Abstract
Pyroptosis plays a key role in the death of cells including cardiomyocytes, and it is associated with a variety of cardiovascular diseases. However, the role of pyroptosis-related genes (PRGs) in hypertrophic cardiomyopathy (HCM) is not well characterized. This study aimed to identify key biomarkers and explore the molecular mechanisms underlying the functions of the PRGs in HCM. The differentially expressed genes were identified by GEO2R, and the differentially expressed pyroptosis-related genes (DEPRGs) of HCM were identified by combining with PRGs. Enrichment analysis was performed using the “clusterProfiler” package of the R software. Protein-protein interactions (PPI) network analysis was performed using the STRING database, and hub genes were screened using cytoHubba. TF-miRNA coregulatory networks and protein-chemical interactions were analyzed using NetworkAnalyst. RT-PCR/WB was used for expression validation of HCM diagnostic markers. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and Western Blot (WB) were used to measure and compare the expression of the identified genes in the cardiac hypertrophy model and the control group. A total of 20 DEPRGs were identified, which primarily showed enrichment for the positive regulation of cytokine production, regulation of response to biotic stimulus, tumor necrosis factor production, and other biological processes. These processes primarily involved pathways related to Renin-angiotensin system, Adipocytokine signaling pathway and NF-kappa B signaling pathway. Then, a PPI network was constructed, and 8 hub genes were identified. After verification analysis, the finally identified HCM-related diagnostic markers were upregulated gene protein tyrosine phosphatase non-receptor type 11 (PTPN11), downregulated genes interleukin-1 receptor-associated kinase 3 (IRAK3), and annexin A2 (ANXA2). Further GSEA analysis revealed these 3 biomarkers primarily related to cardiac muscle contraction, hypertrophic cardiomyopathy, fatty acid degradation and ECM − receptor interaction. Moreover, we also elucidated the interaction network of these biomarkers with the miRNA network and known compounds, respectively. RT-PCR/WB results indicated that PTPN11 expression was significantly increased, and IRAK3 and ANXA2 expressions were significantly decreased in HCM. This study identified PTPN11, IRAK3, and ANXA2 as pyroptosis-associated biomarkers of HCM, with the potential to reveal the development and pathogenesis of HCM and could be potential therapeutic targets.
Keywords: bioinformatics, biomarkers, hypertrophic cardiomyopathy, pyroptosis-related genes
1. Introduction
Hypertrophic cardiomyopathy (HCM) is an inherited cardiovascular disease with left ventricular hypertrophy, myocardial hypercontraction, cardiomyocyte disturbance, and fibrosis.[1] It is one of the most critical reasons behind sudden cardiac death in children and adolescents.[2] Echocardiography shows that the incidence of HCM in adults is estimated to be 0.2%,[3] and considering the clinical and genetic characteristics (including the patient family members), the prevalence reaches 0.5%.[4] However, the pathophysiology of HCM is complex, and the clinical manifestations and prognosis are diverse, warranting successful diagnosis only when the cardiac morphology changes and function failure worsens. In addition, drug-mediated and surgical therapies are the most important treatment methods for HCM. However, these methods primarily play a role in reducing clinical symptoms, and to date, treatment plans for the subsequent developmental stages of the disease are lacking. Currently, the signaling pathways and regulatory networks underlying the pathogenesis of HCM are yet to be fully understood.[5,6] Therefore, more in-depth exploration and identification of potential genes and biological pathways underlying the pathogenesis of HCM is crucial for diagnosis and treatment.
Pyroptosis, a novel form of programmed cell death, is characterized by cell swelling, numerous bubble-like protrusions, rupture of the plasma membrane, the release of cellular contents and inflammatory factors, and subsequent final activation of an inflammatory response.[7] The balance between cell formation and death is essential for the cardiovascular system to maintain normal structure and function. Pyroptosis is a critical innate immune response in cardiovascular diseases. An increasing number of studies highlight the indispensable role of pyroptosis plays in various cardiovascular diseases, such as atherosclerosis, diabetic cardiomyopathy, and heart failure.[8–10] Recent studies have also shown that the expression of pyroptosis-related genes is related to cardiac hypertrophy.[11] However, no studies have yet investigated the mechanism of pyroptosis in HCM.
The development of high-throughput sequencing technology has enabled the understanding of diseases gene expression profiles in a more comprehensive and in-depth manner, which is helpful for precise diseases treatment. This study used high-throughput sequencing data to explore the mechanism underlying pyroptosis-related gene functions in HCM, providing new insights for the prevention and treatment of HCM.
2. Material and method
2.1. Data source
We downloaded the microarray expression profiling datasets GSE36961 and GSE89714 from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). GSE36961 was analyzed based on the GPL15389 Illumina HumanHT-12 V3.0 expression beadchip platform, with 145 samples, including 106 HCM ones and 39 healthy controls. GSE89714 contained 5 HCM cases and 4 normal controls, on the GPL1115 Illumina HiSeq 2000 (Homo sapiens) platform. Since the data in this study are from public databases, there is no ethical approval involved.
2.2. Identifying differently expressed pyroptosis-related genes
The differentially expressed genes (DEGs) between HCM and healthy controls in GSE36961 were identified using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) with the inclusion criteria P < .05 and log2|FC|> 0.5. Three hundred eighty-eight pyroptosis-related genes were downloaded from the GeneCards database.[12] Common genes between the DEGs and pyroptosis-related genes were identified as differentially expressed pyroptosis-related genes (DEPRGs). The“ggplot2” package in R was utilized to draw the volcano plots. Heatmap and PCA of DEPRGs were provided in this study by https://www.bioinformatics.com.cn, an online platform for data analysis and visualization.
2.3. GO, KEGG, and DO enrichment analyses of DEPRGs
Gene Ontology (GO) [biological process, cellular component, and molecular function (MF)], and the Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis were performed using the “clusterProfiler” package of R software.[13] The WebGestalt[14] was used for DO enrichment analysis with P < .05 as the significance level.
2.4. Protein-protein interaction (PPI) network
We performed PPI network analysis using the STRING (https://string-db.org/) database to assess protein-protein interactions for DEPRGs. We mapped the DEPRGs onto the PPI network and set an interaction score > 0.4 as a threshold. In addition, we used the plug-in cytoHubba in Cytoscape v3.9.1 software to screen the top ten genes with the highest height values, respectively, by 4 algorithms (MCC, DMNC, MNC, and Degree) and visualized the overlapping hub genes via a Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/).
2.5. Validation of hub genes
The dataset GSE89714 contained 5 HCM and 4 healthy samples and was used as validation set to verify the expression of the hub genes. The statistically significant genes, as the key biomarkers, are instrumental in diagnosing and treating HCM.
2.6. Gene set enrichment analysis (GSEA)
According to the median gene expression, HCM samples were divided into high and low-expression groups. GSEA was performed to identify the significantly enriched relevant pathways.
2.7. TF-miRNA coregulatory network and Protein-chemical interactions
NetworkAnalyst[15] (https://www.networkanalyst.ca/) was used to construct the TF-miRNA coregulatory network of biomarkers and identify their interacting compounds.
2.8. Cell culture
H9c2 cells were purchased from Procell Life Science and Technology Co., Ltd. (Wuhan, China). Cells were cultured in a complete DMEM medium containing 1 g/L glucose and 10% fetal bovine serum (FBS) in an incubator at 37 ° C with 5% CO2.
2.9. Cell viability measurement
Cell viability was determined by 3-(4,5)-dimethylthiahiazo(-z-y1)-2,5-di-phenytetrazoliumromide (MTT, Beijing Solarbio Science & Technology Co., Ltd.) assay. 1 × 104 cells were seeded in 96-well plates in a volume of 100 μL per well. After 24 hours, the medium was discarded, and isoproterenol (Sigma–Aldrich, USA, 0, 2, 5, 10, and 20 μM, dissolved in DMEM containing 1% FBS) was added to the culture for 48 hours. The medium was replaced with a fresh one every 24 hours. The medium was discarded, and each well was washed twice with PBS. MTT (10 μL, 5mg/ml) solution was added to each well of the 96-well plate containing 90 μL medium. After 4 hours of incubation, the medium was discarded, and 150 μL dimethylsulfoxide from Solarbio was added to each well. The absorbance at 490 nm was measured to calculate the percentage of cell viability: Cell viability (%) = (ODIso − ODblank)/(OD control − ODblank) × 100%.
