Abstract
The genetic variation and population structure of gene N (nucleocapsid) and part of gene L (replicase) from 13 eggplant mottle dwarf virus (EMDV) isolates from Spain were evaluated and compared with sequences of EMDV isolates from other countries retrieved from GenBank. Phylogenetic inference of part of gene L showed three main clades, one containing an EMDV isolate from Australia and the other two containing isolates from Iran and Europe, as well as four subclades. EMDV isolates from Spain were genetically very similar and grouped in a subclade together with one isolate from Germany and one from the UK. No new recombination events were detected in addition to one recombination previously reported, suggesting that recombination is rare for EMDV. The comparison of synonymous and non-synonymous rates showed that negative selection played an important role, and only two codons were under positive selection. Genetic differentiation (Fst test), phylogenetic and nucleotide diversity analyses suggest a unique introduction of EMDV to Spain and low gene flow with other countries. In contrast, Greece and Italy showed diverse populations with high gene flow between both.
Keywords: negative-strand RNA virus, Alphanucleorhabdovirus melongenae, phylogeny, recombination, selection, gene flow, nucleotide diversity
1. Introduction
Understanding the genetic variability of virus populations and the factors involved in their evolution is crucial to developing accurate detection and diagnostic tools, implementing efficient disease control strategies and gaining insight into virus epidemiology [1]. The main evolutionary mechanisms shaping the genetic structure and variability of virus populations are mutation, recombination, selection, genetic drift and gene flow or migration [2,3]. RNA viruses have great potential for rapid evolution due to their rapid replication and high mutation rates since RNA replicases lack proofreading activity. Recombination is another source of genetic variation and the emergence of new plant viruses [4]. The genetic variation produced by mutation and recombination is limited by the interplay of selection, genetic drift and gene flow. Natural selection in plant viruses results from the competition among genetic variants differing in some aspects of the life cycle, such as replication, movement between plant cells and transmission to other plants by vectors, so those variants with more reproductive success (fitness) will pass to the next generation. Genetic drift is the change in the frequency of genetic variants in small populations by random chance. Viruses can undergo population bottlenecks or founder events in different life cycle steps such as cell-to-cell movement and transmission by vectors. Gene flow or migration of genetic variants favors genetic uniformity, whereas restricted migration leads to genetic differentiation between populations via selection and genetic drift.
Eggplant mottled dwarf virus (EMDV) has been assigned to the species Alphanucleorhabdovirus melongenae in the genus Alphanucleorhabdovirus, family Rhabdoviridae, order Mononegavirales [5]. The EMDV genome is a single-stranded, negative-sense RNA encapsidated by the nucleocapsid protein (N) and wrapped by a phospholipid membrane forming bacilliform particles. The EMDV genome contains seven open reading frames: N (nucleocapsid), X (unknown function), P (phosphoprotein, a polymerase cofactor), Y (putative movement protein), M (matrix protein, which connects the envelope to the ribonucleocapsid core), G (glycoprotein, which protrudes from the lipid envelope exterior) and L (RNA-dependent RNA polymerase) [5,6].
EMDV has a wide host range and infects important crops (e.g., eggplant, tomato, potato, pepper, cucumber and tobacco), ornamental (e.g., pittosporum, honeysuckle, pelargonium and hibiscus) and wild plants [7]. EMDV is transmitted by the leafhoppers Agallia vorobjevi, Anaceratogallia laevis and A. ribauti [8,9]. It is widespread in the Mediterranean basin and has been detected in Europe: Albania, Azerbaijan, Bulgaria, Croatia, France, Germany, Greece, Italy, Portugal, Slovenia, Spain, Türkiye and the UK; Asia: Afghanistan, Iran, Israel and Jordan; Africa: Algeria, Libya, Morocco and Tunisia; and Oceania: plants imported from Australia in New Zealand [7,10].
The genetic variability of EMDV and some evolutionary factors have been studied by sequence analyses of genes N, X, P, Y, M and G from seven isolates from Greece and one from Cyprus; part of gene L of these isolates from Greece and Cyprus, as well as eight from Italy and five from Spain [11,12]; and the complete sequence of one isolate from Iran [13]. In this work, the nucleotide sequence of gene N and part of gene L in 13 EMDV isolates from four regions of Spain and six hosts were determined and analyzed together with part of the gene L of another 5 Spanish isolates retrieved from GenBank. This work revealed that the population of EMDV in Spain was very homogeneous and had a low migration rate in contrast to those in Italy and Greece. A more comprehensive picture of the genetic variability and evolutionary mechanisms of EMDV globally was obtained by analyzing the entire genome of nine isolates—three from Italy, two from Greece and one each from Iran, Slovenia, Germany and the UK—as well as the N gene from six isolates from Greece and one from Cyprus; part of the L gene of six isolates from Greece, one from Cyprus, two from France and one from Australia; and a Y gene portion from one isolate from Azerbaijan.
2. Results
2.1. Sequencing EMDV Isolates from Spain
We analyzed the N gene (encoding the nucleocapsid) and a portion of the L gene (encoding the RNA-dependent RNA polymerase) to evaluate the genetic variability in EMDV in Spain and other countries. These genomic regions were chosen for three reasons: (i) they are frequently used to study plant virus genetic variability, which facilitates comparison between EMDV and other viruses; (ii) most EMDV sequences in GenBank either corresponded to or contained the selected gene L region, allowing for comparisons of variability in Spain and other countries; and (iii) because both genes, N and L, are separated in the genome, making them useful for recombination analysis.
The complete gene N and a region of gene L (named L1) of 13 EMDV isolates collected in several regions of Spain were amplified by RT-PCR, sequenced and compared with nucleotide sequences of other EMDV isolates retrieved from GenBank (Table 1). The nucleotide sequences were deposited in GenBank under accession numbers OR631742–OR631767.
Table 1.
