Abstract
Bordetella pertussis fimbriae bind to sulfated sugars such as heparin through the major subunit Fim2. The Fim2 subunit contains two regions, designated H1 and H2, which show sequence similarity with heparin binding regions of fibronectin, and the role of these regions in heparin binding was investigated with maltose binding protein (MBP)-Fim2 fusion proteins. Deletion derivatives of MBP-Fim2 showed that both regions are important for binding to heparin. The role of H2 in heparin binding was confirmed by site-directed mutagenesis in which basic amino acids were replaced by alanine. These studies revealed that Lys-186 and Lys-187 are important for heparin binding of MBP-Fim2, whereas Arg-179 is not required. Peptides derived from H1 and H2 (pepH1 and pepH2) also showed heparin binding activity. Using a series of peptides, in each of which a different basic amino acid was substituted for alanine, we demonstrated that the structural requirements for heparin binding differ significantly among pepH1 and pepH2 peptides. A Pepscan analysis of Fim2 revealed regions outside H1 and H2 which bind heparin and showed that not only basic amino acids but also tyrosines may be important for binding to sulfated sugars. A comparison of the heparin binding regions of Fim2 with homologous regions of Fim3 and FimX, two closely related but antigenically distinct fimbrial subunits, showed that basic amino acids and tyrosines are generally conserved. The major heparin binding regions identified in Fim2 are part of epitopes recognized by human antibodies, suggesting that the heparin binding regions are exposed at the fimbrial surface and are immunodominant. Since B. pertussis fimbriae show weak serological cross-reactivity, the differences in primary structure in the heparin binding regions of Fim2, Fim3, and FimX may affect antibody binding but not heparin binding, allowing the bacteria to evade antibody-mediated immunity by switching the fimbrial gene expressed.
Sulfated glycosaminoglycans, such as heparan sulfate, are found in many tissues and cells of mammals (33). In view of their ubiquitous nature, it is perhaps not surprising that many pathogens, including viruses, bacteria, and parasites, colonize host surfaces by binding to sulfated glycosaminoglycans (5). Heparan sulfate and the structurally similar polysaccharide heparin have also been identified as receptors for filamentous hemagglutinin (17) and fimbriae (6), two adhesins of the respiratory pathogen Bordetella pertussis.
B. pertussis produces two serologically distinct fimbriae, designated serotype 2 and 3 fimbriae. Serotype 2 and 3 fimbriae are mainly composed of Fim2 and Fim3 subunits, with molecular weights of 22,500 and 22,000, respectively (1, 15, 18, 19). In addition to these major subunits, the fimbriae contain a single minor fimbrial subunit species (molecular weight, 40,000), designated FimD (35). Both the minor and the major subunits of B. pertussis fimbriae can mediate binding to heparin. Further, FimD also binds to the integrin VLA-5, an interaction which is believed to facilitate invasion of macrophages (11, 12). Studies in animal models (7, 21, 22, 34) have demonstrated the crucial role that B. pertussis fimbriae play in colonization of the respiratory tract.
Proteins binding to heparin contain local accumulations of positively charged amino acid residues bounded by negatively charged amino acid residues (4). Two such regions, designated regions H1 and H2, are found in the Fim2 subunit, and these regions show sequence similarity with heparin binding regions of fibronectin (6). In this work, the roles of regions H1 and H2 in heparin binding were investigated. Further, additional regions of Fim2 able to bind heparin were identified with synthetic peptides.
MATERIALS AND METHODS
Reagents.
Biotinylated heparin was purchased from Sigma Chemical Co. (St. Louis, Mo.); bovine serum albumin (BSA; Boseral PG) was obtained from Organon Teknika, Boxtel, The Netherlands.
Bacterial strains and culture conditions.
The strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown in NZ medium or agar. The conditions for growth of B. pertussis were as described previously (20). Antibiotics were used in the following concentrations: ampicillin, 100 μg/ml, and streptomycin, 300 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant phenotype and/or genotype | Source or reference |
|---|---|---|
| B. pertussis B52 | fim2::SacI fim3::kan Strr | 20, 21 |
| E. coli | ||
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 φ80ΔlacZM15 | Bio-Rad (Gaithersburg, Md.) |
| BL21 | ompT rB− mB−lon mutant | 31 |
| Plasmids | ||
| pRIP250 | pucBM21 with a NotI-ClaI fragment containing fim2 | F. R. Mooi |
| pALTER | Amps Tetr | Promega |
| pAF2 | Derived from pAlter; EcoRI-HindIII fragment containing fim2 | This study |
| pAF2-A179 | Derived from pAF2, containing a mutation at residue 179 of fim2 | This study |
| pAF2-A186 | Derived from pAF2, containing a mutation at residue 186 of fim2 | This study |
| pAF2-AA186-187 | Derived from pAF2, containing a mutation at residues 186 and 187 of fim2 | This study |
| pMAL-cRI | lacIPtac malEΔ2-26-fx lacZ Ampr | New England Biolabs |
| pMBP-Fim2 | pMA1-cRI containing an EcoRI-HindIII fragment with the fim2 gene | This study |
| pMBP-Fim2[179] | Derived from pMBP-Fim2, containing an Arg-Ala mutation at position 179 of fim2 | This study |
| pMBP-Fim2[186] | Derived from pMBP-Fim2, containing a Lys-Ala mutation at position 186 of fim2 | This study |
| pMBP-Fim2[186-187] | Derived from pMBP-Fim2, containing Lys-Ala mutations at positions 186 and 187 of fim2 | This study |
| pMBP-Fim2ΔH1 | Derived from pMBP-Fim2, containing fim2Δ78 | This study |
| pMBP-Fim2ΔH2 | Derived from pMBP-Fim2, containing fim2Δ67 | This study |
DNA techniques.
Unless otherwise stated, standard methods were used for DNA cloning, transformation, plasmid isolation, and agarose gel electrophoresis (28). Restriction enzymes were used according to the instructions provided by the manufacturer (Boehringer Mannheim). DNA sequencing was performed with Applied Biosystems dye terminator kits. Sequence reactions were run on an ABI 373A DNA sequencer (Applied Biosystems).
Construction of mutations in the fim2 gene.
