Abstract
The emergence and continued geographic expansion of arboviruses and the growing number of infected people have highlighted the need to develop and improve multiplex methods for rapid and specific detection of pathogens. Sequencing technologies are promising tools that can help in the laboratory diagnosis of conditions that share common symptoms, such as pathologies caused by emerging arboviruses. In this study, we integrated nanopore sequencing and the advantages of reverse transcription polymerase chain reaction (RT-PCR) to develop a multiplex RT-PCR protocol for the detection of Chikungunya virus (CHIKV) and several orthoflaviviruses (such as dengue (Orthoflavivirus dengue), Zika (Orthoflavivirus zikaense), yellow fever (Orthoflavivirus flavi), and West Nile (Orthoflavivirus nilense) viruses) in a single reaction, which provides data for sequence-based differentiation of arbovirus lineages.
Keywords: nanopore sequencing, arbovirus, Chikungunya virus, orthoflaviviruses, genotyping
1. Introduction
Infectious diseases caused by viruses transmitted by mosquitoes have been making world headlines since arbovirus outbreaks began appearing in large urban areas. An estimated 3.83 billion people are currently living in areas at risk of dengue, and this number is predicted to increase to 6.1 billion people by 2080, which would represent 60% of the global population in 2080 [1]. Other mosquito-borne diseases, such as Zika and Chikungunya fevers, also represent an important threat to the health and economics of populations around the world, mainly in the region of the Americas, where a total of 3.1 million cases of arboviral diseases were reported in 2022, which represented a relative increase of 118.5% from 2021 to 2022 [2].
Many arboviral diseases lead to clinically indistinguishable febrile syndromes, which makes correct diagnosis challenging. For instance, laboratory differentiation of members of the genus Orthoflavivirus (formerly named Flavivirus) [3] using serological methods is limited due to extensive cross-reactivity [4]. Nucleic acid tests, such as quantitative reverse transcription polymerase chain reaction (RT-qPCR) tests, have been extensively used for pathogen detection due to their high specificity and sensitivity [5]; however, available tests cannot provide information for differentiating arboviruses at the level of distinct lineages.
DNA sequencing technology has been proven useful in recent efforts to control infectious disease outbreaks, by providing relevant epidemiological aspects regarding the dynamics of an epidemic [6]. The nanopore sequencing platform is a potential tool for diagnostic purposes due to its cost effectiveness, rapid turnaround time, and portability, which allow it to be employed in the investigation of several viral outbreaks [7,8,9]. Hence, in this study, we integrated nanopore sequencing and the advantages of reverse transcription polymerase chain reaction (RT-PCR) to develop a multiplex RT-PCR protocol for the detection of Chikungunya virus (CHIKV) (genus Alphavirus) and several orthoflaviviruses in a single reaction, which provides data for sequence-based differentiation of arbovirus lineages.
2. Materials and Methods
2.1. Primer Design and Selection
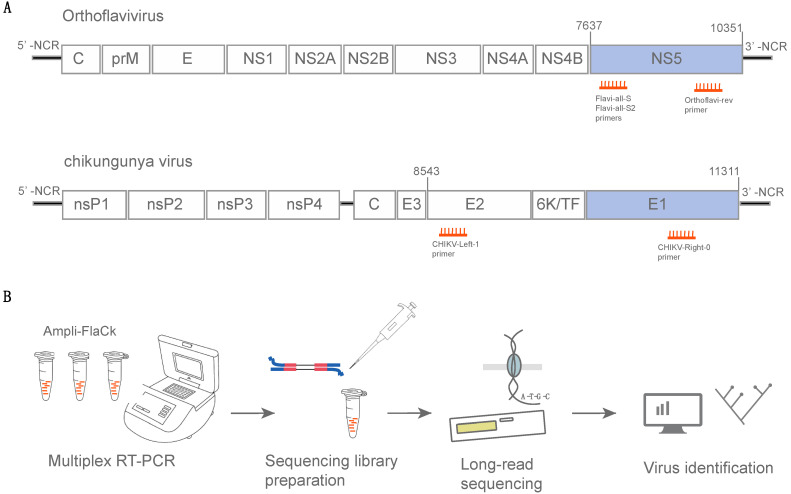
We identified a set of primers, previously designed for real-time quantitative reverse transcription PCR (RT-qPCR) [10], which originally amplified a fragment of 260 bases of a conserved region in the NS5 gene of orthoflaviviruses. To take advantage of the long-read sequencing technology of a MinION device, we performed an alignment of 28 reference sequences of common orthoflaviviruses to design a new reverse primer. When used alongside the forward primers (Flavi-all-S and Flavi-all-S2) from a previously published scheme [10], this new reverse primer enabled the amplification of a fragment of around 1000 bases from the NS5 region.