Eggplant mottled dwarf virus (EMDV) isolates analyzed in this study.
| Isolate | Country | Region | Year | Host | Genes 1 | GenBank |
|---|---|---|---|---|---|---|
| 203/09 | Spain | Valencia | 2009 | Pittosporum tobira | N, L1 | OR631742, OR631755 |
| 80/10 | Spain | Almería | 2010 | Solanum melongena | N, L1 | OR631743, OR631756 |
| 990/11 | Spain | Almería | 2011 | Solanum lycopersicum | N, L1 | OR631744, OR631757 |
| 991/11 | Spain | Almería | 2011 | Cucumis sativus | N, L1 | OR631745, OR631758 |
| 539/12 | Spain | Granada | 2012 | Solanum lycopersicum | N, L1 | OR631746, OR631759 |
| 443/17 | Spain | Granada | 2017 | Solanum melongena | N, L1 | OR631747, OR631760 |
| 478/17 | Spain | Granada | 2017 | Solanum melongena | N, L1 | OR631748, OR631761 |
| 697/17 | Spain | Granada | 2017 | Solanum melongena | N, L1 | OR631749, OR631752 |
| 377/20 | Spain | Granada | 2020 | Solanum melongena | N, L1 | OR631750, OR631763 |
| 593/12 | Spain | Navarra | 2012 | Capsicum annuum | N, L1 | OR631751, OR631764 |
| 1009/11 | Spain | Pontevedra | 2011 | Capsicum annuum | N, L1 | OR631752, OR631765 |
| 464/15 | Spain | Valencia | 2015 | Podranea ricasoliana | N, L1 | OR631753, OR631766 |
| 540/15 | Spain | Zaragoza | 2015 | Pittosporum tobira | N, L1 | OR631754, OR631767 |
| S1 | Spain | Malaga | 2011 | Hibiscus rosa-sinensis | L1 | HG916821 [12] |
| S2 | Spain | Malaga | 2011 | Hibiscus rosa-sinensis | L1 | HG916822 [12] |
| S3 | Spain | Malaga | 2011 | Hibiscus rosa-sinensis | L1 | HG916823 [12] |
| S4 | Spain | Granada | 2011 | Hibiscus rosa-sinensis | L1 | HG916824 [12] |
| S5 | Spain | Almeria | 2013 | Cucumis sativus | L1 | HG916825 [12] |
| SH-eg | Iran | 2011 | Solanum melongena | N, X, P, Y, M, G, L | KC905081 [13] | |
| STR20ST2 | Slovenia | 2020 | Solanum lycopersicum | N, X, P, Y, M, G, L | OL472111 [14] | |
| PV-1127 | Germany | 2014 | Hydrangea macrophylla | N, X, P, Y, M, G, L | OQ847408 [15] | |
| 02923HTS | UK | 2022 | Pittosporum tobira | N, X, P, Y, M, G, L | OQ716555 [16] | |
| Pit-MAIB | Italy | 2008 | Pittosporum tobira | N, X, P, Y, M, G, L | LN680656 [17] | |
| Agapanthus | Italy | 2012 | Agapanthus sp. | N, X, P, Y, M, G, L | KJ082087 [18] | |
| PV-0031 | Italy | 1987 | Solanum melongena | N, X, P, Y, M, G, L | MW854257 | |
| EG1035 | Greece | Thessaloniki | 2009 | Solanum melongena | N, X, P, Y, M, G, L | FR751552 [6] |
| PV-1212 | Greece | Thessaloniki | 2017 | Cucumis sativus | N, X, P, Y, M, G, L | OL584369 |
| EMDVcs | Greece | Katerini | 2008 | Cucumis sativus | N, L1 | HG794532, HG794539 [11,19] |
| EMDVnt | Greece | Thessaloniki | 2006 | Nicotiana tabacum | N, L1 | HG794534, HG794540 [11,19] |
| EMDVsl | Greece | Thessaloniki | 2010 | Solanum lycopersicum | N, L1 | HG794535, HG794543 [11,19] |
| EMDVpit | Cyprus | Nicosia | 2011 | Pittosporum tobira | N, L1 | HG794531, HG794544 [11,19] |
| EMDV-Egg | Greece | 2007 | Hibiscus rosa-sinensis | L1 | AM922322 [19] | |
| EMDVhrs | Greece | Thessaloniki | 2009 | Lonicera japonica | N | HG794541 [11] |
| EMDVlj | Greece | Sifnos island | 2010 | Capparis spinosa | N | HG794542 [11] |
| EMDV-caps | Greece | Santorini | 2005 | Capparis spinosa | N | HG794545 [11] |
| EMDV-capsL | Greece | Rhodes | 2009 | Solanum melongena | L1 | HG794533 [19] |
| EMDVsm | Greece | Thessaloniki | 2010 | Hibiscus rosa-sinensis | L1 | HG794536 [19] |
| Vivaldi | Italy | Emilia-Romagna | 2012 | Solanum tuberosum | L1 | KC760150 [20] |
| C1 | Italy | Calabria | 2010 | Hibiscus rosa-sinensis | L1 | HG916814 [12] |
| C2 | Italy | Calabria | 2010 | Hibiscus rosa-sinensis | L1 | HG916815 [12] |
| SOM-1 | Italy | Campania | 2011 | Solanum melongena | L1 | HG916816 [12] |
| SOM-2 | Italy | Campania | 2011 | Solanum melongena | L1 | HG916817 [12] |
| SOM-3 | Italy | Campania | 2013 | Solanum melongena | L1 | HG916818 [12] |
| SOL-1 | Italy | Campania | 2010 | Solanum lycopersicum | L1 | HG916819 [12] |
| COV-1 | Italy | Emilia-Romagna | 1990 | Codiaeum variegatum | L1 | HG916820 [12] |
| EM170361 | France | 2017 | Solanum lycopersicum | L2 | MN990976 [10] | |
| EM170755 | France | 2017 | Cucumis sativus | L2 | MN990977 [10] | |
| T14_00910 | Australia | 2014 | Hibiscus syriacus | L2 | KJ742827 [21] | |
| AZ15-31 | Azerbaijan | 2015 | Cucumis sativus | Y1 | MG964325 [22] |
1 L1 = position 1090–1965 of gene L, L2 = position 1375–1670 of gene L, Y1 = position 277–865 of gene Y.
EMDV isolates PV-1127 from Germany and 02923HTS from the UK showed the highest nucleotide identities with the Spanish isolates, ranging from 97.8 to 99.3% for gene N and from 97.3 to 99.1% for the genomic region L1 (Supplementary Tables S1 and S2). Comparison of the L1 region of the 13 Spanish isolates sequenced here and that of five Spanish isolates retrieved from GenBank showed that isolates S4 and S5 from the Granada province of Spain were similar to isolate 443/17, also collected in Granada (nucleotide identity > 99.0%). Isolates S1, S2 and S3 from the Spanish province of Malaga showed lower nucleotide identity with the other Spanish isolates (ranging from 96.3 to 97.1%). Isolate 203/09 also differed from the other Spanish isolates (nucleotide identity, 96.9–97.4%). The rest of the Spanish isolates showed more similar sequences (nucleotide identity 98.1–99.5%).