Site-directed mutagenesis of the fim2 gene, performed with the Altered Sites II in vitro mutagenesis kit (Promega), was used to substitute Arg-179, Lys-186, and Lys-187 with alanine (Table 2). A 2.1-kbp EcoRI-HindIII fragment, derived from pRIP250 (Table 1), which contains a complete copy of the fim2 gene, was inserted into the EcoRI-HindIII site of pALTER, resulting in the plasmid pAF2. The following oligonucleotides, derived from the coding strand of fim2, were used to generate mutations in fim2 (the substituted bases are underscored): Arg-179 to Ala, 5′-CCGTCACGATGCGCTACCTGGCCTCC-3′; Lys-186 to Ala, 5′-GCCTCCTACGTCGCAAAGAACGGCGACGTC-3′; and Lys-186–Lys-187 to Ala-Ala, 5′-GCCTCCTACGTCGCAGCGAACGGCGACGTC-3′. After mutagenesis, the fim2 gene was sequenced to determine whether (only) the desired mutation was introduced. The resulting plasmids were designated pAF2-A179, pAF2-A186, and pAF2-AA186, respectively. Fusions between the malE gene encoding the maltose binding protein (MBP) and the wild-type and mutated fim2 genes, respectively, were constructed to enable the isolation of Fim2 subunits for binding studies. For the construction of the malE-fim2 fusions, the part of the fim2 gene coding for the mature fimbrial subunit was amplified with Pfu polymerase (Stratagene) by using the oligonucleotides CGGCGGAattCGCGCGGCACCATC and GAATCTGGATCcCTAGGGGTAGACCACG. To facilitate subsequent cloning, EcoRI and BamHI sites (underscored) were included. Lowercase letters indicate mismatches. Plasmids containing the wild-type (pRIP250) or mutated fim2 (pAF2 series) gene were used as the template for the PCR. After PCR, the 550-bp product was digested with EcoRI and BamHI and inserted into EcoRI-BamHI-digested pMAL-cRI. The resulting plasmids contained an in-frame gene fusion between the 5′ end of the MBP gene and the 3′ end of fim2 (Table 2). All constructs were verified by DNA sequencing.
TABLE 2.
Heparin binding to MBP-Fim2 fusion proteinsa
| Protein designation | Amino acid sequence | Amt of heparin binding
|
|
|---|---|---|---|
| OD450b | %c | ||
| 27 45 72 165 192 207 | |||
| MBP-Fim2 | isefDDGT/../EDPSGPNHTKVVQLPKISKNALKANGDQ/../PEVQTGGTSRTVTMRYLASYVKKNGDVE/../YP | 1.4 ± 0.03 | 100 |
| H1 H2 | |||
| MBP-Fim2ΔH1 | isefDDGT/../ED------------------------DQ/../PEVQTGGTSRTVTMRYLASYVKKNGDVE/../YP | 0.1 ± 0.1 | 7 |
| MBP-Fim2ΔH2 | isefDDGT/../EDPSGPNHTKVVQLPKISKNALKANGDQ/../PE-----------------------DVE/../YP | 0.8 ± 0.1 | 57 |
| MBP-Fim2[179] | isefDDGT/../EDPSGPNHTKVVQLPKISKNALKANGDQ/../PEVQTGGTSRTVTMAYLASYVKKNGDVE/../YP | 1.7 ± 0.1 | 121 |
| MBP-Fim2[186] | isefDDGT/../EDPSGPNHTKVVQLPKISKNALKANGDQ/../PEVQTGGTSRTVTMRYLASYVAKNGDVE/../YP | 0.9 ± 0.1 | 64 |
| MBP-Fim2[186-187] | isefDDGT/../EDPSGPNHTKVVQLPKISKNALKANGDQ/../PEVQTGGTSRTVTMRYLASYVAANGDVE/../YP | 0.2 ± 0.03 | 14 |
| MBP | isef-------------------------------------------------------------------------- | 0.2 ± 0.1 | 14 |
Protein designations are shown on the left and only the relevant parts of the amino acid sequences are shown. Omitted segments are indicated by “/../”. The H1 and H2 regions, which show sequence similarity with the heparin binding regions of fibronectin, are underlined. Dashes indicate deleted regions; lowercase letters indicate the MBP-derived amino acids. Substitutions are double underlined. Numbers above the MBP-Fim2 sequence refer to amino acid positions in Fim2.
Proteins were adsorbed on microtiter plates, and the ability of biotinylated heparin to bind to the adsorbed proteins was determined at a concentration of 10 μg of biotinylated heparin per ml as described in Materials and Methods. Shown are the mean ELISA readings at OD450 of eight independent experiments (± standard deviation). In all experiments, the differences in binding relative to MBP-Fim2 were significant (P ≤ 0.002).
Percentage of binding of heparin at 10 μg/ml was calculated as follows: [(OD450 MBP-mutated Fim2)/(OD450 MBP-Fim2)] × 100.
Plasmid pMBP-Fim2 was used to delete two regions of fim2, designated H1 and H2 (Table 2). Region H1, coding for amino acids 21 to 44 of the mature Fim2 subunit, was deleted by digesting pMBP-Fim2 with AocI and NaeI. Subsequently, the AocI site was filled in by using T4 polymerase, and the molecule was recircularized with T4 DNA ligase. The resultant plasmid was designated pMBP-Fim2ΔH1 (Table 2). Region H2, coding for amino acids 141 to 163 of the mature Fim2 subunit, was deleted by digesting pMBP-Fim2 with PinAI and AatII. After the single-stranded extensions were filled in, the DNA was recircularized with T4 DNA ligase. The resultant plasmid was designated pMBP-Fim2ΔH2 (Table 2). The fim2 genes in pMBP-Fim2ΔH1 and pMBP-Fim2ΔH2 were sequenced to determine that the required deletion was obtained.
Production and purification of MBP-Fim2 fusion proteins.
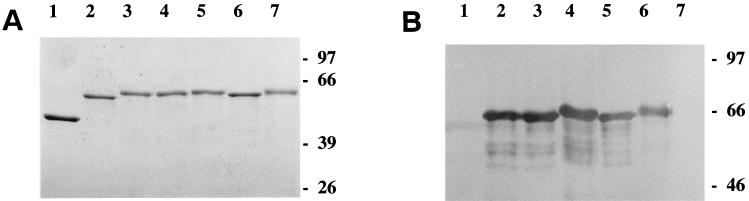
E. coli BL21 containing the appropriate plasmids was used for production of MBP and MBP-Fim2 fusion proteins. Induction of the fusion protein was performed at 37°C by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside to bacterial cultures grown to an optical density at 600 nm (OD600) of 1.0. After 3 h of induction, the cells were harvested by centrifugation at 13,000 × g for 20 min and resuspended in 20 mM NaCl–1 mM EDTA–20 mM Tris-HCl (pH 7.4). To break the cells, they were frozen (−20°C), thawed, and subsequently sonicated four times for 30 s (Branson sonifier; 50% output). NaCl was added to a final concentration of 200 mM, and the insoluble fraction was removed by centrifugation at 26,000 × g for 30 min. The fusion protein was purified from the soluble fraction by affinity chromatography on amylose columns. Fusion proteins were eluted with maltose, and aliquots of each fraction were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1 [14]). Protein-containing fractions were pooled and frozen at −20°C or used immediately in binding assays. Between 7 and 10 mg of each protein fraction was purified from 500 ml of cultured cells.
