Abstract
The bacterial artificial chromosome (BAC) cloning system is capable of stably propagating large, complex DNA inserts in Escherichia coli. As part of the Mycobacterium tuberculosis H37Rv genome sequencing project, a BAC library was constructed in the pBeloBAC11 vector and used for genome mapping, confirmation of sequence assembly, and sequencing. The library contains about 5,000 BAC clones, with inserts ranging in size from 25 to 104 kb, representing theoretically a 70-fold coverage of the M. tuberculosis genome (4.4 Mb). A total of 840 sequences from the T7 and SP6 termini of 420 BACs were determined and compared to those of a partial genomic database. These sequences showed excellent correlation between the estimated sizes and positions of the BAC clones and the sizes and positions of previously sequenced cosmids and the resulting contigs. Many BAC clones represent linking clones between sequenced cosmids, allowing full coverage of the H37Rv chromosome, and they are now being shotgun sequenced in the framework of the H37Rv sequencing project. Also, no chimeric, deleted, or rearranged BAC clones were detected, which was of major importance for the correct mapping and assembly of the H37Rv sequence. The minimal overlapping set contains 68 unique BAC clones and spans the whole H37Rv chromosome with the exception of a single gap of ∼150 kb. As a postgenomic application, the canonical BAC set was used in a comparative study to reveal chromosomal polymorphisms between M. tuberculosis, M. bovis, and M. bovis BCG Pasteur, and a novel 12.7-kb segment present in M. tuberculosis but absent from M. bovis and M. bovis BCG was characterized. This region contains a set of genes whose products show low similarity to proteins involved in polysaccharide biosynthesis. The H37Rv BAC library therefore provides us with a powerful tool both for the generation and confirmation of sequence data as well as for comparative genomics and other postgenomic applications. It represents a major resource for present and future M. tuberculosis research projects.
Cosmid clones (2, 16) have played a crucial role in the Mycobacterium tuberculosis H37Rv genome sequencing project. However, problems such as underrepresentation of certain regions of the chromosome, unstable inserts, and the relatively small insert size complicated the production of a comprehensive set of canonical cosmids representing the entire genome.
In an attempt to obtain complete coverage of the genome with a minimal overlapping set of clones, a bacterial artificial chromosome (BAC) library of M. tuberculosis was constructed by using the vector pBeloBAC11 (12), which combines a simple phenotypic screen for recombinant clones with the stable propagation of large inserts (21). The BAC cloning system is based on the Escherichia coli F factor, whose replication is strictly controlled and thus ensures stable maintenance of large constructs (23). BACs have been widely used for cloning of DNA from various eucaryotic species (6, 12, 15, 24, 25). In contrast, to our knowledge this report describes the first attempt to use the BAC system for cloning bacterial DNA.
A central advantage of the BAC cloning system over cosmid vectors is that the F plasmid is present in only one or a maximum of two copies per cell, reducing the potential for recombination between DNA fragments and, more importantly, avoiding the lethal overexpression of cloned bacterial genes. However, the presence of the BAC as just a single copy means that plasmid DNA has to be extracted from a large volume of culture to obtain sufficient DNA for sequencing, and we describe here a simplified protocol to achieve this.
Forty-seven cosmids chosen from the integrated map of the 4.4-Mb circular chromosome (18) were shotgun sequenced during the initial phase of the H37Rv genome sequence project. The sequences of these clones were used as landmarks in the construction of a minimally overlapping BAC map. Comparison of the sequence data from the termini of 420 BAC clones allowed us to establish a minimal overlapping BAC map and to fill in the existing gaps between the sequence of cosmids. As well as using the BAC library for genomic mapping and sequencing, we also tested the system in comparative genomic experiments to uncover differences between two closely related mycobacterial species. As shown in a previous study (17), M. tuberculosis, M. bovis, and M. bovis BCG exhibit a high level of global genomic conservation, but certain polymorphic regions were also detected. Therefore, it was of great interest to find a reliable, easy, and rapid way to exactly localize polymorphic regions in mycobacterial genomes by using selected BAC clones. This approach was validated by determining the exact size and location of the polymorphisms in the genomic region of DraI fragment Z4 (18), taking advantage of the availability of an appropriate BAC clone covering the polymorphic region and the H37Rv genome sequence data. This region is located approximately 1.7 Mb from the origin of replication.
(Portions of this work were presented at the American Society for Microbiology conference Tuberculosis: Past, Present, and Future, July 8 to 12, 1997, Copper Mountain, Colo.)
MATERIALS AND METHODS
DNA preparation.
Preparation of M. tuberculosis H37Rv DNA in agarose plugs was conducted as previously described (7, 18). Plugs were stored in 0.2 M EDTA at 4°C and washed three times in 0.1% Triton X-100 buffer prior to use.
BAC vector preparation.
pBeloBAC11 was kindly provided by H. Shizuya, Department of Biology, California Institute of Technology (Pasadena, Calif.). Preparation was carried out as described by Woo et al. (24).
Partial digestion with HindIII.
Partial digestion was carried out on plugs, each containing approximately 10 μg of high-molecular-weight DNA, after three 1-h equilibration steps in 50 ml of 1× HindIII digestion buffer (Boehringer, Mannheim, Germany) plus 0.1% Triton X-100. The buffer was then removed and replaced by ice-cold HindIII enzyme buffer (1 ml/plug) containing 20 U of HindIII (Boehringer). After 2 h of incubation on ice, the plugs were transferred to a 37°C water bath for 30 min. Digestions were stopped by adding 500 μl of 50 mM EDTA (pH 8.0).
Size selection.