2.2. Phylogenetic Relationships among Worldwide EMDV Isolates
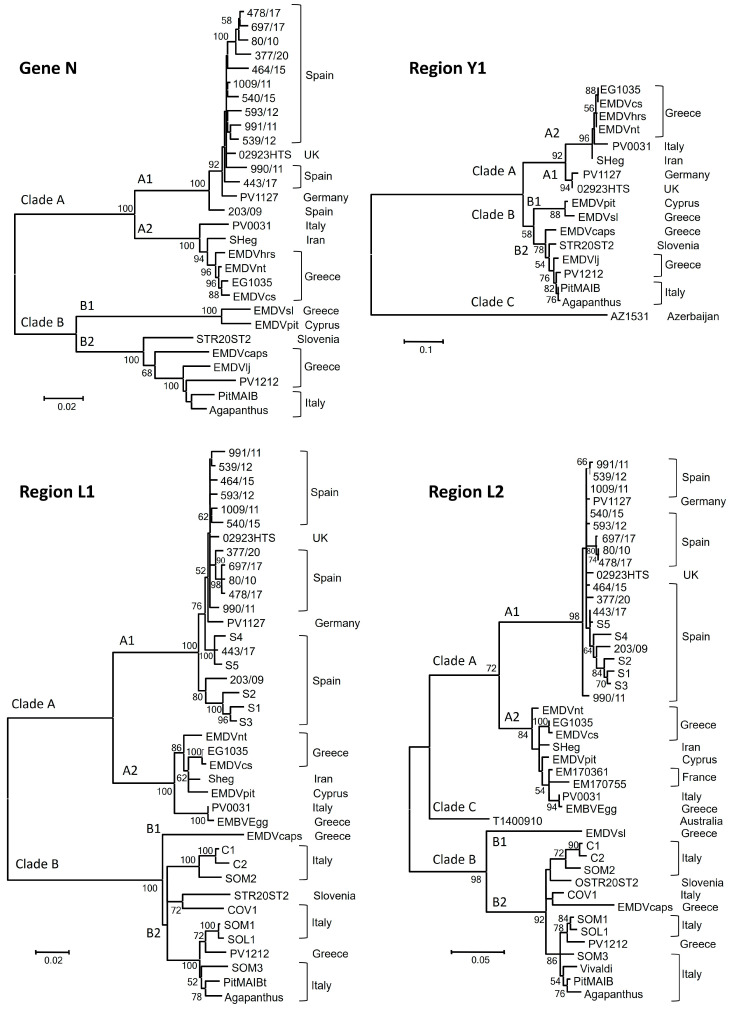
Phylogenetic analysis of the N gene of 29 EMDV isolates from eight countries showed two main clades, A and B, each divided into two subclades, A1, A2, B1 and B2 (Figure 1). Subclade A1 was composed of all isolates from Spain, one isolate from Germany and another from the UK, forming a homogenous and undifferentiated group (the minimum nucleotide identity between isolate pairs was 97.3%). Subclade A2 was composed of isolates from Iran, Italy and Greece. The minimum nucleotide identity within subclade A2 was 97.1%, whereas nucleotide identity between subclades A1 and A2 varied from 91.7 to 92.9%. Subclade B1 had two isolates, one from Greece and another from Cyprus (nucleotide identity of 97.2%), and B2 was composed of isolates from Italy, Greece and Slovenia. The minimum nucleotide identity within subclade B2 was 94.1%, and nucleotide identities between subclades B1 and B2 ranged from 89.0 to 89.6%. Nucleotide identity between clades A and B ranged from 85.3 to 87.6%.
Figure 1.
Unrooted maximum likelihood phylogenetic trees of four EMDV genomic regions: gene N, Y1 (gene Y, nucleotide position 277–865), L1 (gene L, position 1090–1965) and L2 (gene L, position 1375–1670) of EMDV isolates from different countries (see Table 1). Horizontal bars are proportional to evolutionary distances, and bootstrap values greater than 50% are in the nodes. Clades A, B and C and subclades A1, A2, B1 and B2 are indicated in the corresponding branches.
Phylogenetic trees of the L1 genomic region (876 nt) of 39 isolates from eight countries and the L2 region (296) of 44 isolates from 10 countries (Figure 1) had the same topology with high values of bootstrap for the external nodes, but L1 showed better resolution in the external nodes. The phylogenetic tree of L2 comprised three main clades: A, B and C. Clades A and B and their divisions (A1, A2, B1 and B2) contained isolates in the same clades and subclades of the phylogenetic tree of gene N. Clade C contained the EMDV isolate T14_00910 from Australia. Nucleotide identities were 81.4–86.4% between clades A and B, 84.5–88.9% between A and C and 84.5–87.4% between B and C. Subclade A1 contained 1 isolate from Germany, 1 from the UK, the 13 Spanish isolates sequenced here and another 5 Spanish isolates retrieved from GenBank. Subclade A2 contained isolates from Iran, Greece, Italy and France and the isolate EMDVpit from Cyprus that was in the subclade B1 for gene N. Clades A1 and A2 had minimum nucleotide identities of 96.3% and 95.6%, respectively, and the nucleotide identity between these two clades ranged from 87.5 to 91.2%. Subclade B1 had only isolate, EMDVsl from Greece, and subclade B2 contained isolates from Slovenia, Italy and Greece. The minimum nucleotide identity within clade B2 was 90.5%, and that between B1 and B2 ranged from 85.8 to 88.5%.
The phylogenetic tree of a 588 nt region of gene Y (named Y1) in 17 EMDV isolates from eight countries showed three main clades: A, B and C (Figure 1). Clades A and B and the subdivisions A1, A2, B1 and B2 contained the same isolates as the equivalent clades for the N gene and the L1 and L2 genomic regions. Nucleotide identities within each subclade were higher than 94.0%, 91.3–92.9% between subclades A1 and A2 and 88.6–90.1% between B1 and B2. Clade C contained the isolate AZ15-31 from Azerbaijan, showing high divergence from the other isolates (nucleotide identities between 74.4 and 76.4%, whereas clade A and B had nucleotide identities ranging from 83.3 to 88.8%. Blastn analysis showed that isolate AZ15-31 had nucleotide identities of 82.6% with tomato alphanucleorhabdovirus 1 (TARV1) and 78.0% with physostegia chlorotic mottle virus (PhCMoV), both being distinct species belonging to the genus Alphanucleorhabdovirus.
The phylogenic trees showed no correlation between genetic distances and hosts. For example, for gene N, the isolate EMDVcs from cucumber was genetically close to the isolate EG1035 from eggplant and genetically distant to another cucumber isolate, PV1212. Another example is for the genomic region L2, where the isolate EM170361 from tomato was close to the eggplant isolate EG1035 and the cucumber isolate EMDVcs but distant from another tomato isolate, Sol1, which was close to the cucumber isolate PV1212.
