Abstract
The characterization of natural fungal diversity impacts our understanding of ecological and evolutionary processes and can lead to novel bioproduct discovery. Russula and Lactarius, both in the order Russulales, represent two large genera of ectomycorrhizal fungi that include edible as well as toxic varieties. Based on morphological and phylogenetic analyses, including nucleotide sequences of the internal transcribed spacer (ITS), the 28S large subunit of ribosomal RNA (LSU), the second largest subunit of RNA polymerase II (RPB2), the ribosomal mitochondrial small subunit (mtSSU), and the translation elongation factor 1-α (TEF1-α) gene sequences, we here describe and illustrate two new species of Russula and one new species of Lactarius from southern China. These three new species are: R. junzifengensis (R. subsect. Virescentinae), R. zonatus (R. subsect. Crassotunicatae), and L. jianyangensis (L. subsect. Zonarii).
Keywords: Basidiomycota, Russulales, Russula, Lactarius, new species, morphological and phylogenetic analyses
1. Introduction
Fungi classified within the order Russulaceae include members of some of the most significant ectomycorrhizal genera found in almost all forest ecosystems, spanning across temperate, subtropical, and tropical regions [1]. The proliferation and ecological relevance of Russulaceae are evident in the lush forests of southern China and various other regions dispersed throughout South Asia. This fungal assemblage plays a pivotal role in mycorrhizal associations, contributing significantly to the vitality and sustainability of forest ecosystems [2,3]. Russulaceae establish intimate associations with various host plants [4,5]. In addition, these fungi have significant medicinal, nutritional, and bioremediation value, including as resources for novel drug discovery [6,7,8]. Fujian, a coastal province in southern China, is surrounded by mountains on three sides and the sea on the other. The subtropical monsoon climate in this region results in relatively warm, short winters and long, rainy summers compared to northern China [9,10]. Botanically, Fujian is positioned at the southernmost end of the Sino-Japanese Floristic Region and faces Taiwan across the sea, the latter of which belongs to the Indo-Malay Region [11]. The main mountain ranges in Fujian include the Wuyi, Shanling, Jiufeng, and Tailao (elevation of ~200–2158 m), which house subtropical evergreen broadleaf forests, mixed coniferous and broadleaf forests, and South Asian tropical rainforests. The major tree species in Fujian comprise Masson’s pine, bamboo groves, willow trees, banyans, and camphor [12], among which the Chinese fir, Chinese yew, Fujian pine, and Chinese swamp cypress are indigenous to Fujian. The diverse and unique local tree species found in Fujian, coupled with the warm and humid climate, are likely important factors conducive to the proliferation of their associated Russulaceae fungi.
The genus Russula Pers. (Russulaceae, Russulales, Basidiomycota) was established by Persoon in 1796 [13]. Members of this genus often constitute crucial components of forest ecosystems via their extensive associations with plants, and also likely as a food source (their fruiting bodies/mushrooms) for a variety of animals [14]. Indeed, a number of Russula species are globally recognized as edible fungi [15] and have displayed promising (biopharmaceutical) properties with respect to possessing anticancer and antioxidative activities [16,17]. The morphological classification system for Russula is characterized by brightly colored fragile caps, brittle context, amyloid warty spores, abundant sphaerocysts in a heteromerous trama, an absence of latex, and simple-septate hyphae [18,19]. The documented number of species cataloged within the genus Russula currently surpasses > 2000, with their fruiting bodies encompassing a vast array of variations in color, morphology, and anatomical characteristics. However, due to the substantial variability exhibited within this taxonomic group, many species still pose considerable challenges in terms of their accurate identification and differentiation. This complexity underscores the likelihood of new species awaiting detection and classification through attempts to compare molecular phylogenetic reconstruction with modern infrageneric classification [20]. Consequently, challenges persist in differentiating and taxonomically categorizing Russula species within fungal surveys and ethnopharmacological investigations [21].
The genus Lactarius also belongs to the family Russulaceae in the order Russulales [22]. In traditional classification, all species that exude latex (or “milk”, hence the term “milkcap” fungi) were grouped under the genus Lactarius. Buyck et al. [22,23] separated Lactifluus and L. furcatus Coker from the genus Lactarius, establishing a new genus named Lactarius sensu novo, which is mainly classified into three subgenera: L. subg. Lactarius, L. subg. Plinthogalus (Burl.) Hesler & A.H. Sm., and L. subg. Russularia (Fr.) Kauffman. Although the exploration of extensive fungal resources has led to the identification of several dozen Russulaceae species across various regions [24,25,26,27], investigations concerning this genus in southern China remain inadequately addressed, with the continual discovery of new species [28,29].
During an exploration aimed at delineating the diversity and geographical distribution of Russula in China, a series of intriguing samples was gathered within Fujian province, China. These isolates displayed characteristics that did not correspond to any known species within the genus. Employing both morphological and molecular phylogenetic analyses, we identify three new species within the Russulaceae family. We present detailed descriptions of these newfound species, complemented by illustrations elucidating their distinctive morphological attributes.
2. Materials and Methods
2.1. Collections and Morphological Analyses
Fresh fruiting bodies of two unknown (putative members of the Russula) mushrooms were collected from the Junzifeng National Nature Reserve, and one from Jiufeng Mountain, Jianyang (putative member of the Lactarius), in the Fujian Province, China, in August 2021. These specimens were collected during field expeditions focused on fungi. Images of the fresh fruiting bodies were captured using a Canon (Tokyo, Japan) EOS 6D Mark II camera. The meticulous documentation of their macroscopic attributes involved the careful examination of fresh samples in their natural diurnal environment. Comprehensive records encompassing macroscopic characteristics and habitat specifics were meticulously collated from collection records and accompanying visual documentation, adhering to the conventions of mycological taxonomic research. To ensure the permanent preservation of specimens, one crucial step was a dehydration process, during which the specimens underwent desiccation within a drying oven set at 45 °C. This meticulous procedure persisted until the moisture content of the fruiting bodies was diligently reduced to below 10%, ensuring their suitability for long-term storage. Microsections of dried specimens were stained with a mixture of 5% potassium hydroxide (KOH) and 1% Congo red. A detailed illustration of the structure and ornamentation of the spores was carried out using a scanning electron microscope (ZEM15C, ZEPTOOLS, Tongling, China). Microscopic features were observed using a Leica microscope (DM2500, Wetzlar, Germany) at magnifications up to 100×. For the description of basidiospores, 20 basidiospores, in profile view, were measured. The basidium length excludes sterigmata. The notation (a-)b–c(-d) was used to represent the dimensions of basidiospores, where the range ‘b–c’ covered 90% or more of the measured values. ‘a’ and ‘d’ represent the extreme values. An average length/width ratio (Q value) was calculated from 20 spores, along with the standard deviation, reflecting the characteristics of the basidiospores. The specimens were deposited in the Herbarium Mycologicum Academiae Sinicae (HMAS) at the Institute of Microbiology, Chinese Academy of Sciences, with the specimen numbers HMAS 298099, HMAS 298100, HMAS 298101, HMAS 298102, HMAS 298103, and HMAS 298104. Taxonomic information on the new taxa was submitted to MycoBank (http://www.mycobank.org (accessed on 10 January 2024)).
2.2. DNA Extraction, PCR Amplification, and Sequencing
DNA was extracted using the Fungal DNA Mini Kit (OMEGA-D3390, Feiyang Biological Engineering Corporation, Guangzhou, China) following the manufacturer’s protocol. Briefly, 100 mg of starting material (fruiting body) was used for DNA extraction. The amplification of the nucleotide sequences of the internal transcribed spacer (ITS), 28S large subunit regions of ribosomal DNA (LSU), the translation elongation factor 1-α (TEF1-α), the ribosomal mitochondrial small subunit (mtSSU), and second largest RNA polymerase II regions (RPB2) was conducted via polymerase chain reaction (PCR) using the primer pairs: ITS4/ITS5 [30], LROR/LR5 [31], TEF1-α [32], mtSSU [30], and RPB2-6F/RPB2-7cR [33], respectively. The PCR reaction volume was 25 μL, comprising 12.5 μL of 2× Rapid Taq Master Mix (Vazyme, Nanjing, China), 1 μL of each forward and reverse primer (10 μM) (Sangon, Shanghai, China), and 1 μL of template genomic DNA. The reaction mixture was adjusted to a total volume of 25 μL using distilled deionized water. Amplification products were visualized using 1% agarose gel electrophoresis. Sequencing was performed by Fuzhou Tsingke Company (Fuzhou, China) using bidirectional (double-stranded) sequencing.
