Abstract
Apiospora is widely distributed throughout the world, and most of its hosts are Poaceae. In this study, Arthrinium-like strains were isolated from non-Poaceae in the Hainan and Fujian provinces of China. Based on the combined DNA sequence data of the internal transcriptional spacer (ITS), partial large subunit nuclear rDNA (LSU), translation extension factor 1-α gene (TEF1-α) and β-tubulin (TUB2), the collected Apiospora specimens were compared with known species, and three new species were identified. Based on morphological and molecular phylogenetic analyses, Apiospora adinandrae sp. nov., A. bawanglingensis sp. nov. and A. machili sp. nov. are described and illustrated.
Keywords: taxonomy, Ascomycota, multigene phylogeny
1. Introduction
The genus Apiospora (Apiosporaceae, Amphisphaeriales, Sordariomycetes, Ascomycota) was identified and established by Saccardo (1875) with Apiospora montagnei as the type species [1]. The Arthrium-like taxa under Apiosporaceae contains the genus with apiosporous hyaline ascospores and basauxic conidiogenesis [2,3]. Thus, after Ellis (1965), the biological relationship between Apiospora and Arthrinium became widely accepted, and Apiospora was considered a synonym for Arthrinium until the study of Pintos and Alvarado, the former was usually considered the sexual morph and the latter was considered the asexual morph [3,4,5]. During this time, the dual nomenclature was abandoned, and the old name Arthrinium was recommended for unitary nomenclature [6].
The genus Arthrinium was introduced by Kunze and Schmidt (1817) with Ar. caricicola as the type species [7]. In order to clarify the exact status of the Arthrium-like taxa, most species were recollected, and some species were reassigned to different genera by Link (in Willdenow 1824) due to different conidial shapes, but were subsequently reclassified back to Arthrinium by von Höhnel 1925 and Cooke 1954, because of similar conidiophores [8,9,10]. Pintos et al. (2019) published the genetic information of the first model species Ar. caricicola, followed by the phylogenetic analysis of Pintos and Alvarado (2021), which found that Arthrinium formed a separate clade from other sequences of Apiospora, and Apiospora and Arthrinium were phylogenetically distinguished [4,11].
The morphological similarities between the two genera (Apiospora and Arthrinium) make it difficult to determine the boundary between them by morphological methods alone. Morphologically, most of the conidia of Apiospora are nearly spherical in the front and lenticular at the side, and sometimes the conidia develop into pinholes, and the conidia of Arthrinium vary in shape (angular, spherical, curved, boat-shaped, fusiform, polygonal), with some species having thick black intervals [12,13,14,15].
Ecologically, Apiospora was widely distributed in various climatic zones around the world and was found in many species of hosts, whereas Arthrinium was rarely distributed in tropical and subtropical regions and had fewer hosts [13,14]. Species of Apiospora have been found in Poaceae during past studies, while Arthrinium has been found in the family Cyperaceae and Juncaceae [4,15,16,17,18,19,20]. The ecological differences further support the genetic and morphological division of the two genera, and all the evidence is sufficient to support the separation of the two genera in taxonomic status. In addition, the host association and geographical distribution were discussed based on the existing research progress.
Currently, there are 120 records of Arthrinium and 157 records of Apiospora on Index Fungorum (http://www.indexfungorum.org/, accessed on 20 November 2023). In the present study, the authors isolated strains of Arthrinium-like taxa in China. To clarify the taxonomic status, morphological and phylogenetic studies were conducted. Three new species, Apiospora adinandrae sp. nov., A. bawanglingensis sp. nov. and A. machili sp. nov., were identified and classified in Apiospora by multilocus analysis of tandem internal transcribed spacer (ITS), 28S large subunit ribosomal RNA gene (LSU), translation elongation factor 1-alpha gene (TEF1α) and beta-tubulin (TUB) datasets.
2. Materials and Methods
2.1. Isolation and Morphology
2.1.1. Strains Isolation
The four strains were isolated from three host plants in two provinces of China (Table 1). These strains were obtained by tissue separation and the single spore isolation method [21]. Surface sterilization was performed on 5 × 5 mm fragments at the junction of the leaf lesion edge and healthy tissue. The fragments were soaked in 75% alcohol for 90 s, washed with sterile water for 45 s, then immersed in 5% sodium hypochlorite solution for 60 s and finally washed with sterile water 3 times. The sterilized wet fragments were dried on sterilized filter paper and then cultured on potato dextrose agar (PDA: 220 g potato, 20 g agar, 18 g dextrose, 1000 mL sterile water and natural pH) 25 °C for 3 days. Subsequently, morphologically different colonies could be observed on PDA, and the tip of a hyphae with strong growth capacity growing on the edge of the colony was picked and transferred to another Petri dish containing PDA and culture was continued at 25 °C.
Table 1.
Strains associated with host and region in this study.
| Strain | Host | Region |
|---|---|---|
| BW0444 | Indocalamus longiauritus | Hainan Province |
| BW04441 | Indocalamus longiauritus | Hainan Province |
| BW0455 | Indocalamus longiauritus | Hainan Province |
| BW04551 | Indocalamus longiauritus | Hainan Province |
| XG01282B-1 | Adinandra glischroloma | Fujian Province |
| XG01282B-2 | Adinandra glischroloma | Fujian Province |
| XG01175A-4 | Machilus nanmu | Fujian Province |
| XG01175 | Machilus nanmu | Fujian Province |
2.1.2. Morphological Studies
The colonies were observed morphologically on the 7th and 14th day of culture and photographed by digital camera (Canon Powershot G7X, Canon, Tokyo, Japan). A stereomicroscope (Olympus SZX10, OLYMPUS, Tokyo, Japan) and a microscope (Olympus BX53, OLYMPUS, Tokyo, Japan) were used to observe the microscopic morphological characteristics of the colonies. Both microscopes were equipped with high-definition color digital cameras to capture the conidia of fungal structures.
In order to facilitate further research, the four strains in this study were kept in 10% sterilized glycerol or sterile water at 4 °C. Voucher specimens were stowed in two institutions, the Herbarium of the Department of Plant Pathology (HSAUP) of Shandong Agricultural University, Taian, China, and the Herbarium Mycologicum Academiae Sinicae (HMAS) of the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China. Ex-holotype living cultures were stored in the Shandong Agricultural University Culture Collection (SAUCC). All taxonomic information on the new taxa obtained in this paper has been submitted to MycoBank (http://www.mycobank.org, accessed on 20 November 2023).
2.2. DNA Extraction and Amplification
Fungal tissue was obtained on PDA and genomic DNA was extracted from the mycelium. DNA extraction was performed by a magnetic bead kit (OGPLF-400, GeneOnBio Corporation, Changchun, China) and the CTAB method [22,23]. The polymerase chain reaction (PCR) procedure was performed using the primer pairs in Table 2, which contains the entire internal transcriptional spacer (ITS) of the intervening 5.8S rRNA gene, part of the large subunit nrRNA gene (LSU), part of the translation extension factor 1-α gene (TEF1α) and part of the beta-tubulin gene (TUB2).
The polymerase chain reaction was carried out using an Eppendorf Master Thermocycler (Hamburg, Germany). The amplification reaction was performed in a 20 μL reaction system consisting of 10 μL 2 × Hieff Canace® Plus PCR Master Mix (With Dye) (Yeasen Biotechnology, Cat No. 10154ES03, Shanghai, China), the forward and reverse primer (TsingKe, Qingdao, China) each 0.7 μL per 10 μM, and 1.4 μL template genomic DNA, at last replenishing the total volume to 20 μL with distilled deionized water. The PCR products were separated by 1% agarose gel electrophoresis with GelRed added, and the bands were observed under ultraviolet light [24]. Then, we used a Gel Extraction Kit (Cat: AE0101-C) (Shandong Sparkjade Biotechnology Co., Ltd., Ji’nan, China) for gel recovery. The bidirectional sequencing of DNA samples was completed by the Tsingke Company Limited (Qingdao, China), and the resulting sequences were processed by MEGA 7.0 to achieve consistency [25]. The sequences data obtained and used in this article were all uploaded to GenBank (Table S1).
Table 2.
Molecular markers and their PCR primers and programs used in this study.
| Locus | Primers | Sequence (5′–3′) | PCR Cycles | References |
|---|---|---|---|---|
| ITS | ITS5 | GGA AGT AAA AGT CGT AAC AAG G | (95 °C: 30 s, 55 °C: 30 s, 72 °C: 45 s) × 29 cycles | [26] |
| ITS4 | TCC TCC GCT TAT TGA TAT GC | |||
| LSU | LR0R | GTA CCC GCT GAA CTT AAG C | (95 °C: 30 s, 48 °C: 50 s, 72 °C: 1 min 30s) × 35 cycles | [27] |
| LR5 | TCC TGA GGG AAA CTT CG | |||
| TEF1α | EF1-728F | CAT CGA GAA GTT CGA GAA GG | (95 °C: 30 s, 51 °C: 30 s, 72 °C: 1 min) × 35 cycles | [28] |
| EF2 | GGA RGT ACC AGT SAT CAT GTT | |||
| TUB2 | T1 | AAC ATG CGT GAG ATT GTA AGT | (95 °C: 30 s, 56 °C: 30 s, 72 °C: 1 min) × 35 cycles | [29] |
| Bt-2b | ACC CTC AGT GTA GTG ACC CTT GGC |
2.3. Phylogenetic Analyses
The new sequences obtained in present study were all compared in NCBI’s Genbank nucleotide database (https://www.ncbi.nlm.nih.gov/, accessed on 11 November 2023) and all reference sequences for relevant species were downloaded. Multi-sequence analysis was performed using MAFFT 7 online services and the auto policy (http://mafft.cbrc.jp/alignment/server/, accessed on 20 November 2023) to compare the newly generated sequences with other related sequences. To identify isolates at the species level, a phylogenetic analysis of each marker was first performed and then combined (ITS-LSU-TEF1α-TUB2) (See Supplementary File S1).