FIG. 1.
Analysis of MBP-Fim2 fusion proteins by SDS-PAGE (A) and immunoblotting (B). For SDS-PAGE and immunoblotting, respectively, 1 and 0.2 μg of protein was applied to each lane. The immunoblot was incubated with monoclonal antibody 136C6 (20) directed against the Fim2 subunit. Lanes: 1, MBP; 2, MBP-Fim2; 3, MBP-Fim2[179]; 4, MBP-Fim2[186]; 5, MBP-Fim2[186-187]; 6, MBP-Fim2ΔH2; 7, MBP-Fim2ΔH1. Numbers on the right indicate the molecular mass in kilodaltons of the protein standards.
Immunological techniques.
Separated proteins were electroblotted onto a nitrocellulose membrane (BA 83; Schleicher & Schuell, Dassel, Germany). The blots were blocked for 1 h with PTBE (0.5% fat-free milk powder, 0.1% BSA, 0.1% Tween 20 in phosphate-buffered saline [PBS]) and were subsequently incubated with a 1:100 dilution of polyclonal anti-Fim2 serum KO7096, a 1:100 dilution of monoclonal antibody 136C6 (25), or a 1:10,000 dilution of polyclonal anti-MBP serum (Biolabs) in PTBE for 1 h. The blots were washed with PBS containing 0.1% Tween 20 and subsequently incubated with a 1:500 dilution of alkaline phosphatase swine anti-rabbit or rabbit anti-mouse immunoglobulin G (heavy and light chain) conjugate (Dako A/S, Glostrup, Denmark) in PTBE buffer for 1 h. The blots were developed in a solution of nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate (Sigma) in dimethyl formamide.
The ability of polyclonal sera, directed against native or denatured fimbriae, to react with MBP-Fim2 was determined by enzyme-linked immunosorbent assay (ELISA). Microtiter plates (EIA/RIA; Costar) were coated overnight with MBP-Fim2, MBP, and fimbriae at concentrations of 5 μg/ml in PBS. The plates were washed with running tap water and blocked by incubation with PBSTB (PBS [pH 7.0], 1% BSA [Boseral PM; Organon Teknika], 0.05% Tween 20) for 1 h at room temperature. Reactivity with anti-fimbria sera was determined by incubation with serial dilutions of the polyclonal rabbit antibodies KO7094 and KO7096 raised against native fimbriae and fimbrial subunits, respectively. The plates were then washed and incubated with goat anti-rabbit conjugate. Finally, the wells were washed and incubated with a peroxidase substrate (0.4 mM 3,3,5,5′-tetramethylbenzidine [Sigma] and 0.009% H2O2 in 110 mM sodium acetate buffer [pH 5.5]) and, after stopping the reaction by adding 3 M H2SO4, the OD450 was determined.
Binding of biotinylated heparin to proteins and peptides.
Microtiter plates were coated with MBP-Fim2 fusion proteins or MBP overnight at room temperature at a concentration of 5 μg/ml in PBS. H1- and H2-derived synthetic peptides (see below) were applied at a concentration of 20 μg/ml for 2 h at 37°C. After being coated, the plates were washed four times with PBS containing 0.05% Tween 20 (PBST) in a 96-plate washer (SLT 96PW; Proton-Wilson) and then blocked by incubation with heat-treated PBSB (1% BSA in PBS) for 1 h at room temperature. Heat treatment of BSA consisted of incubation at 56°C for 60 min and filtering the solution through a 0.22-μm-pore-size filter (23). Plates were incubated overnight with biotinylated heparin (Sigma) at room temperature, washed four times with PBST, and incubated with a streptavidin-peroxidase conjugate (1:1,000 dilution; Amersham) for 2 h at 37°C. Finally, the plates were washed four times with PBST, and 100 μl of a peroxidase substrate (0.4 mM 3,3,5,5′-tetramethylbenzidine [Sigma] and 0.009% H2O2 in 110 mM sodium acetate buffer [pH 5.5]) was added to each well. After 5 min the reaction was stopped by adding 50 μl of 3 M H2SO4, and the OD450 was determined. The percentage of binding for proteins and peptides (Tables 2 and 3) was determined at a concentration of biotinylated heparin of 10 μg/ml as follows: [(OD450 of mutant derivatives of MBP-Fim2) (OD450 of MBP-Fim2)] × 100. A biotinylated heparin concentration of 10 μg/ml was found to be sufficient for saturation of binding.
TABLE 3.
Heparin binding to pepH1- and pepH2-derived peptidesa
| Peptide | Amino acid sequence | Net charge | OD450b | % Bindingc |
|---|---|---|---|---|
| 47 70 | ||||
| | ______ | | +5 | 1.21 ± 0.2 | 100 | |
| pepH1 | PSGPNHTKVVQLPKISKNALKANG | |||
| pepH1a | ....................A... | +4 | 0.20 ± 0.1 | 17 |
| pepH1b | ................A....... | +4 | 0.2 ± 0.04 | 17 |
| pepH1c | .............A.......... | +4 | 0.12 ± 0.02 | 10 |
| pepH1d | ...........A............ | +5 | 0.40 ± 0.05 | 33d |
| pepH1e | .......A................ | +4 | 0.12 ± 0.01 | 10 |
| pepH1f | .....A.................. | +4 | 0.12 ± 0.05 | 10 |
| pepH1g | .....A.A.....A..A...A... | 0 | 0.12 ± 0.03 | 10 |
| pepH1h | ....................ANK. | +5 | 1.33 ± 0.2 | 110 |
| 167 189 | ||||
| | ______ | | ||||
| pepH2 | VQTGGTSRTVTMRYLASYVKKNG | +4 | 1.82 ± 0.3 | 100 |
| pepH2a | ....................A.. | +3 | 1.69 ± 0.2 | 93 |
| pepH2b | ...................A... | +3 | 1.74 ± 0.05 | 96 |
| pepH2c | ..............A........ | +3 | 1.76 ± 0.2 | 97 |
| pepH2d | ............A.......... | +3 | 1.78 ± 0.2 | 98 |
| pepH2e | .......A............... | +3 | 1.76 ± 0.2 | 97 |
| pepH2f | ...................AA.. | +2 | 1.46 ± 0.2 | 80 |
| pepH2g | .......A....A......AA.. | 0 | 1.45 ± 0.2 | 80 |
| pepH2h | ...................NGKK | +4 | 1.92 ± 0.4 | 105 |
pepH1- and pepH2-derived peptides were adsorbed on microtiter plates, and the binding of biotinylated heparin was assayed as described in Materials and Methods. Dots indicate identical amino acids. Basic amino acids are indicated in boldface. Numbers refer to the positions of residues in the Fim2 subunit. Regions with a spacing of 6 or 12 residues between basic amino acids are indicated by lines above and below the sequence.