The partially digested DNA was subjected to contour-clamped homogeneous electric field (CHEF) electrophoresis on a 1% agarose gel, using a DR III apparatus (Bio-Rad, Hercules, Calif.) in 1× Tris-acetate-EDTA buffer at 13°C, with a ramp from 3 to 15 s at 6 V/cm for 16 h. Agarose slices from 25 to 75 kb, 75 to 120 kb, and 120 to 180 kb were excised from the gel and stored in Tris-EDTA at 4°C.
Ligation and transformation.
Agarose slices containing fractions from 25 to 75 kb, 75 to 120 kb, and 120 to 180 kb were melted at 65°C for 10 min and digested with Gelase (Epicentre Technologies, Madison, Wis.), using 1 U per 100 μl of gel slice. Then 25 to 100 ng of the size-selected DNA was ligated to 10 ng of HindIII-digested, dephosphorylated pBeloBAC11 in a 1:10 molar ratio, using 10 U of T4 DNA ligase (New England Biolabs, Beverly, Mass.) at 16°C for 20 h. Ligation mixtures were heated at 65°C for 15 min and then drop-dialyzed against Tris-EDTA, using VS 0.025 mM membranes (Millipore, Bedford, Mass.). Fresh electrocompetent E. coli DH10B cells (20) were harvested from 200 ml of a mid-log (optical density at 550 nm of 0.5) culture grown in SOB medium. Cells were washed three times in ice-cold water and finally resuspended in ice-cold water to a cell density of 1011 cells/ml (optical density at 550 nm of 150). One microliter of the ligation mix was used for electroporation of 30 μl of electrocompetent E. coli DH10B in an Easyject Plus electroporator (Eurogentec, Seraing, Belgium), with settings of 2.5 kV, 25 μF, and 99 Ω, in 2-mm-wide electroporation cuvettes. After electroporation, cells were resuspended in 600 μl of SOC medium, allowed to recover for 45 min at 37°C with gentle shaking, and then plated on LB agar containing chloramphenicol (12.5 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 50 μg/ml), and isopropyl-β-d-thiogalactopyranoside (IPTG; 25 μg/ml). The plates were incubated overnight, and recombinants (white colonies) were picked manually to 96-well plates. Each clone was inoculated three times (twice with 200 μl and once with 100 μl of 2YT-chloramphenicol 12.5 μg/ml per clone) and incubated overnight. One of the microtiter plates, containing 100 μl of culture per well, was maintained as a master plate at −80°C after 100 μl of 80% glycerol was added to each well, while minipreps (19) were prepared from the remaining two plates to check for the presence of inserts. Clones containing inserts were then designated Rv clones and repicked from the master plate to a second set of plates for storage of the library at −80°C.
Preparation of DNA for sizing, direct sequencing, and comparative genomics.
A modified Birnboim and Doly protocol (5) was used for extraction of plasmid DNA for sequencing purposes. Each Rv clone was inoculated into a 50-ml Falcon polypropylene tube containing 40 ml of 2YT medium with 12.5 μg of chloramphenicol per ml and grown overnight at 37°C with shaking. Cells were harvested by centrifugation and stored at −20°C. The frozen pellet was resuspended in 4 ml of solution A (50 mM glucose, 10 mM EDTA, 25 mM Tris [pH 8.0]), and 4 ml of freshly prepared solution B (0.2 M NaOH, 0.2% sodium dodecyl sulfate) was then added. The solution was gently mixed and kept at room temperature for 5 min before addition of 4 ml of ice-cold solution C (3 M sodium acetate [pH 4.7]). Tubes were kept on ice for 15 min and centrifuged at 10,000 rpm for 15 min. After isopropanol precipitation, the DNA pellet was dissolved in 600 μl of RNase solution (15 mM Tris HCl [pH 8.0], 10 μg of RNase A per ml). After 30 min at 37°C, the DNA solution was extracted with chloroform-isoamyl alcohol (24:1) and precipitated from the aqueous phase with isopropanol. The DNA pellet was then rinsed with 70% ethanol, air dried, and dissolved in 30 μl of distilled water. In general, DNA prepared by this method was clean and concentrated enough to give good-quality results by automatic sequencing (at least 300 bp of sequence). For a few DNA preparations, an additional polyethylene glycol (PEG) precipitation step was necessary, which was performed as follows. The 30 μl of DNA solution was diluted to 64 μl, mixed gently, and precipitated with 16 μl of 4 M NaCl and 80 μl of 13% PEG 8000. After 30 min on ice, the tubes were centrifuged at 4°C, and the pellet was carefully rinsed with 70% ethanol, air dried, and diluted in 20 μl of distilled water.
Sizing of inserts.
Insert sizes were determined by pulsed-field gel electrophoresis (PFGE) after cleavage with DraI (Promega). DNA (100 to 200 ng) was DraI cleaved in a 20-μl total reaction volume as recommended by the manufacturer and then loaded onto a 1% agarose gel and migrated, using a pulse of 4 s for 15 h at 6.25 V/cm at 10°C on an LKB-Pharmacia CHEF apparatus. Mid-range and low-range PFGE markers (New England Biolabs) were used as size standards. Insert sizes were estimated after ethidium bromide staining of gels.
Direct sequencing.
For each sequencing reaction 7 μl of BAC DNA (300 to 500 ng), 2 μl of primer (2 μM), 8 μl of reaction mix of a Taq DyeDeoxy Terminator cycle sequencing kit (Applied Biosystems), and 3 μl of distilled water were used. After 26 cycles (96°C for 30 s; 56°C for 15 s; 60°C for 4 min) in a thermocycler (MJ Research Inc., Watertown, Mass.), DNA was precipitated with 70 μl of 70% ethanol–0.5 mM MgCl2, centrifuged, rinsed with 70% ethanol, dried, and dissolved in 2 μl of formamide-EDTA buffer. SP6 and T7 samples of 32 BAC clones were loaded onto 64-lane, 6% polyacrylamide gels, and electrophoresis was performed on a model 373A automatic DNA sequencer (Applied Biosystems) for 12 to 16 h. The sequences of oligonucleotides used as primers are shown in Table 1.