Phylogenetic analysis of multi-labeled data was performed based on Bayesian inference (BI) and maximum likelihood (ML) algorithms. Both ML and BI were run on the CIPRES Science Gateway portal (https://www.phylo.org/, accessed on 20 November 2023) or offline software (The ML was operated in RaxML-HPC2 on XSEDE v8.2.12 and BI analysis was operated in MrBayes v3.2.7a with 64 threads on Linux) [30,31,32,33,34,35]. For ML analyses, the default parameters were used and 1000 rapid bootstrap replicates were run with the GTR+G+I model of nucleotide evolution, and the BI analysis was performed using a fast bootstrap algorithm with an automatic stop option. The BI analyses included 2 million generations of sixty-four parallel threads with the stop rule options and 100 generations of sampling frequency. The burn-in score was set to 25% and posterior probabilities (PP) were determined from the remaining trees. All resulting trees were plotted using FigTree v. 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree, accessed on 20 November 2023), or ITOL: Interactive Tree Of Life (https://itol.embl.de/, accessed on 21 November 2023) [36], and the layout of the trees was produced in Adobe Illustrator CC 2019.
3. Results
3.1. Phylogenetic Analyses
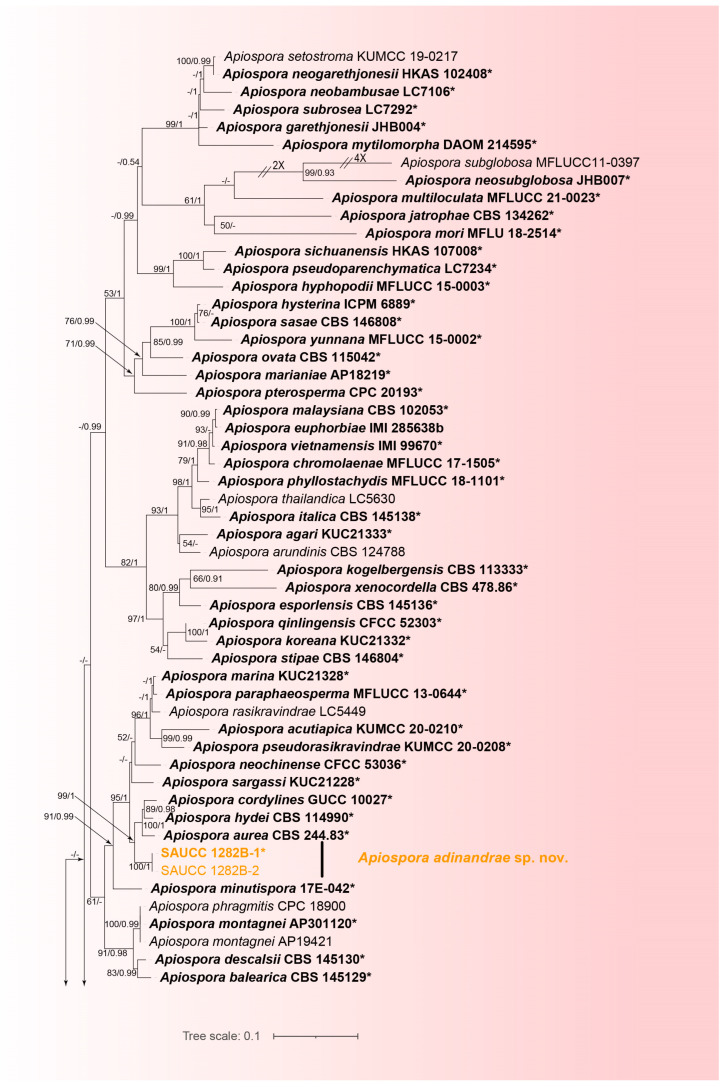
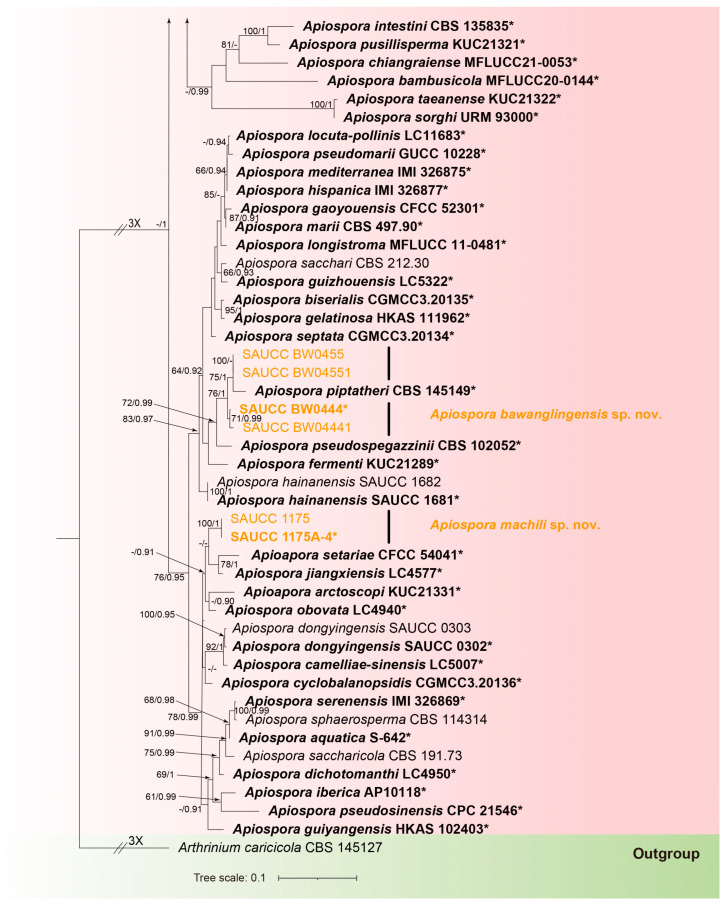
Phylogenetic analysis was performed on 99 isolates representing Apiospora species, of which 97 isolates were considered to be the ingroup and 2 strains of Arthrinium caricicola (CBS 145127) were used as the outgroup. The final alignment contained 2843 concatenated characters, viz. 1–838 (ITS), 839–1674 (LSU), 1675–2293 (TEF1α), 2294–2843 (TUB2), 1619 were constant, 421 were variable and parsimony-uninformative and 803 were parsimony-informative. The topology of the ML tree confirms the tree topology obtained from Bayesian inference; therefore, only the ML tree is presented (Figure 1). The alignment has 1456 distinct alignment patterns. The proportion of gaps and completely undetermined characters in this alignment: 33.53%. The estimated base frequencies were as follows: A = 0.234029, C = 0.250395, G = 0.257923, T = 0.257653; substitution rates AC = 1.257520, AG = 3.156093, AT = 1.159778, CG = 0.983356, CT = 4.511962 and GT = 1.000000; gamma distribution shape parameter α = 0.263195. Final ML optimization likelihood: −25299.135520. The GTR+I+G model was proposed for ITS, LSU, TEF1α and TUB2. BI analysis of these four tandem genes was performed over 3,400,000 generations in 68,002 trees. The first 17,000 trees representing the burn-in phase of the analysis are discarded, while the remaining trees are used to calculate the posterior probability in the majority rule consensus tree (Figure 1; first value: PP > 0.90 shown). The alignment embodied a total of 1469 unique site patterns (ITS: 475, LSU: 204, TEF1α: 440, TUB2: 350).
Figure 1.
A maximum likelihood tree showing the phylogenetic relationships of Apiospora inferred from the ITS, LSU, TEF1α and TUB2 sequences, and the roots on Arthrinium caricicola (CBS 145127). The Bayesian inference posterior probability (left, BIPP ≥ 0.90) and the maximum likelihood bootstrap value (right, MLBV ≥ 50%) are shown as BIPP/ML above the nodes. Strains marked with a star “*” and bolded represented are ex-types or ex-holotypes. Strains from the present study are in orange. The scale in the bottom middle indicates 0.1 substitutions per site. In order to make the layout of the evolutionary tree beautiful, some branches are shortened by two diagonal lines (“/”) with the number of times.
In our phylogenetic analyses, 97 strains of Apiospora were identified as a monophyletic clade (Figure 1). Among them, 8 strains produced three new species lineages, Apiospora adinandrae (SAUCC 1282B-1, SAUCC 1282B-2) closely related to A. aurea (CBS 244.83), A. hydei (CBS 114990) and A. cordylines (GUCC 10027) with full support (99% MLBV and 1.0 BIPP); A. bawanglingensis (SAUCC BW0444 and SAUCC BW04441) closely related to A. piptatheri (CBS 145149) with good support (1.0 BIPP and 76% MLBV); and A. machili (SAUCC 1175A-4) forming a separate single-species lineage. The present study revealed three species, viz. Apiospora adinandrae sp. nov., A. bawanglingensis sp. nov. and A. machili sp. nov.
