Abstract
This study aimed to investigate the genomic and epidemiological features of a methicillin-resistant Staphylococcus aureus sequence type 1 (MRSA ST1) strain associated with caprine subclinical mastitis. An S. aureus strain was isolated from goat’s milk with subclinical mastitis in Paraiba, Northeastern Brazil, by means of aseptic procedures and tested for antimicrobial susceptibility using the disk-diffusion method. Whole genome sequencing was performed using the Illumina MiSeq platform. After genome assembly and annotation, in silico analyses, including multilocus sequence typing (MLST), antimicrobial resistance and stress-response genes, virulence factors, and plasmids detection were performed. A comparative SNP-based phylogenetic analysis was performed using publicly available MRSA genomes. The strain showed phenotypic resistance to cefoxitin, penicillin, and tetracycline and was identified as sequence type 1 (ST1) and spa type 128 (t128). It harbored the SCCmec type IVa (2B), as well as the lukF-PV and lukS-PV genes. The strain was phylogenetically related to six community-acquired MRSA isolates (CA-MRSA) strains associated with human clinical disease in North America, Europe, and Australia. This is the first report of a CA-MRSA strain associated with milk in the Americas. The structural and epidemiologic features reported in the MRSA ST1 carrying a mecA-SCCmec type IVa suggest highly complex mechanisms of horizontal gene transfer in MRSA. The SNP-based phylogenetic analysis suggests a zooanthroponotic transmission, i.e., a strain of human origin.
Keywords: antimicrobial resistance genes, goat milk, mecA, MRSA, plasmids, virulence genes, whole genome sequencing
1. Introduction
Although methicillin-resistant Staphylococcus aureus (MRSA) can be commonly found in the skin and nostrils of healthy individuals [1], an increasing number of infections associated with community-acquired and livestock-acquired MRSA strains have been reported in recent decades worldwide [2,3,4]. According to the World Health Organization [5], MRSA ranks as a high-priority organism for studies involving novel treatment options.
In dairy animals, MRSA-associated intra-mammary infections have been associated with frequent antimicrobial therapeutic failures, resulting in significant economic losses due to reduced production and compromised milk quality [6,7,8]. The tolerance to increased antimicrobial levels, especially β-lactams drugs, is related to the versatility of these bacteria in acquiring antimicrobial resistance and virulence genes by horizontal gene transfer mechanisms [2]. Therefore, mobile genetic elements (MGEs), such as genomic islands, bacteriophages, pathogenicity islands, insertion sequences, transposons, chromosomal cassettes, and plasmids, play a key role in the dissemination of resistance and virulence genes, contributing to the survival and plasticity of staphylococci [2,9,10]. Although antimicrobial resistance, virulence, and toxin-encoding genes contributing to the adaptation and rapid dissemination of staphylococci in different environments are commonly carried in plasmids [11,12], there is a lack of studies addressing this MGE in staphylococci.
Considering the global relevance of MRSA as a human pathogen and the emerging importance of animals and animal-derived products as sources of MRSA to humans, we reported herein an in-depth genomic investigation of a community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) strain associated with subclinical caprine mastitis in Northeastern Brazil.
2. Materials and Methods
The S. aureus isolate, originally identified as SA31, was cultured from an udder half-milk sample aseptically collected from a lactating goat in Northeastern Brazil, according to the protocol of the National Mastitis Council [13]. We used a blood agar base (Oxoid, ThermoFisher, Waltham, MA, USA) supplemented with 5% defibrinated sheep blood for conventional microbiological isolation. The isolated was identified as S. aureus by means of Gram staining and biochemical testing, including catalase, oxidase, and tube coagulase. After confirmation by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-TOF MS) (Bruker Daltonics, Bremen, Germany), the isolate was transferred into tryptic soy broth (TSB, BD), supplemented with 3.5 mg/L of cefoxitin, and incubated at 37 °C for 18 h. Then, 100 µL aliquot of inoculum was streaked onto mannitol salt agar (MSA, BD), also supplemented with cefoxitin (3.5 mg/L), and incubated at 37 °C for 18 h [14]. Antimicrobial susceptibility was determined using the disk-diffusion method according to the guidelines of the Clinical and Laboratory Standards Institute [15] using the following antibiotics and their respective concentrations: cefoxitin (FOX, 30 µg), ciprofloxacin (CIP, 5 µg), clindamycin (CLI, 2 µg), chloramphenicol (CHL-30 µg), erythromycin (ERY, 15 µg), gentamicin (GEN, 10 µg), oxacillin (OXA, 1 µg), penicillin (PEN, 10U), trimethoprim/sulfamethoxazole (SXT, 23.75/1.25 µg), and tetracycline (TET, 30 µg). S. aureus US400 reference strain was used as positive control. For storage, five copies of the isolate were kept at −70 °C in broth heart infusion (BHI, BD) supplemented with 40% glycerol.