For the detection of the Chikungunya virus’s nucleic acid, we selected a pair of primers previously published for nanopore sequencing [11] and redirected these primers to compose the primer scheme for the optimized protocol described in this study.
We evaluated the specificity of the Chikungunya primers in silico using the National Center for Biotechnology Information (NCBI) Primer-BLAST tool against the NCBI nucleotide collection. However, assessment of the orthoflavivirus primers using the Primer-BLAST tool was not feasible due to its limited ability to accept ambiguity letters beyond “N” within the primer sequences. Consequently, we employed an alternative tool, MFEprimer v.3.0 [11], to conduct in silico checks on both the CHIKV and the orthoflavivirus primers for primer-specific amplification against the NCBI virus database.
2.2. Virus and Clinical Specimen Selection
We used virus stocks of Chikungunya virus (CHIKV), as well as Orthoflavivirus denguei (DENV) (dengue virus types 1–4), Orthoflavivirus zikaense (Zika virus; ZIKV), Orthoflavivirus flavi (yellow fever virus; YFV), and Orthoflavivirus nilense (West Nile virus; WNV) viruses passaged in C6/36 cells, in L-15 medium kindly provided by the Public Health Laboratory of Minas Gerais state (Fundação Ezequiel Dias) and by the Flavivirus Laboratory of the Oswaldo Cruz Foundation-Rio de Janeiro. Clinical specimens (serum) were provided as residual samples from the epidemiological surveillance routines of the Brazilian Central Public Health Laboratories (LACEN) in Minas Gerais, Rio Grande do Norte, Acre, and Amazonas states. Clinical samples were also obtained from genomic surveillance activities carried out in the Public Health Laboratory of Uruguay and in Ceará state (Universidade de Fortaleza, Brazil). This study was approved by the Pan American Health Organization/World Health Organization (PAHO/WHO) and by the Research Ethics Committee of the Universidade Federal de Minas Gerais, with approval No. 32912820.6.1001.5149. Personal information was de-identified to minimize the risk of the unintended disclosure of the identity of individuals.
Viral RNA, taken from both culture supernatant and clinical specimens, was extracted using the chemagic Viral DNA/RNA 300 Kit H96 (PerkinElmer, Wellesley, MA, USA) according to the manufacturer’s instructions. Extracted RNA was subjected to molecular diagnosis using RT-qPCR to confirm the presence of viral RNA in the samples. Obtained cycle threshold (Ct) values were then used as proxy indicators for the amount of viral genetic material in the screened samples. RT-qPCR tests were performed using the Applied Biosystems 7500 (Thermo Fisher Scientific Inc., Sunnyvale, CA, USA) following previously described protocols [12,13,14,15] for the detection of YFV, ZIKV, CHIKV, and DENV.