3.2. Taxonomy
3.2.1. Apiospora adinandrae X.Y. Liu, Z.X. Zhang and X.G. Zhang, sp. nov. (Figure 2)
MycoBank—No: 850667;
Etymology—The epithet adinandrae pertains to the generic name of the host plant Adinandra glischroloma;
Type—Wuyishan National Forest Park, Fujian Province, China, on diseased leaves of Adinandra glischroloma, 15 October 2022, X.Y. Liu (HMAS 352657, holotype), ex-holotype living culture SAUCC 1282B–1;
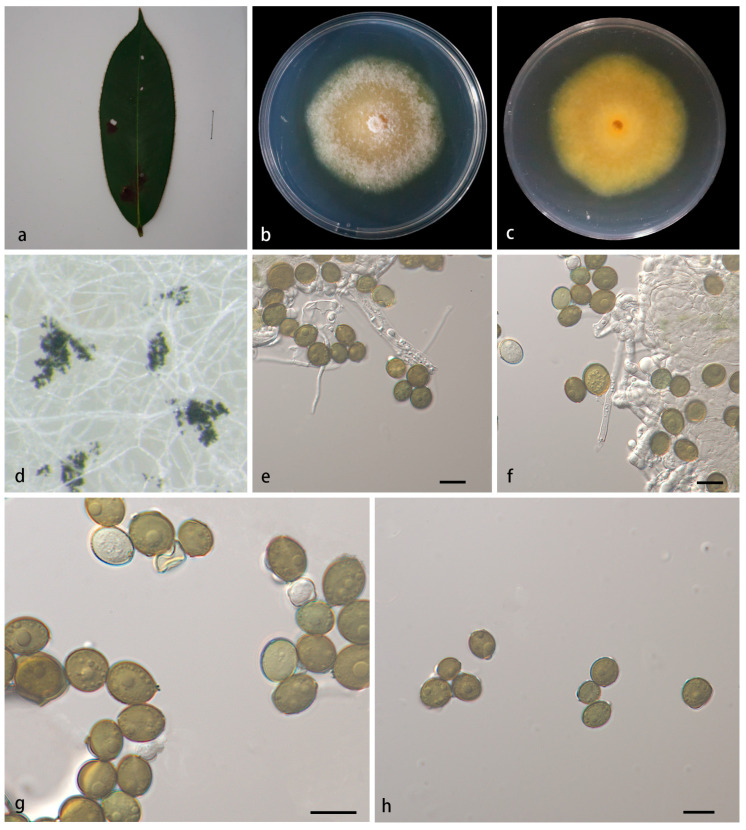
Description—On PDA, hyphae 2.5–4.0 μm in diameter, branched, hyaline and septate. Sexual morph: Undetermined. Asexual morph: Conidiophore reduced to conidiogenous cells, aggregated in clusters on hyphae. Conidiogenous cells are 3.5–6.0 × 2.0–3.5 μm, hyaline becoming pale green, polyblastic, cylindrical, septate, verrucose, flexuous. Conidia smooth, rounded to ovoid, globose to subglobose, green to pale brown, 7.5–12.4 × 6.3–10.5 μm, mean ± SD = 9.1 ± 1.0 × 8.1 ± 1.1 μm. See Figure 2;
Culture characteristics—PDA, colonies concentrically spreading with subcircular margin, flat, abundant white aerial mycelium. In reverse, the sites where mycelium is abundant appear pale yellow, and the sites with sparse mycelium appear yellow. After seven days of incubation at 25 °C, the colony diameter reached 57.5–65.5 mm and the growth rate was 8.21–9.35 mm/day;
Additional specimen examined—China, Fujian Province: Wuyishan National Forest Park, on diseased leaves of Adinandra glischroloma, 15 October 2022, X.Y. Liu, HSAUP 1282B–2, living culture SAUCC 1282B–2;
Notes—Phylogenetic analyses based on ITS-LSU-TEF1α-TUB2 rDNA sequences showed that Apiospora adinandrae sp. nov. formed an independent clade which is closely related to A. aurea (CBS 244.83), A. hydei (CBS 114990) and A. cordylines (GUCC 10027). The base-pair comparison of ITS, LSU, TEF1α and TUB2 sequences, respectively, showed 1.01%, 0.48%, 6.81% and 3.62% differences between A. adinandrae (SAUCC 1282B-1) and A. aurea (CBS 244.83); showed 1.18%, 0.36%, 6.79% and 4.75% differences between A. adinandrae and A. hydei (CBS 114990); and showed 1.04%, 0%, 6.47% and 4.97% differences between A. adinandrae and A. cordylines (GUCC 10027);
Morphologically, A. adinandrae differs from A. aurea, A. hydei and A. cordylines in conidiophore, conidiogenous cells and conidia. The conidiophore of A. adinandrae usually reduced to conidiogenous cells, while both A. aurea and A. cordylines have brown transverse septa conidiophore. The conidiogenous cells of A. adinandrae hyaline become pale green, while the conidiogenous cells of A. aurea, A. hydei and A. cordylines become colorless to brown. A. aurea has dark brown conidia (10.0–30.0 × 10.0–15.0 μm) and sterile cells of a different shape than the conidia. The conidia of A. hydei are brown, roughened, globose in surface view (15.0–22.0 × 10.0–14.0 μm). The conidia of A. cordylines brown, smooth to finely roughened, subglobose (15.0–19.0 × 12.5–18.5 μm). The conidia of A. adinandrae are smaller in size (7.5–12.4 × 6.3–10.5 μm) and more oval in shape than the conidia of A. aurea, A. hydei and A. cordylines [4]. For details, see Table 3.
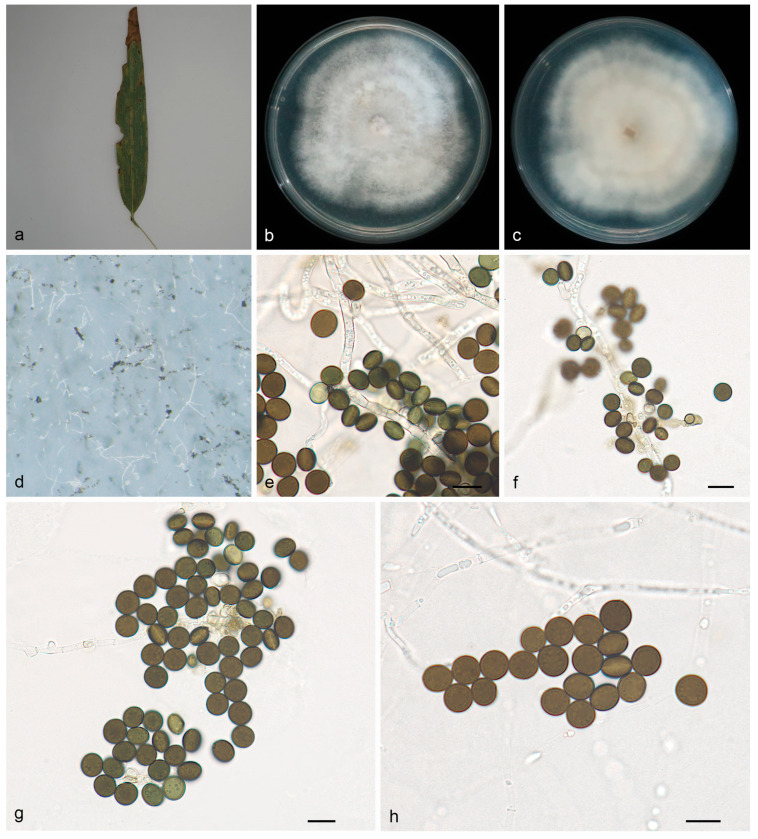
Figure 2.
Apiospora adinandrae (HMAS 352657, holotype) (a) diseased leaf of Adinandra glischroloma, (b,c) colonies after 14 days on PDA ((b), obverse; (c), reverse), (d) colony overview, (e,f) conidia with conidiogenous cells, (g,h) conidia. Scale bars: 10 μm (e–h).