TABLE 1.
Primers used for PCRs and sequencing
| Primer | Sequence |
|---|---|
| Vector-specific primers for DOP-PCR, 1st amplification step | |
| SP6-BAC1 | AGT TAG CTC ACT CAT TAG GCA |
| T7-BAC1 | GGA TGT GCT GCA AGG CGA TTA |
| Vector-specific primers (direct sequencing, nested primers for 2nd DOP-PCR step) | |
| SP6 Mid | AAA CAG CTA TGA CCA TGA TTA CGC CAA |
| T7-Belo2 | TCC TCT AGA GTC GAC CTG CAG GCA |
| Degenerate primers | |
| Deg2 | TCT AGA NNN NNN TCC GGC |
| Deg3 | TCT AGA NNN NNN GGG CCC |
| Deg4 | CGT TTA AAN NNN NWA GGC CG |
| Deg6 | GGT ACT AGT NNN NNW TCC GGC |
| Primers used for amplification of M. bovis DNA in polymorphic chromosomal region of Rv58 | |
| Primer 1 | ACG ACC TCA TAT TCC GAA TCC C |
| Primer 2 | GCA TCT GTT GAG TAC GCA CTT CC |
PCR with degenerate oligonucleotide primers (DOP-PCR).
As an alternate procedure we used partially degenerate oligonucleotides in combination with vector-specific (SP6 or T7) primers to amplify insert ends of BAC clones, following a previously published protocol for P1 clones (13). The degenerate primers Deg2, Deg3, Deg4, and Deg6 (Table 1) gave the best results for selected amplification of insert termini.
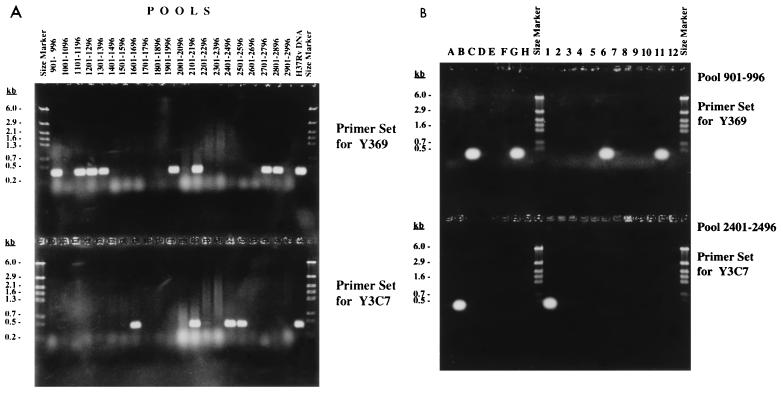
Screening by pooled PCR.
To identify particular clones in the library which could not be detected by random end sequencing of the 400 BAC clones, PCR screening of DNA pools was performed. Primers were designed for regions of the chromosome where no BAC coverage was apparent, using cosmid or H37Rv whole-genome shotgun sequences. Primers were designed to amplify approximately 400 to 500 bp. Ninety-six-well plates containing 200 μl of 2YT-chloramphenicol (12.5 μg/ml) per well were inoculated with 5 μl of −80°C glycerol stock cultures each from the master plates and incubated overnight. The 96 clones of each plate were pooled by taking 20 μl of culture from each well, and this procedure was repeated for 31 plates. Pooled cultures were centrifuged, the pellets were resuspended in sterile water, boiled for 5 min, and centrifuged, and the supernatants were kept for PCRs. As an initial screening step, the 31 pools of a total of 2,976 BACs, representing about two-thirds of the library, were tested for the presence of a specific clone, using appropriate PCR primers. PCR was performed with 10 μl of supernatant, 5 μl of assay buffer [100 mM β-mercaptoethanol, 600 mM Tris HCl (pH 8.8), 20 mM MgCl2, 170 mM (NH4)2SO4], 5 μl of dimethyl sulfoxide, 5 μl of deoxynucleoside triphosphates (20 mM), 5 μl of water, 10 μl of primer (2 μM), 10 μl of inverse primer (2 μM), and 0.2 U of Taq DNA polymerase (Boehringer). Thirty-two cycles of PCR (95°C for 30 s; 55°C for 1 min 30 s; 72°C for 2 min) were performed after an initial denaturation at 95°C for 1 min. An extension step at 72°C for 5 min finished the PCR. If a pool of 96 clones yielded an appropriate PCR product (Fig. 1A), subpools were made to identify the specific clone. Subpools representative for lane A of a 96-well plate were made by pooling clones 1 to 12 from lane A into a separate tube. Subpools for lanes B to H were made in the same way. In addition, subpools of each of the 12 rows (containing eight clones each) were made, so that for one 96-well plate, 20 subpools were obtained. PCR with these 20 subpools identified the specific clone (Fig. 1B, bottom). If more than one specific clone was present among the 96 clones of one plate (Fig. 1B, top), additional PCRs had to be performed with the possible candidates (data not shown).
FIG. 1.
PCR screening for unique BAC clones with specific primers for two selected genomic regions of the H37Rv chromosome, using 21 pools representating 2,016 BACs (A) and sets of 20 subpools from selected positive pools (B).
Genomic comparisons.