2.3. Recombination between EMDV Variants
Several sets of nucleotide sequences were analyzed with seven recombination detection methods implemented in the package RDP4 [23]. Analysis of the complete genome (13163 nt) or each gene individually (N, X, P, Y, M, G and L) of the nine EMDV isolates with the genome completely sequenced (isolates SH-eg, STR20ST2, PV-1127, 02923HTS, Pit-MAIB, Agapanthus, PV-0031, EG1035 and PV-1212, see Table 1) showed no recombination with any of these methods. Also, no recombination was found after analyzing the N gene of 29 EMDV isolates or two regions of gene L: L1 (876 nt) of 39 EMDV isolates and L2 (296 nt) of 44 EMDV isolates.
Finally, the sequences of N and L1 were concatenated for each isolate separately for the 13 Spanish isolates sequenced here, and isolates EMDVcs, EMDVnt and EMDVpit, along with equivalent sequences which were prepared from the nine isolates with the complete genome sequenced. Thus, we generated sequences composed of gene N and the region L1 from 25 EMDV isolates to use as input. The seven methods performed using the RDP4 package detected one recombination event for isolate EMDVpit involving an ancestor of isolate STR20ST2 as the major parental and an SH-eg ancestor as the minor parental. The phylogenetic trees confirmed this recombination event since EMDVpit was placed in different clades depending on the genomic region analyzed. Thus, isolates EMDVpit and STR20ST2 are in clade B and SH-eg is in clade A in the phylogenetic trees of genes N and Y, while in the phylogenetic tree of gene L, EMDVpit and SH-eg are in clade A and STR20ST2 is in clade B (Figure 1). The nucleotide identity of EMDVpit was higher with SH-eg (98.2%) than with STR20ST2 (85.0%) for gene L, whereas the opposite occurred for genes N and Y (Table 2). This result suggests that the breaking point could exist between the genes Y and L. This recombination even was detected previously in the gene G, but only the minor parental was found [11].
Table 2.
Nucleotide identity between the recombinant isolate EMDVpit and the parental isolates SH-eg and STR20ST2.
| Isolates | N | Y | L |
|---|---|---|---|
| SH-eg/STR20ST2 | 87.6 | 86.7 | 85.2 |
| SH-eg/EMDVpit | 85.9 | 84.4 | 98.2 |
| STR20ST2/EMDVpit | 89.2 | 90.1 | 85.0 |
2.4. Genetic Variation, Selection and Coevolution in Different EMDV Genes
Analysis of the nine completely sequenced EMDV isolates with the genome showed differences in genetic variability between genes. Nucleotide diversity was similar for genes N, P, M and G and slightly higher for genes X and Y (Table 3). Surprisingly, gene Y had the highest nucleotide diversity and the lowest proportion of segregating sites. The ratio between non-synonymous and synonymous rates (dN/dS) was less than one for every gene, indicating negative selection, ranging from 0.010 for gene Y to 0.123 for gene X. Evaluation of selection at individual codons (amino acids) showed that around 10% of the amino acids were under negative (purifying) selection, and amino acid positions 449 of gene N and 10 of gene X were under positive (diversifying) selection.
Table 3.
Population genetics parameters of different coding regions of EMDV.
| Gene | S | π | dN | dS | dN/dS | N | NSe | Pos | Neg |
|---|---|---|---|---|---|---|---|---|---|
| N | 0.209 | 0.167 | 0.012 | 0.443 | 0.027 | 1431 | 299 | 1 (449) | 140 |
| X | 0.320 | 0.225 | 0.072 | 0.587 | 0.123 | 294 | 94 | 1 (10) | 24 |
| P | 0.226 | 0.170 | 0.021 | 0.437 | 0.048 | 885 | 200 | 0 | 86 |
| Y | 0.189 | 0.241 | 0.005 | 0.485 | 0.010 | 864 | 163 | 0 | 112 |
| M | 0.228 | 0.178 | 0.017 | 0.474 | 0.036 | 756 | 172 | 0 | 96 |
| G | 0.223 | 0.163 | 0.012 | 0.501 | 0.023 | 1848 | 412 | 0 | 231 |
| L | 0.224 | 0.179 | 0.013 | 0.495 | 0.028 | 5841 | 1309 | 0 | 761 |
S = proportion of segregating (polymorphic) sites. π = nucleotide diversity (mean nucleotide differences per site between sequence pairs, dN = average number of non-synonymous substitutions per non-synonymous site. dS = average number of synonymous substitutions per synonymous site. N = number of nucleotide sites of each gene. NSe = number of segregating sites. Pos = number of codons under positive selection (between parentheses are the codon positions). Neg = number of codons under negatives selection.
Finally, we analyzed coevolution among different amino acid sites within each EMDV protein for EMDV isolates with the complete genome sequenced. Only positions 190 and 376 of N and 57 and 86 of X were coevolving. However, analysis of gene N from 29 EMDV isolates showed six coevolving amino acid pairs: 68–157, 169–193, 218–423, 228–442, 240–432 and 381–399. Analysis of L1 showed ten coevolving amino acid pairs: 377–541, 393–402, 406–413, 417–614, 422–624, 441–535, 519–541, 521–589, 568–651 and 628–654 (position relative to L1).
2.5. Genetic Differentiation of EMDV Populations
Genetic differentiation between EMDV populations, determined by the gene flow (migration) rate, was estimated by calculating the nucleotide diversity within and between geographical regions and using the statistic Fst.
The nucleotide diversity of EMDV within Italy or Greece was high (>0.100) and about ten times lower than that within Spain (Table 4). Comparison between countries showed nucleotide diversities in the same order as those within Italy or Greece. Fst values suggest high gene flow between Italy and Greece and low gene flow between both countries and Spain.
Table 4.
Genetic diversity and differentiation of EMDV populations in three Mediterranean countries.
| Gene | N | Italy | Greece | Spain | |
|---|---|---|---|---|---|
| N | 3 | Italy | 0.166 ± 0.015 | ||
| 8 | Greece | 0.148 ± 0.012 (−0.068) | 0.177 ± 0.014 | ||
| 13 | Spain | 0.187 ± 0.015 (0.524) | 0.203 ± 0.017 (0.487) | 0.019 ± 0.002 | |
| L1 | 10 | Italy | 0.110 ± 0.013 | ||
| 6 | Greece | 0.239 ± 0.036 (0.277) | 0.206 ± 0.031 | ||
| 18 | Spain | 0.285 ± 0.046 (0.688) | 0.206 ± 0.026 (0.475) | 0.025 ± 0.003 |
Gene = Genomic regions analyzed: gene N (1431 nt) and L1, a segment of gene L (876 nt). N = number of isolates per country and genomic region. N values < 5 are too low to represent a country and should be interpreted with caution. Nucleotide diversity estimated as mean nucleotide distance between sequence pairs using the substitution model T92 + G, where G = 0.27. Nucleotide diversity values within countries are in bold on the diagonal, and diversity values between countries are below the diagonal. Fst values are between parentheses.