Total DNA was extracted using a commercial extraction kit (Power Soil, Qiagen, Hilden, Germany) following the manufacturer’s protocol. After DNA integrity analysis in agarose gel and fluorometric quantification (Qubit 2.0, Life Technologies, Carlsbad, CA, USA), genomic libraries were prepared using the Nextera XT DNA Library Preparation kit (Illumina, San Diego, CA, USA). DNA fragment sizes were evaluated using a capillary electrophoresis system (Fragment Analyzer, Agilent, Waldbronn, Germany). Paired-end sequencing was performed in Illumina MiSeq using a v3 kit (2 × 150 cycles). The quality of the reads was checked using FastQC version 0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ accessed on 2 September 2023). The genome was de novo assembled using Unicycler [16]. Genomic-based predictions and functional annotations were performed using the PATRIC server [17].
The prediction of SA31 as a human pathogenic strain was performed by PathogenFinder 1.1 [18]. MLST 2.0 [19] was used for in silico multilocus sequence typing (MLST), and spaTyper 1.0 [20] for spa-typing was performed in silico using the Center for Genomic Epidemiology (CGE) server (https://cge.cbs.dtu.dk/services/ accessed on 3 September 2023). Virulence factors were screened using the Virulence Factors of Pathogenic Bacteria (VFDB) platform [21]. Antimicrobial resistance determinants were investigated using ResFinder 4.1 [22]. SCCmecFinder 1.2 [23] was used to identify the staphylococcal chromosomal cassette. Plasmids were detected by Plasmidfinder 2.1 [24] and manually curated in Geneious (v.9.0.5).
A comparative SNP-based phylogenetic analysis was performed using publicly available MRSA genomes using Isolates Browser (https://www.ncbi.nlm.nih.gov/pathogens/isolates accessed on 5 September 2023). AMRFinderPlus [25] was used for the detection of antimicrobial resistance determinants, stress response, and virulence genes.
3. Results and Discussion
Staphylococcus aureus SA31 was phenotypically resistant to β-lactams (FOX, OXA, PEN) and tetracycline (TET), as shown in Table 1 and Table S1 (Supplementary Materials).
Table 1.
Genomic and phenotypic features of a community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) ST1 strain associated with subclinical caprine mastitis in Northeastern Brazil.
| Features | S. aureus Strain 31 |
|---|---|
| Phenotypic antibiotic resistance profile | FOX, OXA, PEN, TET |
| Structural Genomic data | |
| Genome size (bp) | 2,792,445 |
| Contigs number | 9 |
| CDSs | 2614 |
| rRNA | 4 |
| tRNA | 57 |
| % GC | 32.69 |
| Epidemiological Genomic Data | |
| MLST (ST) 1 | ST1 |
| SPATyper 2 | t128 |
| Plasmid 3 | pSA31PB |
| ARG 4,* | blaI, blaR1, blaZ, lmrS, mecA, tet(38) |
| Virulence factors (VFDB) 5 | |
| Adherence | atl, ebh, clfA, clfB, cna, ebp, efb, fnbA, fnbB, icaA, icaB, icaC, icaR, sdrC, sdrD, sdrE, spa |
| Enzyme | sspA, sspB, sspC, hysA, geh, lip, splA, splB, splC, splF, coa, sak, nuc |
| Immune evasion | adsA, scn, sbi |
| Secretion system | esaA, esaB, esaD, esaE, esaG, essA, essB, essC, esxA, esxB, esxC, esxD |
| Toxin | hla, hld, sea, sec, seh, selk, sell, selq, set16, set17, set18, set19, set21, set22, set23, set24, set25, set26, set34, hlgA, hlgB, hlgC, lukD, lukF-PV, lukS-PV |
Legend: FOX—cefoxitin; OXA—oxacillin; PEN—penicillin; TET—tetracycline. * Antibiotic resistance genes. Available in 1 https://cge.cbs.dtu.dk/services/MLST/ (accessed on 5 September 2023), 2 https://cge.cbs.dtu.dk/services/spatyper/ (accessed on 5 September 2023), 3 https://cge.cbs.dtu.dk/services/PlasmidFinder/ (accessed on 5 September 2023); 4 https://cge.cbs.dtu.dk/services/ResFinder/ (accessed on 5 September 2023); 5 http://www.mgc.ac.cn/VFs/main.htm (accessed on 5 September 2023).
