Abstract
A polysaccharide capsule is one of the most important virulence factors for the pathogenic fungus Cryptococcus neoformans. We previously characterized two capsule-associated genes, CAP59 and CAP64. To further dissect the molecular mechanism of capsule synthesis, 16 acapsular mutants induced by 4-nitroquinoline-1-oxide were obtained. The acapsular phenotype of one of these mutants was complemented. The cloned gene was designated CAP60, and deletion of this newly described capsule-associated gene resulted in an acapsular phenotype. The proposed 67-kDa Cap60p contains 592 amino acids and appears to have a putative transmembrane domain close to the N terminus. DNA sequence analysis revealed that CAP60 has similarity to CAP59 at the center portion of its coding regions. Contour-clamped homogeneous electric field blot analysis suggested that these two genes are on the same chromosome. CAP60 and CAP59, however, could not be functionally substituted for each other by direct complementation or by domain swap experiments. In addition, CAP60 is closely linked to a gene which is similar to a cellulose growth-specific gene of Agaricus bisporus, CEL1. Immunogold electron microscopy studies of the epitope-tagged CAP60 gene revealed that Cap60p was primarily localized to the nuclear membrane. Animal model studies indicated that CAP60 is essential for virulence. Thus, CAP60 is required for both capsule formation and virulence.
Cryptococcus neoformans is a pathogenic yeast which produces a thick extracellular polysaccharide capsule. The polysaccharide capsule is a well-recognized virulence factor of C. neoformans (14, 18). Classical recombination analysis has identified several different genetic loci controlling capsule formation (22). Recently, we complemented two previously identified acapsular mutants and cloned two genes, CAP59 and CAP64 (6, 7). Capsule formation requires functional copies of both genes; deletion of either gene results in an acapsular phenotype. CAP59 and CAP64 are not essential genes, and deletion of either one does not interfere with the growth of C. neoformans. However, both genes are essential for virulence in mice, because acapsular strains resulting from gene deletion are unable to produce fatal infections or multiply in vivo and complementation of the acapsular phenotype restores virulence.
Although CAP59 and CAP64 are essential for capsule formation, the biochemical functions of these two genes are not clear. Analysis of DNA sequences did not reveal their functions. Functional analysis of the Cap59p protein, as determined by expressing different regions of CAP59 under control of the C. neoformans GAL7 promoter, indicates that the putative transmembrane domain at the N terminus of Cap59p is required for its ability to complement the cap59 acapsular phenotype (8). In addition, the glycine residue in the center of the gene is important for CAP59 function, because a missense mutation at the Gly324 residue abolished complementation by the GAL7 fusion construct (8).
The CAP59 and CAP64 loci were previously reported to be closely linked (22), but further studies by molecular as well as classical recombinational analysis revealed that they are actually on separate chromosomes: CAP59 is on chromosome I and CAP64 is on chromosome III (7). Several unique features of these two genes have been reported. Both are closely linked to convergently transcribed genes. CAP59 is closely linked to the gene encoding the putative mitochondrial ribosomal L27 protein, and CAP64 is linked to the putative proteasome subunit gene, PRE1. In both cases the distance between the linked genes is under 30 bp. CAP59 contains six introns, and CAP64 contains eight introns.
To further dissect the molecular mechanisms of capsule formation, we isolated more acapsular strains by mutagenesis. In this paper, we describe the isolation and characterization of another capsule-associated gene, CAP60, and our attempt to immunolocalize its product.
MATERIALS AND METHODS
Strains and media.
C. neoformans var. neoformans serotype D wild-type isolates B-3501 (α mating type) and B-3502 (a mating type) have been described before (16). B-4500 is a wild-type congenic strain of B-4476 (17). R748 is a capsule-deficient mutant received from E. S. Jacobson as strain 326 (22). The LP1 strain is an F4 progeny of an ade2 strain, red13B (7). B-4500FO2 is a ura5 auxotroph of B-4500. Strain cap60-17 is an acapsular mutant generated by mutagenesis. cap60-17FO7, which was used for transformations, is a ura5 auxotroph of cap60-17 and was isolated according to the method described previously (19). All strains were maintained on YEPD (1% yeast extract, 2% Bacto Peptone, and 2% dextrose). Minimal medium (YNB) contained 6.7 g of yeast nitrogen base without amino acids (Difco) and 20 g of glucose per liter. 5-Fluoroorotic acid (5-FOA) medium contained 6.7 g of yeast nitrogen base (Difco), 1 g of 5-FOA, 50 mg of uracil, and 20 g of glucose per liter.
Transformation of C. neoformans.
The electroporation method described by Edman and Kwon-Chung was used to transform C. neoformans (12). TYCC111 and CIP3 were stable encapsulated and acapsular transformants, respectively, of cap60-17FO7 which were selected among Ura5+ stable transformants after three transfers on YEPD medium.
Isolation of capsule-deficient strains.
