Abstract
Toxoplasma gondii is an Apicomplexan parasite with a complex life cycle that includes a rapidly dividing asexual stage known as the tachyzoite. The tachyzoite surface has been reported to comprise five major antigens, the most abundant of which is designated SAG1 (for surface antigen 1). At least one of the other four (SAG3) and another recently described minor antigen (SRS1 [for SAG1-related sequence 1]) have previously been shown to be structurally related to SAG1. To determine if further SAG1 homologs exist, we searched a Toxoplasma expressed sequence tag (EST) database and found numerous ESTs corresponding to at least three new genes related to SAG1. Like SAG1, these new SRS genes encode apparently glycosylphosphatidylinositol-anchored proteins that share several motifs and a set of conserved cysteine residues. This family appears to have arisen by divergence from a common ancestor under selection for the conservation of overall topology. The products of two of these new genes (SRS2 and SRS3) are shown to be expressed on the surface of Toxoplasma tachyzoites by immunofluorescence. We also identified strain-specific differences in relative expression levels. A total of 10 members of the SAG1 gene family have now been identified, which apparently include three of the five major surface antigens previously described and one antigen expressed only in bradyzoites. The function of this family may be to provide a redundant system of receptors for interaction with host cells and/or to direct the immune responses that limit acute T. gondii infections.
Toxoplasma gondii possesses the ability to invade and to establish productive infection in almost any nucleated cell. This process involves (i) the initial attachment of the parasite to the host cell and (ii) subsequent invasion of the host through its plasma membrane, with concomitant formation of a parasitophorous vacuole in which the parasite resides and replicates. While these processes have been studied in detail by electron and video microscopy (reviewed in reference 28), the precise molecular events which mediate the initial interaction between the parasite and its host are poorly understood. A goal of our research has therefore been to understand the role of parasite surface antigens in these processes. In particular, we have focused on the major tachyzoite surface antigen, SAG1 (for surface antigen 1), which is one of five major glycosylphosphatidylinositol (GPI)-anchored antigens revealed by surface-labeling studies of laboratory reference strains (6, 30, 32). The genes encoding three of these have previously been cloned: SAG1, or P30 (4); SAG2, or P22 (23); and SAG3, or P43 (5). The latter study indicated that SAG1 and SAG3 are structurally related, with 24% overall amino acid identity. In particular, both proteins contain 12 cysteine residues and a number of short peptides which align directly in the primary sequence and suggest a common overall topology. The identification of an additional gene which encodes a protein related to SAG1 (10) suggested that the parasite may have a number of surface antigens with this common structure. We therefore set out to identify further potential homologs of SAG1 in a Toxoplasma expressed sequence tag (EST) database (1, 18). We report a further two members (plus a fragment of a third) and show that two, at least, are expressed on the tachyzoite surface. The implications of these findings for the function of SAG1 and the other SRS (for SAG1-related sequence) genes are discussed.
MATERIALS AND METHODS
Host cells and parasites.
Tachyzoites of T. gondii were grown in human foreskin fibroblast monolayers in Dulbecco’s modified Eagle’s medium supplemented with 10% Nu-Serum, 2 mM glutamine, and 20 μg of gentamicin solution per ml at 37°C in a humid 5% CO2 atmosphere. Four T. gondii strains were used in this study: RH (26), CEP (24), PLK (a clonal derivative of ME49 [14]), and the Sag1− mutant ME49 derivative designated B in reference 12.
Searches and analysis.
The libraries used in assembly of the EST database for T. gondii have been described elsewhere (1, 18, 35); summaries of the Toxoplasma sequences with known homologs in GenBank can be found at the Toxoplasma genome web site (http://www.ebi.ac.uk/parasites/toxo/toxpage.html) and at http://cbil.humgen.upenn.edu/toxodb/toxodb.html. Searches of dBEST (Database of Expressed Sequence Tags) for homologs of the known surface antigens SAG1 (GenBank accession no. M23658), SAG3 (GenBank accession no. L21720), SRS1 (GenBank accession no. U77677 [10]), and the SAG1 homolog SAG5 (GenBank accession no. U64517 [29]) were made with the BLAST (Basic Local Alignment Search Tool) suite of programs (at National Center for Biotechnology Information [NCBI]: http://www.ncbi.nlm.nih.gov) and FASTA (at European Bioinformatics Institute: http://www.ebi.ac.uk) via the World Wide Web. Initial identification of SRS sequences was performed by tBLASTn analysis of dBEST (with the NCBI server defaults: expect = 10, matrix = BLOSUM62) with the SAG coding sequences and evaluation of all clones matched according to these parameters. After exclusion of clones yielding low BLAST scores because of overall short or poor-quality sequences, ESTs that could not be assigned to known SAG genes were compared to those in dBEST by using BLASTn and FASTA. This permitted identification of homologous fragments and possible overlaps. The resulting matches were assembled into contigs by using Gelassemble (Wisconsin Package, version 8.0; Genetics Computer Group [GCG], Madison, Wis.). At each stage of reconstruction, the consensus sequences were resubmitted to BLAST analysis to ensure identification of all related sequences. In this way, complete contig assemblies encompassing the entire open reading frames (ORFs) of two distinct members of the T. gondii SAG family, designated SRS2 and SRS3, were obtained. In addition, a single EST corresponding to a further distinct family member (SRS4) was also identified (see Table 2). Alignments of the SRS family were made with ClustalV via the World Wide Web.
TABLE 2.