3.2.2. Apiospora bawanglingensis X.Y. Liu, Z.X. Zhang and X.G. Zhang, sp. nov. (Figure 3)
MycoBank—No: 850661;
Etymology—The specific epithet “bawanglingensis” refers to the Bawangling National Forest Park, where the type was collected;
Type—Bawangling National Forest Park, Hainan Province, China, on diseased leaves of Indocalamus longiauritus, 19 May 2021, X.Y. Liu (HMAS 352654, holotype), ex-holotype living culture SAUCC BW0444;
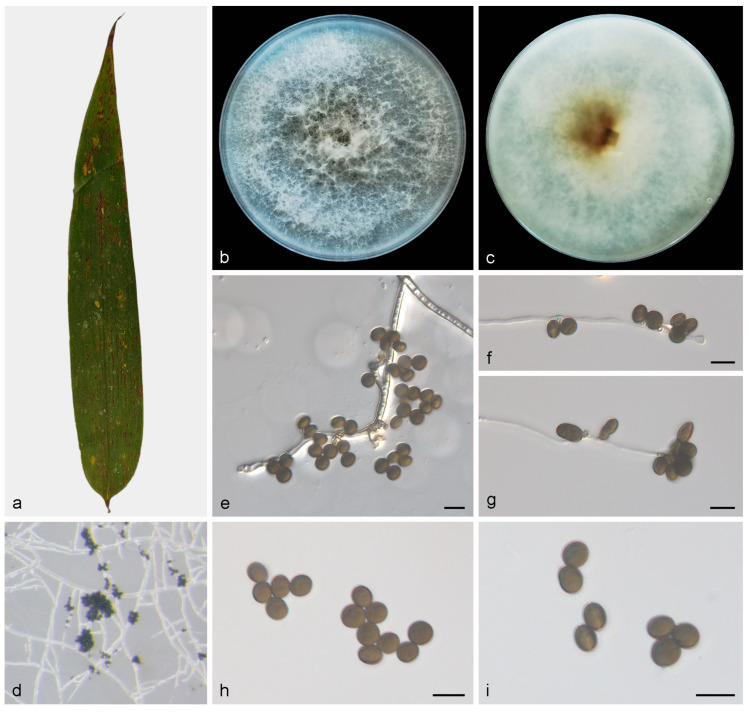
Description—On PDA, hyphae 3.0–3.5 μm in diameter, branched, hyaline and septate. Sexual morph: Undetermined. Asexual morph: Conidiophore reduced to conidiogenous cells, aggregated in clusters on hyphae. Conidiogenous cells dark green, becoming brown, polyblastic, cylindrical, septate, verrucose, flexuous, 5.0–10.0 × 2.0–3.0 μm. Conidia smooth, rounded to ovoid, globose to subglobose, green to dark brown, 6.4–7.7 × 5.3–7.1 μm, mean ± SD = 7.1 ± 0.4 × 6.1 ± 0.5 μm, n = 30. See Figure 3;
Culture characteristics—PDA, colonies concentrically spreading, fluffy, with abundant aerial sparse mycelium, white to cream. In reverse, white, becoming tawny from the center. After seven days of incubation at 25 °C, the colony diameter reached 73.7–82.5 mm and the growth rate was 10.5–11.7 mm/day;
Additional specimen examined—China, Hainan Province: Bawangling National Forest Park, on diseased leaves of Indocalamus longiauritus, 19 May 2021, X.Y. Liu, HSAUP BW04441, living culture SAUCC BW04441;
Notes—Phylogenetic analyses based on ITS-LSU-TEF1α-TUB2 rDNA sequences showed that Apiospora bawanglingensis sp. nov. formed an independent clade which is closely related to A. piptatheri (CBS 145149). The base-pair comparison of ITS, LSU and TEF1α sequences, respectively, showed 2.74%, 0.72% and 8.63% differences between A. bawanglingensis (SAUCC BW0444) and A. piptatheri (CBS 145149);
Morphologically, A. bawanglingensis differs from A. piptatheri in conidiogenous cells and conidia. The conidiogenous cells of A. piptatheri discrete, sometimes branched, measure 6.0–27.0 × 2.0–5.0 µm. Compared to A. piptatheri, A. bawanglingensis has brown, polyblastic, cylindrical, septate, shorter conidiogenous cells (5.0–10.0 × 2.0–3.0 μm). The conidia of A. piptatheri (6.0–8.0 × 3.0–5.0 μm) and A. bawanglingensis (6.4–7.7 × 5.3–7.1 μm) are similar in shape, but the conidia of A. piptatheri with a thin hyaline germ-slit and the conidia of A. bawanglingensis are not observed [4]. For details, see Table 3.
Figure 3.
Apiospora bawanglingensis (HMAS 352654, holotype) (a) diseased leaf of Indocalamus longiauritus, (b,c) colonies after 14 days on PDA ((b), obverse; (c), reverse), (d) colony overview, (e–g) conidia with conidiogenous cells, (h,i) conidia. Scale bars: 10 μm (e–i).
3.2.3. Apiospora machili X.Y. Liu, Z.X. Zhang and X.G. Zhang, sp. nov. (Figure 4)
MycoBank—No: 850665;
Etymology—The epithet machili pertains to the generic name of the host plant Machilus nanmu;
Type—Wuyishan National Forest Park, Fujian Province, China, on diseased leaves of Machilus nanmu, 15 October 2022, X.Y. Liu (HMAS 352656, holotype), ex-holotype living culture SAUCC 1175A–4;
Description—On PDA, hyphae 2.5–3.5 μm in diameter, branched, hyaline and septate. Sexual morph: Undetermined. Asexual morph: Conidiophore reduced to conidiogenous cells, aggregated in clusters on hyphae. Conidiogenous cells pale green, becoming brown, polyblastic, cylindrical, septate, verrucose, flexuous, 6.0–8.0 × 2.5–4.0 μm. Conidia smooth, green to dark brown, 7.1–9.5 × 5.6–8.8 μm, mean ± SD = 8.5 ± 0.6 × 7.7 ± 0.7 μm. In face view, rounded to ovoid, globose to subglobose; in side view, lenticular, with a pale equatorial slit. Sexual morph: Undetermined. See Figure 4;
Culture characteristics—PDA, colonies concentrically spreading with undulate margin, wooly aerial mycelium, flat, ivory. In reverse, the whole is ivory, with a slight yellow in the center. After seven days of incubation at 25 °C, the colony diameter reached 69.7–78.8 mm and the growth rate was 9.9–11.2 mm/day;
Additional specimen examined—Wuyishan National Forest Park, Fujian Province, China, on diseased leaves of Machilus nanmu, 15 October 2022, X.Y. Liu, HSAUP 1175, living culture SAUCC 1175;
Notes—Phylogenetic analyses based on ITS-LSU-TEF1α-TUB2 rDNA sequences showed that Apiospora machili sp. nov. formed an independent clade which is closely related to A. setariae (CFCC 54041) and A. jiangxiensis (LC4577). The base-pair comparison of ITS, LSU, TEF1α and TUB2, respectively, showed 3.40%, 0%, 0% and 0% differences between A. machili (SAUCC 1175A-4) and A. setariae (CFCC 54041), and showed 2.2%, 0.24%, 4.35% and 4.54% differences between A. machili and A. jiangxiensis (LC4577);
Morphologically, A. machili differs from A. setariae and A. jiangxiensis in conidiophore, conidiogenous cells and conidia. A. setariae has erect conidiophore and longer conidiogenous cells (8.0–55.0 × 1.0–3.5 μm), whereas A. machili conidiophore reduced to conidiogenous cells (6.0–8.0 × 2.5–4.0 μm) and the conidiogenous cells polyblastic, cylindrical, flexuous. A. machili differs from A. jiangxiensis by its cylindrical conidiogenous cells (6.0–8.0 × 2.5–4.0 μm), while A. jiangxiensis has conidiogenous cells that are clearly ampulliform (apical neck 2.5–6.0 µm, basal part 3.0–9.0 µm). A. machili, A. setariae and A. jiangxiensis have morphologically similar conidia (7.1–9.5 × 5.6–8.8 μm vs. 7.5–10.5 μm vs. 7.5–10.0 × 4.5–7.0 μm) [4]. For details, see Table 3.
Figure 4.
Apiospora machili (HMAS 352656, holotype) (a) diseased leaf of Machilus nanmu, (b,c) colonies after 14 days on PDA ((b), obverse; (c), reverse), (d) colony overview, (e,f) conidia with conidiogenous cells, (g,h) conidia. Scale bars: 10 μm (e–h).
3.2.4. Apiospora piptatheri Pintos and P. Alvarado, MycoKeys 49: 40. (2019) (Figure 5)
Description—On PDA, hyphae 2.5–4.0 μm in diameter, branched, hyaline and septate. Asexual morph: Conidiophore reduced to conidiogenous cells, aggregated in clusters on hyphae. Conidiogenous cells pale green, becoming brown, polyblastic, cylindrical, septate, verrucose, flexuous, 10.0–15.0 × 3.0–5.0 μm. Conidia smooth, rounded to ovoid, globose to subglobose, green to dark brown, 6.2–7.6 × 4.9–7.2 μm, mean ± SD = 6.9 ± 0.4 × 6.1 ± 0.6 μm. Sexual morph: Undetermined. See Figure 5;
Culture characteristics—PDA, colonies concentrically spreading with irregular margin, abundant fluffy aerial mycelium which can fill a plate, mycelium white to cream. In reverse, the central part is brown to yellow, and gradually lightened outward, from yellow to white. After seven days of incubation at 25 °C, the colony diameter reached 73.7–82.5 mm and the growth rate was 10.5–11.7 mm/day;
Additional specimen examined—Bawangling National Forest Park, Hainan Province, China, on diseased leaves of Indocalamus longiauritus, 19 May 2021, X.Y. Liu, HSAUP BW04551, living culture SAUCC BW04551;
Notes—Phylogenetic analyses based on ITS-LSU-TEF1α rDNA sequences showed that Apiospora piptatheri (SAUCC BW0455) formed an independent clade which is closely related to A. piptatheri (CBS 145149). The base-pair comparison of ITS, LSU and TEF1α showed 0.36%, 0.96% and 56.2% differences between SAUCC BW0455 and CBS 145149, respectively;
Morphologically, strain SAUCC BW0455 and A. piptatheri (CBS 145149) have similar characteristics. The isolates (SAUCC BW0455) are similar to the type strain of A. piptatheri (CBS 145149) in having smooth, subglobose, brown conidia (6.2–7.6 × 4.9–7.2 μm vs. 6.0–8.0 × 3.0–5.0 μm); however, no germ-slit were observed in SAUCC BW0455 [4]. For details, see Table 3.