The log-phase culture of B-4500 was treated with 4-nitroquinoline-1-oxide at 37°C for 30 min to achieve 90% killing. The mutagenized cells were plated on YEPD medium. Yeast cells from colonies with abnormal morphology were examined for the presence of capsules by microscopic examination of India ink slide preparations. Antibody screening of colony blots was performed by standard methods. In brief, a nitrocellulose filter was laid on the plate for 1 min and air dried for 5 min. The filter was washed with a solution containing 50 mM Tris (pH 7.5), 200 mM NaCl, 0.1% Tween 20, and 5% nonfat dry milk; incubated with anti-capsule rabbit antibody; reacted with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) secondary antibody (Bio-Rad Laboratories, Hercules, Calif.); and treated with 4-chloro-1-naphthol and hydrogen peroxide. The reaction was stopped with water.
Preparation and analysis of nucleic acid and proteins.
Genomic DNA isolation and analysis were performed as described previously (6). Random hexamer priming was used to label the DNA probes to specific activities of >108 dpm/μg (13). DNA sequencing was performed by the dideoxy-mediated chain termination method with a Sequenase version 2.0 kit (U.S. Biochemicals, Cleveland, Ohio). Programs of the University of Wisconsin Genetics Computer Group (Madison) were used for analysis of nucleic acid sequences (10).
Total proteins were isolated by glass bead disruption of yeast cells in 20 mM Tris (pH 8.0), 10 mM MgCl2, 1 mM EDTA, 5% glycerol, 1 mM dithiothreitol, 100 mM KCl, 1 mM phenylmethylsulfonyl fluoride, and other proteinase inhibitors. The protein extracts were cleared by centrifugation in a microcentrifuge at 4°C for 30 min, 30 μg of protein was loaded onto a sodium dodecyl sulfate–8% polyacrylamide gel, and the separated proteins were transferred to a polyvinylidene fluoride membrane (Millipore, Bedford, Mass.). The membrane was incubated with anti-hemagglutinin (HA) monoclonal antibody (BAbCO, Richmond, Calif.) followed by secondary antibody, obtained from the Western-Star chemiluminescent detection system (TROPIX, Bedford, Mass.) and used as suggested by the manufacturer.
Construction of plasmids.
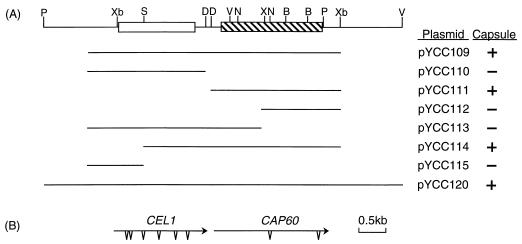
The URA5-containing plasmid, pCIP3, and the ADE2-containing plasmid, pADEΔApa, were received from J. C. Edman (6). To rescue free plasmids from C. neoformans, the genomic DNA from the transformants was digested with NotI, ligated, and transformed into Escherichia coli. For PCR cloning, approximately 50 ng of genomic DNA from the encapsulated transformants of cap60-17FO7 was used along with the primer set flanking the cloning site of pCnTEL1. The PCR was performed with Taqplus DNA polymerase (Stratagene, La Jolla, Calif.) in a 50-μl total reaction volume and allowed to run for 25 cycles of 94°C for 40 s, 60°C for 1 min, and 72°C for 4 min per cycle. To construct pYCC109, the 4.8-kb PCR product was gel isolated (GeneClean II; Bio 101, Vista, Calif.) and cloned into pCIP3. The plasmids pYCC110, pYCC111, pYCC112, pYCC113, pYCC114, and pYCC115 were subclones of pYCC109 in pCIP3 (Fig. 1).
FIG. 1.
Map of CAP60. (A) Restriction enzyme map. Overlapping subclones of pYCC107 were diagrammed. pYCC120 is the reconstituted genomic clone. Plasmids were transformed into cap60-17FO7, and the capsular phenotypes of the resulting transformants were as indicated (+, present; −, absent). Open and hatched boxes represent the coding regions of CEL1 and CAP60, respectively. B, BamHI; D, NdeI; N, NcoI; P, PstI; S, SmaI; X, XhoI; V, EcoRV; Xb, XbaI. (B) Transcriptional direction of CAP60 and CEL1. Arrows indicate the direction of transcription. Triangles represent introns.
To construct a partial library, genomic DNA of B-4500 was digested with XhoI and fractionated on a 0.8% agarose gel. The region from 4 to 6 kb was gel isolated and ligated to the pBluescript vector. The library was screened with the 4.8-kb PCR fragment. Positive clones were isolated, and a clone containing an additional 2.4 kb of the CAP60 3′ flanking region was reconstructed into the pCIP3 vector (pYCC120 [Fig. 1]). The ApaI/EcoRI fragment of pADEΔApa, which contained the functional ADE2 gene, was cloned into the SmaI site of pBluescript to give pYCC76. The 1.5-kb NcoI/BamHI region of pYCC120 was replaced with the 3.0-kb BamHI/EcoRV fragment of the ADE2 gene from pYCC76 to give pYCC122.