2.10. Cell model establishment
The upregulation of ANP and β-MHC expression indicates that the cardiac hypertrophy model has been successfully induced. Cells (1 × 105) in the logarithmic growth phase were seeded in each 35 mm cell culture dish. After 24 hours, cardiomyocytes were randomly divided into 2 groups: control group: given 2 ml DMEM containing 1% FBS for 48 hours; Iso group: given 10 μmol/L Iso in 2 ml DMEM containing 1% FBS for 48 hours. Medium for both groups was changed every 24 hours.
2.11. Real-time quantitative polymerase chain reaction (RT-PCR)
The mRNA levels of ANP, β-MHC, IRAK3, PTPN11, and ANXA2 were detected by RT-PCR. Total RNA was extracted from cells using TRIzol reagent according to the manufacturer instructions. PrimeScriptTM RT reagent kit and TB Green premix (Takara Bio, USA) was used for cDNA synthesis and RT-PCR, respectively. The primer sequences are listed in Table 1. The RT-qPCR data were analyzed by the ΔΔCT method. GAPDH was used as an internal control for normalizing all gene expressions.
Table 1.
qPCR primers.
| Genes | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| ANP | GGCACTTAGCTCCCTCTC | CCCTCAGTTTGCTTTTCA |
| β-MHC | TGGATGCAGACCTCTCCC | TGCTTCTTGCCACCCTTG |
| IRAK3 | TGACACAGAAAACCCCCT | CCAGCATTCCTCACAAGA |
| PTPN11 | GGAAACACAGAGAGAACC | TATCAATCACAATGAACG |
| ANXA2 | GCAGTGTGTGCCACCTCC | GTCCCCTTGCCCTTCATG |
| GAPDH | GGCTCTCTGCTCCTCCCTGT | CGTTCACACCGACCTTCACC |
2.12. Western blotting analysis
After treatment, the medium was removed, and the cells were washed with PBS. Cells were then lysed in cell lysis buffer containing 1% PMSF on ice for 30 minutes and centrifuged at 12 × 103 rpm for 5 minutes. The total protein was quantified with the BCA method. The samples were separated on 10% SDS–PAGE gels and then transferred to polyvinylidene difluoride membranes. The membranes were blocked in 5% skim milk for 2 hours on a shaker and then incubated with primary antibodies such as β-Actin (Beyotime, China), IRAK3(Abcam, ab-8116), PTPN11 (CST, 3397S), ANXA2 (CST, 8235S) overnight at 4 °C. Membranes were washed with TBST and then incubated with corresponding HRP-conjugated secondary antibody for 1 hour at room temperature and detected by ECL detection kit (Bio-Rad, USA).
2.13. Statistical analysis
All data were expressed as mean ± standard deviation (SD). Statistical analysis and graphing were performed using GraphPad prism 9.0. One-way analysis of variance followed by Dunnett post hoc test was used to compare differences between multiple groups and t tests for the differences between the 2 groups. P < .05 was considered to be statistically significant.
3. Results
3.1. Identification of DEPRGs
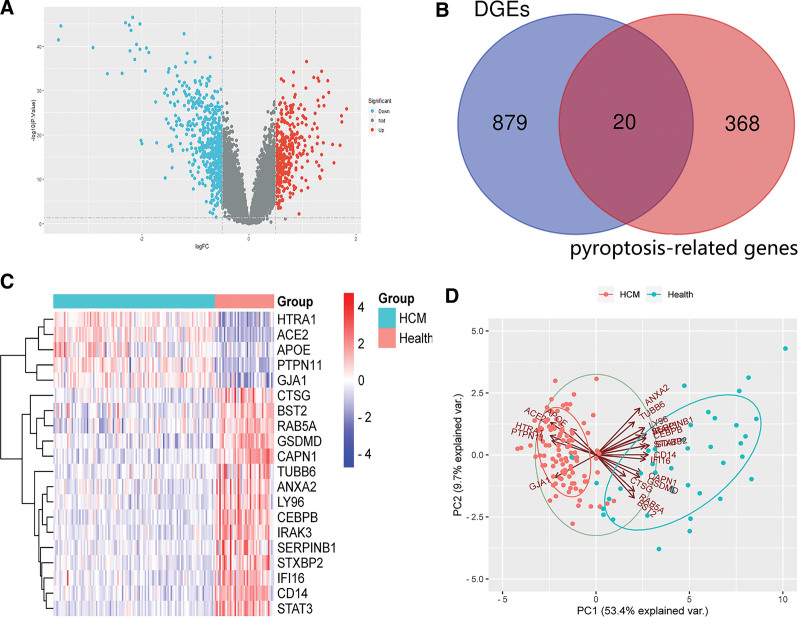
We used the dataset GSE36961 (log2 transformed and normalized) based on the inclusion criteria of |log2FC|>0.5, P < .05. This analysis revealed 899 DEGs (Fig. 1A), including 372 upregulated and 527 downregulated genes. Twenty congruous DEPRGs were identified by integrated bioinformatics analysis (Fig. 1B), including 5 upregulated and 15 downregulated genes, depicted by a heatmap (Fig. 1C). PCA analysis was performed on DEPRGs to clarify the good discrimination between HCM and healthy controls (Fig. 1D).
Figure 1.
Identification of DEPRGs of HCM. (A) The differentially expressed genes of GSE36961. (B) The DEPRGs of HCM. (C) The heat map of the DEPRGs. (D) PCA plots. DEPRGs = differentially expressed pyroptosis-related genes, HCM = hypertrophic cardiomyopathy.
3.2. Function enrichment analyses of the DEPRGs
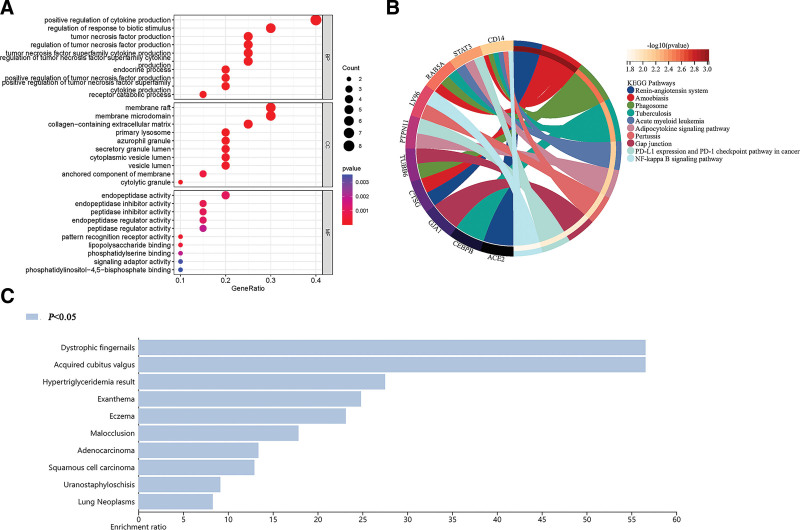
The GO analysis of DEPRGs was performed to reveal their biological functions (Fig. 2A). BP analysis revealed how most DEPRGs were primarily involved in the positive regulation of cytokine production, regulation of response to biotic stimulus, tumor necrosis factor production. Cellular component analysis revealed that most DEPRGs were enriched into the membrane raft, membrane microdomain, and collagen-containing extracellular matrix. MF analysis revealed that the DEPRGs were significantly enriched in endopeptidase activity, peptidase inhibitor activity. KEGG enrichment analysis showed (Fig. 2B) that the DEPRGs were mainly functionally involved in pathways associated with Salmonella infection, Amoebiasis, Renin − angiotensin system, Adipocytokine signaling pathway and NF-kappa B signaling pathway. The WebGestalt online database was used for DO analysis to explore the function of DEPRGs further, which revealed that Dystrophic fingernails, Acquired cubitus valgus, Hypertriglyceridemia result, were the major diseases that the DEPRGs participated in (Fig. 2C).