2.3. Alignment and Phylogenetic Analyses
To construct the phylogenetic tree of Russulaceae, we utilized sequences obtained from six fungal strains and reference sequences for multi-locus phylogenetic analyses which were obtained from Rehner and Buckley [32], Chen et al. [34], Deng et al. [35], Buyck et al. [36], and Roy et al. [37]. The newly generated sequences were screened for similarity through a GenBank BLAST search. The ITS, LSU, mtSSU, RPB2, and TEF1-α sequences were aligned using the MAFFT v. 7.11 online tool (https://mafft.cbrc.jp/alignment/software/ (accessed on 23 December 2023)), followed by manual adjustments in MEGA 7.0. Phylogenetic analyses employed both maximum likelihood (ML) and Bayesian inference (BI) methods. ML analysis was conducted using RaxML-HPC2 on XSEDE v. 8.2.12 via the CIPRES Science Gateway portal, while BI analysis was performed using MrBayes on XSEDE v. 3.2.7a (https://www.phylo.org/ (accessed on 25 December 2023)). The consensus tree was constructed using FigTree v. 1.4.4 and further refined using Adobe Illustrator CS 6.0. Newly generated sequences from this study have been deposited in GenBank. Branches showing ML bootstrap support values (≥70) and Bayesian posterior probability (≥0.90) were considered significantly supported.
3. Results
3.1. Phylogenetic Analyses
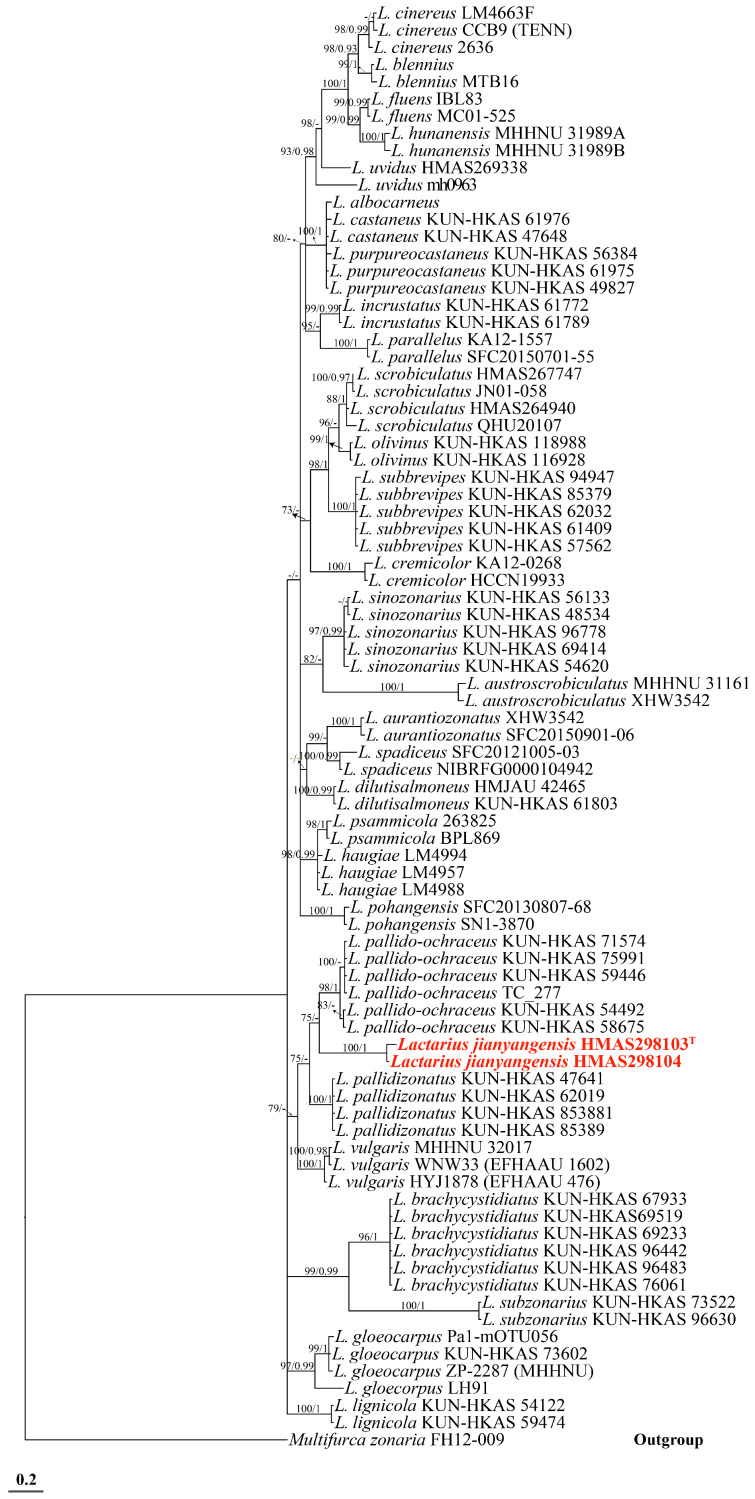
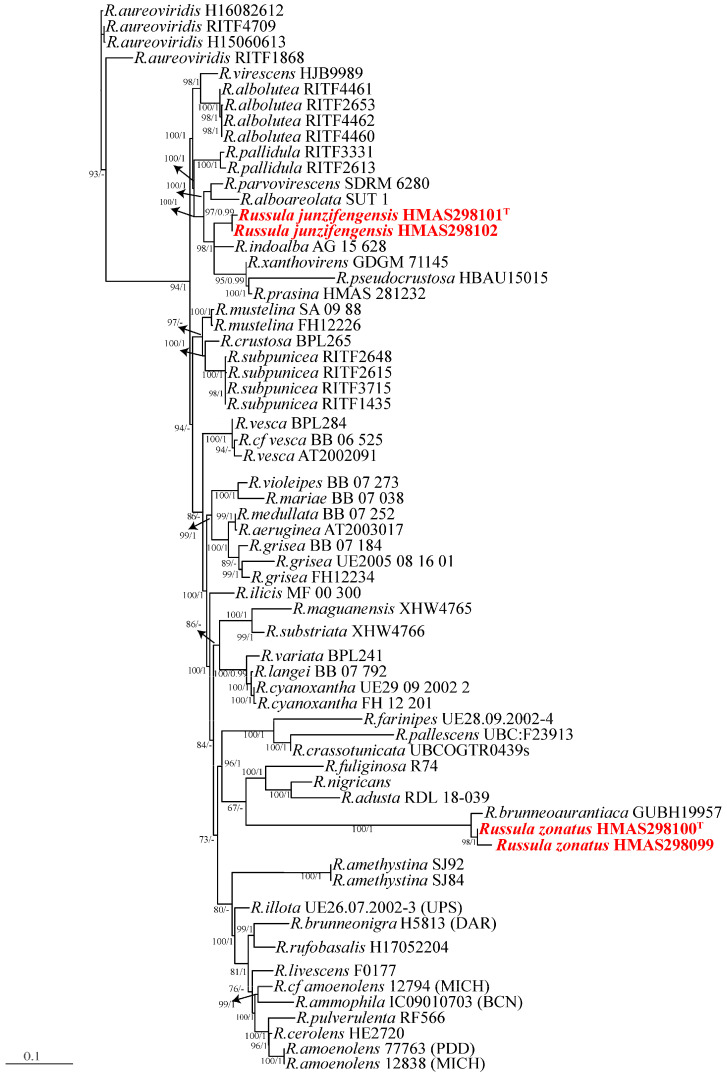
The multi-locus sequence matrix spans a length of 4350 bp. Its dataset comprises 700 bp of ITS, 890 bp of LSU, 1100 bp of TEF1-α, 800 bp of RPB2, and 860 bp of mtSSU. For the multi-locus region, the best substitution model for ITS and RPB2 in the BI analysis is SYM + G4, while for LSU and TEF1-α it is SYM + I + G4, and for mtSSU the best substitution model is GTR + F + G4. A total of 148 sequences, including newly generated ones, were deposited in the GenBank database (Table 1 and Table 2). Based on the foundational rank consistency of the phylogenetic topologies obtained from BI and ML analyses, only the ML trees are depicted in Figure 1 and Figure 2. The resulting phylogenetic trees demonstrate strong support for clades of the four new species in multi-locus phylogenetic analyses. These new species exhibit notable distinctions from known species (Figure 1 and Figure 2). Bootstrap and posterior probability values indicate robust support in multi-locus phylogeny for R. junzifengensis (from subsect. Virescentinae), R. zonatus (from subsect. Virescentinae), and L. jianyangensis (L. subsect. Zonarii), forming a distinct clade.