Peptides were adsorbed on microtiter plates, and the ability of biotinylated heparin to bind to the adsorbed peptides was determined at a concentration of 10 μg of biotinylated heparin per ml as described in Materials and Methods. Shown are the mean ELISA readings at OD450 of eight independent experiments (± standard deviation). In all experiments with pepH1 derivatives, with the exception of pepH1h, the differences in binding relative to pepH1 were significant (P < 0.0001). No significant differences in binding were found with the pepH2 derivatives.
Percent binding at 10 μg of heparin per ml was calculated relative to pepH1 or pepH2 as indicated in footnote c of Table 2.
The difference in binding of pepH1d relative to the other pepH1 derivatives was significant (P < 0.05).
Competition between fimbriae and MBP-Fim2 for binding of heparin.
Biotinylated heparin (660 μg/ml) was incubated for 1 h at 37°C with a serial dilution of fimbriae (0 to 100 μg/ml). Subsequently, the mixtures were added to microtiter plates coated with MBP-Fim2 (1 μg/ml) and allowed to incubate for 2 h. After being washed, the amount of biotinylated heparin that bound to MBP-Fim2 was determined as described above.
Peptides.
Peptides (Table 3) were assembled simultaneously on 10-μmol scale with an automated multiple peptide synthesizer equipped with a 48-column reaction block (AMS 422, ABIMED; Analysen-Technik GmbH, Langenfeld, Germany) and 9-fluorenylmethoxycarbonyl (Fmoc)-protected amino acids. Peptides were prepared as C-terminal amides, starting from 4-(2′,4′-dimethoxyphenyl-Fmoc-aminomethyl)phenoxy resin as described earlier (2, 32).
Pepscan.
Synthesis of peptides on polyethylene rods was carried out according to established Pepscan procedures (9, 10). A complete set of 18 peptides spanning the entire Fim2 protein was synthesized, each consisting of 12 consecutive amino acids that overlap with the previous and with the subsequent peptides by 2 amino acids. Peptide-loaded polyethylene rods were incubated with 80 μg of biotinylated heparin per ml, and heparin binding was detected by immunosorbent assays.
RESULTS
Role of regions H1 and H2 in heparin binding.
Two regions of the Fim2 subunit, designated H1 and H2, show sequence similarity with heparin binding domains of fibronectin (Table 2) (6). The role of these regions in heparin binding was investigated with fim2 derivatives in which the corresponding DNA had been deleted. Initially, the mutated fim2 genes were reintroduced into B. pertussis and attempts were made to isolate the mutated fimbriae. However, it appeared that B. pertussis strains carrying fim2 derivatives with even a single amino acid substitution (see below) produced very small numbers of fimbriae. Presumably, the mutations interfered with the assembly of the subunits. Therefore, the effect of the mutations in fim2 was analyzed with MBP-Fim2 fusion proteins. The two fusion proteins that lacked region H1 or H2 were designated MBP-Fim2ΔH1 and MBP-Fim2ΔH2, respectively (Table 2). The deletions in these hybrid proteins encompassed, respectively, 24 and 23 amino acid residues. After purification, the MBP-Fim2 hybrid proteins and their mutant derivatives were analyzed by SDS-PAGE and immunoblotting (Fig. 1). The fusion proteins revealed the expected molecular size and, with the exception of MBP-Fim2ΔH1, reacted with a Fim2 monoclonal antibody (136C6). The epitope of the monoclonal antibody used may be contained in the region deleted in MBP-Fim2ΔH1.
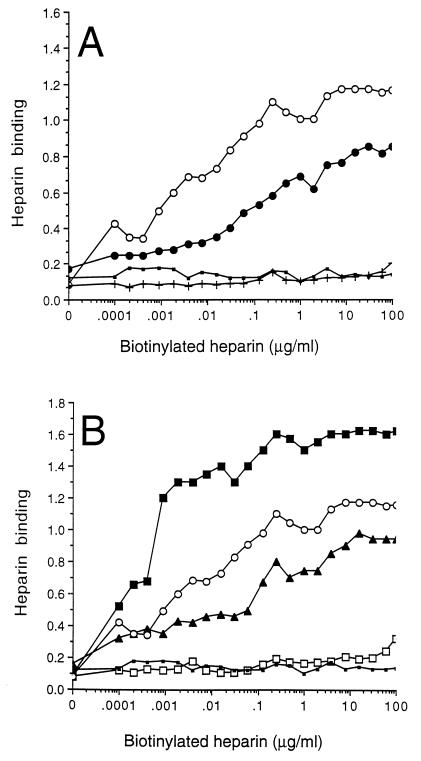
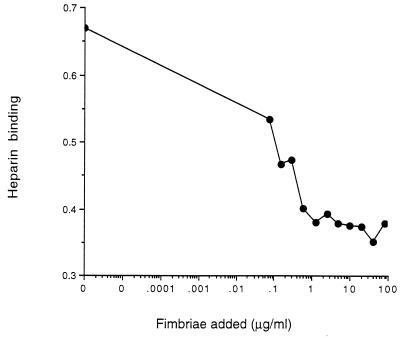
The MBP-Fim2 hybrid protein was immobilized on polystyrene microtiter plates to investigate its ability to bind heparin. Heparin bound to the MBP-Fim2 protein in a dose-dependent manner, and the saturation of binding was observed within the concentration range used (Fig. 2A). In contrast, a very low level of heparin binding was observed to MBP alone, independent of the heparin concentration used (Fig. 2A). Binding of heparin to MBP-Fim2 could be inhibited by fimbriae. At the highest fimbria concentration used (100 μg/ml), the percent inhibition was approximately 50% (Fig. 3). Microtiter plates with MBP-Fim2 (but not with MBP) bound antibodies raised against native fimbriae and fimbrial subunits (not shown). Together, these observations indicate that the MBP-Fim2 fusion protein has retained functional and structural features of the fimbria and could be used to characterize the fimbrial heparin binding site.
FIG. 2.
Binding of biotinylated heparin to MBP-Fim2 fusion proteins. Biotinylated heparin was incubated with immobilized MBP-Fim2 or mutant MBP-Fim2 derivatives harboring deletions (A) or substitutions (B). After being washed, adherent heparin was detected with a streptavidin-peroxidase conjugate. The assay was carried out at least four times, and a representative result is shown. (A) ○, MBP-Fim2; •, MBP-Fim2ΔH2; +, MBP-Fim2ΔH1; ▪, MBP. (B) ▪, MBP-Fim2[179]; □, MBP-Fim2[186-187]; ▴, MBP-Fim2[186]; ○, MBP-Fim2; ▪, MBP.
FIG. 3.