Figure 2.
Function enrichment analyses of DEPRGs (P < .05). (A) GO; (B) KEGG signaling pathway; (C) DO enrichment. DEPRGs = differentially expressed pyroptosis-related genes, GO = Gene Ontology, KEGG = Kyoto Encyclopedia of Genes and Genomes.
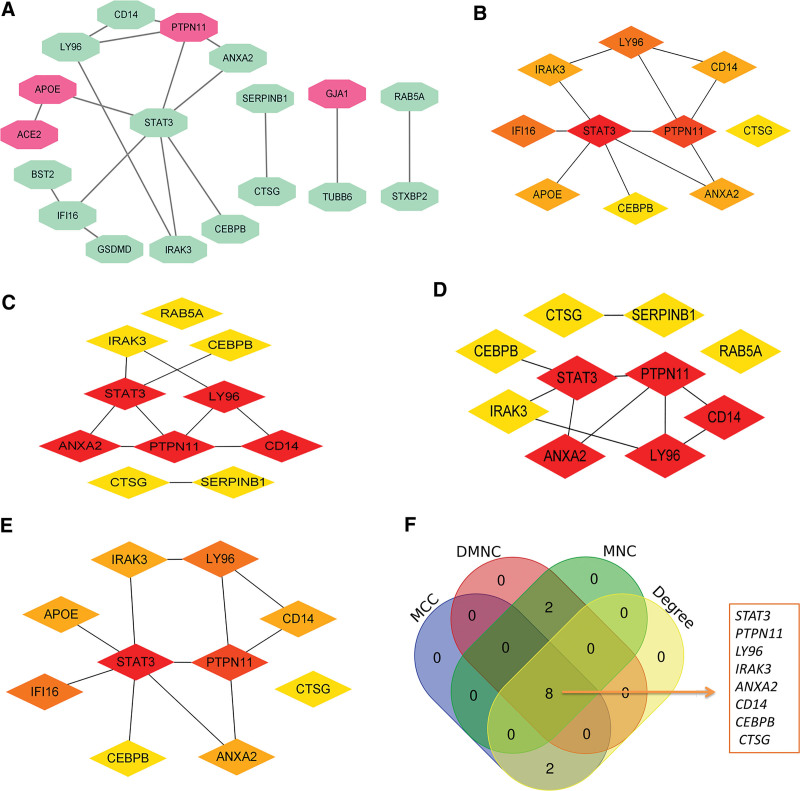
3.3. PPI network analyses of DEPRGs
Based on the network interaction score > 0.4 according to the STRING database, a PPI network with 18 nodes and 17 edges was built. Moreover, among the 18 nodes, 4 nodes were upregulated, and 14 were downregulated (Fig. 3A). The top ten genes were detected via 4 algorithms [MCC (Fig. 3B), DMNC (Fig. 3C), MNC (Fig. 3D), and Degree (Fig. 3E)], respectively, of cytoHubba plug-in in Cytoscape. Finally, the 8 common genes by the 4 algorithms were identified as the hub genes of HCM: one upregulated gene protein tyrosine phosphatase non-receptor type 11 (PTPN11), 7 downregulated genes signal transducer and activator of transcription 3 (STAT3), lymphocyte antigen 96 (LY96), interleukin-1 receptor-associated kinase 3 (IRAK3), annexin A2 (ANXA2), CD14 molecule (CD14), CCAAT enhancer binding protein beta (CEBPB), and cathepsin G (CTSG) (Fig. 3F).
Figure 3.
The PPI network analysis. (A) The PPI network of the DEPRGs. Red indicates upregulated, and green is downregulated. (B) Top 10 nodes detected by MCC. (C) Top 10 nodes detected by DMNC. (D) Top 10 nodes detected by MNC. (E) Top 10 nodes detected by Degree. (F) The 4 algorithms utilized to identify hub genes. DEPRGs = differentially expressed pyroptosis-related genes.
3.4. Gender differences in HCM
We analyzed the GSE36961 dataset with 145 samples, including 19 healthy men and 54 male HCM patients, and 20 healthy women and 52 female HCM patients, to explore whether there are gender differences in the prevalence of HCM. The difference was not statistically significant (χ2 = 0.056, P = .812), suggesting that the prevalence of HCM does not differ between men and women. In addition, we selected HCM patient samples from this dataset, performed differential expression analysis by gender, and found no gender differences in the expression of these 8 hub genes in HCM patients (Table 2).
Table 2.
Eight hub genes in HCM patients grouped by gender.
| Gene symbol | adj.P.Val | P value | logFC |
|---|---|---|---|
| PTPN11 | 0.954 | .213 | 0.073815 |
| IRAK3 | 0.965 | .268 | −0.04424 |
| ANXA2 | 0.981 | .363 | −0.06047 |
| STAT3 | 0.972 | .322 | −0.05204 |
| LY96 | 0.997 | .606 | −0.03469 |
| CD14 | 0.999 | .826 | −0.01925 |
| CEBPB | 0.899 | .0903 | 0.093459 |
| CTSG | 0.999 | .64 | 0.031557 |
3.5. Verification of the hub genes for HCM
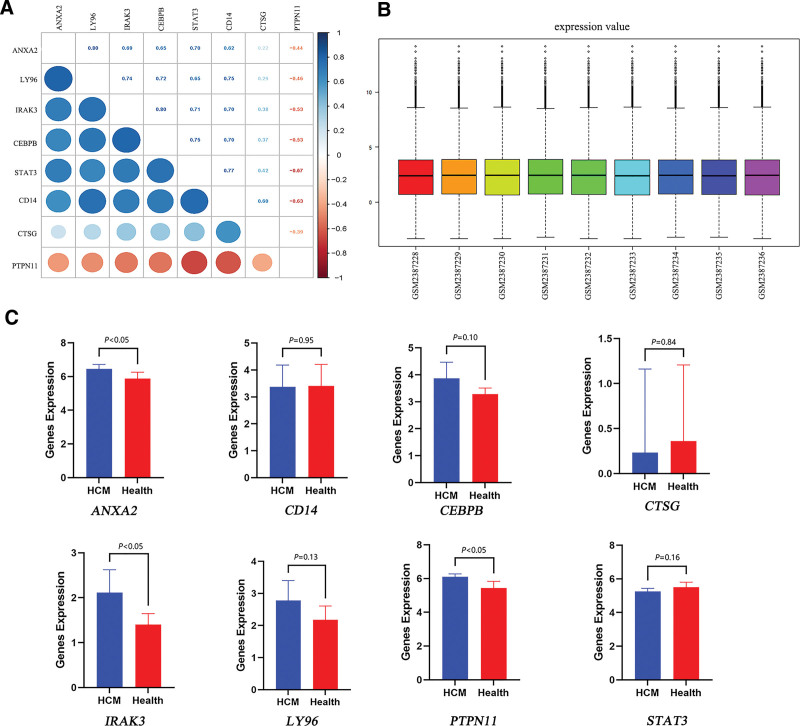
We then performed a correlation analysis on the 8 hub genes to verify their correlation and improve the diagnostic value (Fig. 4A), which revealed a significant association with each other except CTSG, indicating significant shared functional similarities. Analyzing the normalized dataset GSE89714 (Fig. 4B), revealed that only PTPN11, IRAK3, and ANXA2 expressions were statistically significant between the 2 groups, with the expression of PTPN11, IRAK3, and ANXA2 in HCM being higher than that in the healthy group (Fig. 4C). The expression of PTPN11 was consistent with GSE36961, while that of IRAK3 and ANXA2 were opposite. Therefore, further studies are needed to validate the expression of the biomarkers. In short, our results indicated the potential significance of PTPN11, IRAK3, and ANXA2 as essential biomarkers associated with pyroptosis in HCM.
Figure 4.
Validation of hub genes. (A) Heat-map of the correlation analysis between candidate genes. (B) Normalization of GSE89714. (C) Data validation of hub genes by GSE89714.