To study the population structure of EMDV in Spain, four regions were considered: (i) Malaga province, (ii) Granada province, (iii) Almeria province and (iv) the rest, composed of Navarra, Pontevedra, Valencia and Zaragoza provinces. Nucleotide diversities within and between regions were lower than 0.020, except between Malaga and the other Spanish regions, with values around 0.040 (Table 5). Fst values showed two genetically differentiated populations of EMDV in Spain with very low gene flow between them. One was in Malaga province, and the other corresponded to the other provinces of Spain, which showed high gene flow between them.
Table 5.
Genetic diversity and differentiation of EMDV populations in Spain.
| N | Malaga | Granada | Almeria | Rest | |
|---|---|---|---|---|---|
| 3 | Malaga | 0.018 ± 0.003 | |||
| 6 | Granada | 0.039 ± 0.005 (0.520) | 0.016 ± 0.004 | ||
| 4 | Almeria | 0.040 ± 0.005 (0.492) | 0.016 ± 0.002 (−0.094) | 0.019 ± 0.003 | |
| 5 | Rest | 0.038 ± 0.005 (0.472) | 0.019 ± 0.003 (0.046) | 0.019 ± 0.003 (0.011) | 0.018 ± 0.004 |
Genomic regions analyzed: L1, a segment of gene L (876 nt). N = number of isolates per each region of Spain: Malaga, Granada and Almeria provinces, and Rest corresponds to Navarra, Pontevedra, Valencia and Zaragoza provinces. Nucleotide diversity estimated as mean nucleotide distance between sequence pairs using the substitution model T92 + G, where G = 0.27. Nucleotide diversities within Spanish regions are in bold on the diagonal, and diversities between countries are below the diagonal. Fst values are between parentheses.
3. Discussion
EMDV genetic variability was relatively high compared to that of most plant viruses [24,25], which should be considered for detection and disease control [1,26]. Thus, we had to design primers based on conserved sequence stretches for the gene L region since RT-PCR with other primers [12] failed with some Spanish EMDV isolates.
Phylogenetic analyses of genomic regions N, Y1, L1 and L2 were congruent among themselves and with those from previous studies [11,12,21]. These phylogenetic trees show a certain geographic structure since the Australian isolate is in clade C, all Spanish isolates are in subclade A1 and most Italian isolates are in subclade B2. However, Greece contains EMDV isolates in clades A (subclade 2) and B (B1 and B2), and France contains isolates in subclade A2, which are more similar to distant isolates like SH-eg from Iran than isolates located in Spain, Germany and the UK. More isolates from different countries should be analyzed to gain insight into the migration routes of EMDV [27].
Phylogenetic analysis of the Y1 genomic region showed that isolate AZ15-31 from Azerbaijan [22] formed a different clade and was divergent from the other EMDV isolates, with a nucleotide identity of about 75.0%, which is in the same range as EMDV with some other species of the genus Alphanucleorhabdovirus. One of the three criteria to differentiate species within the genus Alphanucleorhabdovirus is that the nucleotide sequence identity of entire genomes must be less than 75.0% [5]. Thus, the entire genome of isolate AZ15-31 should be sequenced to assign it to EMDV or another species of the genus Alphanucleorhabdovirus such as Alphanucleorhabdovirus physostegiae (physostegia chlorotic mottle virus, PhCMoV) or to propose a new species in this genus.
Recombination seems to be uncommon for EMDV since only one recombination event was detected [11]. Homologous recombination is rare in negative-strand RNA viruses [28,29] in contrast to positive-strand RNA viruses [4,30,31]. Recombination events found in negative-strand RNA viruses might be artifacts from mixed infections or laboratory contamination [31]. So, the presence of recombinant variants should be confirmed by analyzing within-plant viral populations [32].
Some EMDV isolates collected in distant places and times showed similar nucleotide sequences. For example, EMDV isolates 1009/11 collected in Spain in 2011 and 02923HTS collected in the UK in 2022 showed 99.3% nucleotide identity. This suggests genetic stability by strong negative selection within a narrow adaptative peak [33] for the phylogenetic subclade A1. Genetic stability has been found in other plant RNA viruses [34,35]. A comparison of synonymous and non-synonymous substitution rates showed that most EMDV genes were subjected to strong negative selection due to functional restrictions. This is frequent for plant viruses [24,36,37] since proteins can have multiple domains and functions in the virus life cycle [38,39]. Only two codons were under diversifying positive selection, which could be result of adaptation to a new host, vector or environment [40]. Positive selection has been detected in other plant viruses, for example, in the substitution of one amino acid (codon) in the movement protein of tomato spotted wilt virus (TSWV), which is an adaptation to overcome resistance in tomato [41].
The low genetic diversity and the low genetic flow of EMDV isolates from Spain with respect to those from Greece and Italy suggest a unique introduction of EMDV to Spain. The EMDV population within Spain was genetically very homogeneous with high gene flow, except in Malaga province, which showed slight divergence and low gene flow with the rest of Spain. EMDV might have undergone genetic drift after a founder or bottleneck event or experienced adaptation to new conditions in Malaga. Further analysis of additional Malaga EMDV isolates from various hosts would be necessary to verify whether this slight divergence is present in other isolates.
In contrast, Greece and Italy had diverse EMDV populations with high gene flow between both. Greece showed the highest diversity with EMDV isolates in clades A and B, so the center of diversification and dispersion might be in Greece or nearby. Some plant viruses could have dispersed and diversified from Middle East [30], where farming originated. More EMDV sequences from these countries and from Middle East would be necessary to test this hypothesis.
4. Material and Methods
4.1. Virus Isolates
We collected 658 plants from different hosts and provinces of Spain from 2009 to 2020 and analyzed them by DAS-ELISA for EMDV (Loewe, Sauerlach, Germany). Only 13 plants were positive for EMDV (Table 1) and were used for RT-PCR, sequencing and sequence analyses.
4.2. Primer Design
Primers were designed with Oligo 7 software [42] from conserved stretches of the nine EMDV isolates with complete genome sequences available (GenBank accessions KC905081, OL472111, OQ847408, OQ716555, LN680656, KJ082087, MW854257, FR751552 and OL584369) to minimize false negatives [1]. Two primer pairs (N1F2/N2bR and N2F2/N1R2) comprising two overlapping regions encompassing gene N and primers PL1a/Pl2B covering a 1053 nt region of gene L were designed (Table 6). An 876 nt region of the PL1a/Pl2B PCR product was selected for nucleotide sequence analysis and was named L1.