For the domain swap experiment, the 0.8-kb PstI/AatII fragment of pYCC14 plasmid containing CAP59 was cloned into the BssHII/PpuMI site of pYCC111 to give pYCC195.
Plasmid pYCC136 was a subclone of pYCC111 containing the carboxyl terminus of Cap60p. The HA epitope (YPYDYPDYA) (28) was inserted in frame at the carboxyl terminus of Cap60p by PCR amplification of pYCC136 as described previously (23). The resulting plasmid (pYCC142) was sequenced to confirm that no errors had been introduced during amplification. The 3′ end of CAP60 in pYCC111 was replaced with the tagged fragment in pYCC142 to generate pYCC145. GETP1 contained three tandem copies of HA pYCC202, as developed by M. Tyers and B. Futcher. The BstXI/XbaI fragment of GETP1 was cloned into pYCC142 to give pYCC198, and the 3′ end of CAP60 in pYCC111 was replaced with the three-HA-tagged fragment in pYCC198 to generate pYCC202.
Protein localization.
The immunofluorescence method used was that described by Pringle et al. (20) with modifications. Cells were fixed in 4% formaldehyde in 50 mM potassium phosphate (pH 6.5) for 2 h at room temperature, washed with 20 mM sodium citrate–1 M sorbitol at pH 5.8, and digested with 10 mg of mureinase (U.S. Biochemicals) per ml in the same buffer at 37°C for 1 h. The fixed cells were attached to polylysine-treated slides and were incubated at room temperature for 1 h with anti-HA antibody in phosphate-buffered saline containing 1 mg of bovine serum albumin per ml. After treatment with fluorescein-conjugated anti-IgG secondary antibody (Boehringer Mannheim, Indianapolis, Ind.), the slides were viewed by immunofluorescence microscopy.
Postembedding immunolabeling procedures were performed by Science Application International Corp. (Frederick, Md.) as described previously (24). Briefly, the procedures were carried out at 4°C to minimize the loss of proteins during the process. Log-phase yeast cells grown in minimal medium were fixed in an equal volume of 8% formaldehyde and 0.2% glutaraldehyde solution overnight at 4°C. The cells were rinsed, dehydrated in graded ethanol, infiltrated in LR Gold resin (Ted Pella, Inc., Redding, Calif.), and allowed to polymerize under UV light in a −20°C cryochamber (Ted Pella, Inc.). Ultrathin sections (50 to 60 nm) were mounted on a 300-mesh nickel grid with a Formvar film. Sections of the grid were blocked with normal goat serum and incubated with anti-HA monoclonal antibody (BAbCO) diluted 1:20 and a 1:100 dilution of 15-nm-diameter colloidal gold-conjugated goat anti-mouse IgG secondary antibody (Amersham Corp., Arlington Heights, Ill.). The thin sections were counterstained with uranyl acetate and lead citrate and observed with an electron microscope (Hitachi H-7000) operated at 75 kV.
Virulence study.
Female BALB/c mice (20 g) were injected in the tail veins with each yeast strain as described previously (6), and the mortality was monitored.
Nucleotide sequence accession number.
The GenBank nucleotide sequence accession numbers for the CAP60 and CEL1 sequences reported in this paper are AF030696 and AF030695, respectively.
RESULTS
Isolation of the CAP60 gene.
Our previous collections of the acapsular mutants were generated by UV irradiation or N-methyl-N′-nitro-N-nitrosoguanidine treatment of B-3501 and B-3502 (15). To generate different types of acapsular mutants, we mutagenized a wild-type strain, B-4500, with 4-nitroquinoline-1-oxide. About 1.8 × 104 colonies that survived after mutagenesis were screened for rough colony morphology, and 38 putative acapsular or hypocapsular strains were obtained. By use of the anti-capsular rabbit antibody, 16 true acapsular strains were obtained, and each was transformed with plasmids containing either CAP59 or CAP64 to complement the acapsular phenotype. Among the 16 strains, 4 were complemented by CAP59 and 3 were complemented by CAP64.
Of the other nine strains, one, cap60-17FO7, was randomly chosen to complement the acapsular phenotype by using the B-4500 DNA constructed in a genomic library of a telomere-based vector (7). Several encapsulated transformants were isolated following electroporation and a two-polymer aqueous-phase treatment to enrich the encapsulated population (6). All the transformants contained free plasmids as determined by hybridizing undigested genomic DNAs with vector sequences (data not shown). These transformants lost the free plasmids and became acapsular when they were grown on nonselective medium. These results indicated that the free plasmids contain the DNA sequence which complements the acapsular mutation. To rescue the free plasmids, DNAs from encapsulated transformants were digested and transformed into E. coli. We failed, however, in several attempts to rescue the plasmids directly from the encapsulated transformants in E. coli. A PCR approach was taken by using primers flanking the cloning site to amplify the DNA responsible for complementation. A 4.8-kb PCR product was obtained, and it was able to complement the acapsular mutation of cap60-17FO7 when the DNA was cloned into a URA5 vector (pYCC109 [Fig. 1]). The PCR DNA product not only complemented cap60-17FO7 but also complemented another three of the nine newly isolated acapsular mutants. In addition, pYCC109 also complemented R-748, which was one of the acapsular mutants previously identified as Cap60 by classical genetic analysis (22). To conform with the nomenclature of capsule genes and in accordance with previous classical mutation analysis, we designated this newly isolated gene CAP60.