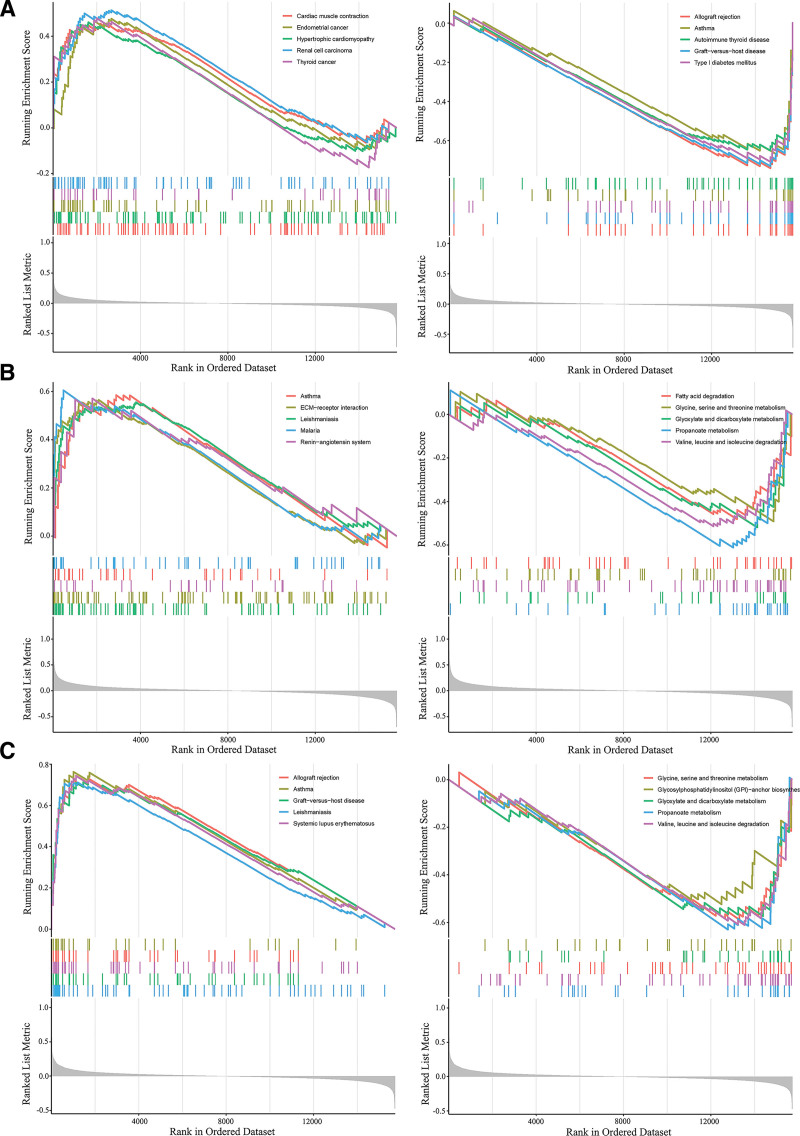
3.6. GSEA of PTPN11, IRAK3, and ANXA2
Taking a cue from above analysis, we chose PTPN11, ANXA2, and IRAK3 for further analysis. GSEA showed that PTPN11 was functionally involved in Asthma, Autoimmune thyroid disease, Cardiac muscle contraction, and Hypertrophic cardiomyopathy, indicating its direct connection with HCM as a critical biomarker (Fig. 5A). ANXA2 and IRAK3 were closely related to the glycine, serine, and threonine metabolism, glyoxylate and dicarboxylate metabolism, Leishmaniasis, propanoate metabolism, valine, leucine, and isoleucine degradation. In addition, ANXA2 was also associated with ECM − receptor interaction, fatty acid degradation, and the renin − angiotensin system (Fig. 5B and C).
Figure 5.
Gene set enrichment analysis. (A) GSEA results of PTPN11. (B) GSEA results of ANXA2. (C) GSEA results of IRAK3.
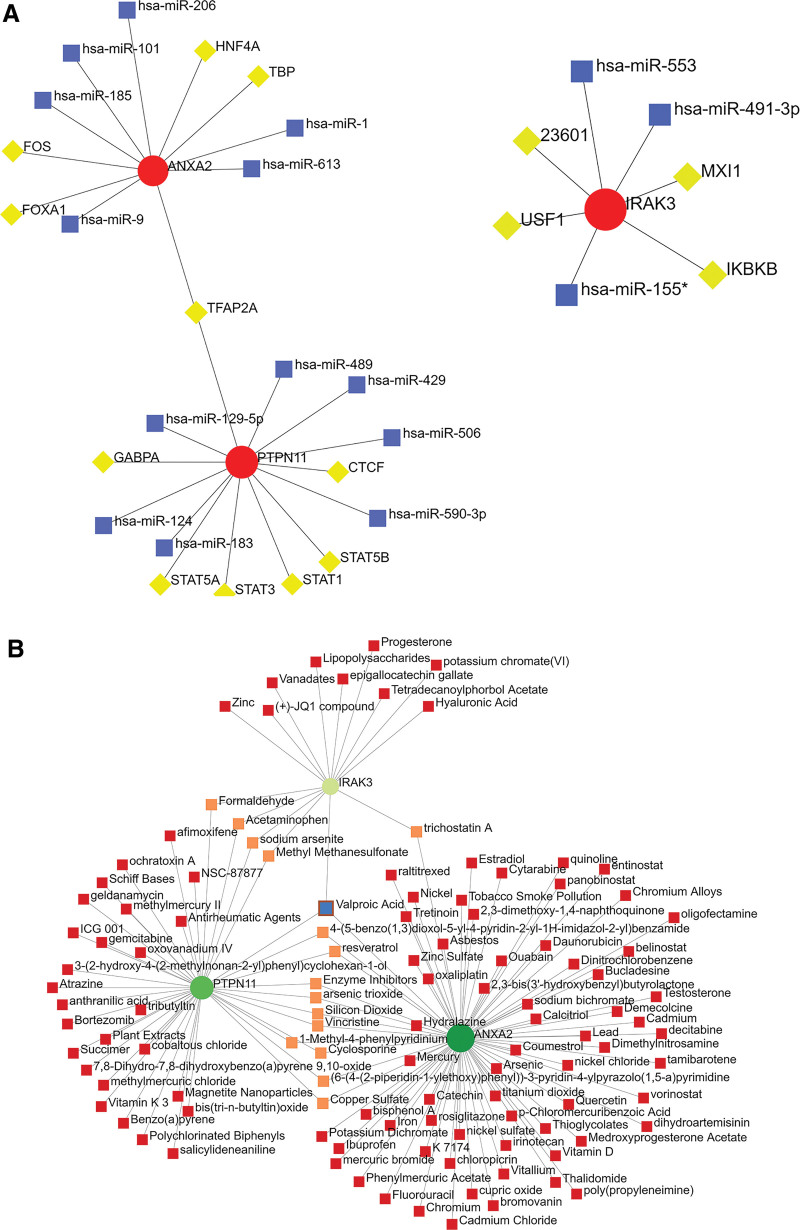
3.7. TF-miRNA coregulatory network and protein-chemical interactions
TF-miRNA coregulatory network showed the interaction of TF genes and miRNAs with the 3 key biomarkers (Fig. 6A). PTPN11 and ANXA2 were co-regulated by transcription factor AP-2 alpha (TFAP2A). In addition, we identified a compound, Valproic Acid, that could interact with the 3 key biomarkers (Fig. 6B). Since these compounds were detected against 3 key biomarkers, they represented common compounds in HCM. The compounds interacting with most key biomarkers are listed in Table 3.
Figure 6.
NetworkAnalyst analysis of hub genes. (A) TF-miRNA coregulatory network. The red nodes represent the key biomarkers, the yellow nodes are TF genes, and the blue nodes indicate miRNA. (B) Protein-chemical interactions. The green nodes represent the key biomarkers. The pink nodes represent compounds that interact in common between 2 key biomarkers. The blue node represent compound that interact with the 3 key biomarkers.
Table 3.