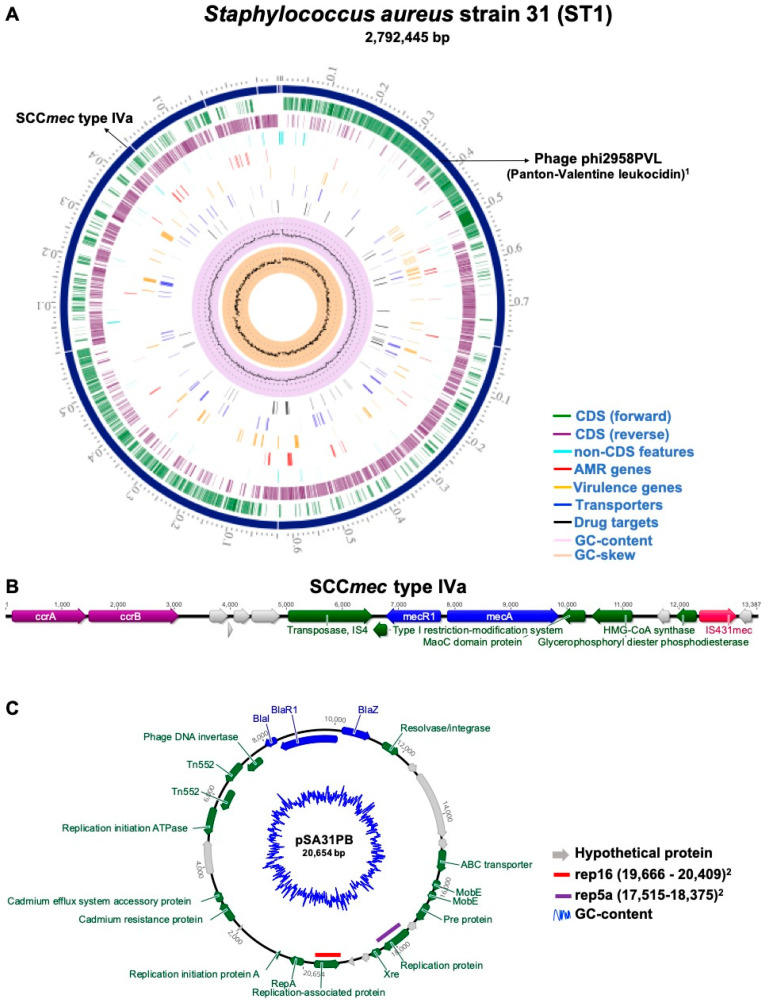
Sequencing generated 7,188,460 reads, averaging 118 bases and a total yield of 848,238,280 bases. A high-quality chromosome composed of contigs with an N50 of 628,879 and 303-fold coverage was assembled. The complete genome contained 2,792,445 bp, a G + C content of 32.69%, with a total of 2614 protein-coding sequences (CDS), 4 rRNA genes, and 57 tRNA genes. The graphical representation of the circular chromosome DNA is shown in Figure 1A.
Figure 1.
Circular genome map of the community-acquired Staphylococcus aureus strain 31 (CA-MRSA ST1) and genomic architecture of pSA31PB plasmid. (A) Genome map of CA-MRSA ST1 strain. From outer to inner ring: contigs (scale-x1Mbp), CDS on the forward strand, CDS on the reverse strand, RNA genes, CDS with homology to known antimicrobial resistance genes, CDS with homology to known virulence factors, GC content, and GC skew. The schematic figure was generated using the Comprehensive Genome Analysis Service in PATRIC server [17]. (B) Schematic representation of the SCCmec Type IVa showing the position of the mec complex, composed of IS431mec, mecA, and intact and truncated regions of the regulatory gene (mecR1), in addition to the ccr complex and the J regions, located between and around the mec and ccr complex. (C) Schematic representation of the 20,654 bp pSA31PB putative plasmid that harbors the blaZ gene and two genes replication rep types: rep16 and rep5a Plasmid figure was generated using Geneious software (v. 9.0.5).
In silico analyses revealed that the SA31 strain belonged to sequence type 1 (ST1) and spa type 128 (t128). It harbored a staphylococcal cassette chromosome mec element (SCCmec) type IVa (2B) carrying the mecA gene (Figure 1B). In addition, according to the pathogenicity prediction analysis, we observed a 97.6% probability of the strain being pathogenic to humans. SCCmec type IV carrying mecA antimicrobial resistance gene is commonly present in CA-MRSA strains [3,26,27,28]. Notably, skin and soft tissue infections caused by CA-MRSA sequence type 1 (ST1) strains carrying SCCmec IV have been reported in several countries [3,27,29]. The presence of SCCmec IVa in S. aureus associated with milk and dairy products [30,31,32,33] suggests a possible route of contamination by direct contact between dairy animals and humans during milking practices (including manual milking) or via the agricultural environment [32].