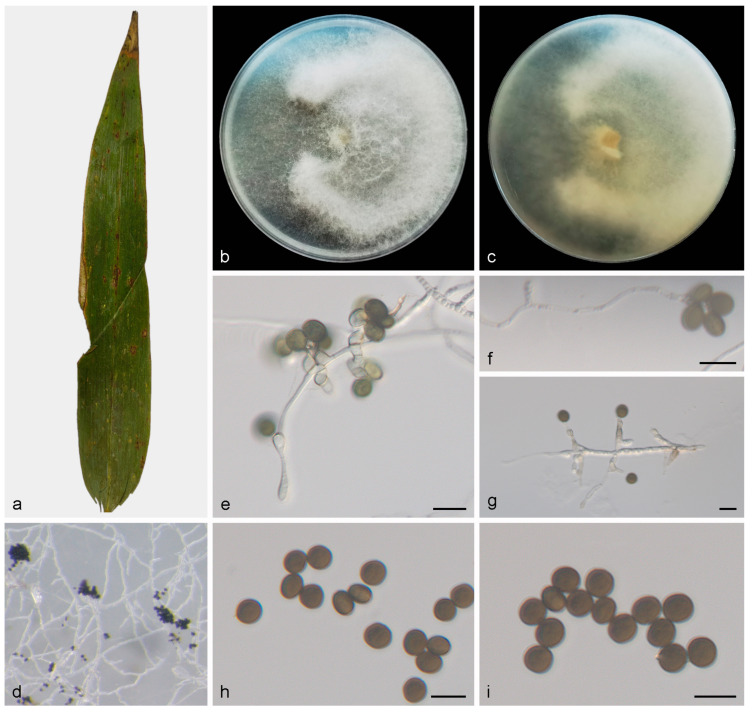
Figure 5.
Apiospora indocalami (HMAS 352655, holotype) (a) diseased leaf of Indocalamus longiauritus (b,c) colonies after 14 days on PDA ((b), obverse; (c), reverse) (d) colony overview (e–g) conidia with conidiogenous cells (h,i) conidia. Scale bars: 10 μm (e–i).
Table 3.
The asexual morphological characters of some Apiospora species.
| Strain | Host | Country | Conidiogenous Cells | Conidia in Surface View | Size (μm) | References |
|---|---|---|---|---|---|---|
| Apiospora descalsii | Ampelodesmos mauritanicus | Spain | solitary on hyphae, ampulliform, hyaline | brown, smooth, guttulate, globose, ellipsoid | (5.0–) 7.0 (–8.0) | [11] |
| A. esporlensis | Poaceae | Spain | polyblastic, aggregated, smooth, hyaline, ampuliform, cylindrical or lageniform | brown, smooth, globose, pale equatorial slit | (8.0–) 9.0–12.0 (–13.0) | [11] |
| A. iberica | Poaceae | Portugal | aggregated or solitary, ampulliform or cylindrical | brown, smooth, globose to ellipsoid | (9.0–) 10.0 (–12.0) | [11] |
| A. italica | Poaceae | Italy | ampulliform, cylindrical or doliiform, hyaline to brown | brown, smooth, globose | (3.0–) 4.0–7.0 (–9.0) × (1.5–) 2.0–3.0 (–5.0) | [11] |
| A. piptatheri | Poaceae | Spain | basauxic, polyblastic, sympodial, cylindrical, discrete, branched, smooth | globose, ellipsoidal, brown, with a thin hyaline germ-slit | 6.0–8.0 × 3.0–5.0 | [11] |
| A. fermenti | seaweed | Korea | - | Globose to elongate, ellipsoid | 7.8–8.8 × 7.2–8.6 | [13] |
| A. hainanensis | Poaceae | China | globose, erect, blastic, branched, aggregated, hyaline, pale brown, smooth | globose to lenticular, longitudinal germ-slit, ellipsoidal, brown, smooth to finely roughened | 5.5–8.5 × 5.0–7.5 | [14] |
| A. dongyingensis | Poaceae | China | globose, erect, blastic, aggregate, hyaline, pale brown, smooth, branched | globose, lenticular, longitudinal germ-slit, elongated, brown, smooth to finely roughened | 8.0–16.5 × 5.5–9.0 | [14] |
| A. acutiapica | Poaceae | China | cylindrical to ampulliform, pale brown |
hyaline apex, smooth, dark equatorial slit | - | [15] |
| A. chiangraiense | Poaceae | Thailand | smooth, monoblastic or polyblastic, aggregated, light brown, cylindrical | aseptate, pale brown to dark brown | 6.5–8.0 × 6.0–8.0 | [15] |
| A. thailandica | Poaceae | Thailand | basauxic, polyblastic, smooth, sympodial, cylindrical, discrete, sometimes branched | globose, occasionally elongated, dark brown, smooth, with a truncate basal scar | 5.0–9.0 × 5.0–8.0 | [16] |
| A. yunnana | Poaceae | China | basauxic, cylindrical, discrete, smooth | lenticular, obovoid, dark brown, smooth, with a truncate basal scar | 17.5–26.5 × 15.5–25.0 | [16] |
| A. camelliae-sinensis | Camellia sinensis | China | erect, aggregated, doliiform to ampulliform, pale brown, smooth | brown, smooth, globose | 9.0–13.5 × 7.0–12.0 | [17] |
| A. dichotomanthi | Dichotomanthus tristaniaecarpa | China | erect, aggregated on hyphae, doliiform to clavate or lageniform, hyaline, pale brown, smooth | brown, smooth to finely roughened, globose, with a longitudinal germ-slit | 9.0–15.0 × 6.0–12.0 | [17] |
| A. guizhouensis | Air | China | erect, aggregated, pale brown, smooth, subglobose, ampulliform or doliiform | dark brown, smooth to finely roughened, globose, germ-slit | 5.0–7.5 × 4.0–7.0 | [17] |
| A. jiangxiensis | Maesa sp. | China | erect, scattered or aggregated, hyaline, smooth, ampulliform | brown, smooth to finely roughened, granular, globose | 7.5–10.0 | [17] |
| A. neobambusae | Poaceae | China | erect, aggregated, hyaline, smooth, doliiform to ampulliform, or lageniform | olivaceous, smooth to finely roughened, subglobose | 11.5–15.5 × 7.0–14.0 | [17] |
| A. obovata | Lithocarpus sp. | China | erect, aggregated, pale brown, smooth, subcylindrical or clavate | dark brown, roughened, globose | 11.0–16.5 | [17] |
| A. pseudoparenchymatica | Poaceae | China | pale yellow, finely roughened, subcylindrical to doliiform | dark brown, smooth, finely guttulate, globose | 13.5–27.0 × 12.0–23.5 | [17] |
| A. subrosea | Poaceae | China | pale brown, smooth, doliiform to subcylindrical | brown, smooth, subglobose or ellipsoidal | 12.0–17.5 × 9.0–16.0 | [17] |
| A. pseudorasikravindrae | Poaceae | China | holoblastic, ampulliform, cylindrical or doliiform, olivaceous | globose | 5.0–10 × 5.5–11.0 | [20] |
| A. agari | Agarum cribrosum | Korea | clusters or solitary, hyaline becoming pale green, cylindrical, ampulliform | smooth to granular, globose to subglobose | (8.5–) 9.0–10.5 × (7.0–) 7.5–8.5 (−9.0) | [37] |
| A. arctoscopi | egg masses of Arctoscopus japonicus | Korea | clusters or solitary, hyaline, cylindrical, ampulliform | brown, smooth to granular, globose to elongate, ellipsoid | (9.5–) 10.0–12.0 (−13) × (7.5–) 8.0–11.0 (−12.0) | [37] |
| A. koreana | egg masses of Arctoscopus japonicus | Korea | aggregated in clusters on hyphae, hyaline, cylindrical | brown, smooth to granular, globose to ellipsoid | (7.5–) 8.0–10 (−11) × (5.5–) 6.5–9.5 (−10) | [37] |
| A. marina | seaweed | Korea | aggregated or solitary, hyaline, erect, ampulliform | globose to elongate ellipsoid, brown, smooth to granular | (9.5–) 10.0–12.0 (−13.0) × (7.5–) 8.0–10.0 | [37] |
| A. pusillisperma | Seaweed | Korea | aggregated, hyaline, cylindrical | brown, smooth to granular, globose | 4.0–6.0 (−6.5) × (3.0–) 3.5–5.0 (−5.5) | [37] |
| A. sargassi | Sargassum fulvellum | Korea | aggregated or solitary, hyaline, basauxic, polyblastic, sympodial, erect, cylindrical | brown, smooth to granular, globose | (8.5–) 9.5–11.0 (−11.5) × (8.0–) 8.5–10 (−11) | [37] |
| A. aquatica | decaying wood | China | erect, aggregated in clusters, smooth, doliform to ampulliform | globose to subglobose, smooth, olivaceous to brown | 9.0–11.0 × 8.0–10.0 | [38] |
| A. bambusicola | Poaceae | Thailand | polyblastic, terminal, cylindrical, smooth, aggregated, light brown | solitary, oval or irregularly round, brown, guttulate, granular | 6.0–8.0 × 6.0–7.8 | [39] |
| A. biserialis | Poaceae | China | integrated, pale brown, doliiform to ampulliform, or lageniform | brown, smooth | 7.0–9.0 | [40] |
| A. cyclobalanopsidis | Cyclobalanopsis glauca | China | aggregated, pale brown, ampulliform or cylindrical | brown, smooth, globose to ellipsoid | 8.0–12.0 | [40] |
| A. septata | Poaceae | China | solitary, integrated, branched, ampulliform, cylindrical, brown | brown, smooth, guttulate, globose | 8.0–11.0 (−13.0) | [40] |
| A. chromolaenae | Poaceae | Thailand | basauxic, broadly filiform to ampulliform, aggregated, hyaline, smooth, elongated, | irregular arrangement, pale brown, smooth, globose | 4.0–6.0 × 4.5–6.5 | [41] |
| A. cordylines | Cordyline fruticosa | China | erect, aggregated into clusters, hyaline, smooth, lageniform | olivaceous, smooth to finely roughened, subglobose, ellipsoid | 15.0–19.0 × 12.5–18.5 | [42] |
| A. gaoyouensis | Poaceae | China | aggregated, smooth, short and wide | brown, smooth, granular, globose to elongate ellipsoid | 5.0–8.0 | [43] |
| A. qinlingensis | Poaceae | China | aggregated in clusters on hyphae, smooth, short | brown, smooth, granular, globose to suborbicular | 5.0–8.0 | [43] |
| A. guiyangensis | Poaceae | China | solitary, integrated, branched, ampulliform, cylindrical, hyaline | brown, smooth, guttulate, globose to ellipsoid |