Properties of members of the SRS homology family
| Gene (alternate protein name) | Surface expressiona | Size by Western blotting (kDa)b | No. of ESTsc
|
GenBank accession no. | Source or reference | |
|---|---|---|---|---|---|---|
| Tachyzoite | Bradyzoite | |||||
| SAG1 (P30) | T | 30 | 103 | 2f | M23658 | 4 |
| SAG3 (P43) | T, B | 43 | 10 | 1 | L21720 | 5 |
| SRS1 | T | 46 | 7 | 0 | U77677 | 10 |
| SRS2 | T | 40–43 | 29 | 0 | AF012276 | This study |
| SRS3 (P35) | T | 35? | 14 | 0 | AF012275 | This study |
| SRS4 | NAd | NA | 1 | 0 | This study | |
| SAG5e | NA | NA | 1 | 0 | U64517 | 29 |
| BSR4 (P36) | B | 36 | 1 | 0 | AF015290 | 16 |
T, tachyzoite; B, bradyzoite (surface expression by immunofluorescence).
Size based on mobility in reducing SDS-PAGE.
Number of essentially perfect matches in 7,074 tachyzoite ESTs and 2,353 bradyzoite ESTs.
NA, not available.
See the text.
Molecular cloning of SRS cDNAs.
The EST coverage of the messages encoding SRS2 and SRS3 was insufficient to allow unambiguous determination of the sequence of the entire ORF. Ambiguities were resolved by complete sequencing of clones following reverse transcription-PCR amplification of the putative ORF and cloning into pcDNA1 (Invitrogen). Briefly, RNA was extracted from T. gondii RH tachyzoites with Ultraspec RNA (Biotecx Lab., Inc.) according to the manufacturer’s instructions. Total RNA was reverse transcribed with an oligo(dT) anchor primer (dTAP) and Superscript II (Gibco-Life Sciences). PCR amplification of the SRS2 ORF from the cDNA template was performed with primers SRS2ORF5 and SRS3ORF3 at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min for 30 cycles. The SRS3 ORF was amplified by the same approach with the SRS3ORF5 and SRSORF3 primer pair. The lengths of the SRS messages were determined by Northern analysis, and the positions of their polyadenylation sites were confirmed by 3′ rapid amplification of cDNA ends with SRS2ORF5 (SRS2) and SRS3ORF5 (SRS3) and the anchor primer AP. Primer sequences for these manipulations are listed in Table 1. All manipulations were performed according to standard protocols (2).
TABLE 1.
Sequences of oligonucleotides used in PCR amplification of SRS2, SRS3, and SRS4 fragments
| Oligonucleotide | Sequencea |
|---|---|
| SRS2ORF3 | 5′-aagctcgagTCAATAGGCAAGTGC-3′ |
| SRS2ORF5 | 5′-ctcggatccAGAATGGCGACGCGTGC-3′ |
| SRS25 | 5′-cgggatccCCTGAAAAGTTTACATGCCGCC-3′ |
| SRS23 | 5′-tgctctagaTTATCCACCTGGTCTTGATCC-3′ |
| SRS3ORF5 | 5′-tgactcgagGAAATGCGCTTACAG-3′ |
| SRS3ORF3 | 5′-cgactcgagCTACACAAGGGAAGA-3′ |
| SRS35 | 5′-gcgtctagaGAGAAGCAGGTAGTGTGTCCT-3′ |
| SRS33 | 5′-acgtctagaTCAGATTGTGAGCTCTACTGT-3′ |
| SRS45 | 5′-gcggaattcTTCGTCGGCTGTGACAACAA-3′ |
| SRS43 | 5′-tgcggatccCGTCTGGACAGCTGAAAATT-3′ |
| dTAP | 5′-ccggaattcggtacctctagat18n-3′ |
| AP | 5′-tctagaggtaccgaattccgg-3′ |
Sequences in lowercase are for cloning purposes and are not derived from Toxoplasma genes.
The single EST clone encoding SRS4 (clone zy52c12 [GenBank accession no. N81407]) was obtained from the EST repository (Genome Systems, Inc.) as λZAPII phage, excised as pBluescript SKII with ExAssist helper phage (Stratagene) according to the manufacturer’s instructions, and sequenced directly with T3 and T7 promoter primers.
Production of recombinant pMAL-SRS fusion proteins and antisera.
An 888-nucleotide (nt) fragment of the SRS2 ORF was amplified from the cDNA template with primers SRS25 and SRS23 and cloned into the BamHI and XbaI sites of pMAL-P2 to yield maltose-binding protein (MBP) fusions (New England Biolabs). In similar fashion, a 660-nt fragment of SRS3 and a 715-nt fragment of SRS4 were amplified with SRS35/33 and SRS45/43, respectively, and cloned into pMAL-P2 at the XbaI site (SRS3) or EcoRI-BamHI sites (SRS4). Expression of MBP-SRS fusion proteins was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h at 37°C and purified from bacterial lysates on amylose resin according to standard protocols (25).
For immunizations, approximately 50 μg of lyophilized fusion protein was resuspended in 500 μl of phosphate-buffered saline (PBS) (pH 7.4) and emulsified with an equal volume of RIBI adjuvant (Immunochem Research, Inc.). BALB/c mice were primed by intraperitoneal injection with 200 μl of each mixture and boosted with an equal amount 14 and 30 days after priming. Blood was collected prior to immunization and after each boost from the tail vein at days 24 and 44.
Western blotting and indirect immunofluorescence.