Top 16 compounds with key biomarker interactions.
| Id | Label | Degree | Betweenness | Gene |
|---|---|---|---|---|
| D014635 | Valproic Acid | 3 | 602.42 | PTPN11/IRAK3/ANXA2 |
| C012589 | trichostatin A | 2 | 353.98 | IRAK3/ANXA2 |
| D015655 | 1-Methyl-4-phenylpyridinium | 2 | 178.47 | IRAK3/ANXA2 |
| C459179 | 4-(5-benzo(1,3)dioxol-5-yl-4-pyridin-2-yl-1H-imidazol-2-yl)benzamide | 2 | 178.47 | IRAK3/ANXA2 |
| C516138 | (6-(4-(2-piperidin-1-ylethoxy)phenyl))-3-pyridin-4-ylpyrazolo(1,5-a)pyrimidine | 2 | 178.47 | IRAK3/ANXA2 |
| C006632 | arsenic trioxide | 2 | 178.47 | IRAK3/ANXA2 |
| D019327 | Copper Sulfate | 2 | 178.47 | IRAK3/ANXA2 |
| D016572 | Cyclosporine | 2 | 178.47 | IRAK3/ANXA2 |
| D004791 | Enzyme Inhibitors | 2 | 178.47 | IRAK3/ANXA2 |
| C059514 | resveratrol | 2 | 178.47 | IRAK3/ANXA2 |
| D012822 | Silicon Dioxide | 2 | 178.47 | IRAK3/ANXA2 |
| D014750 | Vincristine | 2 | 178.47 | IRAK3/ANXA2 |
| D000082 | Acetaminophen | 2 | 69.97 | PTPN11/ANXA2 |
| D005557 | Formaldehyde | 2 | 69.97 | PTPN11/ANXA2 |
| D008741 | Methyl Methanesulfonate | 2 | 69.97 | PTPN11/ANXA2 |
| C017947 | sodium arsenite | 2 | 69.97 | PTPN11/ANXA2 |
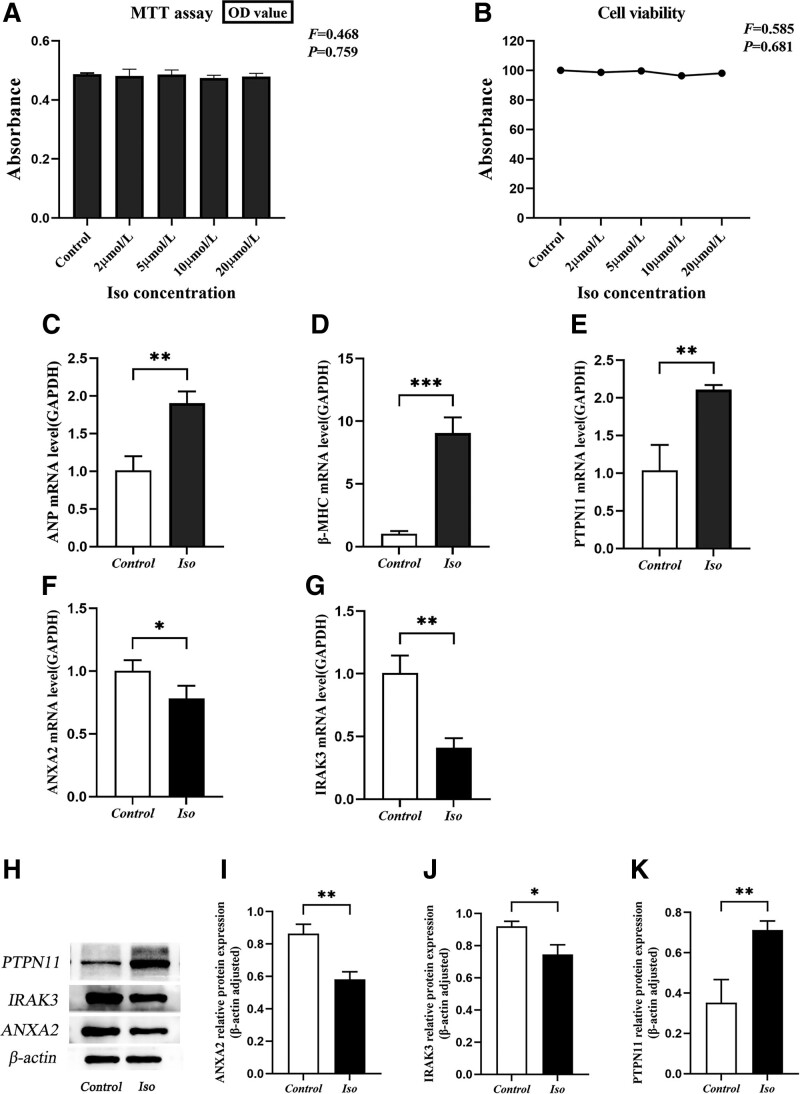
3.8. Effect of isoproterenol on cardiomyocytes viability
Next, we incubated the cells with different concentrations of Iso (0, 2, 5, 10, 20 μmol/L) for 48 hours to determine its cytotoxic effects. Generally, OD values between 0 and 7 indicate that the linear relationship of cell viability is consistent. In this study, the OD values corroborated this notion (Fig. 7A). Assuming that the cell viability of the control group was 100% (0 μmol/L), the MTT assay showed that the concentrations of isoproterenol from 2 to 20 μmol/L failed to significantly affect the viability of H9c2 cells (Fig. 7B). Therefore, gene expression changes in subsequent studies ruled out the cause of cell viability or toxicity. Based on previous studies[16] and our results, 10 μmol/L Iso was chosen to induce H9c2 cardiac hypertrophy.
Figure 7.
The expression levels of PTPN11, IRAK3, and ANXA2 in isoproterenol-induced H9c2 cardiomyocytes. (A) Variations in absorbance values of cells treated with different concentrations of Iso. (B) Iso-treated cell viability. The viability of the control group was considered 100%, and the viability was measured by MTT assay. (C–G) mRNA levels of ANP, β-MHC, PTPN11, ANXA2, IRAK3 in isoproterenol induced H9c2 cardiomyocytes. (H) Representative western blots showing PTPN11, IRAK3, and ANXA2 levels. (I–K) Quantification of the relative changes in PTPN11, IRAK3, and ANXA2 expression. *P < .05, **P < .01 compared with Control group. All values are mean ± SD. MTT = 3-(4,5)-dimethylthiahiazo(-z-y1)-2,5-di-phenytetrazoliumromide.
3.9. The mRNA expression of hypertrophy-related fetal genes and pyroptosis-related genes in isoproterenol-induced H9c2 cardiomyocytes
The mRNA expression of ANP and β-MHC was significantly increased in the Iso (10 µmol/L) group compared to the control group (P < .05), indicating the successful establishment of the cardiac hypertrophy model (Fig. 7C–G). In addition, the mRNA expression of PTPN11 was significantly higher in HCM than control (P < .05), and the mRNA expression of ANXA2 and IRAK3 was lower than control (P < .05).
3.10. The protein expression of PTPN11, IRAK3, and ANXA2 induced by isoproterenol
The protein expression of PTPN11 was significantly increased in the Iso group (P < .05). However, compared with the control group, Iso-induced ANXA2 and IRAK3 protein expression were decreased (P < .05) (Fig. 7H–K).
4. Discussion
This study identified 20 DEPRGs, and the molecular mechanisms underlying the functions of these genes in hypertrophic cardiomyopathy were further explored. GO analysis revealed a robust association of the DEPRGs to positive regulation of cytokine production, regulation of response to biotic stimulus, tumor necrosis factor production, endocrine process receptor, catabolic process, and other biological processes. We hypothesized that these genes might play a key role in HCM pathogenesis by regulating these biological processes. Increasing evidence shows an essential role of proinflammatory cytokines in the treatment and prognosis of cardiovascular diseases.[17] Cytokines produced from different sources may also be of varying importance.[18] The production of tumor necrosis factors also stimulates cardiac hypertrophy.[19] Moreover, the regulation of catabolic processes plays a vital role in the pathophysiology of HCM.[20] On the other hand, our study found that most of these DEPRGs were enriched in the Salmonella infection, Amoebiasis, Renin − angiotensin system, and other related signaling pathways. Previous studies have shown that salmonella infection might lead to left ventricular hypertrophy, causing cardiovascular complications.[21,22] Recent studies have shown that the Renin-angiotensin system plays an essential role in the etiology of cardiomyopathy and can be used as a therapeutic target.[23,24] In addition, PPI network analysis identified 8 hub genes-STAT3, PTPN11, LY96, IRAK3, ANXA2, CD14, CEBPB, and CTSG. Validation analysis of GSE89714 revealed PTPN11, ANXA2, and IRAK3, with statistically significant differential expression, as potential biomarkers of HCM. Furthermore, several compounds interacting with the biomarkers have been identified by NetworkAnalyst. For example, histone deacetylase inhibitor Trichostatin A and valproic acid attenuate cardiac hypertrophy,[25] potentially contributing to the prevention and treatment of HCM.