2.3. Primer Validation using Cultured Viruses and Clinical Specimens
Selected primers were pooled to a concentration of 10 μM and were initially tested using culture isolates of DENV (types 1–4), ZIKV, CHIKV, YFV, and WNV. One-step RT-PCR was performed using the QIAGEN OneStep RT-PCR Kit according to the manufacturer’s instructions, in a total reaction of 25 μL containing 4.7 μL of nuclease-free water, 5 μL of 5x QIAGEN OneStep RT-PCR Buffer, 1 μL of dNTP mix, 6.25 μL of primers (from the pool at 10 μM to a final concentration of 0.5 μM of each primer), 1 μL of QIAGEN OneStep RT-PCR Enzyme Mix, and 7 μL of viral RNA as a template. RT-PCR temperature cycling conditions were as follows: 50 °C for 30 min (reverse transcription); 95 °C for 15 min (initial PCR activation); followed by 40 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 90 s; and lastly 72 °C for 10 min (final extension). The same RT-PCR conditions were applied for the validation of the primers using clinical specimens. All reactions contained no-template controls (NTCs) to assess the extent of cross-contamination between neighboring barcoded samples. Electrophoresis in 1% agarose gel was performed to visualize PCR products alongside a GeneRuler 1 kb Plus DNA Ladder (Thermo Fisher Scientific Inc., Sunnyvale, CA, USA). Amplicon concentrations were then quantified using the Qubit fluorimeter with the Qubit dsDNA HS Assay Kit (Invitrogen, Boston, MA, USA).
2.4. Nanopore Sequencing
Amplicons were purified using 0.8x AMPure XP beads (Beckman Coulter, Brea, CA, USA), and sample concentrations were normalized to an initial input of 10 ng each. After this point, nanopore library preparation (barcode and adapter ligation) followed the protocol previously described by Quick et al. (2017) [11], which used the Ligation Sequencing Kit (SQKLSK109, Oxford Nanopore Technologies, Oxford, UK) and Native Barcoding Expansion (NBD104 and EXPNBD114, Oxford Nanopore Technologies, Oxford, UK) for the multiplexing of 24 samples. Prepared sequencing libraries were loaded on an R9.4 flow cell, and data were collected for 2 h.
2.5. Bioinformatic Analysis for Virus Identification
The raw data (Fast5 files) from the MinION sequencing were basecalled (using the FAST model) and demultiplexed using Guppy v.6.0 (Oxford Nanopore Technologies). Consensus sequence generation, as well as the identification and classification of viral species and lineages [16], were carried out via the Genome Detective software Version 2.41, which uses SPAdes for nanopore single-end reads [17].
The consensus sequences from clinical samples of cases that had tested positive for DENV (types 1–4), CHIKV, ZIKV, and YFV were used to reconstruct the phylogeny of these viruses. Reference sequences were downloaded from NCBI to build separate datasets of partial DENV-2 NS5 (n = 36) and CHIKV envelope (n = 25) genes. After alignment of the sequences generated in this study using MAFFT version 7, these datasets were used to infer the maximum likelihood (ML) phylogenies using IQ-TREE 2.1.1 [18], which also implemented an ultrafast bootstrap (UFBoot) to estimate the statistical support for tree nodes using 10,000 replicates. Distribution charts of reads numbers and RT-qPCR Ct values were built using custom scripts via R studio v.2023.06.1.
3. Results
The sequences of the two previously published forward primers [10] were used alongside the new reverse primer (containing degenerate bases in three positions) designed to amplify a fragment of around 1000 bases from a conserved region in the NS5 gene of orthoflaviviruses (Figure 1A). Additionally, a pair of primers was redirected from a published CHIKV sequencing primer scheme [11] to amplify a fragment of ~1100 bases from the CHIKV envelope gene (Figure 1A). We used an in silico approach to assess the specificity of these selected primers by aligning them against the NCBI virus database, which resulted in 8120 potential amplicons covering 20 different virus species when using the orthoflavivirus primers (Table S2). Performing the same analysis using the CHIKV primers resulted in 584 potential amplicons, all from CHIKV isolates. In addition, a Primer-BLAST analysis returned 6084 BLAST hits for Chikungunya virus genomes (see the Data Availability Statement to access the output from the in silico specificity analysis).
Figure 1.
A multiplex RT-PCR protocol for amplification and sequencing of nucleic acids of Chikungunya virus and orthoflaviviruses (Ampli-FlaCk). (A) Organization of orthoflaviviruses and Chikungunya virus (genus Alphavirus) genomes. Primers (small orange lines) are depicted near their expected binding positions. Genome representations are based on the following references for orthoflaviviruses and Chikungunya virus, respectively: NC_002031 and KP164568.1. (B) Schematic representation of the Ampli-FlaCk workflow.