The MRSA ST1 strain 31 harbored the chromosomal lmrS gene, a multidrug efflux pump mechanism of the Major Facilitator Superfamily (MFS) conferring multidrug resistance in S. aureus strains, including chloramphenicol, erythromycin, and trimethoprim [34]. However, the investigated strain was phenotypically susceptible to these non-β-lactam antibiotics, similar to what has been reported in other MRSA ST1 strains [3,35,36,37]. Previous reports suggest that gene regulators capable of modulating the expression of multiple efflux pumps are involved in lmrS overexpression and, consequently, in the resistance phenotype [38,39]. According to Costa et al. [40], the upregulation of this gene is associated with exposure to sub-inhibitory concentrations of the antimicrobials.
The chromosomal sequence of the CA-MRSA ST1 strain harbored a set of virulence determinants, including lukF-PV and lukS-PV genes encoding the S and F subunits of Panton-Valentine leukocidin (PVL). Moreover, we identified a diversity of enterotoxin-encoding genes (sea, seq, sek, sel, sec2, and seh) and icaC gene involved in the externalization of the nascent polysaccharide [41], as well as the delta-hemolysin (hld), collagen adhesin (cna) and zinc metalloproteinase aureolysin (aur) genes (Table S2—Supplementary Materials). These genes were also identified by VFDB (Table 1).
Manual assembly of the putative plasmid generated a high-quality sequence with only one circular contig of 20,654 bp (pSA31PB). The annotation revealed 27 predicted open reading frames encoding proteins and a 28.37% G + C content. It harbors two encoding replication initiation (rep) genes: rep16 (position 19666..20409; accession number: BX571858) and rep5a (position 17515..18375; accession number: AP003139). Detecting these rep-like sequences in plasmids is crucial as they are useful for staphylococcal plasmid classification [12,42]. In addition, these gene replication initiation sequences can be associated with antibiotic-resistance genes [12,43]. As shown in Figure 1C, pSA31PB co-harbored the stress response cadD gene and a set of antibiotic resistance genes, including the blaZ, associated with β-lactam resistance and usually found as part of the bla operon (blaI, blaR1, and blaZ) which is widely spread among Gram-positive bacteria [44]. The scarcity of characterization of plasmids in MRSA has been previously highlighted [11,12,43,45]. Considering the critical role of plasmids in the successful adaptation of strains, especially in those causing persistent MRSA infections [11,46,47], a better understanding of the genetic context of plasmids is extremely important to understand co-evolutionary events and the dissemination mechanisms of antimicrobial resistance.
The phylogenetic position of SA31 compared to other CA-MRSA genomes performed by the NCBI Isolates Browser tool is shown in Figure 2. SA31 was highly related to six other MRSA strains of human clinical relevance, including a strain recently isolated in England (SAMN08815268) and the ST1-SCCmec IV MRSA strains USA400 (SAMN17703516) and USA400-0051 (SAMN05864218) in the USA. In Brazil, ST1-SCCmec IV isolates emerged as an important healthcare-associated pathogen [28,48,49]. Epidemiological data also demonstrate that CA-MRSA strains predominantly characterized as SCCmec IV have spread throughout the Brazilian territory [29,33,50]. Moreover, they are genetically related to highly virulent CA-MRSA MW2 strains (SAMN03255481; SAMN03255482; SAMD00061104) isolated in the 90s in the USA. The MW2 strain is one of the major pathogens causing community-acquired infections in the Midwestern USA. Several fatal infections were attributed to this strain in the late 1990s [51].
Figure 2.
SNP-cluster tree generated by the NCBI Isolates Browser tool for the community-acquired methicillin-resistant S. aureus Sequence type 1 (CA-MRSA ST1 S) strain SA31 genome. The genome sequence of the CA-MRSA ST1 strain (SA31) causing intramammary subclinical infection in lactating goats in Northeastern Brazil is represented in red.