PTPN11, encoding SHP2 protein, is a classic non-receptor protein tyrosine phosphatase, an essential participant in cell signal transduction, regulating cell growth and proliferation.[26] Aberrant expression of PTPN11 can lead to various diseases, for example, PTPN11 is the most common mutant gene in Noonan syndrome (NS) and LEOPARD syndrome (LS),[27] HCM being one of the characteristics of these 2 diseases. This phenomenon indicates some connection between PTPN11 and HCM. Our GESA results indicated that a high expression of PTPN11 might activate myocardial contraction and hypertrophic cardiomyopathy. Many studies also showed that PTPN11 mutations were a common cause of RASopathy-associated HCM.[28,29] Genotypic-phenotypic correlation studies showed that 8% of SHP2-related NS patients had HCM,[30] and about 70% to 80% of LS cases manifested HCM.[31] The main reason for RASopathy-associated HCM includes PTPN11 mutation leading to abnormal signal transduction and cardiac hypertrophy during cardiac development.[32] Previous studies have shown RAS-mitogen activated protein kinase signal transduction pathway and its upstream and downstream pathways to be upregulated in HCM,[33] wherein PTPN11 mutations critically induce cardiac hypertrophy, suggesting an effective treatment for HCM by blocking this signal pathway.[34] Moreover, our study found a higher expression of PTPN11 in HCM than in the healthy group and demonstrated that the gene was highly expressed in the cardiac hypertrophy model. A recent review elucidated how the overexpression of SHP2 is associated with hereditary developmental diseases.[35] Therefore, we speculated a strong correlation between HCM development and the overexpression of PTPN11. In addition, many studies have highlighted how SHP2 inhibitors play an essential role in cancer treatment. The combination of SHP2 allosteric inhibitors and targeted molecular drugs is a promising therapeutic strategy for cancer.[36] However, whether SHP2 inhibitors help treat HCM or not warrants further research.
ANXA2, a Ca2+-binding multifunctional protein, plays an essential role in various signal pathways and is found in various human tissues and cell types.[37] ANXA2 is associated with cardiovascular diseases; it is overexpressed in heart failure, for which it can be used as a candidate biomarker.[38] Lower ANXA2 expression increases plasma levels of proprotein converting enzyme subtilisin/ kexin 9, increasing the low-density lipoprotein cholesterol levels and the risk of coronary heart disease.[39] Coincidentally, low-density lipoprotein cholesterol is also a prognostic biomarker for HCM.[40] Interestingly, ANXA2 showed a lower expression in the cardiac hypertrophy model, indicating that regulating its expression might reduce the condition and improve cardiac function. Moreover, GSEA results showed that lower expression of ANXA2 protein might inhibit fatty acid degradation, glycine, serine, and threonine metabolism, glyoxylic acid, and dicarboxylic acid metabolism pathways. It has been demonstrated that glycine, serine, and threonine metabolic pathways are differentially regulated in HCM,[41] and targeted amino acid metabolomics can accurately identify HCM.[42] Glycine promotes the biosynthesis of the antioxidant glutathione, reduces mitochondrial oxygen production, and increases adenosine triphosphate production, leading to improved heart function and ventricular remodeling.[43] The protein kinase C (PKC) family of serine/threonine protein kinases has attracted much attention for their roles in cardiac function, and one of its principal members, protein kinase C alpha (PKCα), critically mediates cardiac hypertrophy by regulating kinase 1/2 (ERK-1/2) through extracellular signaling.[44,45] Moreover, the oxidative capacity of fatty acids is known to regulate cardiac hypertrophy.[46] The regulation of the expression of genes related to fatty acid metabolism alleviates cardiac dysfunction and improves cardiac hypertrophy, potentially mediated by activating the sirtuin 3/AMP-activated protein kinase pathway.[47] This suggests that fatty acid degradation may also be a potential mechanism to be therapeutically targeted for HCM treatment. In addition, this study indicated that the lower expression of ANXA2 protein might also activate asthma, ECM-receptor interaction, leishmaniasis, malaria, and renin-angiotensin system (RAS) pathways, all of which might potentially regulate HCM treatment. RAS balances blood pressure and body fluids. Abnormal activation of this system produces the major bioactive hormone angiotensin (Ang) II, which binds to specific receptors and triggers a wide range of biological effects.[48] In this regard, Ang II has been widely used to induce left ventricular hypertrophy,[49,50] and the antagonists of the renin-angiotensin system have been reported to help treat HCM.[51] More importantly, a growing number of studies discussed that activation of the renin-angiotensin-aldosterone system exacerbates left ventricular hypertrophy through increased blood pressure and the direct effects of the active peptides angiotensin-II (AT-II) and aldosterone on cardiomyocytes.[52,53] Renin-angiotensin-aldosterone system blockade has been implicated in having potential therapeutic benefits in patients with HCM.[54] Additionally, ECM-receptor interaction is an essential pathway in cardiac hypertrophy.[55,56] Therefore, we hypothesized that ANXA2 might regulate a potential mechanism that could serve as a therapeutic target for HCM.
IRAK3 is a member of the interleukin-1 receptor-associated kinase (IRAK) family, primarily in monocytes and macrophages, which led to it being named IRAK-M.[57] IRAK-M is a negative regulator of Toll-like receptor signaling and critically regulates inflammation in the innate immune system.[58] Some studies have shown that Nuclear factor-kappaB (NF-kB) activity or the level of inflammatory cytokines such as tumor necrosis factor-α and interleukin-6 (IL-6) increased upon IRAK3 downregulation, indicating how it inhibits excessive inflammatory cytokine production in inflammation.[59,60] NF-kB is a transcription factor that regulates gene expression and regulates inflammation, cell proliferation, and apoptosis.[61] A large number of studies elucidated how the abnormal activation of NF-kB and increased levels of proinflammatory cytokines are involved in the regulatory mechanisms of cardiac hypertrophy,[62,63] and ten years later, the recent studies have evaluated the correlation between NF-kB and the disease state of HCM patients.[64] Myocardial fibrosis in HCM is caused by an inflammatory response.[65] Moreover, tumor necrosis factor-α and IL-6 also play an essential role in the pathogenesis of cardiac hypertrophy.[66,67] Also, reducing inflammatory cytokines is beneficial in attenuating cardiac hypertrophy. Our GSEA results showed that lower expression of IRAK3 inhibits glycine, serine, and threonine metabolism pathway, further demonstrating the importance of IRAK3 in HCM. Therefore, targeting IRAK3 may contribute to developing novel therapeutic drugs for HCM.
5. Conclusion
In summary, this study identified PTPN11, ANXA2, and IRAK3 as essential novel biomarkers of HCM associated with pyroptosis. We also screened the related pathways and compounds, which would play a crucial role in delineating the development and pathogenesis of HCM and provide potential therapeutic targets.
Author contributions
Methodology: Xin Tang.
Supervision: Yan Zhu.
Writing – original draft: Xin Tang.
Writing – review & editing: Xin Tang, Yi Shen, Yun Lu, Wanya He, Ying Nie, Xue Fang, Jinghui Cai, Xiaoyun Si, Yan Zhu.
Abbreviations:
- angiotensin II
- Ang II
- AT-II
- angiotensin-II
- DEPRGs
- differentially expressed pyroptosis-related genes
- DGEs
- differentially expressed genes
- FBS
- fetal bovine serum
- GO
- Gene Ontology
- HCM
- hypertrophic cardiomyopathy
- IL-6
- interleukin-6
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- LS
- LEOPARD syndrome
- MF
- molecular function
- MTT
- 3-(4,5)-dimethylthiahiazo(-z-y1)-2,5-di-phenytetrazoliumromide
- NF-kB
- nuclear factor-kappa B
- NS
- Noonan syndrome
- PKC
- protein kinase C
- PKCα
- protein kinase C alpha
- RAS
- renin-angiotensin system
- SD
- standard deviation
The datasets generated during and/or analyzed during the current study are publicly available.