DNA from the BAC clone Rv58 was digested with the restriction endonucleases EcoRI and PvuII and resolved by agarose gel electrophoresis at low voltage overnight (1.5 V/cm). DNA was transferred via the method of Southern to nitrocellulose membranes (Hybond C Extra; Amersham) according to standard protocols (19) and then fixed to the membranes at 80°C for 2 h. The blot was hybridized with 32P-labeled total genomic DNA from M. tuberculosis H37Rv, M. bovis type strain ATCC 19210, or M. bovis BCG Pasteur. Hybridization was performed at 37°C overnight in 50% formamide hybridization buffer as previously described (18). Results were interpreted from the autoradiograms.
Computer analysis.
Sequence data from the automated sequencer ABI373A were transferred as binary data to a Digital Alpha 200 station or Sun SparcII station and analyzed by using TED, a sequence analysis program from the Staden software package (11). Proofread sequences were compared by using the BLAST programs (1) to the M. tuberculosis H37Rv sequence databases of the Sanger Centre, containing the collected cosmid sequences (TB.dbs) and whole-genome shotgun reads (TB_shotgun_all.dbs) (18a). In addition, local databases containing 1,520 cosmid end sequences and the accumulating BAC end sequences were used to determine the exact locations of end-sequenced BACs on the physical and genetic map. MycDB (4) and public databases (EMBL and GenBank) were also used to compare new sequences, but to a lesser extent. The organization of the open reading frames (ORFs) in the polymorphic region of clone Rv58 was determined by using the DIANA software established at the Sanger Centre.
Nucleotide sequence accession number.
The novel M. bovis sequence of the polymorphic region was deposited under accession no. AJ003103 in the EMBL nucleotide sequence data library.
RESULTS
Construction of a pBeloBAC11 library of M. tuberculosis H37Rv.
Partial HindIII fragments of H37Rv DNA in the size range of 25 to 180 kb were ligated into pBeloBAC11 and electroporated into E. coli DH10B. While cloning of fractions I (25 to 75 kb) and II (75 to 120 kb) gave approximately 4 × 104 transformants (white colonies), cloning of fraction III (120 to 180 kb) repeatedly resulted in empty clones. Parallel cloning experiments using partial HindIII digests of human DNA resulted in stable inserts for all three fractions (data not shown), suggesting that the maximum size of large inserts in BAC clones is strongly dependent on the source of the DNA. Analysis of the clones for the presence of inserts revealed that 70% of the clones had an insert of the appropriate size while the remaining 30% of white colonies represented empty or lacZ′-mutated clones. Size determination of randomly selected, DraI-cleaved BACs via PFGE showed that the insert sizes ranged for the majority of the clones between 40 and 100 kb, with an average size of 70 kb. Clones with inserts of appropriate size were designated with Rv numbers, recultured, and stored at −80°C for further use.
Direct DNA sequence analysis of BACs.
To characterize the BAC clones, we systematically subjected them to insert termini sequencing. Two approaches, direct sequencing of BAC DNA and DOP-PCR, adapted to the high G+C content of mycobacterial DNA, were used. In a first screening phase, 50 BAC clones, designated Rv1 to Rv50, were analyzed by using both methods in parallel. Except for two clones for which the sequences diverged significantly, the sequences obtained by the two methods differed only in length. Sequences obtained directly were on average about 350 bp long, and for 95% of the clones both the SP6 and T7 end sequences were obtained at the first attempt. Sequences obtained by DOP-PCR were mostly shorter than 300 bp. For 40% of the BACs, we obtained only very short amplicons of 50 to 100 bp from one end. In two cases, the sequence obtained with the DOP-PCR differed from the sequences obtained by direct sequencing, and in these cases E. coli or vector sequences were amplified (data not shown). Taking the advantages and disadvantages of both methods into account, we decided to use direct termini sequencing for the systematic determination of the SP6 and T7 end sequences.
Representativity of the library.
After having determined the end sequences of 400 BACs, we observed a certain redundancy. The majority of clones were represented at least three to four times. Maximum redundancy was seen in the vicinity of the unique rrn operon, as 2.5% of the clones carried identical fragments that bridge cosmids Y50 and Y130 (approximate position at 1440 kb [see Fig. 3]). The majority of clones with identical inserts appeared as two variants, corresponding to both possible orientations of the HindIII fragment in pBeloBAC11. This suggests that the redundancy was not the result of amplification during library construction but due to the limited number of possible combinations of partial HindIII fragments in the given size range of 25 to 120 kb. To detect rare BAC clones, a pooled PCR protocol was used. Primers were designed on the basis of the existing cosmid sequences and used to screen 31 pools of 96 BAC clones. When positive PCR products of the correct size were obtained, smaller subpools (of 8 or 12 clones each) of the corresponding pool were subsequently used to identify the corresponding clone (Fig. 1). With this approach, 20 additional BACs (Rv401 to Rv420) were found for the regions where no BACs were found with the initial systematic sequencing approach. The end sequences of these BACs (Rv401 to Rv420) were determined by direct sequencing, which confirmed the predicted location of the clones on the chromosome. A 97% coverage of the genome of H37Rv with BAC clones was obtained. Only one region of ∼150 kb was apparently not represented in the BAC library, as screening of all pools with several sets of specific primers did not reveal the corresponding clone. This was probably due to the fact that HindIII fragments of mycobacterial DNA larger than 110 kb are very difficult to establish in E. coli and that a HindIII fragment of ∼120 kb is present in this region of the chromosome (data not shown).