4. Conclusions
The presence of a CA-MRSA ST1 strain in goat milk, possibly transmitted by cross-contamination due to manual milking practices, indicates that milk and dairy products could eventually play a role in spreading these bacteria. The in-depth characterization of the CA-MRSA ST1 genome associated with mastitis in goat species, including the pSA31PB staphylococcal plasmid, can support future studies addressing the epidemiology, pathogenesis, and evolution of methicillin-resistant Staphylococcus aureus.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/pathogens13010023/s1. Table S1: Annotation data of the genome sequence of the CA-MRSA ST1 strain (SA31) causing intramammary subclinical infection in lactating goat in Northeastern Brazil. Table S2: Annotated resistance and virulence genes identified in the CA-MRSA ST1 strain (SA31) causing intramammary subclinical infection in lactating goat in Northeastern Brazil.
Author Contributions
Conceptualization: C.J.B.O., R.F.C.V. and P.E.N.G.; Data curation: P.C.V., N.M.V.S. and S.P.C.; Formal analysis: P.C.V., E.L.L., M.M.S.S., R.G.F., S.P.C., N.M.V.S. and O.C.F.N.; Funding acquisition: C.J.B.O. and R.F.C.V.; Methodology: P.C.V., E.L.L., M.M.S.S., R.G.F., S.P.C., N.M.V.S. and O.C.F.N.; Investigation: P.C.V., E.L.L., M.M.S.S., R.G.F., S.P.C., N.M.V.S. and O.C.F.N.; Project administration: C.J.B.O. and P.E.N.G., Resources: C.J.B.O., O.C.F.N. and RFVC, P.E.N.G.; Software: E.L.L., M.M.S.S., R.G.F. and S.P.C.; Supervision: C.J.B.O. and P.E.N.G., Validation: E.L.L. and N.M.V.S.; Visualization: P.C.V., E.L.L. and S.P.C.; Writing—original draft preparation: C.J.B.O., E.L.L., R.G.F., O.C.F.N. and P.C.V.; Writing—review and editing: P.E.N.G. and R.F.C.V. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The isolate was obtained from a milk sample, according to the traditional milking practices in private farms. There was no specific animal handling or invasive procedure requiring ethical approval. For the activity of access to Genetic Heritage, the project has been registered in SisGen, in compliance with the provisions of Law No. 13,123/2015 and its regulations. NATIONAL MANAGEMENT SYSTEM OF GENETIC HERITAGE AND ASSOCIATED TRADITIONAL KNOWLEDGE (SISTEMA NACIONAL DE GESTÃO DO PATRIMÔNIO GENÉTICO E DO CONHECIMENTO TRADICIONAL ASSOCIADO) Approval Code: ADC0D0C Approval Date: 24/10/2018.
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw whole-genome sequencing data (Illumina pool-sequencing) have been deposited in the Sequence Read Archive under the accession number (SRR11665884), BioProject (PRJNA593524), BioSample (SAMN14792743). The Fasta file for the assemblies has been deposited in GenBank under the following accession code: JAHHIV000000000.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001. Financeira de Estudos e Projetos (FINEP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Côrtes M.F., Costa M.O., Lima N.C., Souza R.C., Almeida L.G., Guedes L.P.C., Vasconcelos A.T., Nicolás M.F., Figueiredo A.M. Complete genome sequence of community-associated methicillin-resistant Staphylococcus aureus (strain USA400-0051), a prototype of the USA400 clone. Mem. Inst. Oswaldo Cruz. 2017;112:790–792. doi: 10.1590/0074-02760170128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanssen A.-M., Sollid J.U.E. SCCmec in staphylococci: Genes on the move. FEMS Immunol. Med. Microbiol. 2006;46:8–20. doi: 10.1111/j.1574-695X.2005.00009.x. [DOI] [PubMed] [Google Scholar]
- 3.Figueiredo A.M., Ferreira F.A. The multifaceted resources and microevolution of the successful human and animal pathogen methicillin-resistant Staphylococcus aureus. Mem. Inst. Oswaldo Cruz. 2014;109:265–278. doi: 10.1590/0074-0276140016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuny C., Wieler L.H., Witte W. Livestock-associated MRSA: The impact on humans. Antibiotics. 2015;4:521–543. doi: 10.3390/antibiotics4040521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baraldi E., Lindahl O., Savic M., Findlay D., Årdal C. Antibiotic pipeline coordinators. J. Law. Med. Ethics. 2018;46((Suppl. S1)):25–31. doi: 10.1177/1073110518782912. [DOI] [PubMed] [Google Scholar]