Characterization of CAP60.
The minimal region required for complementation of cap60-17FO7 in pYCC109 was determined by overlapping subcloning (Fig. 1), and the smallest clone, pYCC111, was sequenced. A stable Cap+ transformant of cap60-17FO7 (TYCC111) was obtained, and the capsule size was similar to that of the wild-type B-4500 as determined by India ink preparation. The cDNA clones corresponding to the CAP60 gene were isolated, and the sequence was compared to that of the genomic clone. DNA sequence analysis revealed that CAP60 contains two introns and the canonical TATAAA and CAAT sequences upstream of the initiation codon are absent. The proposed Cap60p protein contains 592 amino acids with a calculated molecular mass of 67 kDa and appears to contain a putative transmembrane domain close to the N terminus.
Database searches did not reveal the biochemical function of CAP60. However, CAP60 has some sequence similarity to the CAP59 gene. The proteins encoded by CAP60 and CAP59 have 46% similarity and 22% identity, and most of the conserved regions are in the central portions (Fig. 2). The CAP60 gene, however, could not complement the mutation of cap59 and, likewise, CAP59 could not complement the mutation of cap60. To test if the regions conserved between Cap60p and Cap59p are functionally interchangeable, the coding region for Cap60p from Arg184 to Trp393 was replaced with the coding region for Cap59p from Ile175 to Ile394 (Fig. 2). The resulting construct, pYCC195, was not able to complement the mutation of either cap60 or cap59.
FIG. 2.
Alignment of Cap60p and Cap59p. The alignment was performed with the Bestfit program from the Genetics Computer Group. Arg184 and Trp393 of Cap60p and Ile175 and Ile394 of Cap59p, which delimit the regions used in a domain swap experiment, are underlined. Gly324 (double underlined) is required for Cap59p function. The vertical lines, colons, and periods between the sequences indicate the sequence similarity (10).
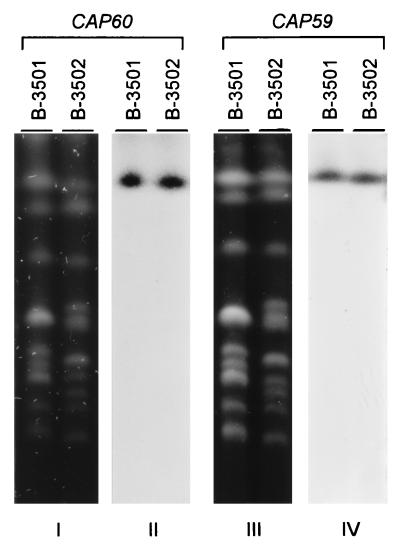
Southern analysis of the contour-clamped homogeneous electric field gel indicated that CAP60 is located on the chromosome I + II doublet, which is similar to the location of CAP59 (Fig. 3). We used a strain, TYCC6, in which the chromosome I + II doublet was resolved (22) and determined that CAP60 is located on chromosome I, just as CAP59 is (data not shown).
FIG. 3.
Chromosomal location of CAP60. The chromosomal DNA was separated by contour-clamped homogeneous electric field gel electrophoresis and stained with ethidium bromide (blots I and III). The gel-separated chromosomal DNA was transferred to a nylon membrane and hybridized with a probe of the 4.8-kb PCR fragment of CAP60 (blot II) or with a probe of CAP59 (blot IV). B-3501 (α mating type) and B-3502 (a mating type) are wild-type strains.
Linkage of CAP60 and CEL1.
During analysis of the CAP60 locus and its flanking region, we found that CAP60 is closely linked to a gene which is immediately upstream and is transcribed in the same direction as CAP60 (Fig. 1). This closely linked gene contains six introns and encodes a putative protein which has a high serine content close to its C terminus. This putative 40-kDa protein has 46% similarity and 25% identity to a cellulose growth-specific gene of Agaricus bisporus (21) (Fig. 4). The CEL1 gene of A. bisporus encodes a protein that has an architecture resembling those of the multidomain fungal cellulases, although the sequence of its putative catalytic core is not matched by any other in the protein and nucleic acid databases (1). The function of the putative CEL1 gene in C. neoformans is not clear. The phenomenon of CAP60 being clustered with a different gene was also observed for the other two capsule-associated genes, CAP59 with L27 and CAP64 with PRE1 (7, 8).
FIG. 4.
Alignment of Cel1p. The alignment was performed with the Bestfit program from the Genetics Computer Group. CN, Cel1p of C. neoformans; AB, Cel1p of A. bisporus. The GenBank accession number for CEL1 of A. bisporus is M86356. The vertical lines, colons, and periods between the sequences indicate the sequence similarity (10).