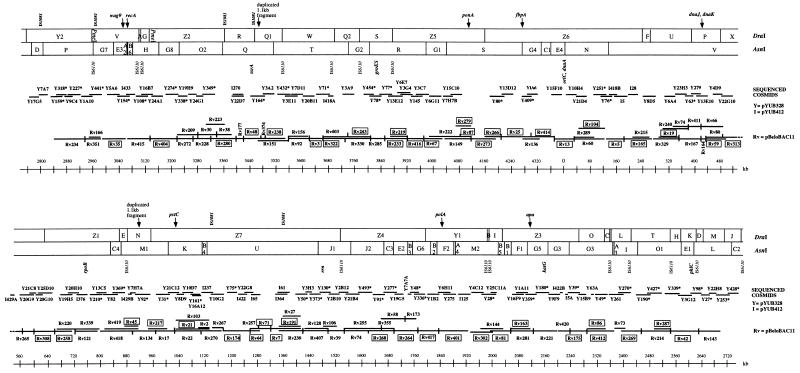
FIG. 3.
Minimal overlapping BAC map of M. tuberculosis H37Rv superimposed on the integrated physical and genetic map established by Philipp et al. (18). Y and I numbers show pYUB328 (2) and pYUB412 (16) cosmids which were shotgun sequenced during the H37Rv genome sequencing project. Y cosmids marked with asterisks were shown in the integrated physical and genetic map (18). Rv numbers show the positions of representative BAC clones relative to sequenced Y and I clones. Rv numbers in boxes represent BACs which were shotgun sequenced at the Sanger Centre.
Establishing a BAC map.
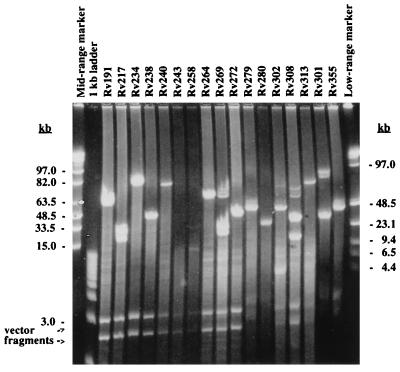
By using all end sequence and shotgun sequence data from the H37Rv genome sequencing project, most of the BAC clones could then be localized by sequence comparison on the integrated map of the chromosome of M. tuberculosis H37Rv (18), and an ordered physical map of the BAC clones was established. PCR with primers from the termini sequences of selected BACs was used for chromosomal walking and confirmation of overlapping BACs (data not shown). The correct order of BACs on the map was also confirmed more recently, using 40,000 whole-genome shotgun reads established at the Sanger Centre. In addition, we performed PFGE of DraI digests of selected BACs (Fig. 2) to see if the approximate fragment size and the presence or absence of DraI cleavage sites in the insert were consistent with the location of the BACs on the physical map (Fig. 3). Comparison of the sequence-based BAC map with the physical and genetic map, established by PFGE and hybridization experiments (18), showed that the two maps were in good agreement. The positions of eight genetic markers previously shown on the physical and genetic map were directly confirmed by BAC end sequence data (Table 2; Fig. 3). The positions of 43 from 47 Y clones (91%) shown on the physical and genetic map, which were later shotgun sequenced, were confirmed by the BAC end sequence and shotgun sequence data. Four clones (Y63, Y180, Y251, and Y253) were located to different positions than previously thought; this discrepancy was found to be due to bookkeeping errors or to chimeric inserts. Their present approximate locations relative to the oriC are shown in Fig. 3: Y63 at 380 kb, Y63A at 2300 kb, Y180 at 2160 kb, Y251 at 100 kb, and Y253 at 2700 kb. A total of 48 BACs, covering regions of the chromosome not represented by cosmids were then shotgun sequenced (9); these are boxed in Fig. 3. No chimeric BACs were found, which is consistent with the observations of other research groups for other BAC libraries (6, 25). The absence of chimeric BACs was of particular importance for the correct assembly of the M. tuberculosis H37Rv sequence. The exact positions of the BAC terminal sequences on the chromosome will be available via the World Wide Web (11a).
FIG. 2.
PFGE gel of DraI-cleaved BAC clones used for estimating the insert sizes of BACs.
TABLE 2.
Identities of genetic markers previously shown on the integrated and genetic map of H37Rv (18) which showed perfect sequence homology with BAC end sequences
| Locus | BAC end sequence | Description of genetic marker | Organism | GenBank accession no. |
|---|---|---|---|---|
| apa | Rv163SP6 | Secreted alanine-proline-rich antigen | M. tuberculosis | X80268 |
| dnaJ, dnaK | Rv164T7 | DnaJ heat shock protein | M. leprae | M95576 |
| fbpA | Rv136T7 | Fibronectin binding protein | M. tuberculosis | M27016 |
| polA | Rv401T7 | DNA polymerase I | M. tuberculosis | L11920 |
| ponA | Rv273T7 | Penicillin binding protein | M. leprae | S82044 |
| pstC | Rv103T7 | Putative phosphate transport receptor | M. tuberculosis | Z48057 |
| recA | Rv415SP6 | Homologous recombination | M. tuberculosis | X58485 |
| wag9 | Rv35SP6 | 35-kDa antigen | M. tuberculosis | M69187 |
Repetitive end sequences.
Repetitive sequences can seriously confound mapping and sequence assembly. In the case of the BAC end sequences, no particular problems with repetitive sequences were observed. Although nine clones with one end in an IS1081 (10) sequence were identified, it was possible to correctly locate their positions on the map by using the sequence of the second terminus. Moreover, these BACs were used to determine the exact locations of IS1081 sequences on the map. Five copies of this insertion sequence, which harbors a HindIII cleavage site, were mapped on the previous physical and genetic map. In contrast, BAC end sequence data revealed an additional copy of IS1081 on the M. tuberculosis H37Rv chromosome. The additional copy was identified by six clones (Rv27, Rv118, Rv142, Rv160, Rv190, and Rv371) which harbored an identical fragment linking Y50 to I364 (Fig. 3, at ∼1380 kb). This copy of IS1081 was not found by previous hybridization experiments, probably because it is located near another copy of IS1081, localized on the same DraI fragment Z7 and AsnI fragment U (Fig. 3, at ∼1140 kb). Furthermore, the position of a copy of IS1081 previously shown in DraI fragment Y1 (Fig. 3, at ∼1840 kb) had to be changed to the region of Y349 (Fig. 3, at ∼3340 kb) according to the end sequences of BAC Rv223. The positions of the four other IS1081 copies were confirmed by the sequence data and therefore remained unchanged. In total six copies of IS1081 were identified in the H37Rv genome, in agreement with the findings of others (10).