Table 1.
Species and specimens of Lactarius used for the molecular phylogenetic analyses.
| Taxon | Voucher | Location | GenBank Accession Number | |||
|---|---|---|---|---|---|---|
| ITS | nrLSU | RPB2 | TEF1 | |||
| L. albocarneus | - | China | KX441117 | KX441364 | KX442105 | - |
| L. aurantiozonatus | HCCN10589 | Korea | MH984993 | MH985118 | MH936950 | - |
| L. aurantiozonatus | SFC20150901-06 | Korea | MH984976 | MH985097 | MH936929 | - |
| L. austroscrobiculatus | MHHNU 31161 | China | OL770185 | - | - | - |
| L. austroscrobiculatus | XHW3542 | China | OL770183 | - | - | - |
| L. blennius | MTB16 | Germany | MN947353 | - | - | - |
| L. blennius | - | Sweden | AY606944 | - | - | - |
| L. brachycystidiatus | KUN-HKAS 67933 | China | MF508951 | - | - | - |
| L. brachycystidiatus | KUN-HKAS 96483 | China | MF508950 | - | - | - |
| L. brachycystidiatus | KUN-HKAS 96442 | China | MF508949 | - | - | - |
| L. brachycystidiatus | KUN-HKAS 69233 | China | MF508948 | - | - | - |
| L. brachycystidiatus | KUN-HKAS 76061 | China | MF508947 | - | - | - |
| L. brachycystidiatus | KUN-HKAS69519 | China | MF508952 | - | - | - |
| L. castaneus | KUN-HKAS 47648 | China | MF508962 | - | - | - |
| L. castaneus | KUN-HKAS 61976 | China | MF508961 | - | - | - |
| L. cinereus | LM4663F | Mexico | FJ348708 | - | - | - |
| L. cinereus | CCB9 (TENN) | USA | MF755272 | - | - | - |
| L. cinereus | 2636 | Canada | KJ705204 | - | - | - |
| L. cremicolor | KA12-0268 | Korea | MH985013 | - | QCH40198 | - |
| L. cremicolor | HCCN19933 | Korea | MH984972 | - | QCH40153 | - |
| L. dilutisalmoneus | HMJAU 42465 | China | MF152847 | - | - | - |
| L. dilutisalmoneus | KUN-HKAS 61803 | China | MF152846 | - | - | - |
| L. fluens | IBL83 | Poland | MZ410712 | - | - | - |
| L. fluens | MC01-525 | Denmark | AJ889961 | - | - | - |
| L. gloecorpus | LH91 | USA | GQ268638 | - | - | - |
| L. gloeocarpus | KUN-HKAS 73602 | China | OL770166 | - | - | - |
| L. gloeocarpus | ZP-2287 (MHHNU) | China | OL770165 | - | - | - |
| L. gloeocarpus | Pa1-mOTU056 | Japan | LC315865 | - | - | - |
| L. haugiae | LM4994 | Mexico | KT583642 | - | AOF41440 | - |
| L. haugiae | LM4988 | Mexico | KT583641 | - | AOF41439 | - |
| L. haugiae | LM4957 | Mexico | KT583640 | - | - | - |
| L. hunanensis | MHHNU 31989B | China | OL770172 | - | - | - |
| L. hunanensis | MHHNU 31989A | China | OL770171 | - | - | - |
| L. incrustatus | KUN-HKAS 61789 | China | MK675285 | - | - | - |
| L. incrustatus | KUN-HKAS 61772 | China | MK675284 | - | - | - |
| L. jianyangensis | HMAS298103T | China | OR835448 | OR826782 | OR915862 | OR887738 |
| L. jianyangensis | HMAS298104 | China | OR835446 | PP033514 | OR915863 | OR887739 |
| L. lignicola | KUN-HKAS 59474 | China | MF508946 | - | - | - |
| L. lignicola | KUN-HKAS 54122 | China | MF508945 | - | - | - |
| L. olivinus | KUN-HKAS 116928 | China | OL770196 | - | - | - |
| L. olivinus | KUN-HKAS 118988 | China | OL770195 | - | - | - |
| L. pallidizonatus | KUN-HKAS 85389 | China | MF508932 | - | - | - |
| L. pallidizonatus | KUN-HKAS 62019 | China | MF508931 | - | - | - |
| L. pallidizonatus | KUN-HKAS 85388 | China | MF508933 | - | - | - |
| L. pallidizonatus | KUN-HKAS 47641 | China | MF508930 | - | - | - |
| L. pallido-ochraceus | KUN-HKAS 71574 | China | MF508942 | - | - | - |
| L. pallido-ochraceus | KUN-HKAS 54492 | China | MF508941 | - | - | - |
| L. pallido-ochraceus | KUN-HKAS 58675 | China | MF508940 | - | - | - |
| L. pallido-ochraceus | TC_277 | China | MW722813 | - | - | - |
| L. pallido-ochraceus | KUN-HKAS 59446 | China | MF508943 | - | - | - |
| L. pallido-ochraceus | KUN-HKAS 75991 | China | MF508939 | - | - | - |
| L. parallelus | SFC20150701-55 | Korea | MH984953 | MH985072 | MH936904 | - |
| L. parallelus | KA12-1557 | Korea | MH984921 | MH985035 | MH936867 | - |
| L. pohangensis | SFC20130807-68 | Korea | MH985018 | MH985143 | MH936975 | - |
| L. pohangensis | SN1-3870 | China | LC622651 | - | - | - |
| L. psammicola | 263825 | USA | MK607513 | - | - | - |
| L. psammicola | BPL869 | USA | KY848507 | - | - | - |
| L. purpureocastaneus | KUN-HKAS 61975 | China | MF508965 | - | - | - |
| L. purpureocastaneus | KUN-HKAS 56384 | China | MF508964 | - | - | - |
| L. purpureocastaneus | KUN-HKAS 49827 | China | MF508963 | - | - | - |
| L. scrobiculatus | QHU20107 | China | OM970920 | - | - | - |
| L. scrobiculatus | HMAS264940 | China | KX441085 | KX441332 | KX442073 | - |
| L. scrobiculatus | JN01-058 | Thailand | KF432968 | - | - | - |
| L. scrobiculatus | HMAS267747 | China | KX441098 | MF893430 | ||
| L. sinozonarius | KUN-HKAS 69414 | China | MF508926 | - | - | - |
| L. sinozonarius | KUN-HKAS 56133 | China | MF508929 | - | - | - |
| L. sinozonarius | KUN-HKAS 48534 | China | MF508928 | - | 0 | - |
| L. sinozonarius | KUN-HKAS 96778 | China | MF508927 | - | - | - |
| L. sinozonarius | KUN-HKAS 54620 | China | MF508925 | - | - | - |
| L. spadiceus | SFC20121005-03 | Korea | MH985021 | MH985146 | MH936978 | - |
| L. spadiceus | NIBRFG0000104942 | Korea | MH984956 | MH985076 | MH936908 | - |
| L. subbrevipes | KUN-HKAS 85379 | China | MF508938 | - | - | - |
| L. subbrevipes | KUN-HKAS 61409 | China | MF508937 | - | - | - |
| L. subbrevipes | KUN-HKAS 57562 | China | MF508936 | - | - | - |
| L. subbrevipes | KUN-HKAS 62032 | China | MF508935 | - | - | - |
| L. subbrevipes | KUN-HKAS 94947 | China | MF508934 | - | - | - |
| L. subzonarius | KUN-HKAS 96630 | China | MF508960 | - | - | - |
| L. subzonarius | KUN-HKAS 73522 | China | MF508959 | - | - | - |
| L. uvidus | mh0963 | Sweden | AY606957 | AF325293 | - | - |
| L. uvidus | HMAS269338 | China | KX441140 | KX441387 | KX442128 | - |
| L. vulgaris | MHHNU 32017 | China | OL770178 | - | - | - |
| L. vulgaris | HYJ1878 (EFHAAU 476) | China | OL770175 | - | - | - |
| L. vulgaris | WNW33 (EFHAAU 1602) | China | OL770173 | - | - | - |
| Multifurca zonaria | FH12-009 | Thailand | KF432960 | - | - | - |
Superscript “T” denotes the type strain of the new species.
Table 2.