Inhibition of biotinylated heparin binding to immobilized MBP-Fim2 by fimbriae. Biotinylated heparin was preincubated with a serial dilution of fimbriae for 1 h at 37°C. The suspension was then added to immobilized MBP-Fim2, and the amount of bound heparin was detected with a streptavidin-peroxidase conjugate. The assay was carried out at least three times, and a representative result is shown.
The ability of mutant derivatives of MBP-Fim2 to bind to heparin was also investigated. To determine whether differences found in heparin binding activity to the MBP-Fim2 hybrids were due to mutations in fim2 per se and not to a difference in binding of the fusion proteins to the polystyrene plates, the efficiency of coating was determined with an anti-MBP serum. All MBP-derived proteins bound equally well to the microtiter wells (not shown). Both MBP-Fim2ΔH1 and MBP-Fim2ΔH2 showed a heparin binding activity lower than that of MBP-Fim2 (i.e., 7 and 57%, respectively [Fig. 2A and Table 2]). The amount of heparin bound by the negative control, MBP, was 14% compared to MBP-Fim2 (Fig. 2A and Table 2). Thus, although both regions H1 and H2 contributed to heparin binding, the presence of region H1 seems to be more essential for heparin binding.
Analysis with synthetic peptides of determinants of regions H1 and H2 that are important for heparin binding.
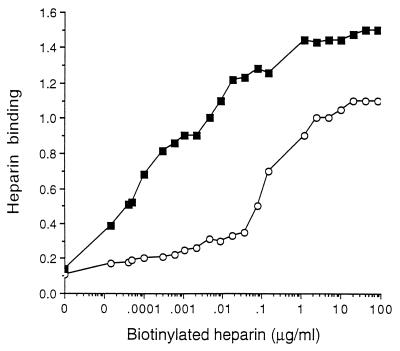
The ability of regions H1 and H2 to bind heparin was further investigated with synthetic peptides derived from Fim2 and immobilized on microtiter plates. Heparin was able to bind the peptides, pepH1 and pepH2, that encompass regions H1 and H2, respectively, in a dose-dependent manner, and saturation of binding was observed within the concentration range used (Fig. 4). pepH2 showed a greater heparin binding activity than pepH1. That pepH2 contains a lower net positive charge than pepH1 (+4 and +5, respectively) suggests that the binding of peptides to the negatively charged heparin is not determined solely by the density of basic amino acids. To study the role of basic amino acids in heparin binding, a series of peptides were synthesized, in each of which a different basic amino acid was substituted for the uncharged amino acid alanine (Table 3). Replacement of only one basic amino acid residue, regardless of its position, reduced heparin binding of pepH1 derivatives by 80 to 90% (Table 3). In contrast, successive replacement of basic amino acids by alanine had only a slight (2 to 7%) effect on heparin binding by pepH2, a finding that is not statistically significant. In fact, pepH2g, a peptide in which all the basic amino acid residues were replaced with alanine, showed only a 20% decrease in heparin binding compared to pepH2. The difference between the two peptides was not statistically significant. Taken together, these experiments showed that the requirements for heparin binding differs among pepH1 and pepH2.
FIG. 4.
Binding of biotinylated heparin to pepH1 (○) and pepH2 (▪). Peptides were immobilized on microtiter plates, and biotinylated heparin was added. The amount of bound heparin was detected with a streptavidin-peroxidase conjugate. The assay was carried out at least four times, and a representative result is shown.
The binding of peptides to heparin is not only dependent on the presence of basic amino acids. It has been shown that nonbasic amino acids, the spatial distribution of residues, and the conformation of the peptide also influence binding affinities (3, 4, 16). To determine whether nonbasic amino acids contributed to the heparin binding of pepH1 and pepH2, Leu-58 and Leu-181, respectively, were substituted with alanine, and the resulting peptides were studied for their heparin binding activity. The substitution of leucine for alanine in pepH1 (as in pepH1d) reduced binding by 67%. The substitution of leucine with alanine in pepH2 (as in pepH2c) did not result in a change in heparin binding activity (Table 3). The differences observed between pepH1d and pepH2c suggest that the conformation of pepH1 is more critical for binding than that of pepH2. This may explain the discrepancy between the relative roles of the H1 and H2 regions as assessed with fusion proteins and peptides. Data obtained with fusion proteins (Fig. 2) suggested that region H1 is more important for heparin binding than region H2, whereas the pepH1 appeared to bind heparin less well than did pepH2 (Fig. 4). Margalit et al. (16) provided evidence that in many regions implicated in heparin binding, two basic amino acids are located 6 or 12 residues apart, depending on whether the regions fold into an α-helix or a β-sheet. Two regions in pepH1 (Lys-54 to Lys-67 and Lys-60 to Lys-67) and pepH2 (Arg-174 to Lys-187 and Arg-179 to Lys-186) meet these criteria (Table 3), and the spacing between these residues was changed. The resulting peptides (pepH1h and pepH2h) bound slightly better (5 to 10%) to heparin than did pepH1 and pepH2 (Table 3). However, the observed differences were not statistically significant. Thus, for both pepH1 and pepH2, a spacing of 6 or 12 residues between basic amino acids does not seem to be crucial for binding to heparin.
Site-specific mutagenesis of region H2.
The role of the basic amino acids located in region H2 in heparin binding was also studied in the more natural context of the MBP-Fim2 fusion protein by replacing basic amino acids with alanine via site-directed mutagenesis. Alanine was chosen as the substitute residue since it is tolerated in both hydrophobic and hydrophilic regions and generally does not alter the conformation of the protein (13). All mutant derivatives of MBP-Fim2 were purified and characterized by SDS-PAGE and immunoblotting (Fig. 1). Substitution of Arg-179 with alanine resulted in a fusion protein (MBP-Fim2[179]) with a significantly higher binding activity (121%) than MBP-Fim2 (Fig. 2B and Table 2). In contrast, substitution of Lys-186 with alanine (as in MBP-Fim2[186]) decreased the heparin binding to 64% and, finally, substitution of Lys-186 and Lys-187 with alanine (as in MBP-Fim2[186-187]) resulted in a decrease in heparin binding activity to 14% (Fig. 2B and Table 2). This demonstrated that the two lysine residues at positions 186 and 187 contribute significantly to heparin binding by MBP-Fim2.
Pepscan analysis.
