Abstract
Purpose
Nascent geographic atrophy (nGA) refers to specific features seen on OCT B-scans, which are strongly associated with the future development of geographic atrophy (GA). This study sought to develop a deep learning model to screen OCT B-scans for nGA that warrant further manual review (an artificial intelligence [AI]-assisted approach), and to determine the extent of reduction in OCT B-scan load requiring manual review while maintaining near-perfect nGA detection performance.
Design
Development and evaluation of a deep learning model.
Participants
One thousand eight hundred and eighty four OCT volume scans (49 B-scans per volume) without neovascular age-related macular degeneration from 280 eyes of 140 participants with bilateral large drusen at baseline, seen at 6-monthly intervals up to a 36-month period (from which 40 eyes developed nGA).
Methods
OCT volume and B-scans were labeled for the presence of nGA. Their presence at the volume scan level provided the ground truth for training a deep learning model to identify OCT B-scans that potentially showed nGA requiring manual review. Using a threshold that provided a sensitivity of 0.99, the B-scans identified were assigned the ground truth label with the AI-assisted approach. The performance of this approach for detecting nGA across all visits, or at the visit of nGA onset, was evaluated using fivefold cross-validation.
Main Outcome Measures
Sensitivity for detecting nGA, and proportion of OCT B-scans requiring manual review.
Results
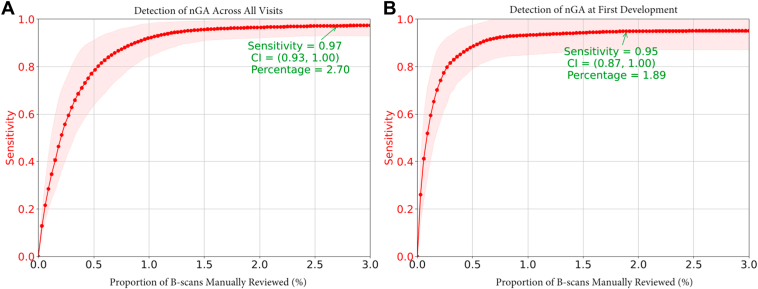
The AI-assisted approach (utilizing outputs from the deep learning model to guide manual review) had a sensitivity of 0.97 (95% confidence interval [CI] = 0.93–1.00) and 0.95 (95% CI = 0.87–1.00) for detecting nGA across all visits and at the visit of nGA onset, respectively, when requiring manual review of only 2.7% and 1.9% of selected OCT B-scans, respectively.
Conclusions
A deep learning model could be used to enable near-perfect detection of nGA onset while reducing the number of OCT B-scans requiring manual review by over 50-fold. This AI-assisted approach shows promise for substantially reducing the current burden of manual review of OCT B-scans to detect this crucial feature that portends future development of GA.
Financial Disclosure(s)
Proprietary or commercial disclosure may be found in the Footnotes and Disclosures at the end of this article.
Keywords: nGA, Deep learning, Multi-instance learning
Geographic atrophy (GA) is a late, vision-threatening complication of age-related macular degeneration (AMD). Once identified on color fundus photography (CFP) or fundus autofluorescence, there is already substantial loss of outer retinal tissue.1 Slowing or preventing the onset of GA in the early stages of AMD is thus needed to prevent irreversible vision loss from this late-stage disease. However, large and lengthy interventional trials2,3 have often been required to detect a clinically meaningful effect of an intervention in the early stages of AMD. The feasibility of such trials could be improved by identifying early signs or predictors of GA onset, so that they could be used to identify high-risk individuals to enrich clinical trial populations. They could also serve as biomarkers of AMD progression, or potentially act as an earlier endpoint in trials aiming to prevent the onset of atrophy.
On OCT imaging, specific features of photoreceptor loss that have been termed nascent GA (nGA)4, 5, 6, 7 have shown promise as one such biomarker for providing a strong predictor of GA onset.7,8 These features include (1) subsidence of the inner nuclear layer (INL) and outer plexiform layer (OPL) and/or (2) a hyporeflective wedge-shaped band within Henle’s fiber layer. A recent study reported that in individuals with bilateral large drusen, eyes that developed nGA showed a 78-fold increased risk of developing GA compared with those that did not have nGA.7 The accurate identification of nGA could thus substantially improve the feasibility of evaluating preventative treatments for the onset of GA.
However, the manual grading of numerous individual B-scans in an OCT volume scan is labor intensive and time consuming, and as such is an expensive undertaking in clinical trials. This is especially the case when a large number of B-scans are acquired per OCT volume scan to provide a high spatial resolution when imaging the retina. This challenge is analogous to one faced in the population-based screening of retinal diseases, where the large number of CFPs requiring assessments has motivated the development of artificial intelligence (AI)-based methods for identifying referable cases.9, 10, 11, 12 Artificial intelligence models can provide a fully autonomous assessment, or they could be used to identify a small subset of cases requiring secondary assessment by human graders. The latter approach could thus provide a means for substantially improving the feasibility of detecting nGA through substantially reducing the number of OCT B-scans requiring manual evaluation.
While deep learning models have recently been developed to automatically detect GA on fundus autofluorescence,13,14 CFP,15,16 and OCT scans,17, 18, 19, 20 such a model for detecting nGA has yet to be developed. This study thus sought to develop a deep learning model that could effectively screen OCT volume scans for the presence of nGA to identify a subset of B-scans for manual assessment, which we describe as “AI-assisted” detection of nGA. We sought to determine what extent of reduction in the number of OCT B-scans could be achieved with this approach while maintaining a near-perfect performance for detecting nGA. We also sought to examine the performance of the deep learning model used in isolation (i.e., a fully automated AI approach).
Methods
This study included a subset of participants21, 22, 23 in the sham treatment arm of the Laser Intervention in the Early Stages of AMD (LEAD) study24,25 (clinicaltrials.gov identifier: NCT01790802) who had ≥ 1 follow-up visit and who did not have nGA at baseline based on the independent masked grading of the OCT imaging data only (as previously described in detail7). The LEAD study was conducted according to the International Conference on Harmonization Guidelines for Good Clinical Practice and the tenets of the Declaration of Helsinki. Institutional review board approval was obtained at all sites and all participants provided written informed consent.