Species and specimens of Russula used for the molecular phylogenetic analyses.
| Taxon | Voucher | Location | GenBank Accession Number | ||||
|---|---|---|---|---|---|---|---|
| ITS | nrLSU | RPB2 | mtSSU | TEF1 | |||
| R. adusta | RDL 18-039 | Belgium | OM833079 | - | - | - | ON015965.1 |
| R. aeruginea | AT2003017 | Sweden | DQ421999 | DQ421999 | - | - | - |
| R. alboareolata | SUT-1 | Thailand | AF345247 | - | - | - | - |
| R. albolutea | RITF4460 | China: Chongqing | - | MW397121 | MW411341 | MW403834 | - |
| R. albolutea | RITF4461 | China: Yunnan | - | MW397122 | MW411342 | MW403835 | - |
| R. albolutea | RITF4462 | China: Yunnan | - | MW397123 | MW411343 | MW403836 | - |
| R. albolutea | RITF2653 | China | MT672478 | MW397120 | MW411340 | MW403833 | - |
| R. amethystina | SJ84 | Pakistan | KT953615 | - | - | - | - |
| R. amethystina | SJ92 | Pakistan | KT953616 | - | - | - | - |
| R. amoenolens | 12838 (MICH) | France | KF245510 | - | - | - | - |
| R. amoenolens | 77763 (PDD) | New Zealand | GU222264 | - | - | - | - |
| R. ammophila | IC09010703 (BCN) | Spain | MK112566 | MK108033 | - | - | - |
| R. aureoviridis | H15060613 | China | KY767808 | - | - | - | - |
| R. aureoviridis | RITF1868 | China | MW397096 | - | - | MW403842 | - |
| R. aureoviridis | H16082612 | China | KY767809 | MK881920 | - | MK882048 | MN617846 |
| R. aureoviridis | RITF4709 | China | MW646980 | MW646992 | - | MW647003 | MW650849 |
| R. brunneoaurantiaca | GUBH19957 | India | OP270714 | - | - | - | - |
| R. brunneonigra | H5813 (DAR) | Australia | EU019945 | - | - | - | - |
| R. cerolens | HE2720 | China | KC505578 | - | - | - | - |
| R. cf. amoenolens | 12794 (MICH) | USA | KF245512 | - | - | - | - |
| R. cf. vesca | BB 06.525 | Mexico | - | KU237465 | KU237751 | KU237309 | - |
| R. crassotunicata | UBCOGTR0439s | Canada | EU597082 | - | - | - | - |
| R. crustosa | BPL265 | USA: Tennessee | - | KT933826 | KT933898 | - | - |
| R. cyanoxantha | UE29.09.2002-2 | France | - | DQ422033 | DQ421970 | - | - |
| R. cyanoxantha | FH 12-201 | Germany | KR364093 | KR364225 | - | - | - |
| R. farinipes | UE28.09.2002-4 | Sweden | DQ421983 | - | - | - | - |
| R. fuliginosa | R74 | CZECH REPUBLIC | HG798529 | - | - | - | - |
| R. grisea | BB 07.184 | Slovakia | - | KU237509 | KU237795 | KU237355 | - |
| R. grisea | UE2005.08.16-01 | Sweden | DQ422030 | DQ422030 | - | - | - |
| R. grisea | FH12234 | Germany | KT934006 | KT933867 | - | - | - |
| R. ilicis | MF 00.300 | Italy | - | KU237595 | KU237880 | KU237443 | - |
| R. illota | UE26.07.2002-3 (UPS) | Sweden | DQ422024 | DQ422024 | DQ421967 | - | - |
| R. indoalba | AG 15-628 | India | KX234820 | - | - | - | - |
| R. junzifengensis | HMAS298101T | China | OR826832 | OR826833 | OR915864 | OR941507 | OR887742 |
| R. junzifengensis | HMAS298102 | China | OR880061 | OR880054 | OR915865 | OR941508 | OR887743 |
| R. langei | BB 07.792 | France | - | KU237510 | KU237796 | KU237356 | - |
| R. livescens | F0177 | China | GU371295 | - | - | - | - |
| R. maguanensis | XHW4765 | China | MH724918 | MH714537 | MH939990 | - | MH939983 |
| R. mariae | BB 07.038 | USA | - | KU237538 | KU237824 | KU237384 | - |
| R. medullata | BB 07.252 | Slovakia | - | KU237546 | KU237832 | KU237392 | - |
| R. mustelina | FH12226 | Germany | - | KT933866 | KT933937 | - | - |
| R. mustelina | SA 09.88 | Slovakia | - | KU237596 | KU237881 | KU237444 | - |
| R. nigricans | - | Germany | AF418607 | - | - | - | - |
| R. pallescens | UBC:F23913 | Canada | KJ146729 | - | - | - | - |
| R. pallidula | RITF2613 | China | - | MH027960 | MH091698 | MW403845 | - |
| R. pallidula | RITF3331 | China | - | MH027961 | MH091699 | MW403846 | - |
| R. parvovirescens | SDRM 6280 | USA | MK532789 | - | - | - | - |
| R. prasina | HMAS 281232 | China | MH454351 | - | - | - | - |
| R. pseudocrustosa | HBAU15015 | China | MT337520 | - | - | - | - |
| R. pulverulenta | RF566 (pers. herb.) | USA | AY061736 | - | - | - | - |
| R. rufobasalis | H17052204 | China | MH168570 | MK881947.1 | - | MK882075.1 | MT085585.1 |
| R. subpunicea | RITF1435 | China: Hunan | - | MW397126 | MW411346 | MW403839 | - |
| R. subpunicea | RITF2615 | China: Hunan | - | MW397127 | MW411347 | MW403840 | - |
| R. subpunicea | RITF3715 | China | MN833635 | MW397124 | MW411344 | MW403837 | - |
| R. subpunicea | RITF2648 | China | MN833638 | MW397125 | MW411345 | MW403838 | - |
| R. substriata | XHW4766 | China | MH724921 | MH714540 | MH939993 | - | MH939986 |
| R. variata | BPL241 | USA | - | KT933818 | KT933889 | - | - |
| R. vesca | BPL284 | USA | KT933978 | KT933839 | - | - | - |
| R. vesca | AT2002091 | Sweden | DQ422018 | DQ422018 | DQ421959 | - | - |
| R. violeipes | BB 07.273 | Slovakia | KU237534 | KU237820 | KU237380 | - | |
| R. virescens | HJB9989 | Belgium | DQ422014 | DQ421955 | - | - | |
| R. xanthovirens | GDGM 71145 | China | MG786056 | - | - | - | - |
| R. zonatus | HMAS298099 | China | OR826839 | OR826846 | OR915866 | OR941505 | OR887740 |
| R. zonatus | HMAS298100T | China | OR880062 | OR880056 | OR915867 | OR941506 | OR887741 |
Superscript “T” denotes the type strain of the new species.
Figure 1.
Phylogeny inferred from Lactarius multigene sequences (nrLSU, ITS, mtSSU, rpb2, and tef1-α) using Bayesian analysis. Support values in normal type are bootstrap support (BS, significant when ≥70%). Values in bold are Bayesian Posterior Probabilities (PP, significant when ≥0.95). The scale bar indicates the number of nucleotide substitutions per site. New species are highlighted in red. Arrows show the support values at the branching points. Superscript “T” denotes the type strain of the new species.
Figure 2.
Phylogeny inferred from Russula multigene sequences (nrLSU, ITS, mtSSU, rpb2, and tef1-α) using Bayesian analysis. Support values in normal type are bootstrap support (BS, significant when ≥70%). Values in bold are Bayesian Posterior Probabilities (PP, significant when ≥0.95). The scale bar indicates the number of nucleotide substitutions per site. New species are highlighted in red. Arrows show the support values at the branching points. Superscript “T” denotes the type strain of the new species.
Lactarius jianyangensis showed the greatest similarity to Lactarius pallido-ochraceus, with an additional 84 sequences from Lactarius collected to construct the tree (Table 1, Figure 1). L. jianyangensis exhibited the highest genetic similarity to L. pallido-ochraceus and clustered with two other species, L. vulgaris and L. pallidizonatus (Figure 1). However, despite clustering with these three species, the substantial phylogenetic distance between L. jianyangensis and the other members of this clade supports its classification as an independent species. Two putative new species within the Russula genus, Russula junzifengensis, formed a strongly supported cluster (BS 100%) and were notably distinct from other known species within the Virescentinae group. Russula junzifengensis clustered together with an unidentified sequence from China (voucher: HMAS250919), which served as the sister clade to R. indoalba, supported by 98% bootstrap support and a posterior probability of 1. R. zonatus clustered alongside R. brunneoaurantiaca and formed a clade sister to R. brunneoaurantiaca with a posterior probability of 1.