Deletion of CAP60 results in the acapsular phenotype.
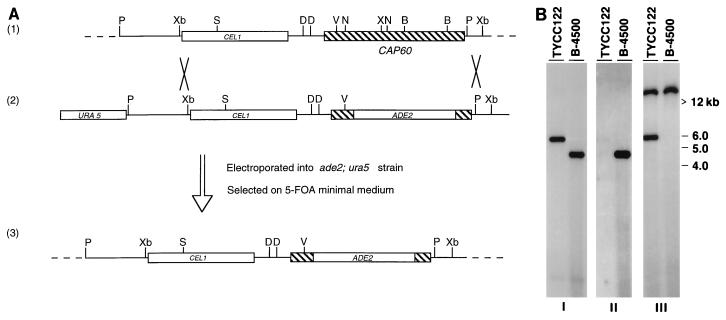
Because of the very low frequency of homologous integration in C. neoformans serotype D strains, a positive-negative selection method has been designed to enrich for the homologous integration event (6). This method required a double crossover at the flanking region of the gene. However, the largest clone rescued by PCR (pYCC109) contained less than 400 bp beyond the stop codon of CAP60 (Fig. 1). To obtain the 3′ flanking region of CAP60, we screened an XhoI-digested partial genomic library. The plasmid pYCC120, which contains the additional 2.4 kb of the CAP60 3′ flanking region, was subsequently constructed and could restore the capsule of cap60-17FO7 (Fig. 1). To delete the CAP60 gene, the 1.5-kb NcoI/BamHI region of pYCC120 was replaced by the ADE2 gene (pYCC122). The resulting plasmid was transformed into an ade2 ura5 strain and plated on 5-FOA medium. Two Ade2+ Ura5− acapsular transformants were isolated. Southern blot analysis was carried out to determine if the acapsular phenotype was derived from a gene replacement event (Fig. 5). The DNA blot was first hybridized with a probe of the 4.8-kb PCR product. The 4.2-kb signal in the wild-type, B-4500, changed to a 5.7-kb signal in the acapsular transformant, TYCC122 (Fig. 5B, blot I). This result suggested an insertion event at the CAP60 locus. The same blot was hybridized with the 1.5-kb NcoI/BamHI region of pYCC120, which was deleted in pYCC122. No hybridization signal was detected in TYCC122, which indicated a deletion event (Fig. 5B, blot II). Finally, the ADE2 gene probe detected a 5.7-kb band in TYCC122 and confirmed that gene replacement occurred at the predicted position (Fig. 5B, blot III). In addition, the greater-than-12-kb signal in TYCC122 represents the native ADE2. Thus, Southern blot analysis confirmed that the acapsular phenotype was generated by deletion of CAP60. In addition, when TYCC122 was transformed with pYCC109, the resulting transformants produced capsules.
FIG. 5.
Deletion of CAP60. (A) Diagram of CAP60 deletion. Strain LP1 (ade2 ura5 CAP60) was transformed with the linearized pYCC122 DNA, and cells were selected on 5-FOA medium. Map 1, chromosomal region containing CAP60; map 2, pYCC122; map 3, deletion of CAP60 resulting from double crossover. The figure is not drawn to scale for simplicity. Hatched box, CAP60 coding region. The CEL1 coding region is represented by the labeled open box. B, BamHI; D, NdeI; N, NcoI; P, PstI; S, SmaI; X, XhoI; V, EcoRV; Xb, XbaI. (B) Southern blot analysis. Genomic DNA of an acapsular transformant (TYCC122) and a capsule-containing strain (B-4500) were digested with XbaI. The membrane was hybridized with the 4.8-kb PCR product (blot I), the 1.5-kb NcoI/BamHI region of pYCC120 (blot II), or the entire pYCC76 plasmid, which contains ADE2 (blot III).
CAP60 is required for virulence.
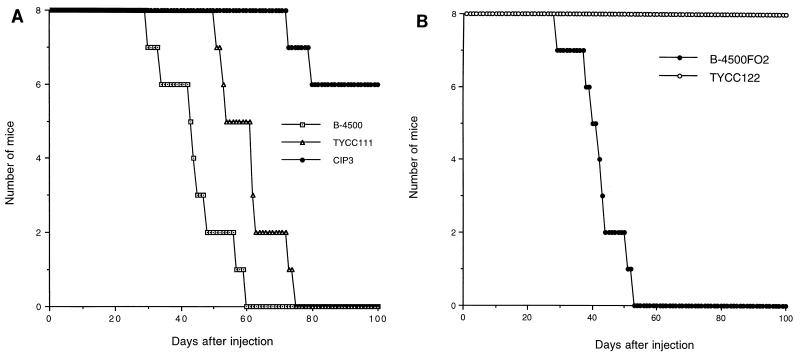
The relationship of virulence and the presence of a capsule has been well established. We anticipated that the acapsular strain created by deletion of CAP60 would be avirulent and that complementation of the acapsular phenotype of cap60 should restore virulence. Animal studies were done to confirm this hypothesis (Fig. 6). The wild-type encapsulated strain (B-4500) and the Cap+ transformant of cap60-17FO7 (TYCC111) caused fatal infections in 100% of inoculated mice within 75 days, although death occurred earlier in mice that received B-4500. The reduction of virulence could have been due to ectopic integration of plasmids in TYCC111, as was the case in studies reported previously (7). Surprisingly, two of the eight mice which received the acapsular transformant of cap60-17FO7 carrying only vector sequence (CIP3) died after 75 to 100 days. Yeast cultures recovered from the brains of these dead mice all produced abundant capsules. Therefore, the mortality in the mice that received CIP3 was due to reversion in the capsule phenotype of the mutant. The virulence of the cap60 deletion mutant (TYCC122) was also compared with the virulence of the isogenic encapsulated strain (B-4500FO2) (Fig. 6B). All mice challenged with B-4500FO2 died within 53 days, whereas the cap60 deletion mutant (TYCC122) failed to produce fatal infection and mice injected with it remained healthy for more than 100 days postinoculation. Thus, these results confirmed that the CAP60 gene is required for C. neoformans to produce fatal infection in mice.