Total lysates from T. gondii tachyzoites were prepared in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (20). Analytical SDS-PAGE and transfer to nitrocellulose filters were performed by standard methods. Filter strips were blocked with 5% nonfat dry milk–0.5% TWEEN 20 in PBS (PBS-M) and incubated with sera or monoclonal antibodies diluted in PBS-M. Bound antibodies were detected with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G and developed by enhanced chemiluminescence (ECL [Amersham]). T. gondii extracts for GPI-specific phospholipase C (GPI-PLC) treatment were prepared as described previously (10). Briefly, washed RH tachyzoites were resuspended in 50 μl of lysis buffer (50 mM Tris [pH 8.0], 5 mM EDTA, 5% Nonidet P-40) and incubated with various amounts of GPI-PLC from Trypanosoma brucei for 1 h at 37°C in the presence of protease inhibitors (pepstatin and phenylmethylsulfonyl fluoride). Cross-reacting determinants (CRDs) of GPI anchors which were unmasked by this treatment were detected with a polyclonal antiserum against CRD of trypanosomal bloodstream-form variant surface glycoprotein (VSG) (20, 27).
Live tachyzoites were prepared for immunofluorescence by passage through a 27-gauge syringe needle to release intact intracellular parasites from host cells. Host cell debris was removed by filtration of the parasite suspension through a 3-μm-pore-diameter filter, and parasites were then washed twice in 2% bovine serum albumin (BSA) in PBS (PBS-BSA). Incubations with primary and fluorescein isothiocyanate (FITC)-conjugated secondary antibodies (Fab) (Cappel) were performed in 100 μl of PBS-BSA at 4°C for 30 min each. Labeled parasites were washed four times with cold PBS after incubations and fixed with 2% formaldehyde for 20 min on ice. The parasites were washed in PBS, dried on coverslips, rehydrated, and embedded with Vectashield (Vector Labs) solution for microscopy. Epifluorescence was detected with an Olympus BX60 microscope.
Nucleotide sequence accession number.
The sequences of the complete genes from the RH strain have been submitted to GenBank under accession no. AF012276 (SRS2) and AF012275 (SRS3).
RESULTS
Identification of ESTs encoding surface antigen homologs.
Identification of SRS sequences was performed by tBLASTn analysis of dBEST (with the NCBI server defaults) with coding sequences of the known SAG1-related genes (SAG1, SAG3, SRS1, and SAG5; SRS1 was previously identified by its proximity to SAG1 in the Toxoplasma genome [10]; SAG5 is a genomic sequence homologous to SAG1 deposited in GenBank by Crisanti et al. [29]) and evaluation of all of the clones that matched according to these parameters. At the time of our initial searches, dBEST included 5,600 ESTs from RH tachyzoites and 1,806 ESTs from ME49 tachyzoites (1). After exclusion of clones yielding low BLAST scores because of overall short or poor-quality sequences, ESTs that could not be assigned to known SAG genes were compared to dBEST by using BLASTn and FASTA, and the resulting matches were assembled into contigs with Gelassemble (GCG). This analysis yielded three previously unidentified genes that we have called SRS2, SRS3, and SRS4.
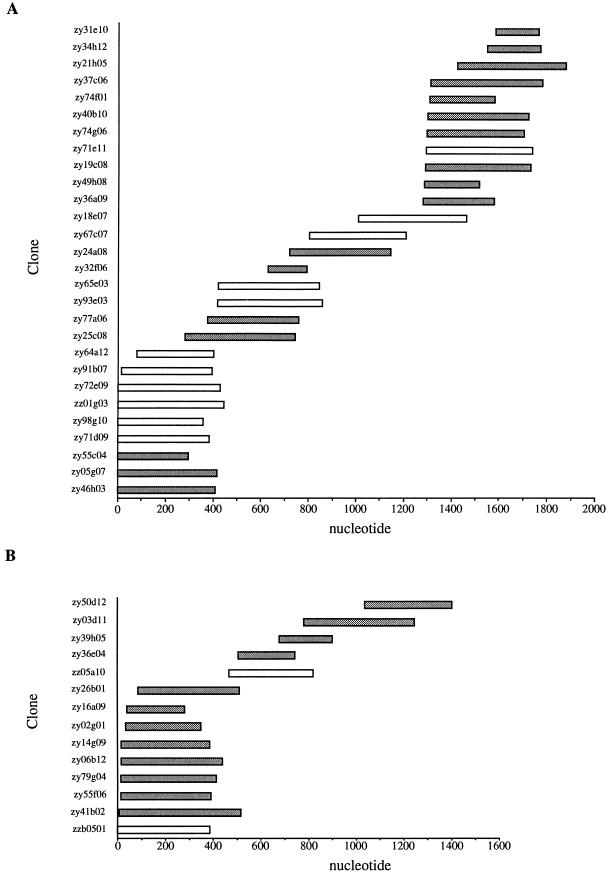
Figure 1A shows the contig assembly of EST sequences corresponding to the SRS2 cDNA derived from the raw sequence data. Although individual EST accessions in dBEST are annotated to indicate regions with high-quality or unambiguous sequences, it was necessary to use the entire sequence to force overlaps in some regions. Manual editing of the consensus in Gelassemble allowed us to derive an ∼1,700-nt contig from a total of 29 overlapping clones from the RH and ME49 tachyzoite EST data sets. The assembled contig was used to design primers to amplify the ORF from the RH strain and obtain the full sequence of the coding region from this strain. The contig assembly indicates that eight clones terminate at approximately the same 5′ position (designated 0 nt) which lies 287 nt upstream of the SRS2 ORF. Although it is possible that this represents a strong stop causing premature termination of reverse transcription, Northern blotting with an SRS2 probe indicated that the mature message for SRS2 is approximately 1.8 kb in length (not shown). Given that the contig created for SRS2 is ∼1.7 kb in length and allowing for polyadenylation of the mRNA, the 5′ termini of these clones appear to converge at or close to the transcriptional start site.