10.0–13.0 × 7.0–10.5 | [44] |
| A. hydei | Trachycarpus fortune | China | aggregated, brown, smooth, subcylindrical to doliiform to lageniform | brown, finely roughened, globose | (15.0–) 17.0–19.0 (–22.0) | [45] |
| A. kogelbergensis | Poaceae | south Africa | aggregated, pale brown, smooth, doliiform to subcylindrical | globose to ellipsoid | 9.0–10.0 × 7.0–8.0 | [45] |
| A. malaysiana | Macaranga hullettii | Malaysia | aggregated, hyaline, pale brown, smooth, doliiform to clavate to ampulliform | brown, smooth, globose | 5.0–6.0 | [45] |
| A. ovata | Poaceae | China | pale brown, smooth, aggregated, ampulliform | broadly ellipsoid, medium brown, finely roughened | 18.0–20.0 | [45] |
| A. pseudosinensis | Poaceae | Netherlands | doliiform or subcylindrical, pale brown, smooth | brown, smooth, ellipsoid, | 8.0–10.0 × 7.0–10.0 | [45] |
| A. pseudospegazzinii | Macaranga hullettii | Malaysia | aggregated, brown, smooth, ampulliform with elongated neck | brown, guttulate, roughened, globose | 8.0–9.0 | [45] |
| A. vietnamensis | Macaranga hullettii | Malaysia | aggregated, pale brown, smooth, doliiform to clavate, ampulliform | aggregated, brown and globose | 5.0–6.0 | [45] |
| A. xenocordella | soil | Austria | aggregated, brown, verruculose, globose to clavate to doliiform, | brown, smooth, guttulate, globose to ellipsoid | (7.0–) 9.0–10.0 (–11.0) | [45] |
| A. hyphopodii | Poaceae | China | basauxic, cylindrical, discrete, with verrucose wall |
globose, dark brown, smooth, truncate scar, with a longitudinal, germ-slit | 4.0–6.0 × 2.0–3.5 | [46] |
| A. locuta-pollinis | honey bee colonies | China | pale brown, smooth, subglobose to ampulliform to doliiform | pale brown with hyaline equatorial rim, smooth, globose | 5.5–9.0 × 4.5–8.0 | [47] |
| A. marianiae | Poaceae | Spain | monoblastic, integrated, terminal, intercalary, cylindrical | brown, solitary | (11.0–) 12.1–13.5 (–18.0) | [48] |
| A. montagnei | Arundo micrantha | Spain | doliiform to lageniform or ampulliform, hyaline | ellipsoidal to ovoid, smooth to finely roughened, with an equatorial germ-slit of paler pigment | (9.0–) 10.3–11.3 (–12.0) | [48] |
| A. minutispora | soil | Korea | erect, ellipsoid to ovoid, hyaline, pale brown to umber in color, and smooth | brown, finely roughened, ellipsoidal to ovoid, thick, solitary or aggregated, irregular dot-like structures | 5.7–8.2 × 4.6–7.0 | [49] |
| A. mori | Morus australis | China | pale yellow, smooth or finely roughened, subcylindrical to doliiform | globose, dark brown, smooth, with a basal scar, occasionally with germ-slit | 4.5–5.5 × 4.0–5.0 | [50] |
| A. neogarethjonesii | Poaceae | China | basauxic, cylindrical, discrete, smooth-walled | globose, dark brown, smooth, with a truncate basal scar | 20–35 × 15–30 | [51] |
| A. paraphaeosperma | Poaceae | Thailand | basauxic, aggregated, hyaline, smooth, elongated, conical | brown, smooth, granular, globose to ellipsoid | 10.0–19.0 | [52] |
| A. phyllostachydis | Poaceae | China | holoblastic, monoblastic, cylindrical, hyaline to pale brown, smooth, thin-walled | globose, irregular, pale brown, guttulate, olive to dark brown, with a germ-slit, smooth | 25.0–35.0 × 20.0–25.0 | [53] |
| A. rasikravindrae | soil | Norway | mononematous, hyaline, straight or flexuous, thin-walled, unbranched, septate, smooth | lenticular, ovoid | 10.0–15.0 × 6.0–10.5 | [54] |
| A. stipae | Poaceae | Spain | aggregated, pale brown, smooth, ampulliform | red-brown, thick-walled, smooth, eguttulate, with lateral germ-slit, often with a pronounced hilum | 6.5–10.5 × 6.0–9.0 | [54] |
| A. sasae | Poaceae | Netherlands | discrete, subcylindrical, subhyaline, proliferating sympodially, smooth to finely verruculose, holoblastic | numerous, aseptate, subglobose, thick-walled, smooth, with a lateral hyaline germ-slit | (16.0–) 17.0–18.0 (–20.0) × (15.0–) 16.0–17.0 (–19.0) | [55] |
| A. setariae | Poaceae | China | erect, hyaline to pale brown, smooth | globose, oval or irregular, brown, guttulate, with a longitudinal germ-slit | 7.5–10.5 | [56] |
| A. setostroma | Poaceae | China | micronematous, holoblastic, monoblastic, hyaline, cylindrical, flexible, discrete, aseptate, smooth | acrogenous, dark brown, obovoid, septate, smooth, multi-guttulate, with a scar | 18–20 × 15–19 | [57] |
| A. sorghi | Poaceae | Brazil | aggregated, hyaline, cylindrical to subcylindrical | brown, smooth, globose, subglobose, with a longitudinal germ-slit | 6.0–8.0 × 6.0–10.0 | [58] |
| A. adinandrae | Adinandra glischroloma | China | pale green, cylindrical, septate, flexuous | smooth, rounded to ovoid, green to dark brown | 7.5–12.4 × 6.3–10.5 | This study. |
| A. bawanglingensis | Poaceae | China | pale green, polyblastic, cylindrical, septate, verrucose, flexuous | smooth, green to dark brown, rounded to ovoid, globose to subglobose, | 6.4–7.7 × 5.3–7.1 | This study. |
| A. piptatheri | Poaceae | China | pale green becoming brown, polyblastic, cylindrical, septate, verrucose, flexuous | smooth, rounded to ovoid, globose, green to dark brown | 6.2–7.6 × 4.9–7.2 | This study. |
| A. machili | Machilus nanmu | China | pale green, becoming brown, polyblastic, cylindrical, septate, verrucose, flexuous, | smooth, rounded to ovoid, globose, green to dark brown, | 7.1–9.5 × 5.6–8.8 | This study. |
Notes: the species information described in this study is marked in bold.
4. Discussion
In this study, three new species, viz., Apiospora adinandrae sp. nov., A. bawanglingensis sp. nov. and A. machili sp. nov., are introduced and described based on their morphological characters and phylogenetic status. A. bawanglingensis was collected from diseased leaves of Indocalamus longiauritus in Bawangling National Forest Park, Hainan Province, China. A. adinandrae and A. machili were collected from Wuyi Mountain, Fujian Province, China; the host of the former was Adinandra glischroloma and the latter was Machilus nanmu.
Phylogenetically, most studies have used phylogenetic analyses of ITS, LSU, TUB2 and TEF1α sequence data to identity Apiospora species. Since the ITS and LSU gene regions are relatively conserved, the TUB2 and TEF1α gene regions have played an important role in the species identification of Apiospora [59]. In the present study, we constructed phylogenetic trees for each gene of ITS, LSU, TEF1α and TUB2 as Supplementary Materials to show the congruence (Figures S1–S4). The phylogenetic analyses showed Apiospora adinandrae sp. nov., A. bawanglingensis sp. nov. and A. machili sp. nov. all as a monophyletic clade (Figure 1), and the comparisons of both ITS and LSU showed small differences. In the A. adinandrae clade, the A. adinandrae is distinguished from A. aurea by 6/590, 4/831, 29/426 and 16/442 nucleotides; from A. hydei by 7/589, 3/831, 29/427 and 21/442; and from A. cordylines by 6/574, 0/831, 28/433 and 22/442 in the ITS, LSU, TEF1α and TUB2 sequences, respectively. In the A. bawanglingensis clade, A. bawanglingensis is distinguished from A. piptatheri by 16/583, 6/829, 39/452 and 0/449 characters, respectively. In the A. machili clade, the A. machili is distinguished from A. setariae by 20/588, 0/830, 0/437 and 0/441 characters and from A. jiangxiensis by 13/588, 2/830, 19/437 and 20/441 in the ITS, LSU, TEF1α and TUB2 sequences, respectively.