- 6.Shrestha A., Bhattarai R.K., Luitel H., Karki S., Basnet H.B. Prevalence of methicillin-resistant Staphylococcus aureus and pattern of antimicrobial resistance in mastitis milk of cattle in Chitwan, Nepal. BMC Vet. Res. 2021;17:239. doi: 10.1186/s12917-021-02942-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khanal S., Boonyayatra S., Awaiwanont N. Prevalence of methicillin-resistant Staphylococcus aureus in dairy farms: A systematic review and meta-analysis. Front. Vet. Sci. 2022;9:947154. doi: 10.3389/fvets.2022.947154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh A.K. A comprehensive review on subclinical mastitis in dairy animals: Pathogenesis, factors associated, prevalence, economic losses and management strategies. CABI Rev. 2022;17:57. doi: 10.1079/cabireviews202217057. [DOI] [Google Scholar]
- 9.Malachowa N., DeLeo F.R. Mobile genetic elements of Staphylococcus aureus. Cell Mol. Life Sci. 2010;67:3057–3071. doi: 10.1007/s00018-010-0389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neyaz L., Rajagopal N., Wells H., Fakhr M.K. Molecular characterization of Staphylococcus aureus plasmids associated with strains isolated from various retail meats. Front. Microbiol. 2020;11:223. doi: 10.3389/fmicb.2020.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baines S.L., Jensen S.O., Firth N., da Silva A.G., Seemann T., Carter G.P., Williamson D.A., Howden B.P., Stinear T.P. Remodeling of pSK1 family plasmids and enhanced chlorhexidine tolerance in a dominant hospital lineage of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2019;63:e02356-18. doi: 10.1128/AAC.02356-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mores C.R., Montelongo C., Putonti C., Wolfe A.J., Abouelfetouh A. Investigation of plasmids among clinical Staphylococcus aureus and Staphylococcus haemolyticus isolates from egypt. Front. Microbiol. 2021;12:659116. doi: 10.3389/fmicb.2021.659116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Middleton J.R., Saeman A., Fox L.K., Lombard J., Hogan J.S., Smith K.L. The National Mastitis Council: A global organization for mastitis control and milk quality, 50 Years and Beyond. J. Mammary Gland. Biol. Neoplasia. 2014;19:241–251. doi: 10.1007/s10911-014-9328-6. [DOI] [PubMed] [Google Scholar]
- 14.Santos S., Saraiva M., Filho A.M., Silva N., De Leon C., Pascoal L., Givisiez P., Gebreyes W., Oliveira C. Swine as reservoirs of zoonotic borderline oxacillin-resistant Staphylococcus aureus ST398. Comp. Immunol. Microbiol. Infect. Dis. 2021;79:101697. doi: 10.1016/j.cimid.2021.101697. [DOI] [PubMed] [Google Scholar]
- 15.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 32nd ed. Clinical & Laboratory Standards Institute; Wayne, PA, USA: 2022. [Google Scholar]
- 16.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wattam A.R., Davis J.J., Assaf R., Boisvert S., Brettin T., Bun C., Conrad N., Dietrich E.M., Disz T., Gabbard J.L., et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017;45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cosentino S., Voldby Larsen M., Møller Aarestrup F., Lund O. PathogenFinder—distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE. 2013;8:e77302. doi: 10.1371/annotation/b84e1af7-c127-45c3-be22-76abd977600f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen M.V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R.L., Jelsbak L., Sicheritz-Pontéen T., Ussery D.W., Aarestrup F.M., et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartels M.D., Petersen A., Worning P., Nielsen J.B., Larner-Svensson H., Johansen H.K., Andersen L.P., Jarløv J.O., Boye K., Larsen A.R., et al. Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2014;52:4305–4308. doi: 10.1128/JCM.01979-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B., Zheng D., Jin Q., Chen L., Yang J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47:D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bortolaia V., Kaas R.S., Ruppe E., Roberts M.C., Schwarz S., Cattoir V., Philippon A., Allesoe R.L., Rebelo A.R., Florensa A.F., et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaya H., Hasman H., Larsen J., Stegger M., Johannesen T.B., Allesøe R.L., Lemvigh C.K., Aarestrup F.M., Lund O., Larsen A.R. SCCmecFinder, a web-based tool for typing of Staphylococcal Cassette Chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere. 2018;3:e00612-17. doi: 10.1128/mSphere.00612-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carattoli A., Zankari E., Garcìa-Fernandez A., Larsen M., Lund O., Voldby Villa L., Møller Aarestrup F., Hasman H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherry N.L., Horan K.A., Ballard S.A., da Silva A.G., Gorrie C.L., Schultz M.B., Stevens K., Valcanis M., Sait M.L., Stinear T.P., et al. An ISO-certified genomics workflow for identification and surveillance of antimicrobial resistance. Nat. Commun. 