Table 6.
Oligonucleotide primers designed for RT-PCR.
| Primer | Sequence | Position 1 | PCR Size 2 |
|---|---|---|---|
| EMDV-N1F2 | ATGACTATTTAAATAAAACCCAACAAC | 181–207 | 914 |
| EMDV-N2bR | AGTGGTGAGGAGCATCTTGTA′ | 1074–1094 | |
| EMDV-N2F2 | TTATTCAAGGCCTAGATACTTACACTG | 922–948 | 846 |
| EMDV-N1R2 | CACACACCACAAACATAGACTAGATAC | 1741–1767 | |
| EMDV-PL1a | ATGGGGGCATCCTATCATAGA- | 8165–8185 | 1053 |
| EMDV-PL2b | GCGACGTACTTTATATCACACACTGTCAT | 9189–9216 |
1 Nucleotide position with respect to the sequence retrieved from GenBank with the accession number LN680656. 2 PCR amplicon size (bp).
4.3. Nucleic Acid Purification, RT-PCR and Sequencing
Total RNAs were extracted from 100 mg of leaf tissues using the silica-capture protocol [43] and stored at −20 °C until use.
RT was performed by denaturing 3 µL of total RNA extracts by heating at 95 °C for 5 min, incubating them on ice for 1 min and adding them to a 10 µL reaction mixture containing 5 mM DTT, 1 mM of each dNTP, first-strand buffer, 0.4 µM of primer EMDV-N1F2 (Table 6), 10 units of Ribolock RNAse Inhibitor (Thermo Fisher Scientific, Waltham, MA, USA) and 40 units of SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA). The reaction mixture was incubated at 50 °C for 10 min and at 80 °C for 10 min to inactivate the reaction. Then, 1 µL of the RT product was added to a 20 µL reaction mixture containing CloneAmp HiFi PCR Premix (Takara, Kusatsu, Shiga, Japan) and 0.2 µM of primers. A Mastercycler epgradient thermal cycler (Eppendorf, Hamburg, Germany)was used with the following thermocycling conditions: denaturation at 98 °C for 1 min; 35 cycles of 98 °C for 10 s, 55 °C for 10 s and 72 °C for 8 s; and an extension step at 72 °C for 1 min. PCR products were analyzed by electrophoresis in 2% agarose gels and visualized with Gel Red (Biotium Inc., Fremont, CA, USA) under UV light. The PCR products were purified using the mi-PCR purification kit (Metabion, Planegg, Germany) and the DNA concentration was estimated with a NanoDrop™ 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Sanger sequencing of the purified RT-PCR products was carried out on an ABI 3730XL capillary sequencer (Applied Biosystems, Waltham, MA, USA) by Eurofins Genomic GATC service (Ebersberg, Germany).
4.4. In Silico Analyses of Nucleotide Sequences
Nucleotide sequences were aligned with the algorithm CLUSTALW [44] implemented in MEGA X software ver. 10.0.5 [45].
Nucleotide identities were estimated from the formula 100 × (1 − p-distance) with MEGA X. Nucleotide diversity was calculated as the mean distance between sequence pairs using the best fitting substitution model based on the lowest Bayesian information criterion (BIC) scores estimated with MEGA X. The T92 + G model was used since it was the first or second model with the lowest BIC for the distinct genomic regions analyzed and was available in further analyses.
Unrooted phylogenetic trees were inferred from the nucleotide sequences with the maximum likelihood method [46], implemented in MEGA X, under the T92 + G substitution model and five discrete gamma rates. The statistical significance of nodes was estimated with 500 bootstrap pseudoreplicates [47].
Possible recombination events between EMDV isolates were analyzed with the algorithms GENECONV, BOOTSCAN, MAXCHI, SISCAN, 3SEQ, LARD and RDP implemented in the RDP4 package [23]. Only recombination events detected by at least five different methods were considered.
Natural selection at the amino acid level was estimated with MEGA X by separately analyzing the rate of non-synonymous (dN) and synonymous (dS) substitutions with the Pamilo–Bianchi–Li method [48]. The rate between dN and dS provides information on the sign and intensity of selection. Selection across the genomic coding regions was studied by estimation of the rates of dN and dS at each codon using the fixed effects likelihood method [49] implemented in the adaptive evolution server Datamonkey 2.0 [50]. Coevolving amino acid sites within each EMDV protein were identified with Spidermonkey-BGM [51] implemented in Datamonkey.
The level of genetic differentiation between populations, determined by gene flow (migration), was estimated with the statistic Fst [52] implemented in the program DNASP v6.12 [53]. Fst values vary between zero, corresponding to genetically undifferentiated populations, and one, indicating genetically isolated populations. An absolute value of Fst > 0.333 suggests infrequent gene flow.
Acknowledgments
The authors thank the Plant Health laboratories of the Autonomous Communities of Spain and Granada La Palma S. Cooperativa for providing plant samples.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants13020250/s1. Supplementary Table S1. Nucleotide identity of the gene N between EMDV isolates from Spain and one isolate from Germany and the UK. Supplementary Table S2. Nucleotide identity of the genomic region L1 (position 1090–1965 of the gene L) between EMDV isolates from Spain and one isolate from Germany and the UK.
Author Contributions
Conceptualization and experimental design, L.R. and L.G.; collection and processing of samples, A.A.-F., E.S.-E. and I.F.-S.-A.; RT-PCR and sequencing, R.T.; nucleotide sequence analyses and primer design, L.R. and R.P.; supervision, L.R.; writing, L.R.; funding acquisition, L.R. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
The 26 nucleotide sequences generated in this work were deposited in GenBank under accession numbers OR631742–OR631767, 3 October 2023.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This research was funded by the Spanish Ministerio de Ciencia e Innovación, grant PID2021-125787OR-C31, co-financed by FEDER.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Rubio L., Galipienso L., Ferriol I. Detection of plant viruses and disease management: Relevance of genetic diversity and evolution. Front. Plant Sci. 2020;11:1092. doi: 10.3389/fpls.2020.01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roossinck M.J. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 1997;35:191–209. doi: 10.1146/annurev.phyto.35.1.191. [DOI] [PubMed] [Google Scholar]
- 3.Moya A., Holmes E.C., González-Candelas F. The population genetics and evolutionary epidemiology of RNA viruses. Nat. Rev. Microbiol. 2004;2:279–288. doi: 10.1038/nrmicro863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chare E.R., Holmes E.C. A phylogenetic survey of recombination frequency in plant RNA viruses. Arch. Virol. 2006;151:933–946. doi: 10.1007/s00705-005-0675-x. [DOI] [PubMed] [Google Scholar]
- 5.International Committee on Taxonomy of Viruses (ICTV) Genus: Alphanucleorhabdovirus. [(accessed on 16 September 2023)]. Available online: https://ictv.global/report/chapter/rhabdoviridae/rhabdoviridae/alphanucleorhabdovirus.