FIG. 1.
EST contig assembly for SRS2 (A) and SRS3 (B). Sequence data from overlapping EST clones (y axis) from RH (shaded bars) and ME49 (open bars) tachyzoites were assembled into contigs with Gelassemble (GCGV8.0).
The ORF of SRS2 encodes a 372-amino-acid protein with predicted signal peptide and GPI anchor addition signals (Fig. 2). Analysis of the signal peptide indicates that this has an unusual structure with two long hydrophobic stretches of ∼18 and ∼22 residues without an intervening methionine residue. As a result, there are two strongly predicted cleavage sites according to the criteria established by Von Heijne (34). The possible significance of this is unclear at present. The apparent GPI anchor signal for SRS2 predicts that the attachment site is in the vicinity of a glycine-serine-alanine sequence 24 to 26 amino acids upstream of the C terminus (33). Removal of the C- and N-terminal processing signals (the latter at the most-C-terminal site predicted by the signal peptide algorithm) would yield a mature peptide consisting of 296 amino acids with a predicted molecular mass of ∼31.6 kDa without further modifications such as N and O glycosylation or the glycolipid anchor itself. Comparison of the putative mature SRS2 sequence with the corresponding SAG1 peptide sequence (by using BESTFIT) indicates that SRS2 shares 32% overall identity and 38% similarity in a central core region (residues 64 to 258 of the complete ORF [Fig. 2]).
FIG. 2.
Amino acid sequence alignment of eight members of the SAG1 family produced by ClustalW. Dashes represent gaps introduced to produce optimal alignment. Amino acids conserved in four or more sequences are boxed, and homologies are shaded when present in two or more sequences. Each allele shown is from the RH strain, except for BSR4, which is from the ME49 strain.
Figure 1B shows the assembly of 14 ESTs from RH and ME49 tachyzoites into a contig of ∼1.4 kb for SRS3. This contig size is in agreement with an ∼1.4-kb SRS3 transcript identified by Northern blotting (not shown). The sequence of the ORF of SRS3 was determined from RH strain cDNA following amplification with the SRS3ORF primer pair. The ORF begins 191 nt downstream from the 5′ end of the contig and encodes 346 amino acids, including a predicted signal peptide and GPI anchor addition signal (Fig. 2). Removal of these two posttranslational processing signals would yield a mature peptide of approximately 281 amino acids with a predicted molecular mass of ∼29.6 kDa, assuming no further posttranslational modification. Comparison of the putative mature SRS3 sequence with the corresponding SAG1 peptide sequence (by using ClustalW) indicates that SRS3 shares 33.3% overall identity and 39.6% similarity in a central core region (residues 61 to 316 of the complete ORF [Fig. 2]).
The single EST corresponding to SRS4 (clone TgEST zy52c12) appears to encode the C-terminal two-thirds of an SRS-type molecule (Fig. 2). At present, we do not know the identity of the N-terminal third, and the characteristics of the complete ORF are unknown. However, SRS4 clearly possesses a hydrophobic consensus GPI anchor addition signal, as is the case with the other SRS sequences. The characteristics of the SRS2, SRS3, and SRS4 gene products are summarized in Table 2. This table also includes summary information on the SAG5 sequence identified by Crisanti et al. (GenBank accession no. U64517 [29]) and the BSR4 (P36) antigen (GenBank accession no. AF015290) identified by Knoll and Boothroyd (16).
Following our initial identification of the SRS genes, a further 2,353 sequences derived from ME49 in vivo bradyzoites were submitted to dBEST (18), thus enabling us to compare expression (in terms of EST numbers) of these sequences between the two stages of the asexual cycle of Toxoplasma. Searches of this data set for sequences corresponding to the SAG1, SAG3, SAG5, and SRS genes revealed only a single EST corresponding to SAG3, which was previously reported to be expressed in this life cycle stage (31), and two ESTs for SAG1 which represent recombinant SAG1 plasmids created in this laboratory and which are a minor contaminant of the cDNA library used for bradyzoite EST sequencing (18). Monoclonal antibodies to SAG1 also fail to react with bradyzoite lysates (31). Thus, failure to detect functional SAG1 ESTs in the bradyzoite data set is consistent with the immunological data indicating its restriction to the tachyzoite stage. The absence of SRS1 to -4 ESTs in the bradyzoite data set, therefore, suggests that these antigens are also not expressed in this stage, but this requires confirmation by direct analysis of in vivo cysts. No new SRS genes were found in this bradyzoite EST data set.
SRS gene products are expressed on the surface of Toxoplasma.
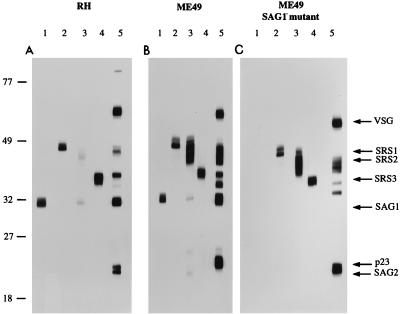
An MBP-SRS2 fusion protein expressed in Escherichia coli and purified by affinity chromatography was used to derive in mice a specific antiserum against SRS2. Western blotting of RH strain lysates with this antiserum (Fig. 3A, lane 3) shows that when reduced, this protein migrates as a weakly staining diffuse band with an apparent mass of 40 to 43 kDa, which overlaps with the SAG3 product. To further examine the relationship between SAG3 and SRS2, we performed Western blots of ME49 lysates on 9% nonreducing SDS-PAGE gels. The SRS2 band has a faster mobility under nonreducing conditions than under reducing conditions, as has also been observed with SAG1 and SAG3 (4, 5). At this resolution, the antiserum to SRS2 appears to react with at least four separate species (Fig. 4). A monoclonal antibody to SAG3 (T41F12) indicated that at least one of the anti-SRS2-reactive species comigrates with this antigen. At present, we lack monospecific reagents which would allow us to discriminate these species and resolve this complex of comigrating antigens. However, Southern blotting (not shown) indicates that SRS2 is encoded by a single gene, and apart from SRS4, no unaccounted for SRS2-like sequences are seen in the EST database. Thus, several of the different species which react with the SRS2 antiserum are most likely to result from posttranslational modifications of one protein.