These primers (a total of five) were pooled to achieve a multiplex RT-PCR protocol for the amplification of the Chikungunya virus’s and the orthoflaviviruses’ nucleic acids (namely Ampli-FlaCk) in a single reaction. Ampli-FlaCk is able to amplify long amplicons, which, combined with nanopore sequencing, can generate long sequences (~1000 bases) from multiple distinct samples. Then, these sequences can be used for the identification and genotyping of the distinct lineages of Chikungunya virus and orthoflaviviruses (Figure 1B). The nucleotide sequence and physical properties of the primers that comprise the Ampli-FlaCk protocol are listed in Table S1.
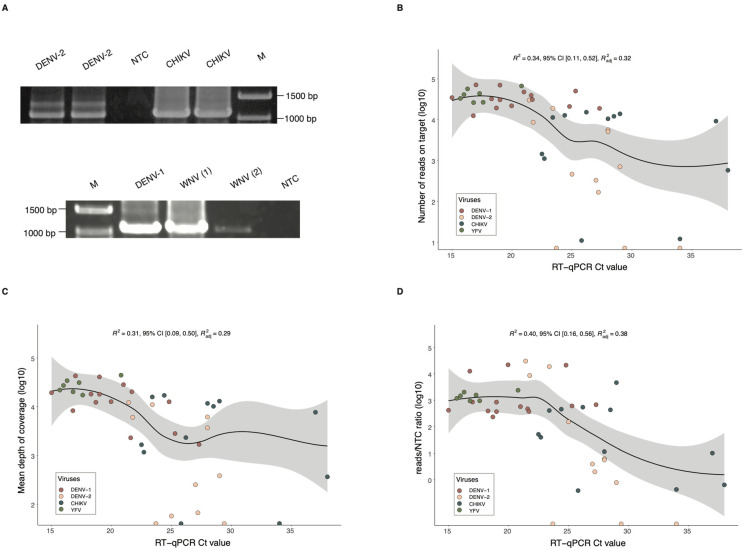
To validate the protocol, the RNA from different cultured viruses (DENV, ZIKV, YFV, WNV) was extracted and used in RT-PCR reactions with the Ampli-FlaCk primers and the OneStep RT-PCR Kit, which allowed both reverse transcription and PCR amplification to occur in the same reaction mix. Gel electrophoresis revealed clearly visible bands corresponding to the expected amplicons for each virus (Figure 2A and Figure S1). Then, the amplicons were barcoded and sequenced using a MinION (nanopore sequencing) device for 2 h. After basecalling, viral reads were analyzed using the Genome Detective Version 2.41 software [17], a user-friendly online tool that performs read mapping, consensus creation, and genotyping of viruses such as ZIKV, DENV, CHIKV, and YFV [16].
Figure 2.
Sequencing performance of the Ampli-FlaCk protocol. (A) Electrophoresis analysis of RT-PCR products from cultured dengue virus type 2 (DENV-2, Ct = 22) and Chikungunya virus (CHIKV, Ct = 33), in duplicates (top image). The bottom image displays electrophoresis results for cultured dengue virus type 1 (DENV-1, Ct = 25) and West Nile virus (WNV, numbers in parentheses represent samples having different Ct values, 1 = 21, and 2 = 33). M = Marker. Images were cropped and converted to grayscale for clarity; see the Data Availability Statement for original, unedited images. (B) Distribution of the number of reads on target and the RT-qPCR Ct values from the clinical specimens (n = 45) used for testing the protocol. (C) Distribution of the mean depth of coverage and the RT-qPCR Ct values from the tested clinical specimens (n = 45). (D) Distribution of the reads/NTC ratio and the RT-qPCR Ct values from the tested clinical specimens (n = 45). (B–D) The dark blue line represents a smooth local regression line (method = LOESS), and the light grey area around the trend line represents the 95% confidence interval.
Sequencing results showed that the protocol can detect all eight viral species and generate nucleotide sequences for the successful discrimination of viral lineages (Table 1). This nanopore library yielded an average of 5682 reads for each sequenced virus, with a mean percentage of 1.75% of off-target reads (reads originating from random crosstalk between neighboring samples). An average of 58.84 reads/NTC ratio was also estimated for this experiment. This ratio indicates the sample’s number of reads on-target-normalized against the number of reads found in the no-template control, and a ratio > 10 was arbitrarily defined to indicate a reliable positive sample for the assigned virus, as previously suggested [19].