- 6.Pappi P.G., Dovas C.I., Efthimiou K.E., Maliogka V.I., Katis N.I. A novel strategy for the determination of a rhabdovirus genome and its application to sequencing of Eggplant mottled dwarf virus. Virus Genes. 2013;47:105–113. doi: 10.1007/s11262-013-0911-5. [DOI] [PubMed] [Google Scholar]
- 7.CABI . Crop Protection Compendium. CAB International; Wallingford, UK: 2021. [(accessed on 16 September 2023)]. Eggplant mottled dwarf virus. Available online: www.cabi.org/cpc. [Google Scholar]
- 8.Della Giustina W., Javoy M., Bansept P., Morel E., Balasse H., Goussard N., Passard C. Les cicadelles du genre Anaceratagallia vectrice du virus responsable de la maladie de la peau de crapaud du concombre. PHM Rev. Hortic. 2000;420:40–43. [Google Scholar]
- 9.Babaie G., Izadpanah K. Vector transmission of eggplant mottled dwarf virus in Iran. J. Phytopathol. 2003;151:679–682. doi: 10.1046/j.1439-0434.2003.00788.x. [DOI] [Google Scholar]
- 10.Desbiez C., Wipf-Scheibel C., Millot P., Berthier K., Girardot G., Gognalons P., Hirsch J., Moury B., Nozeran K., Piry S., et al. Distribution and evolution of the major viruses infecting cucurbitaceous and solanaceous crops in the French Mediterranean area. Virus Res. 2020;286:198042. doi: 10.1016/j.virusres.2020.198042. [DOI] [PubMed] [Google Scholar]
- 11.Pappi P.G., Maliogka V.I., Amoutzias G.D., Katis N.I. Genetic variation of eggplant mottled dwarf virus from annual and perennial plant hosts. Arch. Virol. 2016;161:631–639. doi: 10.1007/s00705-015-2705-7. [DOI] [PubMed] [Google Scholar]
- 12.Parrella G., Greco B. Sequence variation of block III segment identifies three distinct lineages within Eggplant mottled dwarf virus isolates from Italy, Spain and Greece. Acta Virol. 2016;60:100–105. doi: 10.4149/av_2016_01_100. [DOI] [PubMed] [Google Scholar]
- 13.Babaie G., Habibi M.K., Massah A., Dizadji A., Izadinejad L., Simon A. Complete genome sequence and genome analysis of Eggplant mottled dwarf virus-Iranian isolate. J. Phytopathol. 2014;163:331–341. doi: 10.1111/jph.12256. [DOI] [Google Scholar]
- 14.Rivarez M.P.S., Pecman A., Bačnik K., Carvalho Frreira O.M., Vučurović A., Seljak G., Mehle N., Gutierrrez-Aguirre I., Ravnikar M., Kutnjak D. In-depth study of tomato and weed viromes reveals undiscovered plant virus diversity in an agroecosystem. Microbiome. 2023;11:60. doi: 10.1186/s40168-023-01500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menzel W., Winter S., Hamacher J. First report of Eggplant mottled dwarf virus causing flower breaking and vein clearing in Hydrangea macrophylla in Germany. New Dis. Rep. 2016;34:11. doi: 10.5197/j.2044-0588.2016.034.011. [DOI] [Google Scholar]
- 16.Frew L., Hogan C., Andrews K., Fowkes A., Skelton A., Webster G., Dixon M., Conyers C., Adams I., McGreig S., et al. First report of eggplant mottled dwarf virus in Pittosporum tobria in the United Kingdom. J. Plant Pathol. 2023;105:1733. doi: 10.1007/s42161-023-01483-1. [DOI] [Google Scholar]
- 17.Elbeaino T., Kubaa R.A., Tuzlali H.T., Digiaro M. Pittosporum cryptic virus 1: Genome sequence completion using next-generation sequencing. Arch. Virol. 2016;161:2039–2042. doi: 10.1007/s00705-016-2860-5. [DOI] [PubMed] [Google Scholar]
- 18.Zhai Y., Miglino R., Sorrentino R., Masenga V., Alioto D., Pappu H.R. Complete genomic characterization of eggplant mottled dwarf virus from Agapanthus sp. by deep sequencing and de novo assembly. J. Plant Pathol. 2014;96:593–595. [Google Scholar]
- 19.Pappi P.G., Chaintoutis S.C., Dovas C.I., Efthimiou K.E., Katis N.I. Development of one-tube real-time qRT-PCR and evaluation of RNA extraction methods for the detection of eggplant mottled dwarf virus in different species. J. Virol. Methods. 2015;212:59–65. doi: 10.1016/j.jviromet.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Miglino R., Sorrentino R., De Stradis A., Zoina A., Alioto D. Necrotic potato tubers infected by eggplant mottled dwarf virus in Italy. J. Plant Pathol. 2013;95:619–621. [Google Scholar]
- 21.Tang J., Elliott C., Ward L.I., Iqram A. Identification of Eggplant mottled dwarf virus in PEQ Hibiscus syriacus plants imported from Australia. Australas. Plant Dis. Notes. 2014;10:6. doi: 10.1007/s13314-014-0153-y. [DOI] [Google Scholar]
- 22.Desbiez C., Verdin E., Moury B., Lecoq H., Millot P., Wipf-Scheibel C., Mirzayeva S., Sultanova N., Balakishiyeva G., Mammadov A., et al. Prevalence and molecular diversity of the main viruses infecting cucurbit and solanaceous crops in Azerbaijan. Eur. J. Plant Pathol. 2019;153:359–369. doi: 10.1007/s10658-018-1562-0. [DOI] [Google Scholar]
- 23.Martin D.P., Murrell B., Golden M., Khoosal A., Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcıa-Arenal F., Fraile A., Malpica J.M. Variability and genetic structure of plant virus populations. Annu. Rev. Phytopathol. 2001;39:157–186. doi: 10.1146/annurev.phyto.39.1.157. [DOI] [PubMed] [Google Scholar]
- 25.Davino S., Panno S., Iacono G., Sabatino L., D’Anna F., Iapichino G., Olmos A., Scuderi G., Rubio L., Tomassoli L., et al. Genetic variation and evolutionary analysis of Pepino mosaic virus in Sicily: Insights into the dispersion and epidemiology. Plant Pathol. 2017;66:368–375. doi: 10.1111/ppa.12582. [DOI] [Google Scholar]