FIG. 3.
Western blot strips labeled with antisera to SAG1 (lane 1), SRS1 (lane 2), SRS2 (lane 3), SRS3 (lane 4), and CRD (lane 5). Triton X-100 lysates were produced from equal numbers of tachyzoites from RH (A), the ME49 wild type (B), and the ME49 Sag1− mutant (C). Each lysate was treated with trypanosomal GPI-PLC prior to reducing SDS-PAGE. Variant surface glycoprotein (VSG) is a contaminant present in the GPI-PLC preparation. Sizes (kilodaltons) are shown on the left.
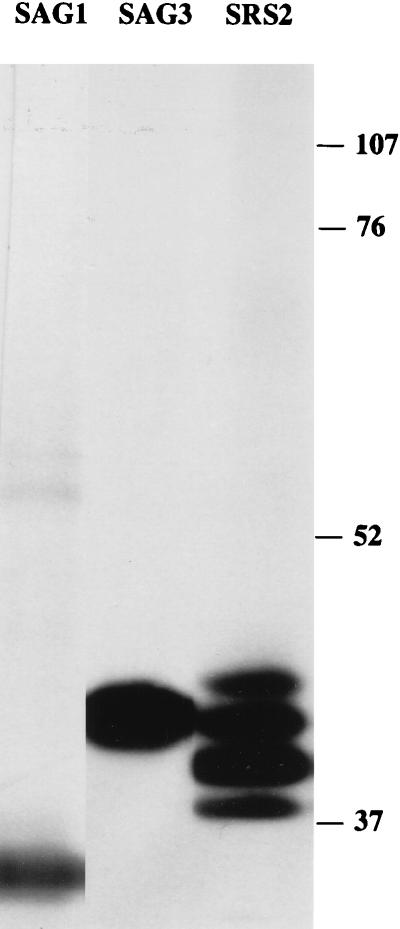
FIG. 4.
Western blot strips of ME49 lysates (without GPI-PLC treatment) labelled with SAG1, SAG3, and SRS2 antisera (from nonreducing SDS-PAGE). Sizes (kilodaltons) are shown on the right.
The SRS2 antiserum also shows limited cross-reactivity with SAG1 on reducing gels (Fig. 3, lane 3) whose identity is evident from the absence of this cross-reacting band in the Sag1− panel (Fig. 3C, lane 3). This cross-reactivity is not evident under nonreducing conditions (Fig. 4). This was further confirmed by fluorescence-activated cell sorter analysis of the Sag1− strain. This strain showed the same levels of fluorescence as its parent strain, ME49, with each of the anti-SRS2 and anti-SAG3 primary antibodies (not shown).
The band reacting with the SRS2 antiserum overlaps in its migration with a weakly reacting band on Western blots of RH lysates probed with antiserum specific for the CRD found in most GPI anchors after cleavage with GPI-PLC. This indicates that as predicted by the hydrophobic C terminus and as shown for SAG1 (19), SRS2 is likely to be GPI anchored.
Western blot analyses were performed with antisera generated to an MBP-SRS3 fusion. The results for RH lysates (Fig. 3A, lane 4) show a strong band migrating at ∼35 kDa. This is the same mobility seen for a major band with the anti-CRD antiserum (Fig. 3A, lane 5) and is presumed to correspond to the previously identified (but uncloned) P35 antigen. The latter antigen was originally defined as a 35-kDa antigen in surface-labeling experiments (6) which reacted with a monoclonal antibody (3F12) raised against Toxoplasma tachyzoites. The 3F12 monoclonal antibody failed to react with the MBP-SRS3 fusion protein produced in E. coli, but it also fails to react with reduced Toxoplasma lysates and therefore recognizes a native, conformational epitope. Thus, while it is likely that SRS3 encodes P35, we currently lack the necessary reagents to determine this definitively.
We examined the approximate relative abundance of each of the SRS products in wild-type RH and ME49 and the Sag1− B mutant of ME49 (12) by Western blotting (Fig. 3A, B, and C, respectively). Comparison of Fig. 3A (RH) and B (ME49) shows that while both strains express SAG1 (lanes 1) and each of the SRS antigens (lanes 2 to 4, corresponding to SRS1, SRS2, and SRS3, respectively), there are differences in the apparent abundance of the SRS antigens between the virulent RH (type I) and avirulent ME49 (type II) strains. In particular, the antiserum to SRS2 gives a strong, broad band corresponding to a 40- to 45-kDa antigen with the ME49 lysate (Fig. 3B, lane 3), which is significantly stronger than that obtained for RH (Fig. 3A, lane 3). These differences are unlikely to be due to differential reactivity of the polyclonal anti-SRS2 antiserum with SRS2 from different strains, since the serum was raised against an RH strain SRS2 fusion protein (and the RH strain SRS2 protein gives a weaker rather than stronger signal). Moreover, the difference in SRS2 antiserum reactivity is reflected in the profile of CRD reactivity obtained with lysates from the two parasite strains (Fig. 3A and B, lane 5).