3.2. Taxonomy
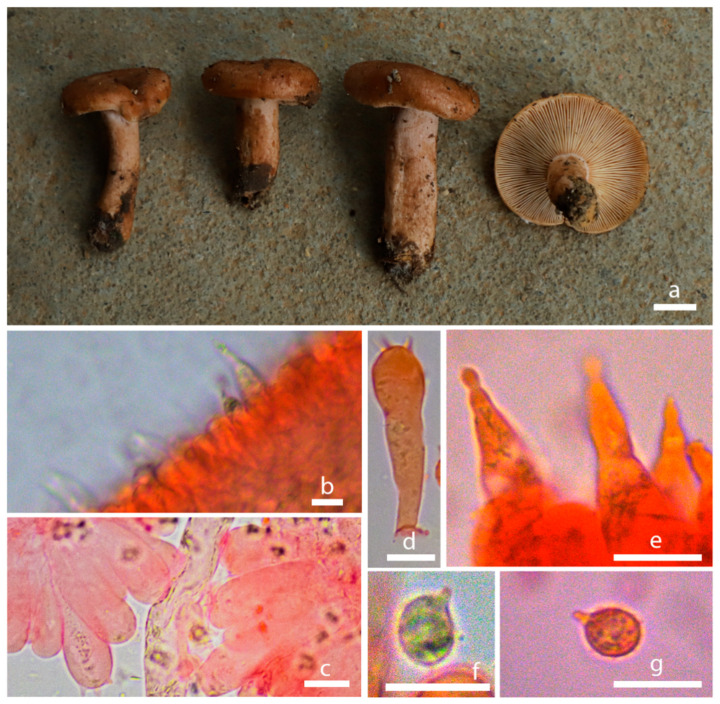
Russula zonatus S. Liu & Jun Z. Qiu, sp. nov. (Figure 3a,b and Figure 4).
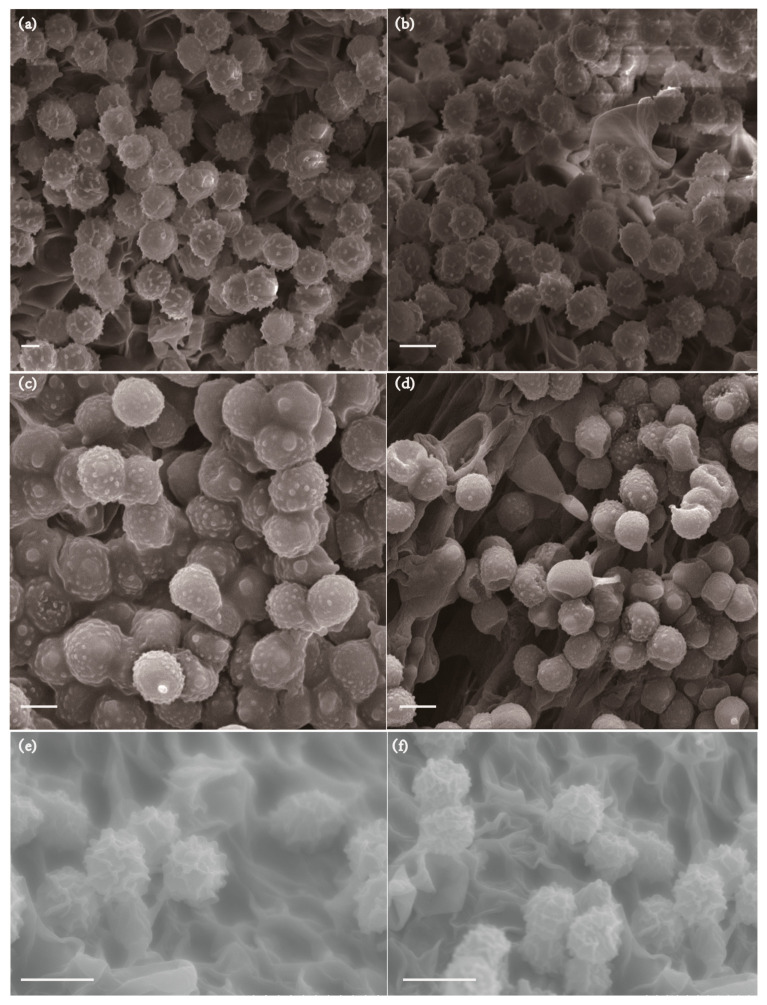
Figure 3.
SEM photos of basidiospores. (a,b) R. zonatus, (c,d) R. junzifengensis, and (e,f) L. jianyangensis. Scale bars: (a–f) = 10 μm.
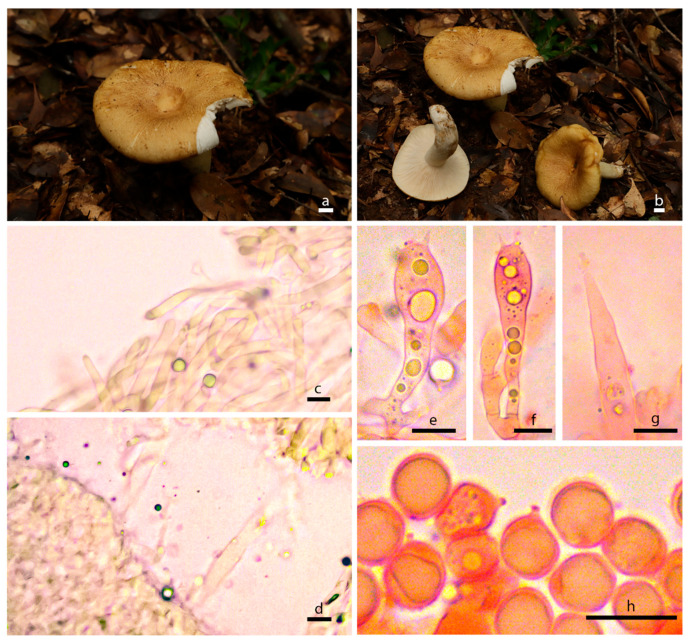
Figure 4.
Morphological characteristics of Russula zonatus (HMAS298100). (a,b) Basidiomata; (c) pileipellis in 5% KOH; (d) lamellae in 5% KOH. (e,f) Basidium in Congo Red reagent; (g) cystidium in Congo Red reagent; (h) basidiospores in Congo Red reagent; bars: (a,b) = 1 cm; (c–h) = 10 µm.
MycoBank: MB 851147.
Etymology: The epithet “zonatus” refers to the morphological feature of ring patterns on the surface.
Holotype: CHINA. Fujian Province, Mingxi County, Xiayang Town, Ziyun Village, in mixed forests, alt. 379 m, 26.34323138° N, 117.45805533° E, 30 August 2021, S. Liu and Jun Z. Qiu (holotype HMAS298100; paratype HMAS298099).
Description: Basidiocarps medium-sized to big. Pileus 7.5–11.9 cm in diam., convex-expanded to infundibuliform with a central depression and slight incurved margin, shallowly infundibuliform when mature, surface glabrous and dry with unclear or golden brown zone lines, no dark brown ring patterns on the surface, brown to grayish brown. Context 3 mm thick, satin white. Lamellae adnate, crowded, marble white, no forking, concolorous with the pileus when fully mature. Stipe 2.5 × 6–3.5 × 10 cm, central, equal, sometimes with fibrils, whitish or sub-concolorous with the pileus.
Basidiospores (6.4) 6.5–7.8 (8) × (5.2) 5.5–6.8 (7) µm (Q = (1.05) 1.06–1.19 (1.23), Q = 1.13 ± 0.05), broadly ellipsoid, with almost isolated warts, plage not amyloid. Basidia 32.5–49 × 10–13 µm, clavate, four-spored. Cystidium common, 30–50 × 5–7.5 µm, fusiform to lanceolate with campulitropal head. Lamella edge sterile, lamellae 8.74 µm thick.
Pileipellis duplex: gelatinous epicutis, 100–230 µm thick, hyphae 2–7 µm wide, hyaline to light yellow in 5% KOH, smooth. Hypodermium well developed, yellowish to lightly yellow intracellular pigment in 5% KOH.
Ecology and distribution: Gregarious in subtropical mixed forests (fagaceous forests or mixed forests with fagaceous trees). Known to inhabit Fujian Province, China.