Table 1.
Summary of sequencing statistics for the cultured viruses used for evaluating the performance of the Ampli-FlaCk protocol.
| Sample | Ct | Number of Reads on Target |
Off-Target Reads (%) | Reads/NTC Ratio | Lineage |
|---|---|---|---|---|---|
| DENV-1 | 25 | 4095 | 0.7 | 67.1 | Genotype I |
| DENV-2 | 22 | 4058 | 0.8 | 66.5 | Genotype III |
| DENV-3 | 19 | 4720 | 1.0 | 77.4 | Genotype V |
| DENV-4 | 28 | 3390 | 2.3 | 55.6 | Genotype II |
| YFV | 28 | 4823 | 1.2 | 79.1 | South America I |
| ZIKV | 25 | 2449 | 1.4 | 40.1 | Asian |
| WNV | 33 | 3967 | 3.9 | 65.0 | Lineage 1A |
| CHIKV | 33 | 17954 | 2.8 | 19.8 * | ECSA |
| NTC | - | 61 | - | - | - |
| Mean (SD) | 5682 (5015.23) | 1.75 (1.14) | 58.84 (20) |
SD = standard deviation; NTC = no-template control; * The CHIKV sample was sequenced using a separate library/flow cell; in this library, 905 reads were detected in the NTC.
Serial dilutions of these eight viruses were also sequenced following the protocol conditions described earlier, and the results allowed us to estimate a preliminary limit of detection for the Ampli-FlaCk protocol, using the dilution’s Ct values as proxy indicators of viral load. The sequencing results showed that the highest Ct values, which generated enough data for reliable detection, ranged from 30 to 37 for dengue viruses, while CHIKV-diluted samples could be positively detected up to a Ct value of 40 (full results are listed in Table S3).
Known positive clinical specimens were selected to validate the Ampli-FlaCk protocol under the same reaction conditions described earlier. Extracted RNA from 14 DENV-1-, 13 DENV-2-, 14 CHIKV-, 1 ZIKV-, and 7 YFV-positive samples was subjected to multiplex RT-PCR treatment and sequenced on the MinION device for up to 6 h to maximize sequencing yield per sample. Analysis of the reads showed that the protocol was able to reliably identify the viral lineage in 83.67% (41/49) of the clinical samples (Table 2 and Table S4). Although some viral reads were detected in 8 samples (16.33%), the reads/NTC ratio for these samples was lower than the threshold to be consider a positive result. The median of 10,229 mapped reads per sample was calculated for the combined sequenced libraries, with long reads of ~1000 bases being recovered from 89.80% (44/49) of the samples. Plotting the number of mapped reads against Ct values, as a proxy indicator for viral load, showed a downward trend in the number of reads as Ct values increased (Figure 2B). As expected, similar trends were also observed in the depth-of-coverage and reads/NTC ratio plots (Figure 2C,D).
Table 2.
Summary of sequencing statistics for the clinical specimens used for evaluating the performance of the Ampli-FlaCk protocol.
| Virus | # of Samples | Ct | # of Reads | % Off-Target Reads | Reads/NTC Ratio | Genotyping | Depth of Coverage 1 | Sequence Length |
|---|---|---|---|---|---|---|---|---|
| DENV-1 | 14 | 19.49 (3.5) | 31,754 (18,341.4) | 0.01 (0.43) | 590.41 (7896.47) | Genotype V | 15,387.15 (12,991.5) | 984.5 (38.3) |
| DENV-2 | 13 | 27.1 (3.6) | 705 (9158.4) | 4.33 (18.17) | 80.10 (10,489.13) | Genotype III | 2282.45 (4641.9) | 1032.5 (109.5) |
| CHIKV | 14 | 26.2 (4.6) | 10,229 (18,977.3) | 3.64 (31.15) | 228.45 (6655.30) | ECSA | 10,946.6 (21,163.4) | 1051.5 (90.5) |
| YFV | 7 | 16.8 (1.7) | 41,143 (15,275.8) | 0.14 (0.04) | 1469.39 (545.57) | South America I | 27,291 (9394.7) | 1044 (49.5) |
| ZIKV | 1 | ND | 62 | 0 | 62 | Asian | 30.4 | 960 |
Median values (standard deviation); ND = not detected; #=number. 1 Mean depth of coverage was estimated using SPAdes implemented via Genome Detective using the following function: read count * read length/contig length.