The asexual morphology of the species in this study is consistent with the basic characteristics of the genus. Morphologically, the biological relationship between Apiospora and Arthrinium has been controversial due to their similar morphological characteristics in having basauxic conidiogenesis [57]. Although some morphological features of Arthrinium species are difficult to observe in Apiospora, such as the conidiophores having black thick septa, and it is still difficult to define the boundary between Apiospora and Arthrinium by the asexual form alone [4,58]. It has been demonstrated that ascogenous Apiospora can reproduce the mycelial asexual state on artificial media such as PDA and MEA [57,60]. Despite this cultural and molecular evidence of asexual to sexual transition, the sexual morphology of Apiospora is still rarely reported [61].
Ecologically, Apiospora is widely distributed in subtropical, tropical, temperate and even cold regions, including Africa, America, Asia, Australia, and Europe, according to reported data [13,15]. As endophytes, plant pathogens and humus, Apiospora is ubiquitous in a wide range of terrestrial environments, such as soil, atmosphere and even marine substrates, but its main hosts are still plants, especially Poaceae [60]. Of all the Apiospora species that have been reported, more than 60% of the hosts are Poaceae, of which about half are from bamboos [59]. The new species A. bawanglingensis in this paper was also collected from Indocalamus longiauritus, which further enriches the diversity of bamboo fungi. Based on the existing statistical data of the USDA fungal database (https://nt.ars-grin.gov/fungaldatabases/, accessed on 10 November 2023.) and the collation of related literature published later on in the genus Apiospora, only 14 records of the genus Apiospora were isolated from woody plants (trees, shrubs, small shrubs), accounting for less than 10% of the total records, and about half of these hosts are Arecaceae. The strains A. adinandrae and A. machili in this study were isolated from Adinandra glischroloma and Machilus nanmu, respectively, which is the first time that Apiospora has been isolated from non-Poaceae hosts in China after Lu, B. (2000) [62].
Based on morphological and phylogenetic analysis, our study identified three new species in which only asexual morphology was found. Apiospora adinandrae sp. nov., A. bawanglingensis sp. nov. and A. machili sp. nov. are morphologically similar to their sister taxa, with significant differences in sequence. A. bawanglingensis was collected from Indocalamus longiauritus. Because most fungi on bamboo were not pathogens and there was no obvious plaque on the host, we believe for the time being that A. bawanglingensis is not a pathogen, and its pathogenicity still needs further study. A. adinandrae and A. machili were isolated from Adinandra glischroloma and Machilus nanmu, respectively. Based on the existing statistical data of the USDA fungal database, this is the first time that Apiospora has been found on Adinandra glischroloma and Machilus nanmu, enriching the host diversity of Apiospora.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof10010074/s1, Table S1. Species and GenBank accession numbers of DNA sequences used in this study. Figure S1. The ITS sequences maximum likelihood tree. Figure S2. The LSU sequences maximum likelihood tree. Figure S3. The TEF1α sequences maximum likelihood tree. Figure S4. The TUB2 sequences maximum likelihood tree.
Author Contributions
Conceptualization, X.L.; methodology, X.L.; software, X.L.; validation, Z.Z.; formal analysis, Z.Z.; investigation, Z.Z.; resources, S.W.; data curation, S.W.; writing—original draft preparation, S.W.; writing—review and editing, Z.Z.; visualization, Z.Z.; supervision, X.Z.; project administration, X.Z.; funding acquisition, X.Z. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable for studies involving humans or animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequences from the present study were submitted to the NCBI database (https://www.ncbi.nlm.nih.gov/, accessed on 20 November 2023) and the accession numbers were listed in Table S1.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by National Natural Science Foundation of China (no. 32300011), Ji’nan City’s ‘New University 20 Policies’ Initiative for Innovative Research Teams Project (202228028), and Key Technological Innovation Program of Shandong Province, China (2022CXGC020710).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Saccardo P.A. Conspectus generum pyrenomycetum italicorum additis speciebus fungorum Venetorum novis vel criticis, systemate carpologico dispositorum. Atti Della Soc. Veneto-Trent. Sci. Nat. 1875;4:77–100. [Google Scholar]
- 2.Hyde K.D., Fröhlich J., Taylor J.E. Fungi from palms. XXXVI. Reflections on unitunicate ascomycetes with apiospores. Sydowia. 1998;50:21–80. [Google Scholar]
- 3.Jiang N., Voglmayr H., Ma C.Y., Xue H., Piao C.G., Li Y. A new Arthrinium-like genus of Amphisphaeriales in China. MycoKeys. 2022;92:27–43. doi: 10.3897/mycokeys.92.86521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pintos Á., Alvarado P. Phylogenetic delimitation of Apiospora and Arthrinium. Fungal Syst. Evol. 2021;7:197–221. doi: 10.3114/fuse.2021.07.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis M.B. Dematiaceous Hyphomycetes. VI. Volume 103. Commonwealth Mycological Institute; Kew, UK: 1965. pp. 1–46. Mycological Papers. [Google Scholar]
- 6.Réblová M., Miller A.N., Rossman A.Y., Seifert K.A., Crous P.W., Hawksworth D.L., Abdel-Wahab M.A., Cannon P.F., Daranagama D.A., De Beer Z.W., et al. Recommendations for competing sexual-asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales) IMA Fungus. 2016;7:131–153. doi: 10.5598/imafungus.2016.07.01.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunze G., Schmidt J.C. Neue Arten. Mykol. Hefte. 1817;1:65–92. [Google Scholar]
- 8.Willdenow C.L. Caroli a Linné Species Plantarum. Volume 6 GC Nauk; Berlin, Germany: 1824. [Google Scholar]
- 9.von Höhnel F.X.R. Über die Gattung Arthrinium Kunze. Mitteilungen aus dem Bot. Inst. Tech. Hochsch. Wien. 1925;2:9–16. [Google Scholar]
- 10.Cooke W.B. The genus Arthrinium. Mycologia. 1954;46:815–822. doi: 10.1080/00275514.1954.12024418. [DOI] [Google Scholar]
- 11.Pintos Á., Alvarado P., Planas J., Jarling R. Six new species of Arthrinium from Europe and notes about A. caricicola and other species found in Carex spp. hosts. MycoKeys. 2019;49:15–48. doi: 10.3897/mycokeys.49.32115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S.J., Zhuang T.H. Binding site of toxic protein from Arthrinium phaeospermum on plasmalemma of hybrid bamboo. J. Zhejiang Univ. Agric. Life Sci. 2012;38:355–361. [Google Scholar]
- 13.Kwon S.L., Cho M., Lee Y.M., Kim C., Lee S.M., Ahn B.J., Lee H., Kim J.J. Two Unrecorded Apiospora Species Isolated from Marine Substrates in Korea with Eight New Combinations (A. piptatheri and A. rasikravindrae) Mycobiology. 2022;1:46–54. doi: 10.1080/12298093.2022.2038857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu R.Y., Li D.H., Zhang Z.X., Liu S.B., Liu X.Y., Wang Y.X., Zhao H., Liu X.Y., Zhang X.G., Xia J.W., et al. Morphological and phylogenetic analyses reveal two new species and a new record of Apiospora (Amphisphaeriales, Apiosporaceae) in China. MycoKeys. 2023;95:27–45. doi: 10.3897/mycokeys.95.96400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian X., Karunarathna S.C., Mapook A., Promputtha I., Xu J., Bao D., Tibpromma S. One New Species and Two New Host Records of Apiospora from Bamboo and Maize in Northern Thailand with Thirteen New Combinations. Life. 2021;11:1071. doi: 10.3390/life11101071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai D., Phookamsak R., Wijayawardene N., Li W., Bhat D., Xu J., Taylor J., Hyde K., Chukeatirote E. Bambusicolous fungi. Fungal Divers. 2017;82:1–105. doi: 10.1007/s13225-016-0367-8. [DOI] [Google Scholar]
- 17.Wang M., Tan X.M., Liu F., Cai L. Eight new Arthrinium species from China. MycoKeys. 2018;34:1–24. doi: 10.3897/mycokeys.34.24221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma R., Kulkarni G., Sonawane M.S., Shouche Y.S. A new endophytic species of Arthrinium Apiosporaceae from Jatropha podagrica. Mycoscience. 2013;55:118–123. doi: 10.1016/j.myc.2013.06.004. [DOI] [Google Scholar]
- 19.Dai D.Q., Jiang H.B., Tang L.Z., Bhat D.J. Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere. 2016;7:1332–1345. doi: 10.5943/mycosphere/7/9/7. [DOI] [Google Scholar]