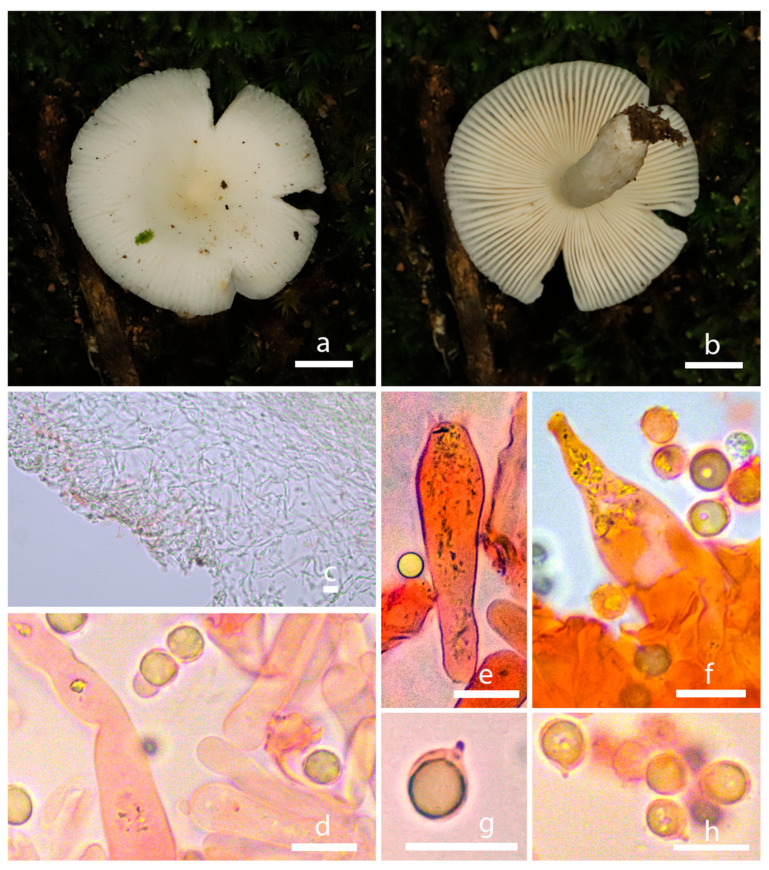
Russula junzifengensis S. Liu & Jun Z. Qiu, sp. nov. (Figure 3c,d and Figure 5).
Figure 5.
Morphological characteristics of Russula junzifengensis (HMAS298101). (a,b) Basidiomata; (c) pileipellis in 5% KOH; (d) lamellae in Congo Red reagent. (e) Basidium in Congo Red reagent; (f) cystidium in Congo Red reagent; (g,h) basidiospores in Congo Red reagent; bars: (a,b) = 1 cm; (c–h) = 10 µm.
MycoBank: MB 851146.
Etymology: Named after the Junzifeng Nature Reserve where the fungus was collected.
Holotype: CHINA. Fujian Province, Mingxi County, Junzifeng Nature Reserve, Xiafang Town, Zhushe Village, in mixed forests, alt. 410 m, 26.56490316° N, 117.03322856° E, 7 August 2021, S. Liu and Jun Z. Qiu (holotype HMAS298101; paratype HMAS298102).
Description: Basidiomata medium-sized, with a diameter of 40–60 mm. Initially hemispherical, later broadly convex to flat with a shallow depression, featuring a sub-transparently striate margin. The lamellae are densely crowded and sometimes slightly decurrent in mature and dry conditions, with a sharp, incurved, and even margin. The surface is glabrous, ranging from dry to slightly glutinous, presenting a satin white appearance, marble white at the center. In the mature stage, the central color turns to a shade of light yellow, pinard yellow, occasionally displaying maize yellow or light orange–yellow to capucine buff. The lamellae are adnate, densely packed, and of yellowish white color, without forking, becoming fragile and matching the pileus color when fully mature. The stipe measures 5.5 × 1.2 cm, central, cylindrical to slightly tapered upwards, rarely becoming subcylindrical to clavate, slightly narrowing towards the base, without an annulus. The stipe is white, appearing yellowish white, smooth in youth, later exhibiting fibrils on the surface. While young, it is full-bodied, eventually becoming hollow. The odor is indistinct.
Basidiospores (5.4) 6–7.5 (7.5) × (5.2) 5.5–6.5 (6.6) µm, [Q = (1.04) 1.05–1.25 (1.25), Q = 1.15 ± 0.06], ellipsoid, composed of small amyloid conical warts, mostly isolated, sometimes fused; indextrinoid, ornamentation small to medium-sized. Basidia 38.5–46.5 × 9–12 µm (smaller), clavate, four-spored, clavate, hyaline to light yellow in 5% KOH. Cystidia, 42 ×13.5 µm (smaller), thin-walled, hyaline in 5% KOH. Pileipellis well developed, separated from the surrounding spherocytes of the context, yellowish to lightly yellow intracellular pigment in 5% KOH.
Ecology and distribution: Gregarious in subtropical mixed forests (solitary or gregarious in Fagaceae forest). Known to inhabit Fujian Province, China.
Lactarius jianyangensis S. Liu & Jun Z. Qiu, sp. nov. (Figure 3e,f and Figure 6).
Figure 6.
Morphological characteristics of Lactarius jianyangensis (HMAS298103). (a) Basidiomata; (b) pileipellis in Congo Red reagent (c) lamellae in Congo Red reagent. (d) Basidium in Congo red reagent; (e) cystidium in Congo red reagent; (f,g) basidiospores in Congo red reagent; bars: (a) = 1 cm; (b–g) = 10 µm.
MycoBank: MB 851149.
Etymology: Named after Jianyang District, where the fungus was collected.
Holotype: CHINA. Fujian Province, Nanping City, Jianyang District, in mixed forests, alt 841 m, 27.34220872° N, 118.16609715° E, 19 August 2021, S. Liu and Jun Z. Qiu (holotype HMAS298103; paratype HMAS298104).
Description: Basidiomata with a small size. Pileus 25–45 mm broad, initially hemispheric, becoming plano-convex and planate when mature, convex with inrolled margin, shallowly infundibuliform when mature, surface greasy when wet, aniline yellow, bittersweet pink, Titian red to agate, sometimes center salmon-orange or Mars yellow with a raw sienna margin, margin glabrous, sub-transparently striate. Context 3–4 mm thick, whitish to brown. The lamella 1–2 mm broad, Mikado orange to cadmium orange when young, xanthine orange, amber brown when mature, concolorous with the pileus when fully mature, sub-crowded to crowded, unequal length and extended. Additionally, the flesh of Lactarius has an aroma. Stipe 35–40 × 7–10 mm, central or tapering downwards, sometimes with longitudinal grooves, surface smooth, greasy, with scattered pits, whitish or sub-concolorous with the pileus, the end of the stipe was slightly enlarged, succulent and hollow, latex white or watery–milky.
Basidiospores (5.99) 6–7.7 (7.8) × (4.8) 4.9–6.3 (6.44) μm, [Q = (1.07) 1.09–1.34 (1.36), Q = 1.20 ± 0.09], broadly ellipsoid, surface has protuberance ridge to reticulate pattern, colorless to hyaline in KOH. Basidia 30.55–39.03 × 8.21–11.32 μm, four-spored, narrowly clavate, colorless to hyaline in KOH, sterigmata 2.84–3.87 μm. Clamp connections abundant in all tissues. The head of pleuromacrocystidia is warping, trama 4.53–4.81 μm.
Ecology and distribution: Gregarious in subtropical fagaceous forests. Known to inhabit Fujian Province, China.
4. Discussion
All Russula and Lactarius species characterized thus far form ectomycorrhizal symbioses with higher plants and trees, and both genera contain cosmopolitan as well as more host-specific members, with both edible and toxic species having been identified [20,38]. Lactarius is characterized by the production of latex, although the genus has now been separated into two (Lactarius and Lactifluus), with an additional separation of several species from Lactarius as well as Russula into Multifurca [39]. Most species of Lactarius form symbioses with broadleaf or coniferous hosts, consistent with their discovery in the Fujian forests of pine.
Russula are distinguished by their bright-colored caps, but do not produce latex and are often characterized by their brittle caps [40]. Due to the difficulty in separating species by their morphological characteristics alone, the modern identification of species within the Russula and Lactarius genera has relied on utilizing the ITS sequence in phylogenetic analysis as the primary molecular method for distinguishing and interpreting these closely related species [41]. However, an overreliance on ITS-based phylogenetic structures can lead to inaccurate subgenus classifications and may overlook the presence of known species, such as the R. queletii complex and the rhodochroa-subsanguinaria complex, often manifested within ITS-based phylogenetics [24]. Additionally, earlier studies focusing on Lactarius species found minimal consistency between Asian Lactarius species and those from other continents. Relying solely on ITS-based phylogenetic analysis and morphological characteristics for Lactarius species’ identification appears insufficient. Hence, the utilization of multi-locus phylogenetic analysis has become the preferred method for revealing the genetic relationships within the Russula and Lactarius genera.