Participants
This study included participants that had bilateral large drusen at baseline (meeting the definition of intermediate AMD based on the Beckman Clinical Classification System of AMD26), and had a best-corrected visual acuity of 20/40 or better in both eyes. Any participant with late AMD, or any other ocular, systemic or neurological disease that could have an impact on the assessment of the retina, were excluded. Late AMD was defined by the presence of neovascular AMD, GA on CFP, or nGA or greater lesion on OCT imaging.21, 22, 23
All participants underwent pupillary dilation, then OCT imaging was performed by obtaining a volume scan covering a 20 × 20° region with 49 horizontal B-scans (25 frames averaged per scan) using the Spectralis HRA + OCT (Heidelberg Engineering). All retinal imaging was performed at the baseline visit and each 6-monthly follow-up visit, for up to 36-months.
Data Labeling
As previously described,7 all OCT volume scans were initially assessed by a senior grader (Z.W.) for the presence of nGA, defined as the presence of subsidence of the INL and OPL, and/or a hyporeflective wedge-shaped band within Henle’s fiber layer.4, 5, 6, 7 Note that the features used to define nGA do not require the presence of choroidal signal hypertransmission ≥ 250 μm with an associated zone of retinal pigment epithelium (RPE) attenuation or disruption ≥ 250 μm (meeting the definition of complete RPE and outer retinal atrophy [cRORA]).27 However, these signs could be present and it is thus possible for nGA lesions seen on OCT imaging to also meet the definition of cRORA. It is also possible that nGA lesions could correspond to GA visible on CFP, but this distinction was not performed (or required) in this study. All visits of any eye deemed to have questionable or definite nGA were then concurrently reviewed by 2 further experienced graders (R.H.G. and Z.W.). For all OCT volume scans labeled as having nGA—which includes lesions that could also meet the definition of complete RPE and cRORA27—the same 2 graders then also concurrently labeled each B-scan for the presence of nGA. Any disagreements were immediately resolved by open adjudication.
Deep Learning Model Development
A multi-instance deep learning model was trained to classify OCT volumes using volume-wise nGA labels. The intermediate logit for each instance is used to identify B-scans requiring manual assessment for the AI-assisted approach described in the next section.
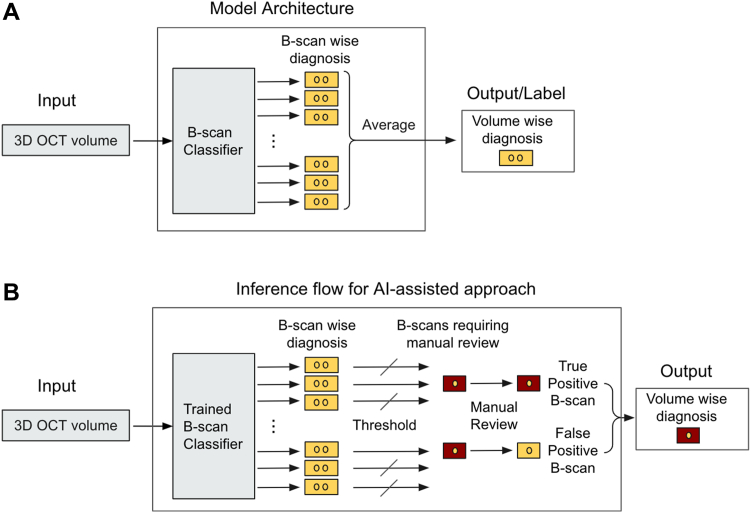
The model architecture is shown in Figure 1A; a late-fusion method with ResNet-1828 backbone was applied for the 3-dimensional OCT volume scans. B-scans from the OCT volume were fed into a B-scan classifier, where they were first preprocessed (resized to 512 × 496, then rescaling the intensity range to [0, 1]). Data augmentation, including small rotations, affine transformations, horizontal flips, and additions of Gaussian noise and blur were randomly applied to improve the invariance of the model to those transformations. The B-scans were then passed through the ResNet-18 backbone pretrained on the ImageNet dataset. The ResNet backbone produces activation maps of 512 × 16 × 16, and a max-pooling layer and an average pooling layer are concatenated to generate a feature vector of 1024. Then a fully connected layer was applied to generate the classification logit for each B-scan. In such a setting, during the training process, the loss from volume label and volume classification (average of B-scan logits) back-propagates and encourages the network to identify as many B-scans that contribute to the final prediction of the presence of nGA at the OCT volume scan level. This framework can thus be considered as a form of simplified multi-instance learning that learns instance labels from only bag labels,29 where the B-scans are the “instances” and OCT volume scans as “bags” (i.e., a set of training instances). During model training, the Adam optimizer was used to minimize focal loss. The L2 weight decay regularization was used to improve the generalization of the model.
Figure 1.
A, Deep learning architecture of the nascent geographic atrophy diagnostic model, which is trained to classify 3-dimensional (3D) OCT volumes. B, The flow for artificial intelligence (AI)-assisted approach where only a small subset of B-scans are identified by the trained B-scan classifier for manual review.
The class imbalance present in our dataset can present a significant challenge to achieving high levels of classification performance, especially for the minority class (OCT volumes with nGA in this study, which was also the positive class), with the deep learning model.30 To minimize the impact of the imbalance in the dataset, we utilized focal loss and over-sampling strategies during the model training. Focal loss has been shown to improve the performance of deep learning models in multiple applications.31, 32, 33 The focal loss down-weights the loss contribution from easy examples in comparison to the traditional cross-entropy loss, and this modification helps the model to focus more on challenging examples, which are typically from the minority class. Over-sampling strategy ensures the model receives sufficient exposure to the positive samples. In addition, we applied transfer learning and the image augmentation techniques during the model training to enhance the performance of the model when dealing with a relatively small dataset.30,34 The data augmentation we performed include rotation and affine transformation to account for typical variations in head positioning when acquiring scans, horizontal flips to remove the impact of eye laterality, and additions of Gaussian noise and blur to simulate scans of different signal-to-noise ratios. Due to the high imbalance and limited size of the dataset, model performance was evaluated on the entire dataset using fivefold cross-validation, with a split at the individual level and stratified based on the presence of nGA to avoid information leak and skewed distribution. The use of cross-validation helps mitigate the impact of random data splits and provides more robust results. We follow the nomenclature for data splitting35 with a training set (64%), a tuning set (16%), and a validation test set (20%). As such, the validation test set was never used for tuning the model. Hyperparameters were tuned optimizing the F1-score on the tuning set to find the best value of learning rate, weight decay, and for early stopping. Thereafter, the model that was trained with the optimal hyperparameter was tested on the validation test set.
