel M, Gentry D R, Hernendez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1458–1496. [Google Scholar]
- 10.Clark A J, Satin L, Chu C C. Transcription of the Escherichia coli recE gene from a promoter in Tn5 and IS50. J Bacteriol. 1994;176:7024–7031. doi: 10.1128/jb.176.22.7024-7031.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper G L. Salmonellosis-infections in man and the chicken: pathogenesis and the development of live vaccines—a review. Vet Bull. 1994;64:123–143. [Google Scholar]
- 12.Craven S E. Altered colonizing ability for the ceca of broiler chicks by lipopolysaccharide-deficient mutants of Salmonella typhimurium. Avian Dis. 1994;38:401–408. [PubMed] [Google Scholar]
- 13.Dorman C. Genetics of bacterial virulence. Oxford, United Kingdom: Blackwell Scientific Publications; 1994. [Google Scholar]
- 14.Ensgraber M, Loos M. A 66-kilodalton heat shock protein of Salmonella typhimurium is responsible for binding of the bacterium to intestinal mucus. Infect Immun. 1992;60:3072–3078. doi: 10.1128/iai.60.8.3072-3078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ensgraber M, Genistsariotis R, Störkel S, Loos M. Purification and characterization of a Salmonella typhimurium agglutinin from gut mucus secretions. Microb Pathog. 1992;12:255–266. doi: 10.1016/0882-4010(92)90044-o. [DOI] [PubMed] [Google Scholar]
- 15a.Genetics Computer Group. Program manual for the Wisconsin package, version 8. Wis: University of Wisconsin, Madison; 1994. [Google Scholar]
- 16.Higgins N P, Hillyard D. Primary structure and mapping of the hupA gene of Salmonella typhimurium. J Bacteriol. 1988;170:5751–5758. doi: 10.1128/jb.170.12.5751-5758.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang P J, Craig E A. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaniga K, Tucker S, Trollinger D, Galan J E. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J Bacteriol. 1995;177:3965–3971. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaniga K, Trollinger D, Galan J E. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitagawa M, Wada C, Yoshioka S, Yura T. Expression of ClpB, an analog of the ATP-dependent protease regulatory subunit in Escherichia coli, is controlled by a heat shock ς factor (ς32) J Bacteriol. 1991;173:4247–4253. doi: 10.1128/jb.173.14.4247-4253.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klose K E, Mekalanos J J. Simultaneous prevention of glutamine synthesis and high-affinity transport attenuates Salmonella typhimurium virulence. Infect Immun. 1997;65:587–596. doi: 10.1128/iai.65.2.587-596.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laskowska E, Kuczynska-Wisnik D, Skorko-Glonek J, Taylor A. Degradation by proteases Lon, Clp and HtrA, of Escherichia coli proteins aggregated in vivo by heat shock; HtrA protease action in vivo and in vitro. Mol Microbiol. 1996;22:555–571. doi: 10.1046/j.1365-2958.1996.1231493.x. [DOI] [PubMed] [Google Scholar]
- 23.Licht T R, Krogfelt K A, Cohen P S, Poulsen L K, Urbance J, Molin S. Role of lipopolysaccharide in colonization of the mouse intestine by Salmonella typhimurium studied by in situ hybridization. Infect Immun. 1996;64:3811–3817. doi: 10.1128/iai.64.9.3811-3817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindquist S, Craig E A. The heat shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 25.MacLachlan R R, Adam S K, Sanderson K E. Cloning, characterization, and DNA sequence of the rfaLK region for lipopolysaccharide synthesis in Salmonella typhimurium LT-2. J Bacteriol. 1991;173:7151–7163. doi: 10.1128/jb.173.22.7151-7163.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manna D, Gowrishankar J. Evidence for involvement of proteins HU and RpoS in transcription of the osmoresponsive proU operon in Escherichia coli. J Bacteriol. 1994;176:5378–5384. doi: 10.1128/jb.176.17.5378-5384.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez R J. Thermoregulation-dependent expression of Yersinia enterocolitica protein 1 imparts serum resistance to Escherichia coli K-12. J Bacteriol. 1989;171:3732–3739. doi: 10.1128/jb.171.7.3732-3739.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mead G C, Impey C S. The present status of the Nurmi concept for reducing carriage of food-poisoning salmonellae and other pathogens in live poultry. In: Smulders F J M, editor. Elimination of pathogenic organisms from meat and poultry. Amsterdam: Elsevier; 1987. pp. 57–77. [Google Scholar]