Here, we employed a combined ITS-nrLSU-RBP2-mtSSU-TEF-1α multi-locus phylogenetic analysis method to support the identification of three species that have been named R. junzifengensis, R. zonatus, and L. jianyangensis. These assignments are based on combined morphological characterizations and molecular multilocus phylogenetic analyses. Russula zonatus appears to be a very common red mushroom in the subtropical-tropical Quercus forests of Fujian and is a member of the subgenus Crassotunicata. Key features for its identification include medium basidiocarps, a convex expanded to infundibuliform pileus with a central depression and slight incurved margin, glabrous and dry surface with indistinct or golden brown zone lines, brown to grayish brown color, very crowded lamellae, moderately ornamented basidiospores with isolated warts, and a subtropical habitat. R. zonatus forms a clade with R. brunneoaurantiaca (with a highest ITS identity of ~99%), R. adusta, and R. nigricans. All these species have a mucilaginous pileus and comparatively large spores and basidia. R. zonatus has a high similarity to R.brunneoaurantiaca’s ITS sequence (0.99%), but they have significant morphological differences. R. zonatus belongs to the subgenus Crassotunicata of the Russula genus and has medium to large basidiocarps, with the convex expanded to the infundibuliform pileus with a central depression and slight incurved margin. It has a glabrous and dry surface with indistinct or golden brown zone lines, a brown to grayish brown color, very crowded lamellae, and moderately ornamented basidiospores with isolated warts. R. zonatus has smaller basidia and cystidia compared to R. brunneoaurantiaca, and, at the macroscopic scale, additional differences are quite obvious, with R. brunneoaurantiaca having a surface that is mucilaginous, brownish orange turning yellowish brown to light brown, and a smooth stipe surface [37].
Russula junzifengensis is characterized by a white or slightly stained white–yellow or yellow pileus, which is broadly convex to flat with a shallow depression, slightly crowded lamellae, medium basidiospores with isolated warts, and a subtropical habitat. This species is similar to R. pseudocrustosa, R. indoalba, and R. xanthovirens, and phylogenetic analysis showed that R. junzifengensis formed a highly supported sister group with R. indoalba, but their ITS sequence similarity is less than 90%. Macromorphologically this species seems to be indistinguishable from R. indoalba; both species have whitish gray basidiomata, a clavate stipe, and ellipsoid basidiospores. But the lamellae of R. junzifengensis are not attached to the stipe and appear with fibrils on the stipe.
The phylogenetic results indicate that L. jianyangensis is closest to L. pallido-ochraceus and L. pallidizonatus. In comparison to L. pallido-ochraceus, both species have few pits on their stipe, a surface that is greasy when wet and basidiospores with reticulate ornamentation. In comparison to L. pallido-ochraceus, the basidiomata of the new species described here has a deeper color, smaller basidia, and narrower pleuromacrocystidia. The ITS similarity between the two species was 94%. Southern China’s Lactarius pallidizonatus X.H. Wang seems to be another closely related species [39]; L. pallidizonatus can be distinguished by its light orange–yellow margin, cream–whitish context, pits near the base on stipes, and basidiospores (80/4/3) (7.0) 7.5–9.0 (9.5) × (5.5) 6.0–7.5 µm [Q = (1.10) 1.12–1.29 (1.35), Q = 1.21 ± 0.05], however, L. pallidizonatus has a lighter color and bigger basidiospores than L. jianyangensis.
5. Conclusions
In this study, we employed a multi-locus phylogenetic analysis method, combined with morphological characteristics, to identify three new fungal species in the Quercus forests of Fujian province, namely R. junzifengensis, R. zonatus, and L. jianyangensis. These new species belong to the Russula and Lactarius genera, which form ectomycorrhizal symbioses with higher plants and trees. We found that relying solely on ITS sequences and morphological characteristics for species identification is insufficient, as it may lead to inaccurate subgenus classifications and the omission of known species. Therefore, we suggest using multi-locus phylogenetic analysis methods to reveal the genetic relationships within the Russula and Lactarius genera, as well as their phylogenetic affinities with species from other geographical regions. Our study provides new data on the fungal diversity and distribution in Fujian province, and also contributes new insights to fungal taxonomy and phylogeny.
Acknowledgments
We would like to thank Ling Wang, Longbin Lin, Huili Pu, Tianyu Tan, Taichang Mu, and Jie Zhao for their help with sample collection.
Author Contributions
Acquisition of funding, Y.C., H.S. and J.Q.; Collection of data, S.L., M.Z., N.O.K., Z.H., J.C., R.C., Y.D., Y.H., Z.W., X.G., C.Y., P.L., W.Z. and H.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All newly generated sequences were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 10 January 2024)). All new taxa were linked with MycoBank (https://www.mycobank.org/ (accessed on 10 January 2024)).
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by the National Natural Science Foundation of China (No. 32270029, U1803232, 31670026), the National Key R & D Program of China (No. 2017YFE0122000), a Social Service Team Support Program Project (No. 11899170165), Science and Technology Innovation Special Fund (No. KFB23084) of Fujian Agriculture and Forestry University, a Fujian Provincial Major Science and Technology Project (No. 2022NZ029017), a Key Project from Fujian Provincial Department of Science and Technology (No. 2020N5005), and the Young and Middle-aged Teacher Education Research Project of Fujian Province (No. JAT210075).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Looney B.P., Meidl P., Piatek M.J., Miettinen O., Martin F.M., Matheny P.B., Labbé J.L. Russulaceae: A new genomic dataset to study ecosystem function and evolutionary diversification of ectomycorrhizal fungi with their tree associates. New Phytol. 2018;218:54–65. doi: 10.1111/nph.15001. [DOI] [PubMed] [Google Scholar]
- 2.Jiang X.M., Li Y.K., Liang J.F., Wu J.R. Russula brunneovinacea sp. nov., from Northeastern China. Mycotaxon. 2018;132:789–797. doi: 10.5248/132.789. [DOI] [Google Scholar]
- 3.Li G.J., Zhao D., Li S.F., Wen H.A. Russula chiui and R. pseudopectinatoides, two new species from Southwestern China supported by morphological and molecular evidence. Mycol. Prog. 2015;14:33. doi: 10.1007/s11557-015-1054-y. [DOI] [Google Scholar]
- 4.Taylor A.F.S., Alexander I.J. Ectomycorrhizal synthesis with an isolate of Russula aeruginea. Mycol. Res. 1989;92:103–107. doi: 10.1016/S0953-7562(89)80103-0. [DOI] [Google Scholar]
- 5.Wang P., Zhang Y., Mi F., Tang X., He X., Cao Y., Liu C., Yang D., Dong J., Zhang K., et al. Recent advances in population genetics of ectomycorrhizal mushrooms Russula spp. Mycology. 2015;6:110–120. doi: 10.1080/21501203.2015.1062810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J.K. Biologically active substances from mushrooms in Yunnan, China. Heterocycles. 2002;57:157–167. doi: 10.3987/REV-01-543. [DOI] [Google Scholar]
- 7.Singh R.S., Walia A.K., Kennedy J.F. Mushroom lectins in biomedical research and development. Int. J. Biol. Macromol. 2020;151:1340–1350. doi: 10.1016/j.ijbiomac.2019.10.180. [DOI] [PubMed] [Google Scholar]
- 8.Ali A., Guo D., Mahar A., Wang P., Shen F., Li R.H., Zhang Z.Q. Mycoremediation of potentially toxic trace elements—A biological tool for soil cleanup: A review. Pedosphere. 2017;27:205–222. doi: 10.1016/S1002-0160(17)60311-4. [DOI] [Google Scholar]
- 9.Chen F., Yuan Y., Wei W., Yu S., Zhang T. Reconstructed Temperature for Yong’an, Fujian, Southeast China: Linkages to the pacific ocean climate variability. Glob. Planet. Chang. 2012;86–87:11–19. doi: 10.1016/j.gloplacha.2012.01.005. [DOI] [Google Scholar]