Given an OCT volume and a trained model, our objective was to identify nGA B-scans. A confidence score of a B-scan containing nGA is estimated as S(l/n) when l > 0 and S(l) when l < 0, where S is the sigmoid function, l is the individual B-scan classification logit, and n is the number of B-scans in a volume. This reverse leaky rectified linear unit operation on logits empirically normalizes the logits to [−1, 1] range, so that the confidence score is distributed more uniformly. B-scans whose confidence scores were higher than a threshold are considered as containing nGA lesions, and the threshold was chosen based on either the sensitivity or specificity of the model from the training and tuning sets (the details on thresholding are described in the next section).
Evaluation of Model Performance
The performance of the approaches utilizing the deep learning model for detecting nGA at the OCT volume scan level was examined in 2 scenarios—first, when evaluating all the OCT volume scans in this study, and second, when evaluating all the OCT volume scans up until the visit of nGA onset (or until the last visit if nGA did not develop in an eye). The former thus included OCT volume scans of subsequent visits following the onset of nGA. The latter was used to evaluate the performance of the approaches within a clinically relevant context—such as within a randomized-controlled trial of an intervention in the early stages of AMD—where it is critical that the approaches accurately identified the onset of nGA, and that they avoid falsely considering that an endpoint has been reached.
Two approaches were examined in this study—a fully automated AI approach and an AI-assisted approach. The former relies entirely on the output of the deep learning model to determine the presence of nGA in an OCT volume, and we thus examined its sensitivity for detecting nGA at sufficiently high levels of specificity (or low false-positive rates) using volume-wise nGA labels. This was achieved by identifying a threshold for the probability output of the model that provides a fixed level of specificity (99%, 98%, and 95%) from the training and tuning sets, and this threshold was then applied in the validation test sets and its performance (sensitivity and specificity) for detecting nGA was determined. For the latter AI-assisted approach (Fig 1B), the same deep learning model was used to identify a subset of OCT B-scans with a higher probability of having nGA lesions from an OCT volume for further manual review. This manual review of the identified subset of B-scans is assumed to provide the same labels of the presence of nGA as the ground-truth (i.e., labels provided by the 2 experienced graders in this study, rather than an assessment from further independent grading). We thus sought to identify a threshold of the B-scan confidence score (described in previous section) in the training and tuning sets that would provide a sensitivity of 99% for the detection of nGA (a near-perfect level of detection), and this threshold was then applied to the validation test sets. The sensitivity of this AI-assisted approach, and the proportion of B-scans requiring manual review, was then determined using the B-scan wise nGA labels. Note that the performance measure of the 2 approaches evaluated, described earlier, differed as each approach has its own specific requirements for it to have clinical utility. The sensitivity of the fully automated AI approach was evaluated at high levels of specificity, which is required to avoid falsely identifying nGA. In contrast, the extent of reduction in the proportion of B-scans requiring manual review by the AI-assisted approach was set to achieve a high level of sensitivity, which is required for a screening tool. The 95% confidence intervals (CIs) of the performance measures of sensitivity and specificity of the fully automated AI approach, and the sensitivity of the AI-assisted approach in the validation test sets, were then calculated using a bootstrap resampling procedure (n = 1000 resamples at the individual level).
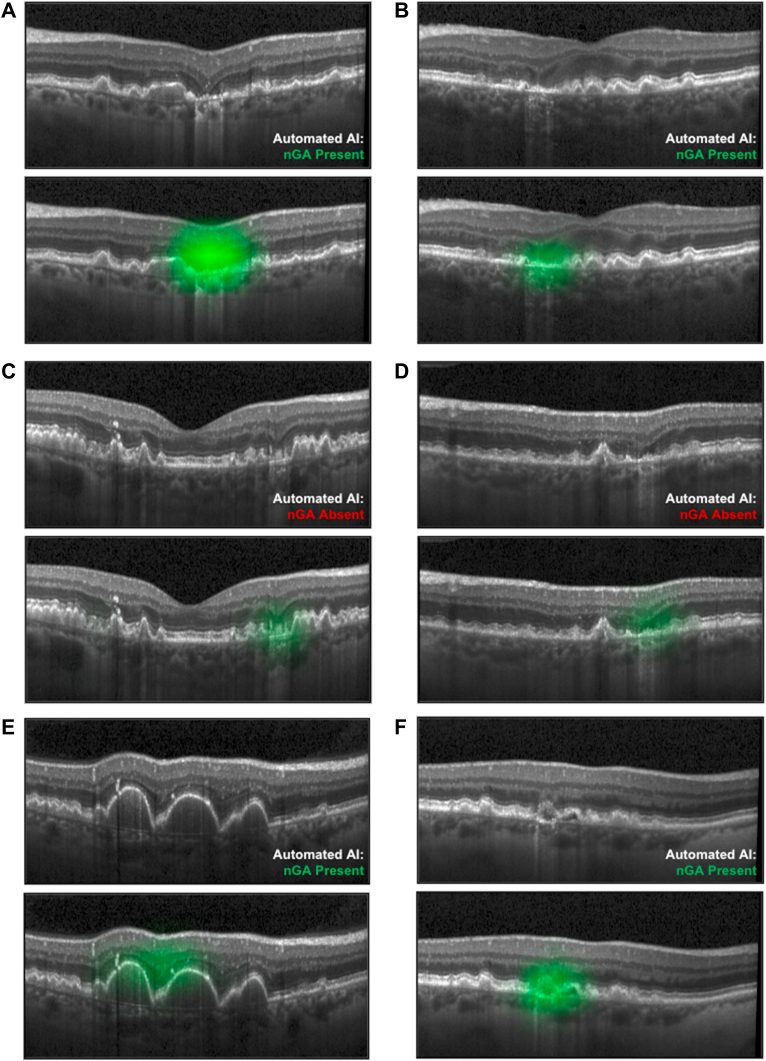
To further illustrate what was learned by the model, we utilized Gradient-weighted Class Activation Mapping technique.36 This method visualizes the regions in the input image that contribute the most to the model’s prediction by computing the gradient of the target class. The saliency map generated by Gradient-weighted Class Activation Mapping helps identify the abnormal regions associated with nGA.