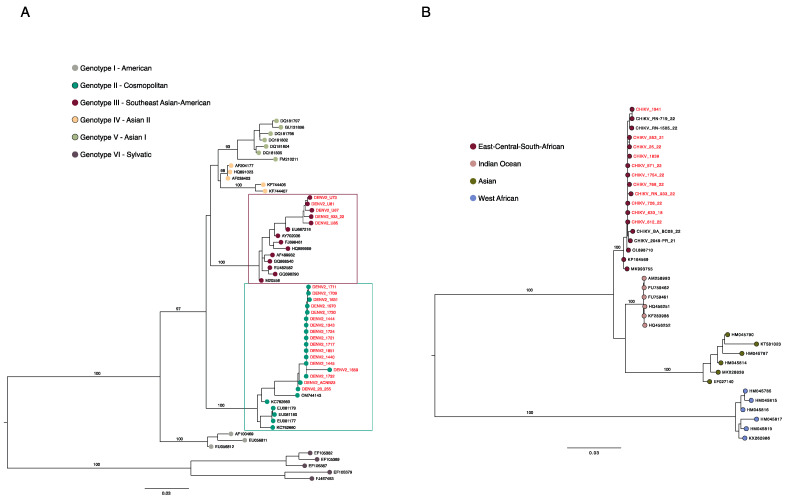
Once its efficacy to recover viral long reads from clinical samples had been established, the Ampli-FlaCk protocol was employed for testing in the field activities of an arbovirus genomic surveillance project carried out in Uruguay (early 2023) [20] and across Brazil in the years 2021 and 2022 [8]. The sequencing of 20 clinical samples collected during those surveillance activities resulted in the recovery and identification of reads of ZIKV, DENV-2, DENV-3, and DENV-4 (Table S5). It is worth mentioning that the protocol was also able to accurately recover reads of a recently detected lineage of DENV-2 in Brazil [21], the genotype II (cosmopolitan). To demonstrate that consensus sequences generated via the Ampli-FlaCk protocol can also be used to reconstruct phylogenetic trees accurately, we performed phylogenetic analyses using DENV-2 and CHIKV sequences from the positive clinical samples used in this study. The maximum likelihood tree showed that the new CHIKV sequences were grouped with other ECSA lineage isolates, while the new DENV-2 sequences were placed either in the genotype II–cosmopolitan clade or in the genotype III clade (Figure 3). Phylogenetic reconstructions were also performed for the other viruses sequenced in this study and can be seen in Supplementary Figures S2 and S3.
Figure 3.
Reconstructed genotyping phylogeny of dengue virus type 2 (Orthoflavivirus denguei) and Chikungunya virus. (A) Maximum likelihood phylogenetic reconstruction of DENV-2 using partial sequences (n = 58) of the NS5 gene, including sequences (n = 22, red tips) generated in this study from clinical samples. (B) Maximum likelihood phylogenetic reconstruction of CHIKV using partial sequences (n = 36) of the envelope gene, including sequences (n = 11, pink tips) generated in this study from clinical samples. Numbers along branches represent Ultrafast bootstrap values (only for the main branches, to maintain clarity).
4. Discussion
DNA sequencing technologies are promising tools that can help in the laboratory detection of emerging and re-emerging pathogens, generating information that can contribute to the fight against epidemics [22,23]. In this study, we developed a protocol for the detection and genotyping of multiple viral species in a single reaction that combines the advantages of the high sensitivity and specificity of multiplex PCR with the portability and speed of data generation provided by nanopore sequencing, which can perform simultaneous sequencing of up to 96 samples in a single experiment. The Ampli-FlaCk protocol also incorporates the benefits of the OneStep RT-PCR Kit, which allows both reverse transcription and PCR amplification to take place in a single reaction [24].