FIG. 6.
Virulence test. Groups of eight mice were injected with about 106 viable cells and monitored to determine mortality. (A) Results for TYCC111, a stable Cap+ transformant of cap60-17FO7; CIP3, a stable Cap− transformant of cap60-17FO7 harboring only the vector sequence; and B-4500, a wild-type strain. P was <0.0004 for the comparison of CIP3 to TYCC111 and B-4500 (Kaplan-Meier analysis). (B) Results for B-4500FO2, a CAP60 ura5 auxotroph, and TYCC122, a cap60 deletion mutant and ura5 auxotroph. P was <0.0001 for the comparison of TYCC122 to B-4500FO2 (Kaplan-Meier analysis).
Localization of Cap60p.
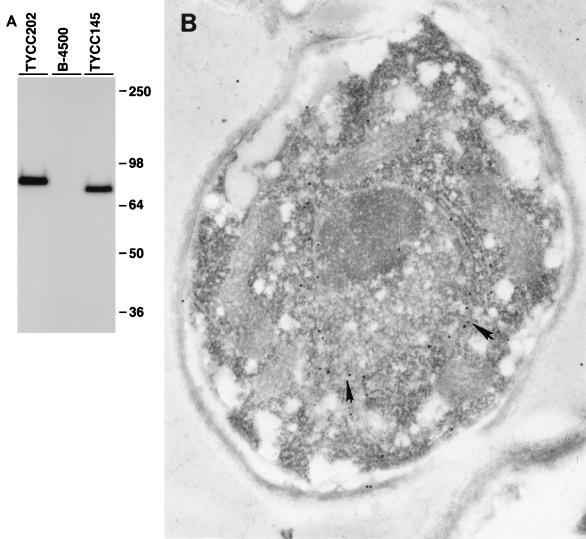
We have employed peptide epitope-tagging methods to identify the cellular location of the CAP60 gene products. The nine-amino-acid epitope of the influenza HA protein (28) was inserted at the carboxy terminus of Cap60p (pYCC145). The resulting plasmid was able to complement the acapsular phenotype of TYCC122. Total proteins were extracted and analyzed by immunoblotting. The size of the protein detected by anti-HA antibody corroborated the predicted molecular weight (Fig. 7A). Immunofluorescence microscopy was used to visualize the HA-tagged Cap60p fusion proteins, but the intensity of fluorescence was not high enough to define the location of Cap60p. A different plasmid, pYCC202, which contained three copies of HA at the C terminus of Cap60p, was constructed and was able to complement the cap60 mutation. The encapsulated transformant containing pYCC202 (TYCC202) produced a protein of the expected size for HA-tagged Cap60p (Fig. 7A). However, the degree of intensity in fluorescence was nearly the same as in TYCC122. Immunogold electron microscopy (EM) was chosen to determine the location of Cap60p. The immunogold labeling appeared to be on the nuclear membrane of TYCC202 (Fig. 7B). Little or no labeling was observed in other places. Control samples without anti-HA antibody showed no labeling. The freeze substitution technique was also used in immunogold EM, and similar results were observed (data not shown).
FIG. 7.
Localization of Cap60p. (A) Immunoblot analysis. Yeast cells were grown in YNB, and total protein extracts were analyzed by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis and incubated with anti-HA antibody and the Western-Star chemiluminescent detection system. B-4500 is a wild-type strain; TYCC145 is an encapsulated transformant containing a plasmid with a single HA-tagged CAP60; and TYCC202 is an encapsulated transformant containing a plasmid with a three-HA-tagged CAP60. (B) Electron micrograph of TYCC202. The yeast cells grown in YNB were fixed in equal volumes of 8% formaldehyde and 0.2% glutaraldehyde. Ultrathin sections (50 to 60 nm) were incubated with anti-HA serum and with 15-nm-diameter colloidal gold-conjugated goat anti-mouse IgG secondary antibody. The thin sections were counterstained with uranyl acetate and lead citrate. The antibodies are associated with the nuclear envelope (arrows). The picture shown is representative of many sections. Magnification, ×3,400.