Results
A total of 280 eyes from 140 participants with bilateral large drusen at baseline that were seen at 6-monthly intervals up to 36 months were included in this study. From a total of 1910 visits, 26 visits were excluded due to the development of neovascular AMD in 9 (3%) eyes. For the remaining 1884 visits, 118 (6%) visits from 40 (14%) eyes were labeled as having nGA. Fig S2 shows our data split strategy.
Performance of the Fully Automated AI Approach
The fully automated AI approach had an overall sensitivity of 0.76 (95% CI = 0.67–0.84) and specificity of 0.98 (95% CI = 0.96–0.99) for detecting nGA when evaluating all OCT volume scans in this study, based on a threshold that achieved 0.99 specificity in the training and tuning sets. Based on the same threshold, the model had a sensitivity of 0.53 (95% CI = 0.40–0.65) for detecting the onset of nGA when evaluating all OCT volume scans up until the visit of nGA development (or until the final visit if nGA did not develop), at a specificity of 0.98 (95% CI = 0.96–0.99). The core performance metrics of the fully automated AI approach for these 2 previously mentioned analyses for other thresholds are summarized in Table 1. Additional performance metrics are summarized in Table S2 and Fig S3.
Table 1.
Performance of the Fully Automated AI Model for Detecting nGA in the Combined Validation Test Sets
| Training and Tuning Specificity∗ | All Visits |
At First Development† |
||
|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | |
| 0.99 | 0.76 (0.67–0.84) | 0.98 (0.96–0.99) | 0.53 (0.40–0.65) | 0.98 (0.96–0.99) |
| 0.98 | 0.86 (0.77–0.93) | 0.97 (0.95–0.99) | 0.75 (0.62–0.87) | 0.97 (0.95–0.99) |
| 0.95 | 0.91 (0.83–0.98) | 0.94 (0.91–0.97) | 0.87 (0.77–0.97) | 0.94 (0.91–0.97) |
All values are presented as the estimate (95% confidence interval). AI = artificial intelligence; nGA = nascent geographic atrophy.
Based on threshold determined from the training and tuning sets.
Including all OCT volume scans up until the visit of nGA development, or until the final visit if nGA did not develop in an eye.
Performance of the AI-Assisted Approach
The AI-assisted approach achieved a sensitivity of 0.97 (95% CI = 0.93–1.00) for detecting nGA when evaluating all OCT volume scans in this study while requiring manual review of only 2.7% of OCT B-scans selected by deep learning model AI from all B-scans in OCT volumes. For the task of detecting the onset of nGA, the AI-assisted approach had a sensitivity of 0.95 (95% CI = 0.87–1.00) when requiring manual review of only 1.9% of the OCT B-scans selected by AI. A plot of the sensitivity against the proportion of OCT B-scans requiring manual review for both scenarios is shown in Figure 4. Since manual review corrects false positive cases, the specificity of AI-assisted approach is by default 1.00.
Figure 4.
Performance of the artificial intelligence-assisted models for detecting nascent geographic atrophy (nGA) at all visits (A), or at the visit when nGA first developed (B), when requiring manual review of a subset of OCT B-scans determined by a deep learning model. CI = confidence interval.
Longitudinally, for the 30 eyes that have ≥ 1 follow up visit after nGA onset, 100% of the AI prediction stays positive consistently. Typical examples are presented in Fig S5.
In addition, we present the probability distributions of B-scans in Fig S6 to provide more insights into the performance of our methodology. All B-scans across the entire dataset were split into 3 groups based on labels of corresponding OCT volumes: non-nGA, nGA onset, and after nGA onset.