Clinical samples were used for assessing the protocol performance, which resulted in viral identification of 83% of the samples. After sequencing, it was evident that the number of mapped reads followed a downward trend when Ct values increased. This negative relationship has been previously reported [25] for whole-genome sequencing of arboviruses from clinical samples and upholds the importance of sampling during the acute period of infections, when a high viral load is present. A Ct value of 37 is considered the limit of detection for dengue samples tested using the Trioplex Real-time RT-PCR Assay (CDC) [26]. Comparatively, the Ampli-FlaCk protocol was able to detect DENV sample dilutions with the highest Ct values ranging from 30 to 37, depending on the serotype, while CHIKV dilutions were detected up to a Ct value of 40. Although these results are encouraging and point to the potential of the protocol for detecting viral genetic material at low concentrations, the small number of samples tested limits our assessment of the effectiveness of the protocol when working with samples with a Ct value above 30. As such, additional testing is needed to determine the exact detection limit of the protocol.
The diagnosis of orthoflavivirus infections using serological methods is challenging due to the limitations imposed by the broad antigenic cross-reactivity among common orthoflaviviruses [27]. The protocol described here was shown to contribute to the successful detection of all orthoflaviviruses tested here by using long nucleotide sequences, which were also used for viral lineage assignments. Although this work did not test other viruses, the protocol has the potential to detect other orthoflavivirus species due to the use of primers with degenerate bases that cover the nucleotide diversity of the NS5 gene, as suggested by in silico analyses.
5. Conclusions
The protocol described in this study allowed us to obtain long reads of ~1000 bases from the NS5 and envelope genes of orthoflaviviruses and CHIKV, respectively. These reads can be used not only for the identification, but also for the classification of viral lineages. The obtained sequences can also be used for the reconstruction of phylogenetic trees and in evolutionary analysis that might help in understanding patterns of viral dispersion during an epidemic. The protocol practicality associated with the portability of the MinION device offers a possibility of using this approach in field studies in areas with limited infrastructure. Furthermore, the ability to wash and reuse the flow cells allows for a reduction in the costs of screening hundreds of samples when considering the protocol’s multiplex approach. Hence, the development of sequencing-based viral detection methods that are integrated into local epidemiological surveillance can be fostered in order to achieve a more anticipatory approach to epidemic prevention and control in public health [28].
Acknowledgments
The authors thank all personnel from the public health central laboratory of each collaborating state, who helped with data collection and assembly.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/v16010023/s1: Table S1. Nucleotide sequence and physical properties of the primers comprising the Ampli-FlaCk protocol. Table S2. List of potential orthoflaviviruses identified during in silico specificity assessment using MFEprimer. Figure S1. Validation of the Ampli-FlaCk primers on cultured viruses. Figure S2. Maximum Likelihood phylogenies reconstructed using the new sequences generated in this study and lineages reference sequences for genotyping. Figure S3. Maximum Likelihood phylogeny of yellow fever virus reconstructed using the new sequences generated in this study and lineages reference sequences for genotyping. Table S3. Sequencing results of serially diluted virus isolates. Table S4. Sequencing and genotyping results of clinical specimens used for validation of the Ampli-FlaCk protocol. Table S5. Sequencing and genotyping results of clinical specimens tested during viral genomic surveillance activities in Brazil and Uruguay. Table S6. List of GenBank access number of the sequences generated from the clinical samples in this study.