DISCUSSION
We have created a new collection of capsule-deficient mutants with 4-nitroquinoline-1-oxide and isolated a new gene, CAP60. Deletion of CAP60 resulted in an acapsular phenotype, and complementation of the mutation restored the capsule. These data show that CAP60 is required for capsule formation. CAP60 is located on the same chromosome as CAP59. Like the clustering of genes in loci of CAP59 and CAP64, CAP60 is closely linked to CEL1, which is similar to a cellulose growth-specific gene of A. bisporus. The results of animal model studies confirmed that a capsule is required for C. neoformans to produce fatal infection in mice. Thus, CAP60, like CAP59 and CAP64, is required for capsule formation and virulence.
Although similarity exists between Cap59p and Cap60p, the gene products cannot functionally substitute for each other by direct complementation or by domain swap experiments. Both Cap59p and Cap60p contain putative transmembrane domains. The one in Cap59p appears to be a signal peptide, which suggests that Cap59p may be secreted, whereas the one in Cap60p is more like a type II transmembrane domain. The possibility of a membrane localization of Cap60p is strengthened by the result of immunogold EM, which localized Cap60p to the nuclear membrane. One potential caveat of the HA epitope-tagging experiment is that insertion of the HA epitope could affect cellular localization, although the resulting construct complemented the acapsular phenotype. The quality of the immunogold EM picture is inferior to those of other fungi which are easy to handle. The cristae of mitochondria were not distinct, and membranes of various organelles appeared to have been disrupted. However, the agreement of the results from many sections of freeze substitution and regular immunogold EM methods strengthened the possibility that the immunogold particle is associated primarily with the nuclear membrane. The reasons for the technical difficulties encountered in the immunogold EM and immunofluorescence methods with C. neoformans are unclear. Difficulties in preservation of cytoplasmic organelles in immunogold EM studies with C. neoformans have also been encountered by other workers (27). Different approaches are needed to explain these difficulties.
The failure experienced in rescuing free plasmids directly from encapsulated transformants of cap60-17FO7 is not uncommon with C. neoformans. In some instances, the modification of the incoming DNA in the transformants was so drastic that the DNA sequence inserted in the cloning site of the telomere vector could not be amplified by use of PCR primers that flank the cloning site (unpublished data). The genomic plasmid library was constructed in a pCnTEL1 vector, which contains telomeres to increase the transformation frequency (11). The telomeres, however, do not prevent the modification of incoming DNA in the transformation of C. neoformans. A vector that can provide more stability may be required to circumvent this problem. Recent isolation of a 1.2-kb DNA fragment from a minichromosome (26) may provide more vector stability for transformation in C. neoformans (26a). Whether this DNA fragment can be used in library construction and prevent the unwanted modifications needs to be tested.
Among the 16 newly isolated acapsular mutants, the acapsular phenotype of 4 strains could be complemented by CAP59, 3 by CAP64, and 4 by CAP60. Thus, close to two-thirds of the easily identifiable acapsular mutants contain mutations which can be complemented by one of the three cloned genes. Although the steps involved in the biosynthetic pathway of capsule formation have not been elucidated, the pathway may involve only a few genes which can dramatically affect the process and result in a clear morphological abnormality—acapsular phenotype. In addition to the truly acapsular mutants, we also obtained 22 hypocapsular mutants. These mutants have a reduced capsule size and a rough colony surface. The major component of capsular polysaccharide is glucuronoxylomannan, which is an α-1,3-d-mannopyranose backbone containing a single β-1,2-linked glucuronate residue on one-third of the mannopyranose residues and varying amounts of xylosylation, depending on the serotype (2–5, 9, 25). It is possible that some of the hypocapsular mutants have defects in the formation of branches or acetylations. Monoclonal antibody or some other reagents which can specifically target the branches or certain acetyl groups may be useful in isolating the genes responsible for these hypocapsule mutations.
ACKNOWLEDGMENTS
We thank J. Cutler for his help with the EM studies and critical review of the manuscript and T. Caesar, L. A. Penoyer, and K. Nagashima for technical assistance.
Part of this work was supported by NIAID grant 5 R01-AI24912 to J. Cutler.