Examples of Findings in This Study
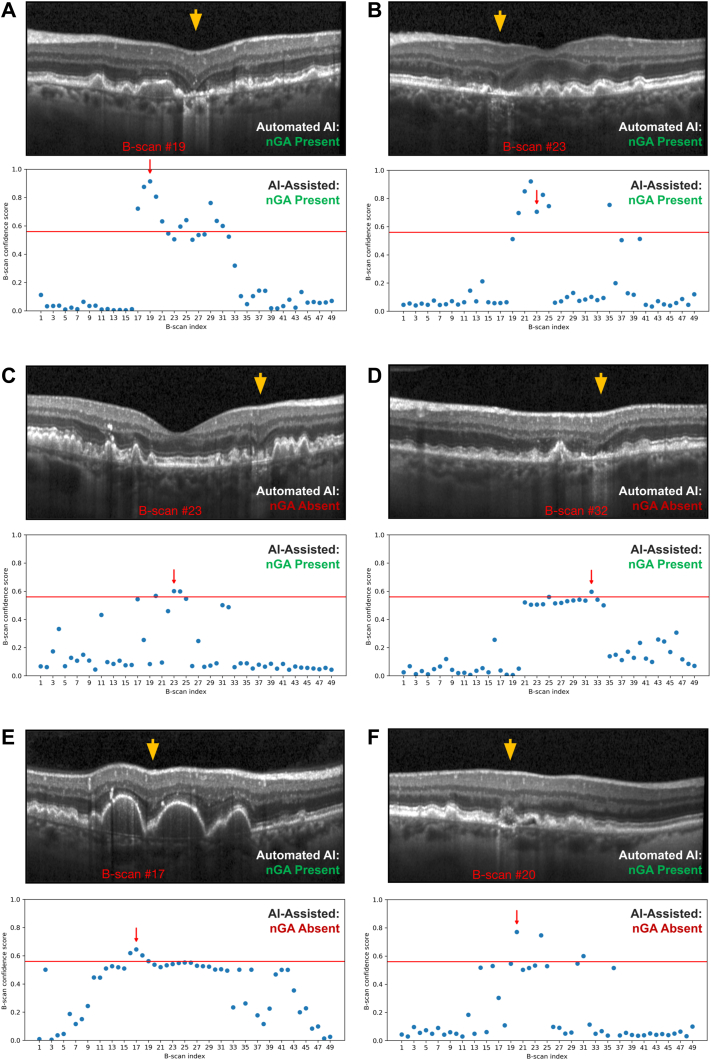
Six examples of cases are presented in Figure 7 to illustrate the findings of this study. The saliency maps generated by Gradient-weighted Class Activation Mapping are presented in Figure 8 to provide visual insights of the model’s prediction. The first 2 cases (Fig 7A, B) represent cases with nGA that were correctly identified with both the fully automated AI (based on a threshold that provided a 0.98 specificity in the validation test set) and AI-assisted approaches. The next 2 cases (Fig 7C, D) represent cases where the fully automated AI approach did not consider nGA to be present in the OCT volume scan, but ≥ 1 B-scans were identified as requiring manual review and then confirmed to have nGA based on the AI-assisted approach. The final 2 cases (Fig 7E, F) represent cases where the fully automated AI approach had incorrectly considered nGA to be present, while B-scans identified as requiring manual review with the AI-assisted approach in these cases were then deemed not to have nGA. For the first of these 2 cases (Fig 7E), the fully automated AI approach may likely have considered subsidence of the OPL and INL to be present between 2 large regions of drusen-associated RPE elevations (that elevated the OPL and INL, thus giving the impression that subsidence occurred between these 2 elevated regions). For the second of these 2 cases (Fig 7F), it is unclear what the fully automated AI approach would have considered in its identification of this B-scan as having nGA. It is possible that the deep learning model learned about other OCT features that are often, but not always, present in regions of nGA—such as choroidal signal hypertransmission, RPE disruption or attenuation, or disruption of the external limiting membrane or ellipsoid zone—and considered these in its assessment of nGA presence in this B-scan.
Figure 7.
Examples of cases in this study, with a selected OCT B-scan shown on the top of each case (with region of interest indicated by the orange arrow), and a plot of the confidence scores of having nascent geographic atrophy (nGA) at each of the 49 B-scans from the OCT volume scan shown on the bottom (with the selected B-scan shown on the top indicated by the red arrow, and the threshold for identifying B-scans requiring manual review that resulted in a 0.98 specificity shown as the horizontal red line). The first 2 cases (A and B) represent cases where the fully automated artificial intelligence (AI) model considered the volume scan to have nGA, and where the AI-assisted approach also identified nGA as being present. The next 2 cases (C and D) show examples where the fully automated AI model did not consider nGA as being present, but the manual review of selected B-scans with the AI-assisted approach led to the correct detection of nGA. The 2 final cases (E and F) represent cases deemed to have nGA by the fully automated AI model, that were correctly identified as not having nGA following manual review of the selected B-scans with the AI-assisted approach. The orange arrows indicate the region of interest in (A and B), they are nGA lesions located by manual review and also highlighted by AI (Gradient-weighted Class Activation Mapping, see Fig 4); in (C and D), they are nGA lesions located by manual review only. In (E and F), they are suspicious nGA lesions suggested by AI, but rejected by human review.
Figure 8.
Gradient-weighted Class Activation Mapping saliency map for the cases presented in Figure 7. The saliency is overlaid as green. AI = artificial intelligence; nGA = nascent geographic atrophy
Discussion
Our study demonstrated that an AI-assisted approach for detecting nGA—whereby a deep learning model identifies selected OCT scans for manual evaluation—reduces the number of B-scans requiring manual review by more than 50-fold (i.e., only 1.9% of OCT B-scans required manual review) while maintaining a near-perfect level of detection of nGA onset. In contrast, the fully automated AI approach (relying solely on the output of the deep learning model) correctly detects approximately half of the eyes with nGA at its onset at a sufficiently high specificity of 0.98. These findings underscore the value of an AI-assisted approach for substantially reducing the burden of manual assessments in the detection of nGA.
This is the first study, to our knowledge, that has developed a deep learning model for detecting nGA on OCT imaging, performed in a longitudinal cohort of individuals with bilateral large drusen at baseline. A recent study15 developed a deep learning model to detect the presence of GA on CFPs of > 4500 individuals who were also reviewed longitudinally in the Age-Related Eye Disease Study. When evaluating nearly 60 000 CFPs of eyes that spanned the entire spectrum of the disease (from not having AMD to having advanced AMD), their model had a sensitivity of 0.69 for detecting GA at a 0.98 specificity, with the performance varying based on the GA area present. For instance, it had a sensitivity of 0.21 for detecting definite GA lesions with a total area that was smaller than a 354-μm diameter circle (being twice the size of the minimum diameter used to define GA37), but a sensitivity of 0.78 for detecting GA lesions with a total area larger than half a disc area.15 These findings underscore the challenge of detecting early signs of atrophic changes generally, which is noteworthy when considering the task of detecting nGA on OCT imaging in this study, as these features represent early signs that portend the onset of GA.4, 5, 6, 7 This challenge of detecting early signs of atrophic changes is also reflected in the findings of this study, where we observed a lower performance of the deep learning model for detecting nGA at its first onset compared to its detection across all study visits. These findings are unsurprising as the features used to define nGA often become more pronounced over time as the atrophic lesion progresses, likely accounting for the improved performance of the deep learning model for detecting nGA across all visits in this study compared to the detection of nGA onset. Another study17 developed a deep learning model to detect the presence of GA (which included cRORA) on OCT imaging, using data from the Age-Related Eye Disease Study 2 Ancillary Spectral-Domain OCT Study. This study included 1284 OCT volume scans from 311 participants, where a quarter of the scans had GA and the remaining three-quarters did not. The deep learning model had a sensitivity of 0.83 for detecting GA on OCT imaging at a specificity of 0.95,17 which was similar to the sensitivity of 0.86 for detecting the onset of nGA across all visits with the deep learning model in this study at a higher specificity of 0.97. A fully automated deep learning model has recently been developed to segment and quantify GA lesions on OCT.19 The model was developed using data from routine clinical care and the validation results show that the model achieved comparable performance with human specialists. Another recent study showed that a deep learning model developed using fundus autofluorescence images and OCT en face images predicted the GA growth rate more effectively than a benchmark model that included parameters such as the baseline GA area, distance of the lesion(s) from the fovea, and lesion contiguity (e.g., unifocal or multifocal).14 This model was developed and evaluated using data from the phase III Chroma and Spectri trials and evaluated in independent test sets, and underscores the value of deep learning models for identifying eyes at higher risk of faster GA progression. These studies collectively highlight the promising role of deep learning models for monitoring and predicting GA progression, and also motivate the exploration of using deep learning models in nGA detection.