- 10.Yin Y., Gemmer M., Luo Y., Wang Y. Tropical cyclones and heavy rainfall in Fujian province, China. Quat. Int. 2010;226:122–128. doi: 10.1016/j.quaint.2010.03.015. [DOI] [Google Scholar]
- 11.Liu Y., Xu X., Dimitrov D., Pellissier L., Borregaard M.K., Shrestha N., Su X., Luo A., Zimmermann N.E., Rahbek C., et al. An updated floristic map of the world. Nat. Commun. 2023;14:2990. doi: 10.1038/s41467-023-38375-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yue Y., Zheng Z., Huang K., Chevalier M., Chase B.M., Carré M., Ledru M.P., Cheddadi R. A Continuous record of vegetation and climate change over the past 50,000 years in the Fujian province of eastern subtropical China. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2012;365–366:115–123. doi: 10.1016/j.palaeo.2012.09.018. [DOI] [Google Scholar]
- 13.Wisitrassameewong K., Park M.S., Lee H., Ghosh A., Das K., Buyck B., Looney B.P., Caboň M., Adamčík S., Kim C., et al. Taxonomic revision of Russula subsection Amoeninae from South Korea. MycoKeys. 2020;75:1–29. doi: 10.3897/mycokeys.75.53673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G.J., Dong Z., Li S.F., Wen H.A. Recent research progress of Russula (Russulales, Agaricomycetes): A review. Mycosystema. 2015;34:821–848. doi: 10.13346/j.mycosystema.150085. [DOI] [Google Scholar]
- 15.Vera M., Adamčík S., Adamčíková K., Hampe F., Caboň M., Manz C., Ovrebo C., Piepenbring M., Corrales A. Morphological and genetic diversification of Russula floriformis, sp. nov., along the Isthmus of Panama. Mycologia. 2021;113:807–827. doi: 10.1080/00275514.2021.1897377. [DOI] [PubMed] [Google Scholar]
- 16.Khatua S., Paloi S., Acharya K. An untold story of a novel mushroom from Tribal Cuisine: An Ethno-Medicinal, taxonomic and pharmacological approach. Food Funct. 2021;12:4679–4695. doi: 10.1039/D1FO00533B. [DOI] [PubMed] [Google Scholar]
- 17.Panda M.K., Das S.K., Mohapatra S., Debata P.R., Tayung K., Thatoi H. Mycochemical composition, bioactivities, and phylogenetic placement of three wild edible Russula species from Northern Odisha, India. Plant Biosyst. 2021;155:1041–1055. doi: 10.1080/11263504.2020.1813829. [DOI] [Google Scholar]
- 18.Looney B.P., Manz C., Matheny P.B., Adamčík S. Systematic revision of the Roseinae clade of Russula, with a focus on eastern North American taxa. Mycologia. 2022;114:270–302. doi: 10.1080/00275514.2021.2018881. [DOI] [PubMed] [Google Scholar]
- 19.Noffsinger C., Cripps C.L. Systematic analysis of Russula in the North American Rocky Mountain alpine zone. Mycologia. 2021;113:1278–1315. doi: 10.1080/00275514.2021.1947695. [DOI] [PubMed] [Google Scholar]
- 20.Paloi S., Kumla J., Karunarathna S.C., Lumyong S., Suwannarach N. Taxonomic and phylogenetic evidence reveal two new Russula species (Russulaceae, Russulales) from Northern Thailand. Mycol. Prog. 2023;22:72. doi: 10.1007/s11557-023-01921-5. [DOI] [Google Scholar]
- 21.Miller S.L., Buyck B. Molecular phylogeny of the genus Russula in Europe with a comparison of modern infrageneric classifications. Mycol. Res. 2002;106:259–276. doi: 10.1017/S0953756202005610. [DOI] [Google Scholar]
- 22.Buyck B., Hofstetter V., Eberhardt U., Verbeken A., Kauff F. Walking the thin line between Russula and Lactarius: The dilemma of Russula subsect. Ochricompactae. Fungal Divers. 2008;28:15–40. [Google Scholar]
- 23.Buyck B., Hofstetter V., Verbeken A., Walleyn R. Proposal to conserve Lactarius Nom. Cons. (Basidiomycota) with a conserved type. Taxon. 2010;59:295–296. doi: 10.1002/tax.591031. [DOI] [Google Scholar]
- 24.Li G.J., Liu T.Z., Li S.M., Zhao S.Y., Niu C.Y., Liu Z.Z., Xie X.J., Zhang X., Shi L.Y., Guo Y.B., et al. Four new species of Russula subsection Sardoninae from China. J. Fungi. 2023;9:199. doi: 10.3390/jof9020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou H., Cheng G.Q., Wang Q.T., Guo M.J., Zhuo L., Yan H.F., Li G.J., Hou C.L. Morphological characteristics and phylogeny reveal six new species in Russula Subgenus Russula (Russulaceae, Russulales) from Yanshan Mountains, North China. J. Fungi. 2022;8:1283. doi: 10.3390/jof8121283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S.H., Li G.J., Phurbu D., He M.Q., Zhang M.Z., Zhu X.Y., Li J.X., Zhao R.L., Cao B. Four new species of Russula from the Xizang Autonomous Region and other provinces of China. Mycology. 2023;14:1–28. doi: 10.1080/21501203.2023.2265667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G.J., Li S.M., Buyck B., Zhao S.Y., Xie X.J., Shi L.Y., Deng C.Y., Meng Q.F., Sun Q.B., Yan J.Q., et al. Three new Russula Species in Sect. Ingratae (Russulales, Basidiomycota) from Southern China. MycoKeys. 2021;84:103–139. doi: 10.3897/mycokeys.84.68750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen B., Song J., Liang J., Li Y. Two new species of Russula Subsect. Virescentinae from Southern China. Mycol. Prog. 2021;20:993–1005. doi: 10.1007/s11557-021-01716-6. [DOI] [Google Scholar]
- 29.Li F., Deng Q.L. Three new species of Russula from South China. Mycol. Prog. 2018;17:1305–1321. doi: 10.1007/s11557-018-1447-9. [DOI] [Google Scholar]
- 30.White T.J., Bruns T., Lee S., Taylor J. PCR Protocols. Elsevier; Amsterdam, The Netherlands: 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics; pp. 315–322. [Google Scholar]
- 31.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rehner S.A., Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1- sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y.J., Hall B.D. Body plan evolution of Ascomycetes, as inferred from an RNA polymerase II phylogeny. Proc. Natl. Acad. Sci. USA. 2004;101:4507–4512. doi: 10.1073/pnas.0400938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen B., Song J., Chen Y., Zhang J., Liang J. Morphological and phylogenetic evidence for two new species of Russula Subg. Heterophyllidia from Guangdong province of China. MycoKeys. 2021;82:139–157. doi: 10.3897/mycokeys.82.64913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng L.S., Kang R., Zeng N.K., Yu W.J., Chang C., Xu F., Deng W.Q., Qi L.L., Zhou Y.L., Fan Y.G. Two new Inosperma (Inocybaceae) species with unexpected muscarine contents from tropical China. MycoKeys. 2021;85:87–108. doi: 10.3897/mycokeys.85.71957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buyck B. One step closer to unravelling the origin of Russula: Subgenus Glutinosae subg. nov. Mycosphere. 2020;11:285–304. doi: 10.5943/mycosphere/11/1/6. [DOI] [Google Scholar]
- 37.Roy N., Beypih J., Tanti B., Dutta A.K. Russula brunneoaurantiaca, a novel taxon of Russula subg. Crassotunicata from west Bengal, India, with morpho-molecular analysis and scanning electron microscopy. Microsc. Res. Tech. 2023;86:1–7. doi: 10.1002/jemt.24463. [DOI] [PubMed] [Google Scholar]
- 38.Shi S.F., Wang X.H., Bau T. Three new species of Lactarius (Russulaceae, Russulales) from Northeast China. Mycoscience. 2018;59:206–217. doi: 10.1016/j.myc.2017.11.001. [DOI] [Google Scholar]
- 39.Wang X.H. Seven new species of Lactarius subg. Lactarius (Russulaceae) from Southwestern China. Mycosystema. 2017;36:1463–1482. doi: 10.13346/j.mycosystema.170155. [DOI] [Google Scholar]
- 40.Lee H., Park M.S., Jung P.E., Eimes J.A., Seok S.J., Lim Y.W. Re-evaluation of the taxonomy and diversity of Russula section Foetentinae (Russulales, Basidiomycota) in Korea. Mycoscience. 2017;58:351–360. doi: 10.1016/j.myc.2017.04.006. [DOI] [Google Scholar]
- 41.Adamčík S., Looney B., Caboň M., Jančovičová S., Adamčíková K., Avis P.G., Barajas M., Bhatt R.P., Corrales A., Das K., et al. The quest for a globally comprehensible Russula language. Fungal Divers. 2019;99:369–449. doi: 10.1007/s13225-019-00437-2. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All newly generated sequences were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 10 January 2024)). All new taxa were linked with MycoBank (https://www.mycobank.org/ (accessed on 10 January 2024)).