In addition, a 1,165-bp-long sequence containing a HindIII site was found in two copies in the genome of H37Rv in different regions. The end sequences of BAC clones Rv48 and Rv374 covering cosmid Y164, as well as Rv419 and Rv45 that cover cosmid Y92, had perfect identity with the corresponding parts of this 1,165-bp sequence (Fig. 3, at ∼3480 and ∼900 kb). Analysis of the sequence did not reveal any homology with insertion sequences or other repetitive elements. However, as each of the two locations showed appropriate BAC coverage, chimerism of the sequenced cosmids Y164 and Y92 can be ruled out as the probable cause.
Using BAC clones in comparative genomics.
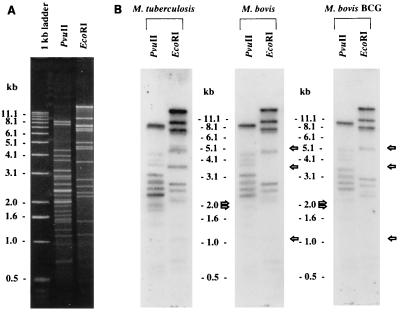
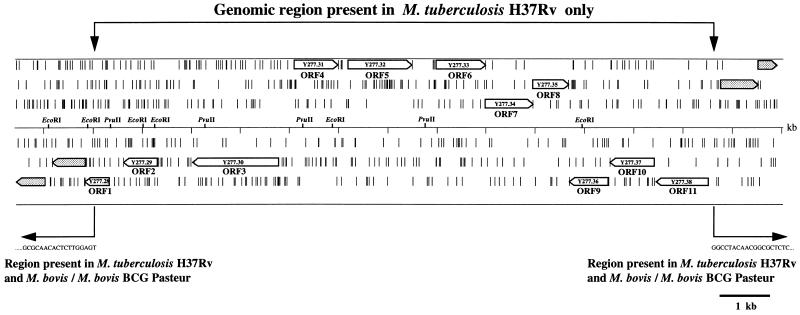
The minimal overlapping set of BAC clones represents a powerful tool for comparative genomics. For example, with each BAC clone containing on average an insert of 70 kb, it should be possible to cover a 1-Mb section of the chromosome with 15 BAC clones. Restriction digests of overlapping clones can then be blotted to membranes and probed with radiolabeled total genomic DNA from, for example, M. bovis BCG Pasteur. Restriction fragments that fail to hybridize with the M. bovis BCG Pasteur DNA must be absent from its genome, hence identifying polymorphic regions between M. bovis BCG Pasteur and M. tuberculosis H37Rv. The results of such an analysis with clone Rv58 (Fig. 3, at ∼1680 kb) are shown here. This clone covers a previously described polymorphic genomic region between M. tuberculosis and M. bovis BCG strains (17). EcoRI and PvuII digests from clone Rv58, fixed on nitrocellulose membranes, were hybridized with 32P-labeled total genomic DNA from M. tuberculosis H37Rv, M. bovis (ATCC 19120), and M. bovis BCG Pasteur. The results of this analysis are shown in Fig. 4, where it is clear that several restriction fragments from clone Rv58 failed to hybridize with genomic DNA from either M. bovis or M. bovis BCG Pasteur. On the basis of the various missing restriction fragments, a restriction map of the polymorphic region was established and compared to the H37Rv sequence data. The localization of the polymorphism could therefore be estimated, and appropriate oligonucleotide primers (Table 1) were selected for the amplification and sequencing of the corresponding region in M. bovis. The alignment of M. bovis and M. tuberculosis H37Rv sequences showed that 12,732 bp were absent from the chromosomal region of the M. bovis type strain and M. bovis BCG Pasteur strain. The G+C content of the polymorphic region is 62.3 mol%, which is similar to the average genome G+C content of the M. tuberculosis genome, hence indicating that this region is not a prophage or other such insertion. Subsequent PCR studies revealed that this segment was also absent from the Danish, Russian, and Glaxo substrains of M. bovis BCG, suggesting that this polymorphism can be used to distinguish M. bovis from M. tuberculosis. Analysis of this sequence showed that 11 putative ORFs, corresponding to ORFs MTCY277.28 to MTCY277.38 (accession no. Z79701 in the EMBL nucleotide sequence data library), are present in M. tuberculosis (Fig. 5). FASTA searches against the protein and nucleic acid databases revealed that the genes of this region may be involved in polysaccharide biosynthesis. Among these putative genes, the highest score was seen with ORF 6 (MTCY277.33), whose putative product shows 51.9% identity with GDP–d-mannose dehydratase from Pseudomonas aeruginosa (accession no. U18320 in the EMBL nucleotide sequence data library) in a 320-amino-acid overlap.
FIG. 4.
Ethidium bromide-stained gel (A) and corresponding Southern blot (B) of EcoRI- and PvuII-digested Rv58 DNA hybridized with 32P-labeled genomic DNA preparations from M. tuberculosis H37Rv, M. bovis ATCC 19210, and M. bovis BCG Pasteur.
FIG. 5.