Our results so far indicated that the H1 and H2 regions of Fim2 are involved in heparin binding. To identify additional heparin binding regions, a Pepscan analysis of the complete Fim2 subunit was performed. Peptides comprising 12 amino acids with an overlap of two residues were synthesized on a solid-phase support. Subsequently, the ability of each peptide to bind biotinylated heparin was determined (Table 4). Of the three peptides (peptides 3, 4, and 5) spanning region H1, one (peptide 4) did not bind to heparin. This was not unexpected, since analyses of H1-derived peptides had shown that small changes abolished heparin binding (Table 3). Peptides 15 and 16 spanned region H2 and both bound heparin, with peptide 16 showing the highest degree of binding. A number of other peptides were also found to bind heparin, most notably peptide 9, which showed the second-highest binding activity of all the peptides analyzed in the Pepscan. Thus, peptide 9 may define a third heparin binding region of Fim2. It is interesting that the two peptides which revealed the highest binding activities, peptides 9 and 16, show structural similarity: they both contain arginine, lysine, and tyrosine residues, and the spacing of the tyrosines is identical in the two peptides. A role of tyrosine in heparin binding was also suggested by the observation that peptide 18, which contains two tyrosines but no basic amino acids, bound heparin. Other peptides which showed significant binding showed some structural similarity in that they contained two basic amino acids (i.e., peptides 3, 5, 6, and 12) and the six most reactive peptides, with the exception of peptide 6, all contain an arginine residue. A single negatively charged amino acid in a peptide did not abolish binding, as shown by peptides 5, 6, 9, and 12. Peptides that showed very little or no binding generally contained two or more negatively charged amino acids and did not contain arginine residues (i.e., peptides 1, 2, 8, 14, and 17).
TABLE 4.
Heparin binding activity of Fim2 analyzed with a Pepscana
| Peptideb (amino acid range) | Amino acid sequencec | OD450 | Rankingd |
|---|---|---|---|
| 1 (27–38) | ddgtivitgtit | 0.298 | 10 |
| 2 (37–48) | itdttcviedps | 0.172 | |
| 3 (47–58) | psgpnHtKvvql | 0.400 | 8 |
| 4 (57–68) | qlpKisKnalKa | 0.182 | |
| 5 (67–78) | KangdqagRtpf | 0.668 | 3 |
| 6 (77–88) | pfiiKlKdcpss | 0.566 | 5/6 |
| 7 (87–98) | sslgngYKaYfe | 0.229 | |
| 8 (97–108) | fepgpttdYstg | 0.207 | |
| 9 (107–118) | tgdlRaYKmvYa | 0.941 | 2 |
| 10 (117–128) | Yatnpqtqlsni | 0.187 | |
| 11 (127–138) | nitaateaqgvq | 0.309 | 9 |
| 12 (137–148) | vqYRisnlndsK | 0.641 | 4 |
| 13 (147–158) | sKitmganeatq | 0.250 | |
| 14 (157–168) | tqqaagfdpevq | 0.238 | |
| 15 (167–178) | vqtggtsRtvtm | 0.566 | 5/6 |
| 16 (177–188) | tmRYlasYvKKn | 0.950 | 1 |
| 17 (187–198) | Kngdveasaitt | 0.163 | |
| 18 (197–207) | ttYvgfsvvYp | 0.419 | 7 |
The ability of overlapping peptides derived from Fim2 to bind biotinylated heparin was assayed as described in Materials and Methods. Numbers in parentheses refer to the positions in the Fim2 subunit.
The position of the peptide within the Fim2 protein is indicated.
Residues which are part of the H1 or H2 region are underlined. Basic residues and tyrosines are in capital letters.
Ranking of peptides with significant binding. Binding was assumed to be significant when the OD450 was two times the background level (i.e., 2 × 0.18).
DISCUSSION
Previously, we identified two regions in Fim2, designated H1 and H2, which show similarity with heparin binding sites of fibronectin. Interestingly, the H1 and H2 regions also display mutual structural similarities (6). MBP-Fim2 fusion proteins in which these regions were deleted confirmed that both regions are involved in binding to heparin. In the fusion protein, the contribution of region H1 seemed to be crucial, since deletion of this region abolished binding to heparin. In contrast, a fusion protein in which region H2 was deleted was still able to bind heparin, albeit less efficiently than did MBP-Fim2, which contained the intact fimbrial subunit. We observed that the MBP-Fim2 protein was only able to bind heparin when it was adsorbed onto a polystyrene surface. No binding was observed when MBP-Fim2 was present in a solution, suggesting that the binding forces between MBP-Fim2 and heparin are weak and that stable binding requires multivalent interactions. Such multivalent interactions occur under natural conditions, since the surface of the bacterium is covered by hundreds of fimbriae, which are composed of hundreds of Fim2 subunits.
The structural requirements of regions H1 and H2 for heparin binding were studied by using synthetic peptides in which basic amino acids were substituted by alanine residues. The peptide derived from region H1 (pepH1) contains five basic amino acids, and all substitutions reduced binding to heparin by 80 to 90%. This suggested that all five basic amino acids are required for heparin binding and/or that the heparin binding ability of pepH1 is sensitive to changes in secondary structure. The latter possibility is consistent with the observation that replacement of a leucine by an alanine reduced the heparin binding activity of pepH1 (by 67%) but not that of pepH2. In contrast, very little effect was observed when the four basic amino acids in pepH2, which was derived from region H2, were replaced by alanine residues. In fact, peptide pepH2h, in which all the basic amino acids were substituted by alanine residues, retained maximum binding activity. This indicated that not only basic amino acids confer the ability to bind to heparin. It has been shown that peptides which bind to heparin are enriched in basic amino acids and tyrosines (3). PepH2 contains two tyrosines, and it seems likely that they are involved in binding of heparin. A role of tyrosines in heparin binding was also suggested by our Pepscan analysis of Fim2. This analysis revealed several regions outside H1 and H2 that were able to bind heparin, and most of these regions contained one or more tyrosines. In fact, peptide 18, which bound heparin (Table 4), contains two tyrosines but no basic amino acids.
Several groups have attempted to define the structural requirements of regions in proteins and peptides that bind heparin. Using sequence alignment, Cardin and Weintraub (4) predicted the presence of two consensus sequence motifs, XBBXBX or XBBBXXBX (where B represents a basic residue and X represents a nonbasic residue), with the capacity to interact with heparin. A larger consensus sequence of 13 residues, i.e., XBBXXBBBXXBBX, was suggested by Sobel et al. (30). However, as with many other heparin binding regions, the sequences of H1 and H2 do not match these three consensus sequences.
Margalit et al. (16) provided evidence that many heparin binding regions contain two basic amino acids which are located about 20 Å apart. Depending on whether the region folds into an α-helix or a β-sheet, this results in a spacing of 12 and 6 residues, respectively, between the basic amino acids. Two regions in pepH1 and pepH2 meet these criteria (Table 3); however, changing the spacing between the basic amino acids defining these regions in pepH1 and pepH2 did not decrease heparin binding. Thus, at least when peptides are used, a spacing of 6 or 12 residues between basic amino acids does not seem to be crucial for heparin binding to regions H1 and H2. Perhaps another type of spatial organization determines the heparin binding capacity of the H1 and H2 regions. The spatial organization and charge distribution of a protein can be represented by helical wheel diagrams (29). The helical wheel diagram of the heparin binding region of apolipoprotein B-100 segregates the basic residues primarily to one side of the helical wheel, forming a region of high-positive-charge density (4). A helical wheel projection of the Fim2 region H1 shows a similar asymmetric distribution of basic residues, with four of five of the basic residues being confined to one side of the helix (not shown). It should be noted, however, that region H1 is predicted to have a β-sheet conformation. A helical wheel representation of region H2 did not show a pronounced segregation of basic residues to one side of the helix.