XT and YS contributed equally to this work.
This research was funded by Doctor Start-up Fund of Affiliated Hospital of Guizhou Medical University (gyfybsky2021-54), Guizhou Science and Technology Support Plan: Qian Science and Technology Cooperation Support ([2020] 4Y231), Guiyang Science and Technology Plan ([2019]9-1-34), and Science and Technology Fund from Guizhou Provincial Health Commission: gzwkj2023-130; Cultivation fund: gyfynsfc[2022]-48.
The authors have no conflicts of interest to disclose.
How to cite this article: Tang X, Shen Y, Lu Y, He W, Nie Y, Fang X, Cai J, Si X, Zhu Y. Identification and validation of pyroptosis-related genes as potential biomarkers for hypertrophic cardiomyopathy: A comprehensive bioinformatics analysis. Medicine 2024;103:4(e36799).
Contributor Information
Xin Tang, Email: tangxin.1009@qq.com.
Yi Shen, Email: 2544137616@qq.com.
Yun Lu, Email: 1017334834@qq.com.
Wanya He, Email: 1719306291@qq.com.
Ying Nie, Email: 3496124270@qq.com.
Xue Fang, Email: 1371193728@qq.com.
Jinghui Cai, Email: 1799186722@qq.com.
Xiaoyun Si, Email: sixiaoyun1@qq.com.
References
- [1].Tuohy CV, Kaul S, Song HK, et al. Hypertrophic cardiomyopathy: the future of treatment. Eur J Heart Fail. 2020;22:228–40. [DOI] [PubMed] [Google Scholar]
- [2].Alexander PMA, Nugent AW, Daubeney PEF, et al. Long-term outcomes of hypertrophic cardiomyopathy diagnosed during childhood: results from a national population-based study. Circulation. 2018;138:29–36. [DOI] [PubMed] [Google Scholar]
- [3].Sen-Chowdhry S, Jacoby D, Moon JC, et al. Update on hypertrophic cardiomyopathy and a guide to the guidelines. Nat Rev Cardiol. 2016;13:651–75. [DOI] [PubMed] [Google Scholar]
- [4].Semsarian C, Ingles J, Maron MS, et al. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol. 2015;65:1249–54. [DOI] [PubMed] [Google Scholar]
- [5].Walsh R, Buchan R, Wilk A, et al. Defining the genetic architecture of hypertrophic cardiomyopathy: re-evaluating the role of non-sarcomeric genes. Eur Heart J. 2017;38:3461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Maron BJ, Maron MS, Maron BA, et al. Moving beyond the sarcomere to explain heterogeneity in hypertrophic cardiomyopathy: JACC review topic of the week. J Am Coll Cardiol. 2019;73:1978–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tan Y, Chen Q, Li X, et al. Pyroptosis: a new paradigm of cell death for fighting against cancer. J Exp Clin Cancer Res. 2021;40:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Qian Z, Zhao Y, Wan C, et al. Pyroptosis in the initiation and progression of atherosclerosis. Front Pharmacol. 2021;12:652963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lu Y, Lu Y, Meng J, et al. Pyroptosis and its regulation in diabetic cardiomyopathy. Front Physiol. 2022;12:791848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chai R, Xue W, Shi S, et al. Cardiac remodeling in heart failure: role of pyroptosis and its therapeutic implications. Front Cardiovasc Med. 2022;9:870924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dai F, Li X, Li X, et al. Caspase-1 abrogates the salutary effects of hypertrophic preconditioning in pressure overload hearts via IL-1β and IL-18. Front Mol Biosci. 2021;8:641585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stelzer G, Rosen N, Plaschkes I, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- [13].Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liao Y, Wang J, Jaehnig EJ, et al. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhou G, Soufan O, Ewald J, et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47:W234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shang L, Pin L, Zhu S, et al. Plantamajoside attenuates isoproterenol-induced cardiac hypertrophy associated with the HDAC2 and AKT/ GSK-3β signaling pathway. Chem Biol Interact. 2019;307:21–8. [DOI] [PubMed] [Google Scholar]
- [17].Arkoumani M, Papadopoulou-Marketou N, Nicolaides NC, et al. The clinical impact of growth differentiation factor-15 in heart disease: a 2019 update. Crit Rev Clin Lab Sci. 2020;57:114–25. [DOI] [PubMed] [Google Scholar]
- [18].Mándi Y, Hogye M, Talha EM, et al. Cytokine production and antibodies against heat shock protein 60 in cardiomyopathies of different origins. Pathobiology. 2000;68:150–8. [DOI] [PubMed] [Google Scholar]
- [19].Ma ZG, Yuan YP, Zhang X, et al. C1q-tumour necrosis factor-related protein-3 exacerbates cardiac hypertrophy in mice. Cardiovasc Res. 2019;115:1067–77. [DOI] [PubMed] [Google Scholar]
- [20].Ranjbarvaziri S, Kooiker KB, Ellenberger M, et al. Altered cardiac energetics and mitochondrial dysfunction in hypertrophic cardiomyopathy. Circulation. 2021;144:1714–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mokhobo KP. Typhoid cardiac involvement. S Afr Med J. 1975;49:55–6. [PubMed] [Google Scholar]
- [22].Filip C, Nicolescu A, Cinteza E, et al. Cardiovascular complications of hemolytic uremic syndrome in children. Maedica (Bucur). 2020;15:305–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rani B, Kumar A, Bahl A, et al. Renin-angiotensin system gene polymorphisms as potential modifiers of hypertrophic and dilated cardiomyopathy phenotypes. Mol Cell Biochem. 2017;427:1–11. [DOI] [PubMed] [Google Scholar]
- [24].Onoue K, Saito Y. New treatment for myocardial diseases. J Cardiol. 2021;77:620–5. [DOI] [PubMed] [Google Scholar]
- [25].Kee HJ, Sohn IS, Nam KI, et al. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation. 2006;113:51–9. [DOI] [PubMed] [Google Scholar]
- [26].Cohen P. The regulation of protein function by multisite phosphorylation--a 25 year update. Trends Biochem Sci. 2000;25:596–601. [DOI] [PubMed] [Google Scholar]
- [27].Tartaglia M, Kalidas K, Shaw A, et al. PTPN11 mutations in Noonan syndrome: molecular spectrum, genotype-phenotype correlation, and phenotypic heterogeneity. Am J Hum Genet. 2002;70:1555–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chen H, Li X, Liu X, et al. Clinical and mutation profile of pediatric patients with RASopathy-associated hypertrophic cardiomyopathy: results from a Chinese cohort. Orphanet J Rare Dis. 2019;14:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang J, Chandrasekhar V, Abbadessa G, et al. In vivo efficacy of the AKT inhibitor ARQ 092 in Noonan Syndrome with multiple lentigines-associated hypertrophic cardiomyopathy. PLoS One. 2017;12:e0178905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sznajer Y, Keren B, Baumann C, et al. The spectrum of cardiac anomalies in Noonan syndrome as a result of mutations in the PTPN11 gene. Pediatrics. 2007;119:e1325–31. [DOI] [PubMed] [Google Scholar]
- [31].Limongelli G, Pacileo G, Marino B, et al. Prevalence and clinical significance of cardiovascular abnormalities in patients with the LEOPARD syndrome. Am J Cardiol. 2007;100:736–41. [DOI] [PubMed] [Google Scholar]
- [32].Lauriol J, Cabrera JR, Roy A, et al. Developmental SHP2 dysfunction underlies cardiac hypertrophy in Noonan syndrome with multiple lentigines. J Clin Invest. 2016;126:2989–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shimada YJ, Raita Y, Liang LW, et al. Comprehensive proteomics profiling reveals circulating biomarkers of hypertrophic cardiomyopathy. Circ Heart Fail. 2021;14:e007849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gelb BD, Tartaglia M. RAS signaling pathway mutations and hypertrophic cardiomyopathy: getting into and out of the thick of it. J Clin Invest. 2011;121:844–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yuan X, Bu H, Zhou J, et al. Recent advances of SHP2 inhibitors in cancer therapy: current development and clinical application. J Med Chem. 2020;63:11368–96. [DOI] [PubMed] [Google Scholar]