Western blotting of lysates from the type III avirulent strain, CEP, with anti-MBP-SRS2 and anti-CRD antisera yielded patterns indistinguishable from those obtained for ME49 (not shown). SRS2 is the most likely candidate for the 40-kDa surface antigen identified by Ware and Kasper (36) using antisera specific to ME49 and CEP strains and which apparently failed to react with antisera from RH. The clear difference between the virulent and avirulent strains with respect to SRS2 expression indicated by Western blots is partially reflected in a more-than-twofold-greater relative abundance (11 of 1,806 [0.61%]) of SRS2 ESTs in the ME49 data set relative to RH (15 of 5,600 [0.27%]). The lack of perfect correlation between abundance of the gene product and EST number has been found for a number of other proteins in Toxoplasma (18).
In addition to the striking differences in apparent SRS2 levels, the CRD profiles for the RH and ME49 strains show another, as yet unidentified, antigen migrating between SAG1 and SRS3 which is more abundant in ME49 than in RH (Fig. 3, lanes 5). The results therefore reveal clear strain-specific differences in the CRD profiles and the apparent abundance of SRS antigens between the virulent RH (type I) and avirulent ME49 (type II) strains. The same result is apparent for the type III strain CEP (not shown).
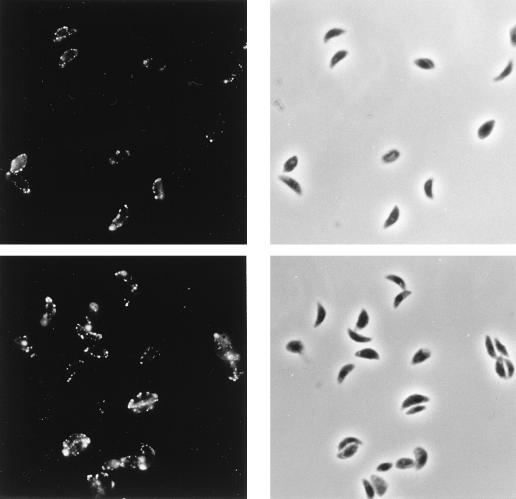
An indirect immunofluorescence assay (Fig. 5, upper panels) shows that the surface of live Toxoplasma tachyzoites can be labeled with antiserum to MBP-SRS2. Antiserum to MBP-SRS3 reveals a similar pattern of surface staining (Fig. 5, lower panels). The mottled appearance of parasite staining results from the fact that the parasites were stained prior to fixation and reflects partial capping during staining. Postfixation staining yields a characteristic circumferential staining pattern (not shown). These data further support our conclusion that SRS2 and SRS3 are indeed expressed on the surface of tachyzoites. As yet, our attempts to derive an antiserum against an MBP-SRS4 fusion protein which reacts with Toxoplasma lysates have been unsuccessful. As such, we cannot be sure that this gene product is expressed.
FIG. 5.
Surface immunofluorescence (left) and phase-contrast microscopy (right) of RH tachyzoites following live staining with antisera to SRS2 (top) and SRS3 (bottom) and fixation with paraformaldehyde. Preimmune sera failed to stain T. gondii tachyzoites (not shown).
DISCUSSION
Analysis of the Toxoplasma EST database has identified at least three new genes (SRS2 to -4) transcribed in tachyzoites which share structural homology with SAG1 and SAG3. Further members of this gene family have also been identified (SAG5, SAG5.1, and SAG5.2 [GenBank accession no. U64517, AF013968, and AF017268, respectively]), although their patterns of expression have not been reported. These results indicate that the surface of T. gondii tachyzoites is more complex than previously thought, displaying considerably more than the five major surface antigens originally identified in surface-labeling experiments. At the same time, the parasite surface appears simpler than previously believed, in the sense that most of the antigens now appear to be members of a single family, with only SAG2 clearly belonging to a separate class (23) (although the P23 antigen has yet to be cloned). T. gondii bradyzoites appear to express at least two members of the SRS family (SAG3 and BSR4 [P36]), in addition to at least two structurally unrelated GPI-anchored antigens, designated SAG4 (P18) (22) and SAG4.2 (GenBank accession no. AF015715 [17]).
The SRS family members show a conserved architecture, with conservation of cysteines and hydrophobicity patterns arguing that they are under selective pressure to maintain a particular secondary structure. Their deduced amino acid sequences show a hydrophobic COOH terminus, which strongly suggests that all SRSs are GPI anchored, as has been demonstrated for SAG1 (20, 32). Consistent with this, antiserum to the CRD of GPI anchors detects antigens comigrating with each of the SRS proteins.
The role of this expanding family of surface antigens is unclear. Despite reports that indicate that antisera to SAG1 partly inhibit invasion (9, 19), it is clear that Sag1− mutants derived by chemical mutagenesis can invade and replicate effectively both in vitro and in vivo (13), albeit at reduced efficiency compared to that of the parental ME49 strain. This implies that the parasite has evolved additional means to enable it to fulfill this function. SAG1 is also highly immunogenic, and humoral and cellular responses of the host against this protein are capable of limiting acute infection by clearing Toxoplasma tachyzoites (3, 15). In doing so, SAG1 drives the infection by selection in favor of the encysted bradyzoite form. This fosters the long-term survival of the parasite in a viable host. SAG1 is therefore critical to the parasite in several different ways. Recent observations suggest that SAG3 may also play a significant role in host cell attachment and invasion: SAG3− mutants obtained by targeted gene disruption are 1,000-fold less virulent and invade host cells with 50% of the efficiency of their wild-type counterparts (30). It is possible that SRS3 and the other family members play roles similar to those of SAG1 and SAG3. The existence of such redundant systems may also account for the ability of Toxoplasma to invade a wide variety of host cells.