While the performance of our fully automated deep learning approach would only miss the onset of nGA in 1 in 8 eyes at the visit of its development at a 6% false-positive rate, this rate increases up to missing almost 1 in 2 eyes at a 2% false-positive rate. Such a level of specificity may be necessary in certain settings to avoid falsely considering an eye to have developed a very high-risk feature for the subsequent development of GA.7,8 The AI-assisted approach evaluated in this study therefore provides an alternative strategy for avoiding such false-positives through incorporating manual review. We demonstrated in this study that it could enable near-perfect detection of nGA onset (a sensitivity of 0.95) while requiring only < 2% of the OCT B-scans to be manually reviewed. This AI-assisted approach is analogous to the one taken to tackle the population-based screening of eye diseases using deep learning models to identify a subset of referable cases for further assessment.9, 10, 11, 12 This approach is especially useful in settings with a low prevalence or incidence of the outcome of interest, and it is thus especially relevant in the longitudinal assessment of individuals with the early stages of AMD. For instance, the OCT volume scans at the time of onset from the 40 eyes where nGA developed represents only 2% of all the OCT volume scans over time in this study up until the point of nGA development or until the last visit if it did not develop. Note that since the AI-assisted approach was tuned to achieve near-perfect levels of sensitivity for detecting nGA, its sensitivity would remain unchanged when applied in cohorts with a different prevalence of nGA, although the proportion of B-scans identified as requiring manual review would be expected to be higher in a cohort with a higher prevalence of nGA than in this study (and vice versa).
The deep learning model developed in this study could thus be useful for application in clinical trials of the early stages of AMD, where it could be used to help reduce the burden of manual grading to identify the onset of nGA if it is used as a study endpoint (as was the case in the LEAD study25,26). It could also be used to help automatically screen for individuals with nGA to potentially target them for inclusion in treatment trials aiming to slow progression to GA. This deep learning model could also be used as a clinical decision-support tool for screening for the presence of nGA, in order to guide risk stratification and counseling patients.38
A key limitation of this study is the lack of an independent dataset from a similar population of individuals with intermediate AMD and a matching imaging protocol, to validate the model, which necessitated instead the use of fivefold cross-validation. During each round of the fivefold cross-validation, the validation test set was strictly reserved for evaluation purposes only. It was not used for model training or tuning, ensuring that the evaluation results remain unbiased and accurately reflect the model's performance on unseen data. It is essential to emphasize that the findings of this study serve as proof-of-principle and we encourage further external validation once more relevant datasets become available in the future. Another limitation is that only B-scans from 1 type of scan protocol of only 1 OCT device was used, and thus the generalizability of the findings of this study to OCT scans obtained from other devices remains to be determined. The performance of the model would likely be improved through including a larger cross-sectional sample of eyes with and without nGA, and this would be especially important if the model is to be used as a fully automated means for detecting nGA. However, a strength of this study is that the model was trained on a clinically important label, namely nGA, which we have previously shown in this dataset to be associated with a 78-fold increased rate of GA development and that explained 91% of the variance in the time to develop GA.7 We have also more recently shown that the development of nGA is more strongly associated with GA development than the presence or development of incomplete RPE and outer retinal atrophy, which is why we focused on the development of a deep learning model to screen for nGA in this study.39 Nonetheless, we have previously shown that there was 1 case in this cohort where nGA was not detected prior to, or when, GA developed (as this case was characterized by having an unusual RPE detachment).39 Note finally that the deep learning model in this study was only developed and tested in eyes with non-neovascular AMD, but future studies could extend the findings of this study to develop a deep learning model that could be used to identify OCT B-scans potentially with signs of neovascular AMD or nGA that require manual review with an AI-assisted approach. Such a model would have an even greater level of clinical utility, as both neovascular and atrophic AMD changes are important clinical endpoints when evaluating disease progression in the early stages of AMD. Regarding clinical utility, as with any other computer-aided methods, there might be confirmation bias, i.e., human reviewers may tend to grade the AI-identified cases as positive. In this study, we only evaluated the proposed AI-assisted approach using the already annotated B-scans. The findings of this study are thus only considered proof-of-principle but it encourages further prospective examination.
While these findings suggest that there may be atypical cases that do not conform to the more common transition from nGA to GA, the strong association between nGA with GA onset still underscores the value of accurately identifying nGA on OCT scans. As such, replication of the promising findings of this study in the future is now warranted to establish the clinical applicability of the approaches developed.
In conclusion, this study demonstrated that a deep learning model developed for an AI-assisted approach for detecting the onset of nGA could help to dramatically reduce the number of OCT B-scans requiring manual review by over 50-fold while maintaining near-perfect sensitivity. This approach has the potential to substantially reduce the current manual burden of reviewing OCT B-scans in both clinical trials and practice for detecting nGA.