The role of basic amino acids in region H2 was also studied with MBP-Fim2 proteins in which basic residues were substituted by alanine by site-specific mutagenesis. This approach showed that Lys-186 and Lys-187 were important for binding (Table 2 and Fig. 2B). Substitution of Lys-186 by alanine (as in MBP-Fim2[186]) decreased binding to heparin by 36%, whereas substitution of Lys-186 and Lys-187 by alanine (as in MBP-Fim2[186-187]) completely abolished binding to heparin. In contrast, replacement of Arg-179 by alanine (as in MBP-Fim2[179]) increased the binding activity of the MBP-Fim2 fusion protein by 21%. The finding that MBP-Fim2[186-187] did not bind to heparin was unexpected, since an MBP-Fim2 fusion protein (i.e., MBP-Fim2ΔH2), in which the region containing the two lysine residues (i.e., region H2) had been deleted, still bound heparin. Possibly, the deletion of H2 exposed novel heparin binding sites in Fim2. The results with MBP-Fim2[186-187] also do not concur with results obtained with peptides derived from region H2, which showed that removal of the two lysine residues did not significantly affect heparin binding. These discrepancies may be attributed to the different conformations of the region investigated in the fusion protein and peptides, respectively. Also, because of the greater flexibility of peptides, their binding to heparin may be less affected by substitutions.
In addition to Fim2, B. pertussis produces the closely related subunit Fim3 (19), which can also bind to heparin (8). Further, B. pertussis contains a silent gene for a third subunit, designated fimX (24). The three genes were probably derived from an ancestral gene by duplication (18), and it seems likely that they are functionally equivalent. The three Fim subunits have diverged substantially, presumably under pressure of the host immune response. We compared the heparin binding region of Fim2 with homologous regions in Fim3 and FimX to identify conserved features important for heparin binding (Fig. 5). In general, the comparison reveals that basic residues and tyrosines tend to be conserved and that arginine, lysine, and histidine residues are interchangeable. In the H1 region, the spacing between the basic residues differs among the Fim sequences, thereby confirming studies with pepH1 which showed that a strict spacing between basic residues is not important in the heparin binding of region H1.
FIG. 5.
Comparison of the heparin binding sites of Fim2 with homologous regions in Fim3 and FimX. Dots indicate residues conserved in all three sequences. Dashes indicate gaps introduced to increase the number of matches. Regions in Fim2 implicated in heparin binding regions are underlined, and amino acids (arginine, lysine, histidine, and tyrosine) presumed to be important for heparin binding are in boldface. Fim2 and Fim3 epitopes recognized by human sera (37) are boxed.
Recently, Williamson and Matthews (37) mapped epitopes present in the Fim2 and Fim3 subunits that reacted with sera from patients infected with, or vaccinated against, whooping cough. Remarkably, the four major heparin binding regions of Fim2 identified in this work are part of the identified epitopes (Fig. 5). It seems reasonable to assume that fimbrial antibodies induced after infection are mainly directed against surface- exposed epitopes. Thus, these observations suggest that the heparin binding regions are exposed at the fimbrial surface and are immunodominant. Antibodies raised against purified serotype 2 and 3 fimbriae show weak cross-reactivity (26, 27), and the observed variation in the heparin binding regions might therefore affect antibody binding but not heparin binding. B. pertussis is able to switch the expression of the fim2 and fim3 genes on and off at random by small mutations in the promoter region (36). This fimbrial phase variation allows the bacteria to evade antibodies which interfere with binding of fimbriae to sulfated glycosaminoglycans.
ACKNOWLEDGMENTS
This work was supported by Dutch Organization for Scientific Research (NWO) grant number 901-14-135.
We thank Jan Poolman and Betsy Kuipers for providing monoclonal antibodies.
REFERENCES
- 1.Ashworth L A, Dowsett A B, Irons L I, Robinson A. The location of surface antigens of Bordetella pertussis by immuno-electron microscopy. Dev Biol Stand. 1985;61:143–151. [PubMed] [Google Scholar]
- 2.Brugghe H F, Timmermans H A, Van Unen L M, Ten Hove G J, Van de Werken G, Poolman J T, Hoogerhout P. Simultaneous multiple synthesis and selective conjugation of cyclized peptides derived from a surface loop of a meningococcal class 1 outer membrane protein. Int J Pept Protein Res. 1994;43:166–172. doi: 10.1111/j.1399-3011.1994.tb00518.x. [DOI] [PubMed] [Google Scholar]
- 3.Caldwell E E, Nadkarni V D, Fromm J R, Linhardt R J, Weiler J M. Importance of specific amino acids in protein binding sites for heparin and heparan sulfate. Int J Biochem Cell Biol. 1996;28:203–216. doi: 10.1016/1357-2725(95)00123-9. [DOI] [PubMed] [Google Scholar]
- 4.Cardin A D, Weintraub H J R. Molecular modelling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 5.Fuller A O. Microbes and the proteoglycan connection. J Clin Invest. 1994;93:460. doi: 10.1172/JCI116992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geuijen C A W, Willems R J L, Mooi F R. The major fimbrial subunit of Bordetella pertussis binds to sulfated sugars. Infect Immun. 1996;64:2657–2665. doi: 10.1128/iai.64.7.2657-2665.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geuijen, C. A. W., R. J. L. Willems, M. Bongaerts, J. Top, H. Gielen, and F. R. Mooi. The role of the Bordetella pertussis minor fimbrial subunit, FimD, in colonization of the mouse respiratory tract. Infect. Immun. 65:4222–4228. [DOI] [PMC free article] [PubMed]
- 8.Geuijen, C. A. W., and F. R. Mooi. Unpublished data.