REFERENCES
- 1.Armesilla A L, Thurston C F, Yague E. CEL1: a novel cellulose binding protein secreted by Agaricus bisporus during growth on crystalline cellulose. FEMS Microbiol Lett. 1994;116:293–299. doi: 10.1111/j.1574-6968.1994.tb06718.x. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharjee A K, Bennett J E, Glaudemans C P. Capsular polysaccharides of Cryptococcus neoformans. Rev Infect Dis. 1984;6:619–624. doi: 10.1093/clinids/6.5.619. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharjee A K, Kwon-Chung K J, Glaudemans C P. On the structure of the capsular polysaccharide from Cryptococcus neoformans serotype C-II. Mol Immunol. 1979;16:531–532. doi: 10.1016/0161-5890(79)90081-6. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharjee A K, Kwon-Chung K J, Glaudemans C P. Capsular polysaccharides from a parent strain and from a possible, mutant strain of Cryptococcus neoformans serotype A. Carbohydr Res. 1981;95:237–248. doi: 10.1016/s0008-6215(00)85580-9. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharjee A K, Kwon-Chung K J, Glaudemans C P. The major capsular polysaccharide of Cryptococcus neoformans serotype B. Carbohydr Res. 1992;233:271–272. doi: 10.1016/s0008-6215(00)90942-x. [DOI] [PubMed] [Google Scholar]
- 6.Chang Y C, Kwon-Chung K J. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol Cell Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang Y C, Penoyer L A, Kwon-Chung K J. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect Immun. 1996;64:1977–1983. doi: 10.1128/iai.64.6.1977-1983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang Y C, Wickes B L, Kwon-Chung K J. Further analysis of the CAP59 locus of Cryptococcus neoformans: structure defined by forced expression and the description of a new ribosomal protein gene. Gene. 1995;167:179–183. doi: 10.1016/0378-1119(95)00640-0. [DOI] [PubMed] [Google Scholar]
- 9.Cherniak R, Reiss E, Turner S H. A galactoxylomannan antigen of Cryptococcus neoformans serotype A. Carbohydr Res. 1982;103:239–250. [Google Scholar]
- 10.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edman J C. Isolation of telomerelike sequences from Cryptococcus neoformans and their use in high-efficiency transformation. Mol Cell Biol. 1992;12:2777–2783. doi: 10.1128/mcb.12.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edman J C, Kwon-Chung K J. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol Cell Biol. 1990;10:4538–4544. doi: 10.1128/mcb.10.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 14.Fromptling R A, Shadomy H K, Jacobson E S. Decreased virulence in stable acapsular mutants of Cryptococcus neoformans. Mycopathologia. 1982;79:23–29. doi: 10.1007/BF00636177. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson E S, Ayers D J, Harrell A C, Nicholas C C. Genetic and phenotypic characterization of capsule mutants of Cryptococcus neoformans. J Bacteriol. 1982;150:1292–1296. doi: 10.1128/jb.150.3.1292-1296.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon-Chung K J. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia. 1976;68:821–833. [PubMed] [Google Scholar]
- 17.Kwon-Chung K J, Edman J C, Wickes B L. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon-Chung K J, Rhodes J C. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect Immun. 1986;51:218–223. doi: 10.1128/iai.51.1.218-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon-Chung K J, Varma A, Edman J C, Bennett J E. Selection of ura5 and ura3 mutants from the two varieties of Cryptococcus neoformans on 5-fluoroorotic acid medium. J Med Vet Mycol. 1992;30:61–69. [PubMed] [Google Scholar]
- 20.Pringle J R, Adams A E, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 21.Raguz S, Yague E, Wood D A, Thurston C F. Isolation and characterization of a cellulose-growth-specific gene from Agaricus bisporus. Gene. 1992;119:183–190. doi: 10.1016/0378-1119(92)90270-y. [DOI] [PubMed] [Google Scholar]
- 22.Still C N, Jacobson E S. Recombinational mapping of capsule mutations in Cryptococcus neoformans. J Bacteriol. 1983;156:460–462. doi: 10.1128/jb.156.1.460-462.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stringer M A, Timberlake W E. dewA encodes a fungal hydrophobin component of the Aspergillus spore wall. Mol Microbiol. 1995;16:33–44. doi: 10.1111/j.1365-2958.1995.tb02389.x. [DOI] [PubMed] [Google Scholar]
- 24.Tobin G J, Nagashima K, Gonda M A. Immunologic and ultrastructural characterization of HIV pseudovirions containing Geg and Env precursor proteins engineered in insect cells. Methods Companion Methods Enzymol. 1996;10:208–218. doi: 10.1006/meth.1996.0096. [DOI] [PubMed] [Google Scholar]
- 25.Turner S H, Cherniak R, Reiss E, Kwon C K. Structural variability in the glucuronoxylomannan of Cryptococcus neoformans serotype A isolates determined by 13C NMR spectroscopy. Carbohydr Res. 1992;233:205–218. doi: 10.1016/s0008-6215(00)90932-7. [DOI] [PubMed] [Google Scholar]
- 26.Varma A, Kwon-Chung K J. Formation of a minichromosome in Cryptococcus neoformans as a result of electroporative transformation. Curr Genet. 1994;26:54–61. doi: 10.1007/BF00326305. [DOI] [PubMed] [Google Scholar]
- 26a.Varma, A., and K. J. Kwon-Chung. Unpublished data.
- 27.Vartivarian S E, Reyes G H, Jacobson E S, James P G, Cherniak R, Mumaw V R, Tingler M J. Localization of mannoprotein in Cryptococcus neoformans. J Bacteriol. 1989;171:6850–6852. doi: 10.1128/jb.171.12.6850-6852.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]