Organization of the ORFs in the 12.7-kb genomic region present in M. tuberculosis H37Rv but not present in M. bovis ATCC 19210 and M. bovis BCG Pasteur. Arrows show the direction of transcription of the putative genes. Positions of EcoRI and PvuII restriction sites are shown. Vertical dashes represent stop codons. The 11 ORFs correspond to the ORFs MTCY277.28 to MTCY277.38 (accession no. Z79701 in the EMBL nucleotide sequence data library). The junction sequences flanking the polymorphic region are shown.
DISCUSSION
Radical measures are required to prevent the grim predictions of the World Health Organization for the evolution of the global tuberculosis epidemic in the next century becoming a tragic reality. The powerful combination of genomics and bioinformatics is providing a wealth of information about the etiologic agent, M. tuberculosis, that will facilitate the conception and development of new therapies. The start point for genome sequencing was the integrated map of the 4.4-Mb circular chromosome of the widely used, virulent reference strain, M. tuberculosis H37Rv, and appropriate cosmids were subjected to systematic shotgun sequence analysis at the Sanger Centre.
The finding that some regions of the chromosome were apparently underrepresented in cosmids and further problems such as the presence of a small number of chimeric or unstable inserts have complicated the production of a minimal overlapping cosmid map of the genome. To finish the sequence of the whole genome and to confirm the topology of the sequence assembly, another, more representative library was needed. Therefore, we decided to construct a BAC library of M. tuberculosis H37Rv to overcome these problems. The success of this approach depended on whether the resulting BAC clones could maintain large mycobacterial DNA inserts. There are various reports describing the successful construction of a BAC library for eucaryotic organisms (6, 12, 15, 24, 25) where inserts of up to 725 kb (25) were cloned and stably maintained in the E. coli host strain. Here, it is shown that the BAC system can also be used for mycobacterial DNA, as 70% of the clones contained inserts with sizes of 25 to 104 kb. Attempts to clone larger mycobacterial DNA fragments into pBeloBAC11 failed; cloning of partial HindIII digests of fraction III in the size range of 120 to 180 kb resulted in empty clones, probably as the result of lethal overexpression of certain genes or to the particular procedure which is necessary to lyse the very resistant cell walls of mycobacteria when one is preparing chromosomal DNA in agarose plugs. The upper insert size limit of ∼110 kb was apparently the reason why one region of the chromosome, in the size range of ∼150 kb, was not covered by BACs. Sequence analysis and PFGE mapping revealed that in this genomic region HindIII sites were 120 kb apart. As this region is almost completely covered by Y clones (Fig. 3, from ∼2700 to ∼2850 kb), no further attempts to clone the 120-kb fragment and the neighboring small fragments were made. Nevertheless, through the combined use of systematic end sequencing and PCR screening, we established a canonical collection of BACs that represents 97% of the genome (Fig. 3), and 48 BACs were subsequently shotgun sequenced.
The identification of genomic differences possibly responsible for phenotypic variations in different mycobacterial species continues to be an attractive research objective. The stability and fidelity of maintenance of the clones in the BAC library represent ideal characteristics for such projects. As we show in this report, BACs can be allied with conventional hybridization techniques for refined analyses of genomes from different mycobacterial species. Having established a reliable procedure to screen for genomic polymorphisms, we can now conduct these comparisons on a more systematic basis, using representative BACs throughout the chromosome and genomic DNA from a variety of mycobacterial species. As another approach to display genomic polymorphisms, we have started to use selected H37Rv BACs for “molecular combing” experiments in combination with fluorescent in situ hybridization (3, 14). With such techniques, we will be able to explore the genome of M. tuberculosis for further polymorphic regions.
The 12.7-kb segment that is present in M. tuberculosis and absent in M. bovis contains 11 putative ORFs, identified on the basis of their codon usage (Fig. 5). Although the ORFs do not show high sequence similarity with other sequences in the EMBL and GenBank databases, the FASTA scores of some of the putative gene products indicate a low similarity with proteins involved in polysaccharide biosynthesis. It is not immediately evident what phenotypic difference between human and bovine strains of tubercle bacilli this region may encode. The diagnostic tests used to differentiate between human and bovine strains exploit the fact that human strains can respire with nitrate, can produce niacin, and are resistant to thiophene-2-carboxylic acid hydrazide, while bovine strains show the opposite phenotypes. Furthermore, bovine tubercle strains can infect humans whereas human strains are not pathogenic for bovines. Complementation studies of M. bovis BCG with the deleted region from M. tuberculosis are under way, with the aim of identifying the function of this 12.7-kb region.
For diagnostic purposes, this 12.7-kb deletion should allow a rapid PCR screening of tubercle isolates to identify whether they are bovine or human strains. The primers listed in Table 1 flank the deleted region and give a 722-bp amplicon in M. bovis or M. bovis BCG strains but a fragment of 13,453 bp in M. tuberculosis that is practically impossible to amplify under the same PCR conditions. More importantly, assuming that some of the gene products from this region represent proteins with antigenic properties, it could be possible to develop a test that can reliably distinguish between the immune response induced by vaccination with M. bovis BCG vaccine strains and infection with M. tuberculosis.
The alliance of such BAC-based approaches to the advances in comparative genomics by the availability of an increased number of complete genomes and the rapid increase of well-characterized gene products in the public databases will allow an exhaustive analysis of the mycobacterial genome. Since 97% of the chromosome is covered by the BAC library, it will also play an important role in other postgenomic applications, such as in mycobacterial gene expression studies where the canonical set of BACs could be used as a matrix for hybridization studies. Probing such matrices with cDNA probes prepared from total mRNA will uncover genetic loci induced or repressed under different physiological conditions (8, 22). As such, the H37Rv BAC library represents a fundamental resource for present and future research projects, particularly as the various clones from this library are freely available to the scientific community upon request.
ACKNOWLEDGMENTS
We are particularly grateful to Gilles Thomas for his interest and kind support.