- 29.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neidhardt F C, VanBogelen R A. Heat shock response. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1334–1345. [Google Scholar]
- 32.Ohkubo S, Yamaguchi K. A suppressor of mutations in the region adjacent to the iterons of pSC101 ori. J Bacteriol. 1997;179:2089–2091. doi: 10.1128/jb.179.6.2089-2091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porter S B, Curtiss R., III Effect of inv mutations on Salmonella virulence and colonization in 1-day-old white leghorn chicks. Avian Dis. 1997;41:45–57. [PubMed] [Google Scholar]
- 34.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate: carbohydrate phosphotransferase systems. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1149–1174. [Google Scholar]
- 35.Reitzer L J. Sources of nitrogen and their utilization. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 380–390. [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sanderson K E, Hessel A, Liu S-L, Rudd K E. The genetic map of Salmonella typhimurium, edition VIII. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1903–1999. [Google Scholar]
- 38.Sasakawa C, Yoshikawa M. A series of Tn5 variants with various drug resistance markers and suicide vector for transposon mutagenesis. Gene. 1987;56:283–288. doi: 10.1016/0378-1119(87)90145-4. [DOI] [PubMed] [Google Scholar]
- 39.Schnaitman C A, Klena J D. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993;57:655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 137–139. [Google Scholar]
- 41.Simon R, Priefer U, Puhler A. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 42.Smith H W. Observations on the flora of the alimentary tract of animals and factors affecting its composition. J Pathol Bacteriol. 1965;89:95–122. [PubMed] [Google Scholar]
- 43.Smith H W, Tucker J F. The effect of antibiotic therapy on the faecal excretion of Salmonella typhimurium by experimentally infected chickens. J Hyg. 1975;75:275–292. doi: 10.1017/s0022172400047306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith H W, Tucker J F. The virulence of Salmonella strains for chickens; their excretion by infected chickens. J Hyg Cambridge. 1980;84:479–488. doi: 10.1017/s0022172400027017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soerjadi A S, Rufner R, Snoeyenbos G H, Weinack O M. Adherence of Salmonella and native gut microflora to the gastrointestinal mucosa of chicks. Avian Dis. 1982;26:576–584. [PubMed] [Google Scholar]
- 46.Squires C L, Pedersson S, Ross B M, Squires C. ClpB is the Escherichia coli heat shock protein F84.1. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka Y, Katsube Y. Infectivity of Salmonella typhimurium for mice in relation to fimbriae. Jpn J Vet Sci. 1978;40:671–681. doi: 10.1292/jvms1939.40.671. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka Y, Katsube Y, Mutoh T, Imaizumi K. Fimbriae of Salmonella typhimurium and their role in mouse intestinal colonisation of the organism. Jpn J Vet Sci. 1981;43:51–62. doi: 10.1292/jvms1939.43.51. [DOI] [PubMed] [Google Scholar]
- 49.Thorns C J, Turcotte C, Gemmel C G, Woodward M J. Studies into the role of the SEF14 fimbrial antigen in the pathogenesis of Salmonella enteritidis. Microb Pathog. 1996;20:235–246. doi: 10.1006/mpat.1996.0022. [DOI] [PubMed] [Google Scholar]
- 50.Van Dyk T K, Reed T R, Vollmer A C, LaRossa R A. Synergistic induction of the heat shock response in Escherichia coli by simultaneous treatment with chemical inducers. J Bacteriol. 1995;177:6001–6004. doi: 10.1128/jb.177.20.6001-6004.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J. 1995;14:1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamanaka K, Mitani T, Ogura T, Niki H, Hiraga S. Cloning, sequencing, and characterization of multicopy suppressors of a mukB mutation in Escherichia coli. Mol Microbiol. 1994;13:301–312. doi: 10.1111/j.1365-2958.1994.tb00424.x. [DOI] [PubMed] [Google Scholar]
- 53.Youderian P, Sugiono P, Brewer K L, Higgins N P, Elliot T. Packaging specific segments of the salmonella chromosome with locked-in Mud-P22 prophages. Genetics. 1988;118:581–592. doi: 10.1093/genetics/118.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]