2023;14:60. doi: 10.1038/s41467-022-35713-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma X.X., Ito T., Tiensasitorn C., Jamklang M., Chongtrakool P., Boyle-Vavra S., Daum R.S., Hiramatsu K. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 2002;46:1147–1152. doi: 10.1128/AAC.46.4.1147-1152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Côrtes M.F., Botelho A.M.N., Almeida L.G.P., Souza R.C., Cunha O.d.L., Nicolás M.F., Vasconcelos A.T.R., Figueiredo A.M.S. Community-acquired methicillin-resistant Staphylococcus aureus from ST1 lineage harboring a new SCCmec IV subtype (SCCmec IVm) containing the tetK gene. Infect. Drug Resist. 2018;11:2583–2592. doi: 10.2147/IDR.S175079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrade M.M., Luiz W.B., da Silva Oliveira Souza R., Amorim J.H. The history of methicillin-resistant Staphylococcus aureus in Brazil. Can. J. Infect. Dis. Med. Microbiol. 2020;2020:1721936. doi: 10.1155/2020/1721936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuenck R.P., Nouér S.A., Winter C.d.O., Cavalcante F.S., Scotti T.D., Ferreira A.L.P., Marval M.G.-D., dos Santos K.R.N. Polyclonal presence of non-multiresistant methicillin-resistant Staphylococcus aureus isolates carrying SCCmec IV in health care-associated infections in a hospital in Rio de Janeiro, Brazil. Diagn. Microbiol. Infect. Dis. 2009;64:434–441. doi: 10.1016/j.diagmicrobio.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Alba P., Feltrin F., Cordaro G., Porrero M.C., Kraushaar B., Argudín M.A., Nykäsenoja S., Monaco M., Stegger M., Aarestrup F.M., et al. Livestock-associated methicillin resistant and methicillin susceptible Staphylococcus aureus sequence Type (CC)1 in European farmed animals: High genetic relatedness of isolates from Italian cattle herds and humans. PLoS ONE. 2015;10:e0137143. doi: 10.1371/journal.pone.0137143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cortimiglia C., Bianchini V., Franco A., Caprioli A., Battisti A., Colombo L., Stradiotto K., Vezzoli F., Luini M. Short communication: Prevalence of Staphylococcus aureus and methicillin-resistant S. aureus in bulk tank milk from dairy goat farms in Northern Italy. J. Dairy. Sci. 2015;98:2307–2311. doi: 10.3168/jds.2014-8923. [DOI] [PubMed] [Google Scholar]
- 32.Carfora V., Giacinti G., Sagrafoli D., Marri N., Giangolini G., Alba P., Feltrin F., Sorbara L., Amoruso R., Caprioli A., et al. Methicillin-resistant and methicillin-susceptible Staphylococcus aureus in dairy sheep and in-contact humans: An intra-farm study. J. Dairy. Sci. 2016;99:4251–4258. doi: 10.3168/jds.2016-10912. [DOI] [PubMed] [Google Scholar]
- 33.de Carvalho S.P., de Almeida J.B., Andrade Y.M., da Silva L.S., Chamon R.C., dos Santos K.R., Marques L.M. Molecular characteristics of methicillin-resistant Staphylococcus aureus isolates from hospital and community environments in northeastern Brazil. Braz. J. Infect. Dis. 2019;23:134–138. doi: 10.1016/j.bjid.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Floyd J.L., Smith K.P., Kumar S.H., Floyd J.T., Varela M.F. LmrS is a multidrug efflux pump of the major facilitator superfamily from Staphylococcus aureus. Antimicrob. Agents Chemother. 2010;54:5406–5412. doi: 10.1128/AAC.00580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tristan A., Ferry T., Durand G., Dauwalder O., Bes M., Lina G., Vandenesch F., Etienne J. Virulence determinants in community and hospital meticillin-resistant Staphylococcus aureus. J. Hosp. Infect. 2007;65((Suppl. S2)):105–109. doi: 10.1016/S0195-6701(07)60025-5. [DOI] [PubMed] [Google Scholar]
- 36.Carvalho K.S., Mamizuka E.M., Filho P.P.G. Methicillin/Oxacillin-resistant Staphylococcus aureus as a hospital and public health threat in Brazil. Braz. J. Infect. Dis. 2010;14:71–76. doi: 10.1016/S1413-8670(10)70014-3. [DOI] [PubMed] [Google Scholar]