- [36].Song Y, Wang S, Zhao M, et al. Strategies targeting protein tyrosine phosphatase SHP2 for cancer therapy. J Med Chem. 2022;65:3066–79. [DOI] [PubMed] [Google Scholar]
- [37].Sharma MC. Annexin A2 (ANX A2): an emerging biomarker and potential therapeutic target for aggressive cancers. Int J Cancer. 2019;144:2074–81. [DOI] [PubMed] [Google Scholar]
- [38].Chugh S, Ouzounian M, Lu Z, et al. Pilot study identifying myosin heavy chain 7, desmin, insulin-like growth factor 7, and annexin A2 as circulating biomarkers of human heart failure. Proteomics. 2013;13:2324–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fairoozy RH, Cooper J, White J, et al. Identifying low density lipoprotein cholesterol associated variants in the Annexin A2 (ANXA2) gene. Atherosclerosis. 2017;261:60–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jansen M, Algül S, Bosman LP, et al. Blood-based biomarkers for the prediction of hypertrophic cardiomyopathy prognosis: a systematic review and meta-analysis. ESC Heart Fail. 2022;9:3418–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shimada YJ, Batra J, Kochav SM, et al. Difference in metabolomic response to exercise between patients with and without hypertrophic cardiomyopathy. J Cardiovasc Transl Res. 2021;14:246–55. [DOI] [PubMed] [Google Scholar]
- [42].Guo L, Wang B, Zhang F, et al. Novel biomarkers identifying hypertrophic cardiomyopathy and its obstructive variant based on targeted amino acid metabolomics. Chin Med J (Engl). 2022;135:1952–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Padrón-Barthe L, Villalba-Orero M, Gómez-Salinero JM, et al. Activation of serine one-carbon metabolism by calcineurin Aβ1 reduces myocardial hypertrophy and improves ventricular function. J Am Coll Cardiol. 2018;71:654–67. [DOI] [PubMed] [Google Scholar]
- [44].Braz JC, Bueno OF, De Windt LJ, et al. PKC alpha regulates the hypertrophic growth of cardiomyocytes through extracellular signal-regulated kinase1/2 (ERK1/2). J Cell Biol. 2002;156:905–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Marrocco V, Bogomolovas J, Ehler E, et al. PKC and PKN in heart disease. J Mol Cell Cardiol. 2019;128:212–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Previs MJ, O’Leary TS, Morley MP, et al. Defects in the proteome and metabolome in human hypertrophic cardiomyopathy. Circ Heart Fail. 2022;15:e009521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Xu M, Xue RQ, Lu Y, et al. Choline ameliorates cardiac hypertrophy by regulating metabolic remodelling and UPRmt through SIRT3-AMPK pathway. Cardiovasc Res. 2019;115:530–45. [DOI] [PubMed] [Google Scholar]
- [48].Sparks MA, Crowley SD, Gurley SB, et al. Classical renin-angiotensin system in kidney physiology. Compr Physiol. 2014;4:1201–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zhou Q, Wei SS, Wang H, et al. Crucial role of ROCK2-mediated phosphorylation and upregulation of FHOD3 in the pathogenesis of angiotensin II-induced cardiac hypertrophy. Hypertension. 2017;69:1070–83. [DOI] [PubMed] [Google Scholar]
- [50].Okabe K, Matsushima S, Ikeda S, et al. DPP (Dipeptidyl Peptidase)-4 inhibitor attenuates Ang II (Angiotensin II)-induced cardiac hypertrophy via GLP (Glucagon-Like Peptide)-1-dependent suppression of Nox (Nicotinamide Adenine Dinucleotide Phosphate Oxidase) 4-HDAC (Histone Deacetylase) 4 Pathway. Hypertension. 2020;75:991–1001. [DOI] [PubMed] [Google Scholar]
- [51].Cooper RM, Raphael CE, Liebregts M, et al. New developments in hypertrophic cardiomyopathy. Can J Cardiol. 2017;33:1254–65. [DOI] [PubMed] [Google Scholar]
- [52].Akhtar H, Al Sudani H, Hussein M, et al. Effects of renin-angiotensin-aldosterone system inhibition on left ventricular hypertrophy, diastolic function, and functional status in patients with hypertrophic cardiomyopathy: a systematic review. Cureus. 2022;14:e26642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].van der Merwe L, Cloete R, Revera M, et al. Genetic variation in angiotensin-converting enzyme 2 gene is associated with extent of left ventricular hypertrophy in hypertrophic cardiomyopathy. Hum Genet. 2008;124:57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Philipson DJ, DePasquale EC, Yang EH, et al. Emerging pharmacologic and structural therapies for hypertrophic cardiomyopathy. Heart Fail Rev. 2017;22:879–88. [DOI] [PubMed] [Google Scholar]
- [55].Meng X, Cui J, He G. Bcl-2 is involved in cardiac hypertrophy through PI3K-Akt pathway. Biomed Res Int. 2021;2021:6615502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Chen Y, Zhou J, Wei Z, et al. Identification of circular RNAs in cardiac hypertrophy and cardiac fibrosis. Front Pharmacol. 2022;13:940768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wesche H, Gao X, Li X, et al. IRAK-M is a novel member of the Pelle/interleukin-1 receptor-associated kinase (IRAK) family. J Biol Chem. 1999;274:19403–10. [DOI] [PubMed] [Google Scholar]
- [58].Kobayashi K, Hernandez LD, Galán JE, et al. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110:191–202. [DOI] [PubMed] [Google Scholar]
- [59].Zhou H, Yu M, Fukuda K, et al. IRAK-M mediates Toll-like receptor/IL-1R-induced NFκB activation and cytokine production. EMBO J. 2013;32:583–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Nguyen TH, Turek I, Meehan-Andrews T, et al. Analysis of interleukin-1 receptor associated kinase-3 (IRAK3) function in modulating expression of inflammatory markers in cell culture models: a systematic review and meta-analysis. PLoS One. 2020;15:e0244570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Pai P, Sukumar S. HOX genes and the NF-κB pathway: a convergence of developmental biology, inflammation and cancer biology. Biochim Biophys Acta Rev Cancer. 2020;1874:188450. [DOI] [PubMed] [Google Scholar]
- [62].Tang L, Yao F, Wang H, et al. Inhibition of TRPC1 prevents cardiac hypertrophy via NF-κB signaling pathway in human pluripotent stem cell-derived cardiomyocytes. J Mol Cell Cardiol. 2019;126:143–54. [DOI] [PubMed] [Google Scholar]
- [63].Koivisto E, Jurado Acosta A, Moilanen AM, et al. Characterization of the regulatory mechanisms of activating transcription factor 3 by hypertrophic stimuli in rat cardiomyocytes. PLoS One. 2014;9:e105168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Pelliccia F, Limongelli G, Rosano GMC, et al. Nuclear factor-kappa B predicts long-term clinical outcome in patients with hypertrophic cardiomyopathy: 10-year follow-up study. Eur J Prev Cardiol. 2022;29:e108–11. [DOI] [PubMed] [Google Scholar]
- [65].Kuusisto J, Kärjä V, Sipola P, et al. Low-grade inflammation and the phenotypic expression of myocardial fibrosis in hypertrophic cardiomyopathy. Heart. 2012;98:1007–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Yokoyama T, Nakano M, Bednarczyk JL, et al. Tumor necrosis factor-alpha provokes a hypertrophic growth response in adult cardiac myocytes. Circulation. 1997;95:1247–52. [DOI] [PubMed] [Google Scholar]
- [67].Zhao L, Cheng G, Jin R, et al. Deletion of interleukin-6 attenuates pressure overload-induced left ventricular hypertrophy and dysfunction. Circ Res. 2016;118:1918–29. [DOI] [PMC free article] [PubMed] [Google Scholar]