- 26.Ferrer R.M., Luis-Arteaga M., Guerri J., Moreno P., Rubio L. Detection and identification of species of the genus Fabavirus by RT-PCR with a single pair of primers. J. Virol. Meth. 2007;144:156–160. doi: 10.1016/j.jviromet.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Davino S., Willemsen A., Panno S., Davino M., Catara A., Elena S.F., Rubio L. Emergence and Phylodynamics of Citrus tristeza virus in Sicily, Italy. PLoS ONE. 2013;8:e66700. doi: 10.1371/journal.pone.0066700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chare E.R., Gould E.A., Holmes E.C. Phylogenetic analysis reveals a low rate of homologous recombination in negative-sense RNA viruses. J. Gen. Virol. 2003;84:2691–2703. doi: 10.1099/vir.0.19277-0. [DOI] [PubMed] [Google Scholar]
- 29.Han G.Z., Worobey M. Homologous recombination in negative sense RNA viruses. Viruses. 2011;3:1358–1373. doi: 10.3390/v3081358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velasco L., Salem N., Willemsen A., Lapidot M., Mansour A.N., Rubio L., Galipienso L. Genetic variation and evolutionary forces shaping Cucumber vein yellowing virus populations: Risk of emergence of virulent isolates in Europe. Plant Pathol. 2016;65:847–856. doi: 10.1111/ppa.12465. [DOI] [Google Scholar]
- 31.Bentley K., Evans D.J. Mechanisms and consequences of positive-strand RNA virus recombination. J. Gen. Virol. 2018;99:1345–1356. doi: 10.1099/jgv.0.001142. [DOI] [PubMed] [Google Scholar]
- 32.Vives M.C., Rubio L., Sambade A., Mirkov T.E., Moreno P., Guerri J. Evidence of multiple recombination events between two RNA sequence variants within a Citrus tristeza virus isolate. Virology. 2005;331:232–237. doi: 10.1016/j.virol.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 33.Wright S. The roles of mutation, inbreeding, crossbreeding and selection in evolution. Proc. Sixth Intl. Cong. Genet. 1932;1:356–366. [Google Scholar]
- 34.Albiach-Martí M.R., Mawassi M., Gowda S., Satyanarayana T., Hilf M.E., Shanker S., Almira E.C., Vives M.C., Lopez C., Guerri J., et al. Sequences of Citrus tristeza virus separated in time and space are essentially identical. J. Virol. 2000;74:6856–6865. doi: 10.1128/JVI.74.15.6856-6865.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elvira-González L., Rubio L., Galipienso L. Geographically distant isolates of the persistent southern tomato virus (STV) show very low genetic diversity in the putative coat protein gene. Virus Genes. 2020;56:668–672. doi: 10.1007/s11262-020-01785-x. [DOI] [PubMed] [Google Scholar]
- 36.Ferrer R.M., Ferriol I., Moreno P., Guerri J., Rubio L. Genetic variation and evolutionary analysis of Broad bean wilt virus 2. Arch. Virol. 2011;156:1445–1450. doi: 10.1007/s00705-011-0990-3. [DOI] [PubMed] [Google Scholar]
- 37.Rubio L., Guerri J., Moreno P. Genetic variability and evolutionary dynamics of viruses of the family Closteroviridae. Front. Microbiol. 2013;4:151. doi: 10.3389/fmicb.2013.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Callaway A., Giesman-Cookmeyer D., Gillock E.T., Sit T.L., Lommel S.A. The multifunctional capsid proteins of plant RNA viruses. Annu. Rev. Phytopathol. 2001;39:419–460. doi: 10.1146/annurev.phyto.39.1.419. [DOI] [PubMed] [Google Scholar]
- 39.Plisson C., Drucker M., Blanc S., German-Retana S., Le Gall O., Thomas D., Bron P. Structural characterization of HC-Pro, a plant virus multifunctional protein. J. Biol. Chem. 2003;278:23753–23761. doi: 10.1074/jbc.M302512200. [DOI] [PubMed] [Google Scholar]
- 40.LaTourrette K., Garcia-Ruiz H. Determinants of Virus Variation, Evolution, and Host Adaptation. Pathogens. 2022;11:1039. doi: 10.3390/pathogens11091039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.López C., Aramburu J., Galipienso L., Soler S., Nuez F., Rubio L. Evolutionary analysis of tomato Sw-5 resistance-breaking isolates of Tomato spotted wilt virus. J. Gen. Virol. 2011;92:210–215. doi: 10.1099/vir.0.026708-0. [DOI] [PubMed] [Google Scholar]
- 42.Rychlik W. OLIGO 7 primer analysis software. Methods. Mol. Biol. 2007;402:35–60. doi: 10.1007/978-1-59745-528-2_2. [DOI] [PubMed] [Google Scholar]
- 43.Foissac X., Svanella-Dumas L., Gentit P., Dulucq M.J., Candresse T. Polyvalent detection of fruit tree Tricho, Capillo and Foveavirus by nested RT-PCR using degenerate and inosine containing primers (PDO RT-PCR) Acta Hortic. 2001;550:37–43. doi: 10.17660/ActaHortic.2001.550.2. [DOI] [Google Scholar]
- 44.Higgins D., Thompson J., Gibson T. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nei M., Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press; Oxford, UK: 2000. [Google Scholar]
- 47.Efron B., Halloran E., Holmes S. Bootstrap confidence levels for phylogenetic trees. Proc. Natl. Acad. Sci. USA. 1996;93:13429–13434. doi: 10.1073/pnas.93.23.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pamilo P., Bianchi N.O. Evolution of the Zfx and Zfy genes: Rates and interdependence between the genes. Mol. Biol. Evol. 1993;10:271–281. doi: 10.1093/oxfordjournals.molbev.a040003. [DOI] [PubMed] [Google Scholar]
- 49.Kosakovsky Pond S.L., Frost S.D.W. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 50.Weaver S., Shank S.D., Spielman S.J., Li M., Muse S.V., Kosakovsky Pond S.L. Datamonkey 2.0: A modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 2018;35:773–777. doi: 10.1093/molbev/msx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poon A.F.Y., Lewis F.I., Frost S.D.W., Kosakovsky Pond S.L. Spidermonkey: Rapid detection of co-evolving sites using Bayesian graphical models. Bioinformatics. 2008;24:1949–1950. doi: 10.1093/bioinformatics/btn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weir B.S., Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 53.Rozas J., Ferrer-Mata A., Sanchez-DelBarrio J.C., Guirao-Rico S., Librado P., Ramos-Onsins S.E., Sanchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 26 nucleotide sequences generated in this work were deposited in GenBank under accession numbers OR631742–OR631767, 3 October 2023.