- 37.Macedo-Viñas M., Conly J., Francois P., Aschbacher R., Blanc D.S., Coombs G., Daikos G., Dhawan B., Empel J., Etienne J., et al. Antibiotic susceptibility and molecular epidemiology of Panton-Valentine leukocidin-positive meticillin-resistant Staphylococcus aureus: An international survey. J. Glob. Antimicrob. Resist. 2014;2:43–47. doi: 10.1016/j.jgar.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Truong-Bolduc Q.C., Wang Y., Chen C., Hooper D.C. Transcriptional regulator TetR21 controls the expression of the Staphylococcus aureus LmrS efflux pump. Antimicrob. Agents Chemother. 2017;61:10–1128. doi: 10.1128/AAC.00649-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephen J., Salam F., Lekshmi M., Kumar S.H., Varela M.F. The major facilitator superfamily and antimicrobial resistance efflux pumps of the ESKAPEE pathogen Staphylococcus aureus. Antibiotics. 2023;12:343. doi: 10.3390/antibiotics12020343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costa S.S., Viveiros M., Amaral L., Couto I. Multidrug efflux pumps in Staphylococcus aureus: An update. Open Microbiol. J. 2013;7:59–71. doi: 10.2174/1874285801307010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arciola C.R., Campoccia D., Ravaioli S., Montanaro L. Polysaccharide intercellular adhesin in biofilm: Structural and regulatory aspects. Front. Cell Infect. Microbiol. 2015;5:7. doi: 10.3389/fcimb.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen L.B., Garcia-Migura L., Valenzuela A.J., Løhr M., Hasman H., Aarestrup F.M. A classification system for plasmids from enterococci and other Gram-positive bacteria. J. Microbiol. Methods. 2010;80:25–43. doi: 10.1016/j.mimet.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 43.McCarthy A.J., Lindsay J.A. The distribution of plasmids that carry virulence and resistance genes in Staphylococcus aureus is lineage associated. BMC Microbiol. 2012;12:104. doi: 10.1186/1471-2180-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shore A.C., Deasy E.C., Slickers P., Brennan G., O’Connell B., Monecke S., Ehricht R., Coleman D.C. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2011;55:3765–3773. doi: 10.1128/AAC.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shahkarami F., Rashki A., Rashki Ghalehnoo Z. Microbial susceptibility and plasmid profiles of methicillin-resistant Staphylococcus aureus and methicillin-susceptible S. aureus. Jundishapur. J. Microbiol. 2014;7:e16984. doi: 10.5812/jjm.16984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwong S.M., Ramsay J.P., Jensen S.O., Firth N. Replication of staphylococcal resistance plasmids. Front. Microbiol. 2017;8:2279. doi: 10.3389/fmicb.2017.02279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vestergaard M., Frees D., Ingmer H. Antibiotic resistance and the MRSA problem. Microbiol. Spectr. 2019;7:10–1128. doi: 10.1128/microbiolspec.GPP3-0057-2018. [DOI] [PubMed] [Google Scholar]
- 48.Silva-Carvalho M.C., Bonelli R.R., Souza R.R., Moreira S., dos Santos L.C.G., Conceição M.d.S., Junior S.J.d.M., Carballido J.M., Rito P.N., Vieira V.V., et al. Emergence of multiresistant variants of the community-acquired methicillin-resistant Staphylococcus aureus lineage ST1-SCCmecIV in 2 hospitals in Rio de Janeiro, Brazil. Diagn. Microbiol. Infect. Dis. 2009;65:300–305. doi: 10.1016/j.diagmicrobio.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 49.Caboclo R.M.F., Cavalcante F.S., Iorio N.L.P., Schuenck R.P., Olendzki A.N., Felix M.J., Chamon R.C., dos Santos K.R.N. Methicillin-resistant Staphylococcus aureus in Rio de Janeiro hospitals: Dissemination of the USA400/ST1 and USA800/ST5 SCCmec type IV and USA100/ST5 SCCmec type II lineages in a public institution and polyclonal presence in a private one. Am. J. Infect. Control. 2013;41:e21–e26. doi: 10.1016/j.ajic.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 50.Romero L.C., de Souza da Cunha M.L.R. Insights into the epidemiology of community-associated methicillin-resistant Staphylococcus aureus in special populations and at the community-healthcare interface. Braz. J. Infect. Dis. 2021;25:101636. doi: 10.1016/j.bjid.2021.101636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baba T., Takeuchi F., Kuroda M., Yuzawa H., Aoki K.-I., Oguchi A., Nagai Y., Iwama N., Asano K., Naimi T., et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw whole-genome sequencing data (Illumina pool-sequencing) have been deposited in the Sequence Read Archive under the accession number (SRR11665884), BioProject (PRJNA593524), BioSample (SAMN14792743). The Fasta file for the assemblies has been deposited in GenBank under the following accession code: JAHHIV000000000.