- 9.Geysen H M, Meloen R H, Barteling S J. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc Natl Acad Sci USA. 1984;81:3998–4002. doi: 10.1073/pnas.81.13.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geysen H M, Meloen R H, Barteling S J. Small peptides induce antibodies with a sequence and structural requirement for binding antigen comparable to antibodies raised against the native protein. Proc Natl Acad Sci USA. 1985;82:178–182. doi: 10.1073/pnas.82.1.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazenbos W L W, van den Berg B M, Geuijen C A W, Mooi F R, van Furth R. Binding of FimD on Bordetella pertussis to very late antigen-5 on monocytes activates complement receptor type 3 via protein tyrosine kinase. J Immunol. 1995;155:3972–3978. [PubMed] [Google Scholar]
- 12.Hazenbos W L W, Geuijen C A W, van den Berg B M, Mooi F R, van Furth R. Binding of Bordetella pertussis to human monocytes: role of FimD. J Infect Dis. 1995;171:924–929. doi: 10.1093/infdis/171.4.924. [DOI] [PubMed] [Google Scholar]
- 13.Klann A G, Hull R A, Palzkill T, Hull S I. Alanine-scanning mutagenesis reveals residues involved in binding of pap-3 encoded pili. J Bacteriol. 1994;176:2312–2317. doi: 10.1128/jb.176.8.2312-2317.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Livey I, Duggleby C J, Robinson A. Cloning and nucleotide sequence analysis of the serotype 2 fimbrial subunit gene of Bordetella pertussis. Mol Microbiol. 1987;1:203–209. doi: 10.1111/j.1365-2958.1987.tb00513.x. [DOI] [PubMed] [Google Scholar]
- 16.Margalit H, Fischer N, Ben-Sasson S A. Comparative analysis of structural defined heparin binding sequences reveals a distinct spatial distribution of basic residues. J Biol Chem. 1993;268:19228–19231. [PubMed] [Google Scholar]
- 17.Menozzi F D, Gantiez C, Locht C. Interaction of Bordetella pertussis filamentous hemagglutinin with heparin. FEMS Microbiol Lett. 1991;78:59–64. doi: 10.1111/j.1574-6968.1991.tb04417.x. [DOI] [PubMed] [Google Scholar]
- 18.Mooi F R. Bordetella pertussis fimbriae. In: Klemm P, editor. Fimbriae: adhesion, genetics, biogenesis, and vaccines. Boca Raton, Fla: CRC Press, Inc.; 1994. [Google Scholar]
- 19.Mooi F R, ter Avest A, van der Heide H G J. Structure of the Bordetella pertussis gene coding for the serotype 3 fimbrial subunit. FEMS Microbiol Lett. 1990;66:327–332. doi: 10.1016/0378-1097(90)90307-c. [DOI] [PubMed] [Google Scholar]
- 20.Mooi F R, van der Heide H G, ter Avest A R, Welinder K G, Livey I, van der Zeijst B A M, Gaastra W. Characterization of fimbrial subunits from Bordetella species. Microb Pathog. 1987;2:473–484. doi: 10.1016/0882-4010(87)90054-4. [DOI] [PubMed] [Google Scholar]
- 21.Mooi F R, van der Heide H G J, Walvoort H C, Brunings H, Jansen W H, Guinee P A M. The analysis of Bordetella pertussis fimbrial mutants in a rabbit model. In: Ron E Z, Rottem S, editors. Microbial surface components and toxins in relation to pathogenesis. New York, N.Y: Plenum Publishing Corp.; 1991. pp. 69–76. [Google Scholar]
- 22.Mooi F R, Jansen W H, Brunings H, Gielen H, van der Heide H G J, Walvoort H C, Guinee P A M. Construction and analysis of Bordetella pertussis mutants defective in the production of fimbriae. Microb Pathog. 1992;12:127–135. doi: 10.1016/0882-4010(92)90115-5. [DOI] [PubMed] [Google Scholar]
- 23.Ortegia-Barria E, Pereira M E. A novel T. cruzi heparin-binding protein promotes fibroblast adhesion and penetration of engineered bacteria and trypanosomes into mammalian cells. Cell. 1991;67:411–421. doi: 10.1016/0092-8674(91)90192-2. [DOI] [PubMed] [Google Scholar]
- 24.Pedroni P, Riboli B, de Ferra F, Grandi G, Toma S, Aricò B, Rappuoli R. Cloning of a novel pilin-like gene from Bordetella pertussis: homology to the fim2 gene. Mol Microbiol. 1988;2:539–543. doi: 10.1111/j.1365-2958.1988.tb00061.x. [DOI] [PubMed] [Google Scholar]
- 25.Poolman J T, Kuipers B, Vogel M L, Hamstra H J, Nagel J. Description of a hybridoma bank towards Bordetella pertussis toxin and surface antigens. Microb Pathog. 1990;8:377–382. doi: 10.1016/0882-4010(90)90024-k. [DOI] [PubMed] [Google Scholar]
- 26.Preston N W. Essential immunogens in human pertussis: the role of fimbriae. Dev Biol Stand. 1985;61:137–141. [PubMed] [Google Scholar]
- 27.Robinson A, Gorringe A R, Funnel S G P, Fernandez M. Serospecific protection of mice against intranasal infection with Bordetella pertussis. Vaccine. 1989;7:321–324. doi: 10.1016/0264-410x(89)90193-x. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schiffer M, Edmunson A B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967;7:121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobel M, Soler D F, Kermode J C, Harris R B. Localization and characterization of a heparin binding domain peptide of human von Willebrand factor. J Biol Chem. 1992;267:8857–8862. [PubMed] [Google Scholar]
- 31.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high level of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 32.Wiertz E J H J, van Gaans-vdBrink J A M, Gausepohl H, Prochnicka-Chalufour A, Hoogerhout P, Poolman J T. Identification of T cell epitopes occurring in a meningococcal class 1 outer membrane protein using overlapping peptides assembled with simultaneous multiple peptide synthesis. J Exp Med. 1992;176:79–88. doi: 10.1084/jem.176.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wight T N, Heingard D, Hascall V C. Proteoglycan structure and function. In: Hay E G, editor. Cell biology of extracellular matrix. 2nd ed. New York, N.Y: Plenum Publishing Corp.; 1991. pp. 45–78. [Google Scholar]
- 34.Willems R J L. Genetic and functional studies on Bordetella pertussis fimbriae. Ph.D. dissertation. Utrecht, The Netherlands: Utrecht University; 1993. [Google Scholar]
- 35.Willems R J L, Geuijen C, van der Heide H G J, Matheson M, Robinson A, Versluis L F, Ebberink R, Theelen J, Mooi F R. Isolation of a putative fimbrial adhesin from Bordetella pertussis and the identification of its gene. Mol Microbiol. 1993;9:623–634. doi: 10.1111/j.1365-2958.1993.tb01722.x. [DOI] [PubMed] [Google Scholar]
- 36.Willems R J L, Paul A, van der Heide H G J, ter Avest A, Mooi F R. Fimbrial phase variation in Bordetella pertussis: a novel mechanism for transcriptional regulation. EMBO J. 1990;9:2803–2809. doi: 10.1002/j.1460-2075.1990.tb07468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williamson P, Matthews R. Epitope mapping of the Fim2 and Fim3 proteins of Bordetella pertussis with sera from patients infected with or vaccinated against whooping cough. FEMS Immunol Microbiol. 1996;13:169–178. doi: 10.1016/0928-8244(95)00123-9. [DOI] [PubMed] [Google Scholar]