Funding for this work was provided by the Wellcome Trust. Additional funding was provided by the Association Française Raoul Follereau and the Institut Pasteur. S. V. Gordon received a Wellcome International Prize Travelling Research Fellowship.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Balasubramanian V, Pavelka M S, Jr, Bardarov S S, Martin J, Weisbrod T R, McAdam R A, Bloom B R, Jacobs W R., Jr Allelic exchange in Mycobacterium tuberculosis with long linear recombination substrates. J Bacteriol. 1996;178:273–279. doi: 10.1128/jb.178.1.273-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bensimon A, Simon A, Chiffaudel A, Croquette V, Heslot F, Bensimon D. Alignment and sensitive detection of DNA by a moving interface. Science. 1994;265:2096–2098. doi: 10.1126/science.7522347. [DOI] [PubMed] [Google Scholar]
- 4.Bergh S, Cole S T. MycDB: an integrated mycobacterial database. Mol Microbiol. 1994;12:517–534. doi: 10.1111/j.1365-2958.1994.tb01039.x. [DOI] [PubMed] [Google Scholar]
- 5.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai L, Taylor J F, Wing R A, Gallagher D S, Woo S S, Davis S K. Construction and characterization of a bovine bacterial artificial chromosome library. Genomics. 1995;29:413–425. doi: 10.1006/geno.1995.9986. [DOI] [PubMed] [Google Scholar]
- 7.Canard B, Cole S T. Genome organization of the anaerobic pathogen Clostridium perfringens. Proc Natl Acad Sci USA. 1989;86:6676–6680. doi: 10.1073/pnas.86.17.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuang S, Daniels D L, Blattner F R. Global regulation of gene expression in Escherichia coli. J Bacteriol. 1993;175:2026–2036. doi: 10.1128/jb.175.7.2026-2036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole S T, Brosch R, Eiglmeier K, Garnier T, Gordon S V, Churcher C, Harris D, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Holroyd S, Gentles S, Jagels K, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Parkhill J, Quail M, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J, Taylor K, Whitehead S, Barrell B G. Genome sequence of Mycobacterium tuberculosis H37Rv. Microb Comp Genomics. 1997;2:174. [Google Scholar]
- 10.Collins D M, Stephens D M. Identification of an insertion sequence, IS1081, in Mycobacterium bovis. FEMS Microbiol Lett. 1991;67:11–15. doi: 10.1016/0378-1097(91)90435-d. [DOI] [PubMed] [Google Scholar]
- 11.Dear S, Staden R A. Sequence assembly and editing program for the efficient management of large projects. Nucleic Acids Res. 1991;19:3907–3911. doi: 10.1093/nar/19.14.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Gas, S., and T. Garnier. MycDB: an integrated mycobacterial database, release 4.23. www.pasteur.fr/mycDB/
- 12.Kim U J, Birren B W, Slepak T, Mancino V, Boysen C, Kang H L, Simon M I, Shizuya H. Construction and characterization of a human bacterial artificial chromosome library. Genomics. 1996;34:213–218. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y G, Whittier R F. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- 14.Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, Vanslegtenhorst M, Wolfe J, Povey S, Beckmann J S, Bensimon A. Dynamic molecular combing—stretching the whole human genome for high-resolution studies. Science. 1997;277:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- 15.Misumi D J, Nagle D L, McGrail S H, Dussault B J, Jr, Smutko J S, Chen H, Charlat O, Duyk G M, Ebeling C, Baldini L, Carlson G A, Moore K J. The physical and genetic map surrounding the Lyst gene on mouse chromosome. Genomics. 1997;40:147–150. doi: 10.1006/geno.1996.4549. [DOI] [PubMed] [Google Scholar]
- 16.Pavelka M S, Jr, Jacobs W R., Jr Biosynthesis of diaminopimelate, the precursor of lysine and a component of peptidoglycan, is an essential function of Mycobacterium smegmatis. J Bacteriol. 1996;178:6496–6507. doi: 10.1128/jb.178.22.6496-6507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philipp W J, Nair S, Guglielmi G, Lagranderie M, Gicquel B, Cole S T. Physical mapping of Mycobacterium bovis BCG Pasteur reveals differences from the genome map of Mycobacterium tuberculosis H37Rv and from M. bovis. Microbiology. 1996;142:3135–3145. doi: 10.1099/13500872-142-11-3135. [DOI] [PubMed] [Google Scholar]
- 18.Philipp W J, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom B R, Jacobs W R, Jr, Cole S T. An integrated map of the genome of the tubercle bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Rajandream, M.-A. Microbial genomes: Mycobacterium tuberculosis. http://www.sanger.ac.uk/Projects/M_tuberculosis/.
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Sheng Y, Mancino V, Birren B. Transformation of Escherichia coli with large DNA molecules by electroporation. Nucleic Acids Res. 1995;23:1990–1996. doi: 10.1093/nar/23.11.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shizuya H, Birren B, Kim U J, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trieselmann B A, Charlebois R L. Transcriptionally active regions in the genome of the archaebacterium Haloferax volcanii. J Bacteriol. 1992;174:30–34. doi: 10.1128/jb.174.1.30-34.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willets N, Skurray R. Structure and function of the F-factor and mechanism of conjugation. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C: American Society for Microbiology; 1987. pp. 1110–1133. [Google Scholar]
- 24.Woo S S, Jiang J, Gill B S, Paterson A H, Wing R A. Construction and characterization of a bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 1994;22:4922–4931. doi: 10.1093/nar/22.23.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmer R, Gibbins A M V. Construction and characterization of a large-fragment chicken bacterial artificial chromosome library. Genomics. 1997;42:217–226. doi: 10.1006/geno.1997.4738. [DOI] [PubMed] [Google Scholar]