The different abundances of SRS2 in RH and ME49 parasites may provide an interesting lead to unravel the function of these antigens in infection. The results from Western blotting and indirect immunofluorescence assay indicated that all three of the reference strains we analyzed (corresponding to types I, II, and III, respectively) expressed all members of the SRS gene family at least at the population level, although they do so at different levels. In particular, the highly virulent type I strain RH expresses little SRS2, whereas this protein is highly abundant in the avirulent type II (ME49) and type III (CEP) strains. The significance of this correlation with virulence is unknown.
It is unclear whether the family of SAG1-related sequences represents a system for antigenic variation. To date, we have not observed individual parasites in any population that fail to express a given antigen (17) (Fig. 5), which argues against the extreme sort of antigenic variation observed in T. brucei or Plasmodium falciparum (7, 21). One way to explore this further would be to examine SRS sequences in non-culture-adapted strains. The protective nature of humoral responses against SAG1 suggests at least that modern avirulent Toxoplasma strains lack the capacity to switch or to select tachyzoites expressing alternative surface antigens. An alternative paradigm emerging in parasite immunology (8, 11) is that coexpressed antigens with variant T-cell epitopes can function to inhibit or modulate T-cell responses. It is possible that the expanding family of Toxoplasma surface antigens functions in a similar manner. Such a function may play a key role in creation of the immunological milieu which favors differentiation to the bradyzoite form and the establishment of long-term infections.
ACKNOWLEDGMENTS
We thank Eduardo Ortega, Serge Bonnefoy, Laura Knoll, Michael Grigg, and other members of the Boothroyd Laboratory for help with this project; Nahid Madhani (Palo Alto DVA Sequencing Laboratory) for automated sequencing; and Stan Tomavo, Furio Spano, and David Roos for helpful discussions and communication of data prior to publication.
This work was supported by grants from the NIH (AI21423, AI30230, and AI41014). A.H. was supported by fellowships from the Swiss National Science Foundation and the Roche Research Foundation.
REFERENCES
- 1.Ajioka J, Boothroyd J C, Brunk B P, Hehl A, Hillier L, Manger I D, Overton G C, Marra M, Roos D, Wan K L, Waterston R, Sibley L D. Sequencing of ESTs from the protozoan parasite Toxoplasma gondii: efficient identification of genes and identification of phylogenetically restricted sequences of the Apicomplexa. Genome Res. 1998;8:18–28. doi: 10.1101/gr.8.1.18. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. 1 to 3. New York, N.Y: John Wiley and Sons, Inc.; 1995. [Google Scholar]
- 3.Bulow R, Boothroyd J C. Protection of mice from fatal Toxoplasma gondii infection by immunization with P30 antigen in liposomes. J Immunol. 1991;147:3496–3500. [PubMed] [Google Scholar]
- 4.Burg J L, Perelman D, Kasper L H, Ware P L, Boothroyd J C. Molecular analysis of the gene encoding the major surface antigen of Toxoplasma gondii. J Immunol. 1988;141:3584–3591. [PubMed] [Google Scholar]
- 5.Cesbron-Delauw M F, Tomavo S, Beauchamps P, Fourmaux M P, Camus D, Capron A, Dubremetz J F. Similarities between the primary structures of two distinct major surface proteins of Toxoplasma gondii. J Biol Chem. 1994;269:16217–16222. [PubMed] [Google Scholar]
- 6.Couvreur G, Sadak A, Fortier B, Dubremetz J F. Surface antigens of Toxoplasma gondii. Parasitology. 1988;97:1–10. doi: 10.1017/s0031182000066695. [DOI] [PubMed] [Google Scholar]
- 7.Cross G A M. Cellular and genetic aspects of antigenic variation in trypanosomes. Annu Rev Immunol. 1990;8:83–110. doi: 10.1146/annurev.iy.08.040190.000503. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert S C, Plebanski M, Gupta S, Morris J, Cox M, Aidoo M, Kwiatkowski D, Greenwood B M, Whittle H C, Hill A V S. Association of malaria parasite population structure, HLA, and immunological antagonism. Science. 1998;279:1173–1177. doi: 10.1126/science.279.5354.1173. [DOI] [PubMed] [Google Scholar]
- 9.Grimwood J, Smith J E. Toxoplasma gondii: the role of a 30-kDa surface protein in host cell invasion. Exp Parasitol. 1992;74:106–111. doi: 10.1016/0014-4894(92)90144-y. [DOI] [PubMed] [Google Scholar]
- 10.Hehl A B, Krieger T, Boothroyd J C. Identification and characterization of SRS1, a Toxoplasma gondii surface antigen upstream of and related to SAG1. Mol Biochem Parasitol. 1997;89:271–282. doi: 10.1016/s0166-6851(97)00126-6. [DOI] [PubMed] [Google Scholar]
- 11.Kahn S J, Wleklinski M. The surface glycoproteins of Trypanosoma cruzi encode a superfamily of variant T cell epitopes. J Immunol. 1997;159:4444–4451. [PubMed] [Google Scholar]
- 12.Kasper L H. Isolation and characterization of a monoclonal anti-P30 antibody resistant mutant of Toxoplasma gondii. Parasite Immunol. 1987;9:433–445. doi: 10.1111/j.1365-3024.1987.tb00521.x. [DOI] [PubMed] [Google Scholar]
- 13.Kasper L H, Khan I A. Role of P30 in host immunity and pathogenesis of Toxoplasma gondii infection. Res Immunol. 1993;144:45–48. doi: 10.1016/s0923-2494(05)80097-5. [DOI] [PubMed] [Google Scholar]
- 14.Kasper L H, Ware P L. Recognition and characterization of stage-specific oocyst/sporozoite antigens of Toxoplasma gondii by human antisera. J Clin Invest. 1985;75:1570–1577. doi: 10.1172/JCI111862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan I A, Ely K H, Kasper L H. A purified parasite antigen (P30) mediates CD8+ T cell immunity against fatal Toxoplasma gondii infection in mice. J Immunol. 1991;147:3501–3506. [PubMed] [Google Scholar]
- 16.Knoll L J, Boothroyd J C. Isolation of developmentally regulated genes from Toxoplasma gondii by a gene trap with the positive and negative selectable marker hypoxanthine-xanthine-guanine phosphoribosyltransferase. Mol Cell Biol. 1998;18:807–814. doi: 10.1128/mcb.18.2.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manger, I. D., A. B. Hehl, and J. C. Boothroyd. Unpublished data.