Acknowledgments
The authors thank the participants and their families who took part in the study, as well as the staff, research coordinators, and investigators at each participating institution. Writing assistance provided by Genentech, Inc.
Manuscript no. XOPS-D-23-00105.
Footnotes
Supplemental material available atwww.aaojournal.org.
Disclosure(s):
All authors have completed and submitted the ICMJE disclosures form.
The authors made the following disclosures:
H.Y.: Shares – F. Hoffmann-La Roche Ltd; Employee – Genentech, Inc.
R.H.G.: Consultant – Roche, Genentech, Inc., Apellis, Novartis, Bayer.
Z.W.: None.
H.C.: Shares – F. Hoffmann-La Roche Ltd; Employee – Genentech, Inc.
S.S.G.: Shares – F. Hoffmann-La Roche Ltd; Employee – Genentech, Inc.
V.S.: Shares – F. Hoffmann-La Roche Ltd; Employee – Genentech, Inc.
M.H.: Shares – F. Hoffmann-La Roche Ltd; Employee – Genentech, Inc.
M.Z.: Shares – F. Hoffmann-La Roche Ltd; Employee – Genentech, Inc.
Supported by Genentech, Inc., United States.
HUMAN SUBJECTS: Human subjects were included in this study. This study included a subset of participants in the sham treatment arm of the Laser Intervention in the Early Stages of AMD (LEAD) study (clinicaltrials.gov identifier: NCT01790802), which was conducted according to the International Conference on Harmonization Guidelines for Good Clinical Practice and the tenets of the Declaration of Helsinki. Institutional Review Board approval was obtained at all sites and all participants provided written informed consent.
No animal subjects were included in this study.
Author Contributions:
Conception and design: Yao, Wu, Zhang
Analysis and interpretation: Yao, Wu, Gao, Guymer, Steffen, Chen, Hejrati, Zhang
Data collection: Yao, Guymer
Obtaining funding: N/A. Study was performed as part of regular employment duties at Genentech, Inc. No additional funding was provided.
Overall responsibility: Zhang
Data Sharing: For up-to-date details on Roche's Global Policy on the sharing of clinical information and how to request access to related clinical study documents, see here: https://www.roche.com/innovation/process/clinical-trials/data-sharing.
Supplementary Data
References
- 1.Holz F.G., Strauss E.C., Schmitz-Valckenberg S., et al. Geographic atrophy: clinical features and potential therapeutic approaches. Ophthalmology. 2014;121:1079–1091. doi: 10.1016/j.ophtha.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 2.Age-Related Eye Disease Study Research Group A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins c and e, beta carotene, and zinc for age-related macular degeneration and vision loss: areds report no. 8. Arch Ophthalmol. 2001;119:1417–1436. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Age-Related Eye Disease Study 2 Research Group Lutein + zeaxanthin and omega-3 fatty acids for age-related macular degeneration. JAMA. 2013;309:2005–2015. doi: 10.1001/jama.2013.4997. [DOI] [PubMed] [Google Scholar]
- 4.Wu Z., Luu C.D., Ayton L.N., et al. Optical coherence tomography defined changes preceding the development of drusen-associated atrophy in age-related macular degeneration. Ophthalmology. 2014;121:2415–2422. doi: 10.1016/j.ophtha.2014.06.034. [DOI] [PubMed] [Google Scholar]
- 5.Wu Z., Luu C.D., Ayton L.N., et al. Fundus autofluorescence characteristics of nascent geographic atrophy in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2015;56:1546–1552. doi: 10.1167/iovs.14-16211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z., Ayton L.N., Luu C.D., et al. Microperimetry of nascent geographic atrophy in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2015;56:115–121. doi: 10.1167/iovs.14-15614. [DOI] [PubMed] [Google Scholar]
- 7.Wu Z., Luu C.D., Hodgson L.A., et al. Prospective longitudinal evaluation of nascent geographic atrophy in age-related macular degeneration. Ophthalmol Retina. 2020;4:568–575. doi: 10.1016/j.oret.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 8.Ferrara D., Silver R.E., Louzada R.N., et al. Optical coherence tomography features preceding the onset of advanced age-related macular degeneration. Invest Ophthalmol Vis Sci. 2017;58:3519–3529. doi: 10.1167/iovs.17-21696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ting D.S.W., Cheung C.Y.-L., Lim G., et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318:2211–2223. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellemo V., Lim Z.W., Lim G., et al. Artificial intelligence using deep learning to screen for referable and vision-threatening diabetic retinopathy in Africa: a clinical validation study. Lancet Digit Health. 2019;1:e35–e44. doi: 10.1016/S2589-7500(19)30004-4. [DOI] [PubMed] [Google Scholar]
- 11.Abràmoff M.D., Lavin P.T., Birch M., et al. Pivotal trial of an autonomous ai-based diagnostic system for detection of diabetic retinopathy in primary care offices. NPJ Digital Medicine. 2018;1:39. doi: 10.1038/s41746-018-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruamviboonsuk P., Tiwari R., Sayres R., et al. Real-time diabetic retinopathy screening by deep learning in a multisite national screening programme: a prospective interventional cohort study. The Lancet Digital Health. 2022;4:e235–e244. doi: 10.1016/S2589-7500(22)00017-6. [DOI] [PubMed] [Google Scholar]
- 13.Treder M., Lauermann J.L., Eter N. Deep learning-based detection and classification of geographic atrophy using a deep convolutional neural network classifier. Graefes Arch Clin Exp Ophthalmol. 2018;256:2053–2060. doi: 10.1007/s00417-018-4098-2. [DOI] [PubMed] [Google Scholar]
- 14.Anegondi N., Salvi A., Cluceru J., et al. Deep learning to predict future region of growth of geographic atrophy from fundus autofluorescence images. Invest Ophthalmol Vis Sci. 2023;64:1117. [Google Scholar]