- 20.Senanayake I.C., Bhat J.D., Cheewangkoon R., Xie N. Bambusicolous Arthrinium Species in Guangdong Province, China. Front. Microbiol. 2020;11:602773. doi: 10.3389/fmicb.2020.602773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z.X., Zhang J., Li D.H., Xia J.W., Zhang X.G. Morphological and Phylogenetic Analyses Reveal Three New Species of Pestalotiopsis (Sporocadaceae, Amphisphaeriales) from Hainan, China. Microorganisms. 2023;11:1627. doi: 10.3390/microorganisms11071627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doyle J.J., Doyle J.L., Brown A.H.D. Chloroplast DNA Phylogenetic Affinities of Newly Described Species in Glycine (Leguminosae: Phaseoleae) Syst. Bot. 1990;15:466–471. doi: 10.2307/2419362. [DOI] [Google Scholar]
- 23.Guo L.D., Hyde K.D., Liew E.C.Y. Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytol. 2000;147:617–630. doi: 10.1046/j.1469-8137.2000.00716.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z.X., Liu R.Y., Liu S.B., Mu T.C., Zhang X.G., Xia J.W. Morphological and phylogenetic analyses reveal two new species of Sporocadaceae from Hainan, China. MycoKeys. 2022;88:171–192. doi: 10.3897/mycokeys.88.82229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White T.J., Bruns T., Lee S., Taylor F.J.R.M., Lee S.H., Taylor L., Shawe-Taylor J. Amplification and direct sequencing of fungal ribosomal rna genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press Inc.; New York, NY, USA: 1990. pp. 315–322. [DOI] [Google Scholar]
- 27.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Donnell K., Kistler H.C., Cigelnik E., Ploetz R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jewell L., Hsiang T. Multigene differences between Microdochium nivale and Microdochium majus. Botany. 2013;91:99–106. doi: 10.1139/cjb-2012-0178. [DOI] [Google Scholar]
- 30.Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. Gatew. Comput. Environ. Workshop. 2010;14:1–8. [Google Scholar]
- 31.Nylander J. MrModeltest V2. Program Distributed by the Author. Bioinformatics. 2004;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [Google Scholar]
- 32.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 34.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 35.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon S.L., Park M.S., Jang S., Lee Y.M., Heo Y.M., Hong J.H., Lee H., Jang Y., Park J.H., Kim C., et al. The genus Arthrinium (Ascomycota, Sordariomycetes, Apiosporaceae) from marine habitats from Korea, with eight new species. IMA Fungus. 2021;12:13. doi: 10.1186/s43008-021-00065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo Z.L., Hyde K.D., Liu J.K., Maharachchikumbura S.S.N., Jeewon R., Bao F.D., Bhat D.J., Lin C.G., Li W.L., Yang J., et al. Freshwater Sordariomycetes. Fungal Divers. 2019;99:451–660. doi: 10.1007/s13225-019-00438-1. [DOI] [Google Scholar]
- 39.Tang X., Goonasekara I.D., Jayawardena R.S., Jiang H.B., Li J.F., Hyde K.D., Kang J.C. Arthrinium bambusicola (Fungi, Sordariomycetes), a new species from Schizostachyum brachycladum in northern Thailand. Biodivers. Data J. 2020;8:e58755. doi: 10.3897/BDJ.8.e58755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng Y., Liu J.J., Lin C.G., Chen Y.Y., Xiang M.M., Liu Z.Y. Additions to the Genus Arthrinium (Apiosporaceae) From Bamboos in China. Front. Microbiol. 2021;12:661281. doi: 10.3389/fmicb.2021.661281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mapook A., Hyde K.D., McKenzie E.H.C., Jones E.B.G., Bhat D.J., Jeewon R., Stadler M., Samarakoon M.C., Malaithong M., Tanunchai B., et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed) Fungal Divers. 2020;101:1–175. doi: 10.1007/s13225-020-00444-8. [DOI] [Google Scholar]
- 42.Chen T.Z., Zhang Y., Ming X.B., Zhang Q. Morphological and phylogenetic resolution of Arthrinium from medicinal plants in Yunnan, including A. cordylines and A. pseudomarii spp. nov. Mycotaxon. 2021;136:183–199. doi: 10.5248/136.183. [DOI] [Google Scholar]
- 43.Jiang N., Li J., Tian C.M. Arthrinium species associated with bamboo and reed plants in China. Fungal Syst. Evol. 2018;2:1–9. doi: 10.3114/fuse.2018.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samarakoon M.C., Hyde K.D., Maharachchikumbura S.S.N., Stadler M., Jones E.B.G., Promputtha I., Suwannarach N., Camporesi E., Bulgakov T.S., Liu J.K. Taxonomy, phylogeny, molecular dating and ancestral state reconstruction of Xylariomycetidae (Sordariomycetes) Fungal Divers. 2022;112:1–88. doi: 10.1007/s13225-021-00495-5. [DOI] [Google Scholar]
- 45.Jiang H.B. Arthrinium setostromum (Apiosporaceae, Xylariales), a novel species associated with dead bamboo from Yunnan, China. Asian J. Mycol. 2019;2:254–268. doi: 10.5943/ajom/2/1/16. [DOI] [Google Scholar]
- 46.Senanayake I.C., Maharachchikumbura S.S.N., Hyde K.D., Bhat D.J., Jones E.B.G., McKenzie E.H.C., Dai D.Q., Daranagama D.A., Dayarathne M.C., Goonasekara I.D., et al. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes) Fungal Divers. 2015;73:73–144. doi: 10.1007/s13225-015-0340-y. [DOI] [Google Scholar]
- 47.Zhao Y.Z. Four new filamentous fungal species from newly-collected and hivestored bee pollen. Mycosphere. 2018;9:1089–1116. doi: 10.5943/mycosphere/9/6/3. [DOI] [Google Scholar]
- 48.Pintos Á., Alvarado P. New studies on Apiospora (Amphisphaeriales, Apiosporaceae): Epitypification of Sphaeria apiospora, proposal of Ap. marianiae sp. nov. and description of the asexual morph of Ap. sichuanensis. MycoKeys. 2022;92:63–78. doi: 10.3897/mycokeys.92.87593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das K., Lee S.Y., Choi H.W., Eom A.H., Cho Y.J., Jung H.Y. Taxonomy of Arthrinium minutisporum sp. nov., Pezicula neosporulosa, and Acrocalymma pterocarpi: New Records from Soil in Korea. Mycobiology. 2020;48:450–463. doi: 10.1080/12298093.2020.1830741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thambugala D.S.T.C., Silva E.G.A.J., Hyde I.P.K.D. Taxonomic and phylogenetic contributions to Celtis formosana, Ficus ampelas, F. septica, Macaranga tanarius and Morus australis leaf litter inhabiting microfungi. Fungal Divers. 2021;48:1–215. [Google Scholar]
- 51.Hyde K.D. Refined families of Sordariomycetes. Mycosphere. 2020;11:305–1059. doi: 10.5943/mycosphere/11/1/7. [DOI] [Google Scholar]
- 52.Hyde K.D., Hongsanan S., Jeewon R., Bhat D.J., McKenzie E.H.C., Jones E.B.G., Phookamsak R., Ariyawansa H.A., Boonmee S., Zhao Q., et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;80:1–270. doi: 10.1007/s13225-016-0373-x. [DOI] [Google Scholar]
- 53.Yang C.L., Xu X.L., Dong W., Wanasinghe D.N. Introducing Arthrinium phyllostachium sp. nov. (Apiosporaceae, Xylariales) on Phyllostachys heteroclada from Sichuan Province, China. Phytotaxa. 2019;406:91–110. doi: 10.11646/phytotaxa.406.2.2. [DOI] [Google Scholar]
- 54.Singh S.M., Yadav L.S., Singh P.N., Hepat R. Arthrinium rasikravindrii sp. nov. from Svalbard, Norway. Mycotaxon. 2013;122:449–460. doi: 10.5248/122.449. [DOI] [Google Scholar]
- 55.Crous P.W., Hernández-Restrepo M., Schumacher R.K., Cowan D.A., Maggs-Kölling G., Marais E., Wingfield M.J., Yilmaz N., Adan O.C.G., Akulov A., et al. New and Interesting Fungi. 4. Fungal Syst. Evol. 2021;7:255–343. doi: 10.3114/fuse.2021.07.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang N., Tian C.M. The holomorph of Arthrinium setariae sp. nov. (Apiosporaceae, Xylariales) from China. Phytotaxa. 2021;483:149–159. doi: 10.11646/phytotaxa.483.2.7. [DOI] [Google Scholar]
- 57.Yuan H.S., Lu X., Dai Y.C., Hyde K.D., Kan Y.H., Kušan I., He S.H., Liu N.G., Sarma V.V., Zhao C.L., et al. Fungal diversity notes 1277–1386: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2020;104:1–266. doi: 10.1007/s13225-020-00461-7. [DOI] [Google Scholar]
- 58.Crous P.W., Groenewald J.Z. A phylogenetic re-evaluation of Arthrinium. IMA Fungus. 2013;4:133–154. doi: 10.5598/imafungus.2013.04.01.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monkai J., Phookamsak R., Tennakoon D.S., Bhat D.J., Xu S., Li Q., Xu J., Mortimer P.E., Kumla J., Lumyong S. Insight into the Taxonomic Resolution of Apiospora: Introducing Novel Species and Records from Bamboo in China and Thailand. Diversity. 2022;14:918. doi: 10.3390/d14110918. [DOI] [Google Scholar]
- 60.Yan H., Jiang N., Liang L.Y., Yang Q., Tian C.M. Arthrinium trachycarpum sp. nov. from Trachycarpus fortunei in China. Phytotaxa. 2019;400:203–210. doi: 10.11646/phytotaxa.400.3.7. [DOI] [Google Scholar]
- 61.Zhao H.J. Bambusicolous fungi in Guangdong, China: Establishing Apiospora magnispora sp. nov. (Apiosporaceae, Amphisphaeriales) based on morphological and molecular evidence. Curr. Res. Environ. Appl. Mycol. 2023;13:1–15. doi: 10.5943/cream/13/1/1. [DOI] [Google Scholar]
- 62.Lu B., Hyde K.D., Ho W.H., Tsui K.M., Taylor J.E., Wong K.M., Zhou Y.D. Checklist of Hong Kong Fungi. Fungal Diversity Press; Hong Kong, China: 2000. p. 207. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences from the present study were submitted to the NCBI database (https://www.ncbi.nlm.nih.gov/, accessed on 20 November 2023) and the accession numbers were listed in Table S1.