- 18.Manger I D, Hehl A, Parmley S, Sibley L D, Marra M, Hillier L, Waterston R, Boothroyd J C. Expressed sequence tag analysis of the bradyzoite stage of Toxoplasma gondii: identification of developmentally regulated genes. Infect Immun. 1998;66:1632–1637. doi: 10.1128/iai.66.4.1632-1637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mineo J R, McLeod R, Mack D, Smith J, Khan I A, Ely K H, Kasper L H. Antibodies to Toxoplasma gondii major surface protein (SAG-1, P30) inhibit infection of host cells and are produced in murine intestine after peroral infection. J Immunol. 1993;150:3951–3964. [PubMed] [Google Scholar]
- 20.Nagel S D, Boothroyd J C. The major surface antigen, P30, of Toxoplasma gondii is anchored by a glycolipid. J Biol Chem. 1989;264:5569–5574. [PubMed] [Google Scholar]
- 21.Newbold C I, Craig A G, Kyes S, Benendt A R, Snow R W, Peshu N, Marsh K. Pfemp1, polymorphism and pathogenesis. Ann Trop Med Parasitol. 1997;91:551–557. doi: 10.1080/00034989760923. [DOI] [PubMed] [Google Scholar]
- 22.Odberg-Ferragut C, Soete M, Engels A, Samyn B, Loyens A, Van Beeumen J, Camus D, Dubremetz J F. Molecular cloning of the Toxoplasma gondii sag4 gene encoding an 18 kDa bradyzoite specific surface protein. Mol Biochem Parasitol. 1996;82:237–244. doi: 10.1016/0166-6851(96)02740-5. [DOI] [PubMed] [Google Scholar]
- 23.Parmley S F, Sgarlato G D, Mark J, Prince J B, Remington J S. Expression, characterization, and serologic reactivity of recombinant surface antigen P22 of Toxoplasma gondii. J Clin Microbiol. 1992;30:1127–1133. doi: 10.1128/jcm.30.5.1127-1133.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfefferkorn E R, Pfefferkorn L C, Colby E D. Development of gametes and oocysts in cats fed cysts derived from cloned trophozoites of Toxoplasma gondii. J Parasitol. 1977;63:158–159. [PubMed] [Google Scholar]
- 25.Riggs P. Expression and purification of maltose-binding protein fusions. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1994. pp. 16.6.1–16.6.14. [DOI] [PubMed] [Google Scholar]
- 26.Sabin A B. Toxoplasmic encephalitis in children. JAMA. 1941;116:801–807. [Google Scholar]
- 27.Shak S, Davitz M A, Wolinsky M L, Nussenzweig V, Turner M J, Gurnett A. Partial characterization of the cross-reacting determinant, a carbohydrate epitope shared by decay accelerating factor and the variant surface glycoprotein of the African Trypanosoma brucei. J Immunol. 1988;140:2046–2050. [PubMed] [Google Scholar]
- 28.Sibley L D. Invasion of vertebrate cells by Toxoplasma gondii. Trends Cell Biol. 1995;5:129–132. doi: 10.1016/s0962-8924(00)88964-3. [DOI] [PubMed] [Google Scholar]
- 29.Spano, F. Personal communication.
- 30.Tomavo S. The major surface proteins of Toxoplasma gondii: structures and functions. Curr Top Microbiol Immunol. 1996;219:45–54. doi: 10.1007/978-3-642-51014-4_4. [DOI] [PubMed] [Google Scholar]
- 31.Tomavo S, Fortier B, Soete M, Ansel C, Camus D, Dubremetz J F. Characterization of bradyzoite-specific antigens of Toxoplasma gondii. Infect Immun. 1991;59:3750–3753. doi: 10.1128/iai.59.10.3750-3753.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomavo S, Schwarz R T, Dubremetz J F. Evidence for glycosyl-phosphatidylinositol anchoring of Toxoplasma gondii major surface antigens. Mol Cell Biol. 1989;9:4576–4580. doi: 10.1128/mcb.9.10.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Udenfriend S, Kodukula K. How glycosylphosphatidylinositol-anchored proteins are made. Annu Rev Biochem. 1995;64:563–592. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- 34.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan K L, Blackwell J M, Ajioka J W. Toxoplasma gondii expressed sequence tags: insight into tachyzoite gene. Mol Biochem Parasitol. 1996;75:179–186. doi: 10.1016/0166-6851(95)02524-3. [DOI] [PubMed] [Google Scholar]
- 36.Ware P L, Kasper L H. Strain-specific antigens of Toxoplasma gondii. Infect Immun. 1987;55:778–783. doi: 10.1128/iai.55.3.778-783.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]