- 15.Keenan T.D., Dharssi S., Peng Y., et al. A deep learning approach for automated detection of geographic atrophy from color fundus photographs. Ophthalmology. 2019;126:1533–1540. doi: 10.1016/j.ophtha.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liefers B., Colijn J.M., González-Gonzalo C., et al. A deep learning model for segmentation of geographic atrophy to study its long-term natural history. Ophthalmology. 2020;127:1086–1096. doi: 10.1016/j.ophtha.2020.02.009. [DOI] [PubMed] [Google Scholar]
- 17.Shi X., Keenan T.D.L., Chen Q., et al. Improving interpretability in machine diagnosis: detection of geographic atrophy in OCT scans. Ophthalmol Sci. 2021;1 doi: 10.1016/j.xops.2021.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang G., Fu D.J., Liefers B., et al. Clinically relevant deep learning for detection and quantification of geographic atrophy from optical coherence tomography: a model development and external validation study. The Lancet Digital Health. 2021;3:e665–e675. doi: 10.1016/S2589-7500(21)00134-5. [DOI] [PubMed] [Google Scholar]
- 19.Mai J., Lachinov D., Riedl S., et al. Clinical validation for automated geographic atrophy monitoring on OCT under complement inhibitory treatment. Sci Rep. 2023;13:7028. doi: 10.1038/s41598-023-34139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogl W.D., Riedl S., Mai J., et al. Predicting topographic disease progression and treatment response of pegcetacoplan in geographic atrophy quantified by deep learning. Ophthalmol Retina. 2023;7:4–13. doi: 10.1016/j.oret.2022.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Wu Z., Bogunovic H., Asgari R., et al. Predicting progression of age-related macular degeneration using optical coherence tomography and fundus photography. Ophthalmol Retina. 2021;5:118–125. doi: 10.1016/j.oret.2020.06.026. [DOI] [PubMed] [Google Scholar]
- 22.Goh K.L., Abbott C.J., Hadoux X., et al. Hyporeflective cores within drusen: association with progression of age-related macular degeneration and impact on visual sensitivity. Ophthalmol Retina. 2022;6:284–290. doi: 10.1016/j.oret.2021.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Goh K.L., Chen F.K., Balaratnasingam C., et al. Cuticular drusen in age-related macular degeneration: association with progression and impact on visual sensitivity. Ophthalmology. 2022;129:653–660. doi: 10.1016/j.ophtha.2022.01.028. [DOI] [PubMed] [Google Scholar]
- 24.Guymer R.H., Wu Z., Hodgson L.A.B., et al. Subthreshold nanosecond laser intervention in age-related macular degeneration: the lead randomized controlled clinical trial. Ophthalmology. 2019;126:829–838. doi: 10.1016/j.ophtha.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 25.Wu Z., Luu C.D., Hodgson L.A.B., et al. Secondary and exploratory outcomes of the subthreshold nanosecond laser intervention randomized trial in age-related macular degeneration: a lead study report. Ophthalmol Retina. 2019;3:1026–1034. doi: 10.1016/j.oret.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Ferris F.L., III, Wilkinson C., Bird A., et al. Clinical classification of age-related macular degeneration. Ophthalmology. 2013;129:844–851. doi: 10.1016/j.ophtha.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadda S.R., Guymer R., Holz F.G., et al. Consensus definition for atrophy associated with age-related macular degeneration on oct: classification of atrophy report 3. Ophthalmology. 2018;125:537–548. doi: 10.1016/j.ophtha.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang F., Jiang M., Qian C., et al. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. IEEE; San Juan, PR: 2017. Residual attention network for image classification; pp. 3156–3164. [Google Scholar]
- 29.Carbonneau M.-A., Cheplygina V., Granger E., et al. Multiple instance learning: a survey of problem characteristics and applications. Pattern Recogn. 2018;77:329–353. [Google Scholar]
- 30.Gosain A., Sardana S. 2017 International Conference on Advances in Computing, Communications and Informatics (ICACCI) IEEE; Udupi, India: 2017. Handling class imbalance problem using oversampling techniques: a review; pp. 79–85. [Google Scholar]
- 31.Lin T.Y., Goyal P., Girshick R., et al. Proceedings of the IEEE International Conference on Computer Vision. IEEE; Cambridge, MA: 2017. Focal loss for dense object detection; pp. 2980–2988. [Google Scholar]
- 32.Mukhoti J., Kulharia V., Sanyal A., et al. Calibrating deep neural networks using focal loss. Adv Neural Inf Process Syst. 2020;33:15288–15299. [Google Scholar]
- 33.Tran G.S., Nghiem T.P., Nguyen V.T., et al. Improving accuracy of lung nodule classification using deep learning with focal loss. J Healthc Eng. 2019;2019 doi: 10.1155/2019/5156416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shorten C., Khoshgoftaar T.M. A survey on image data augmentation for deep learning. J Big Data. 2019;6:1–48. doi: 10.1186/s40537-021-00492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X., Faes L., Kale A.U., et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Health. 2019;1:e271–e297. doi: 10.1016/S2589-7500(19)30123-2. [DOI] [PubMed] [Google Scholar]
- 36.Selvaraju R.R., Cogswell M., Das A., et al. Proceedings of the IEEE international conference on computer vision. IEEE; Venice, Italy: 2017. Grad-cam: visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- 37.Davis M.D., Gangnon R.E., Lee L.Y., et al. The age-related eye disease study severity scale for age-related macular degeneration: areds report no. 17. Arch Ophthalmol. 2005;123:1484–1498. doi: 10.1001/archopht.123.11.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar H., Goh K.L., Guymer R.H., et al. A clinical perspective on the expanding role of artificial intelligence in age-related macular degeneration. Clin Exp Optom. 2022;105:674–679. doi: 10.1080/08164622.2021.2022961. [DOI] [PubMed] [Google Scholar]
- 39.Wu Z., Goh K.L., Hodgson L.A.B., Guymer R.H. Incomplete retinal pigment epithelial and outer retinal atrophy: longitudinal evaluation in age-related macular degeneration. Ophthalmology. 2023;130:205–212. doi: 10.1016/j.ophtha.2022.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.