Author Contributions
Conceptualization: L.C.J.A., M.G. and J.X.; methodology: L.C.J.A., M.G. and J.X.; formal analysis: J.X., M.G., V.F., M.L., C.O. and L.C.J.A.; investigation: J.X., V.F., T.A., F.C.M.I., G.C.P., M.M.D., M.L., E.C., C.O., H.F., N.G., L.O.L., F.K.B., C.M.M.B.d.O., C.C.M.G., D.M.L., E.C.d.O., G.G.d.C.L., I.G., J.M., J.T.N.R., J.A., J.K.B.C., K.G.L., L.B.d.S., L.D., M.C.B.C., M.C.S.U.Z., R.S.M.P., R.B.S., S.K.H., S.F.d.S., S.N.S., T.R., N.M., H.C., A.B., V.B., M.N.C., R.S.M., A.C.P., M.F.d.S. and W.A.J.; resources: J.M.R., L.F., A.R., R.F.d.C.S., C.F.C.d.A., E.L.N.M., M.S.d.O., R.V.d.C., L.C.V.F., A.M.B.d.F. and L.C.J.A.; writing—original draft: J.X.; writing—review and editing: L.C.J.A., M.G. and J.X.; funding acquisition: L.C.J.A., J.M.R., A.R., R.F.d.C.S. and C.F.C.d.A. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This research was reviewed and approved by the Research Ethics Committee of the Universidade Federal de Minas Gerais with approval No. 32912820.6.1001.5149, by the Pan American World Health Organization (No. PAHO-2016-08-0029), by the Oswaldo Cruz Foundation Ethics Committee (CAAE: 45279715.8.0000.0040), and by the Brazilian Ministry of Health (MoH) as part of the arbovirus genomic surveillance efforts.
Informed Consent Statement
Because the serum samples used in this retrospective study were collected originally for diagnostic purposes, patients’ contact and identity information were anonymized and not available to the researchers.
Data Availability Statement
The new sequences generated in this study have been deposited in NCBI GenBank under the accession numbers listed in Supplementary Table S6. Input data used for the phylogenetic analyses are provided in the repository https://doi.org/10.6084/m9.figshare.24270724.v2.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was supported by The Pan American Health Organization (PAHO/WHO), by the Brazilian Ministry of Health grant SCON2021-00180 (Coordenação Geral de Laboratório de Saúde Pública—CGLAB and Coordenação Geral de Vigilância de Arboviroses—CGARB), by the Programa Inova Fiocruz/Oswaldo Cruz Foundation, by the National Institutes of Health USA grant U01 AI151698 for the United World Arbovirus Research Network (UWARN), and in part by the CRP-ICGEB RESEARCH GRANT 2020 Project CRP/BRA20-03, Contract CRP/20/03. J.X. was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Finance Code 001. MG was funded by PON “Ricerca e Innovazione” 2014–2020.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Messina J.P., Brady O.J., Golding N., Kraemer M.U.G., Wint G.R.W., Ray S.E., Pigott D.M., Shearer F.M., Johnson K., Earl L., et al. The current and future global distribution and population at risk of dengue. Nat. Microbiol. 2019;4:1508–1515. doi: 10.1038/s41564-019-0476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan American Health Organization . Annual Arbovirus Bulletin 2022. Regional Office for the Americas of the World Health Organization; Washington, DC, USA: 2023. [Google Scholar]
- 3.Postler T.S., Beer M., Blitvich B.J., Bukh J., de Lamballerie X., Drexler J.F., Imrie A., Kapoor A., Karganova G.G., Lemey P., et al. Renaming of the genus Flavivirus to Orthoflavivirus and extension of binomial species names within the family Flaviviridae. Arch. Virol. 2023;168:224. doi: 10.1007/s00705-023-05835-1. [DOI] [PubMed] [Google Scholar]
- 4.Wilder-Smith A., Gubler D.J., Weaver S.C., Monath T.P., Heymann D.L., Scott T.W. Epidemic arboviral diseases: Priorities for research and public health. Lancet Infect. Dis. 2017;17:e101–e106. doi: 10.1016/S1473-3099(16)30518-7. [DOI] [PubMed] [Google Scholar]
- 5.Kralik P., Ricchi M. A basic guide to real time PCR in microbial diagnostics: Definitions, parameters, and everything. Front. Microbiol. 2017;8:108. doi: 10.3389/fmicb.2017.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maljkovic Berry I., Melendrez M.C., Bishop-Lilly K.A., Rutvisuttinunt W., Pollett S., Talundzic E., Morton L., Jarman R.G. Next generation sequencing and bioinformatics methodologies for infectious disease research and public health: Approaches, applications, and considerations for development of laboratory capacity. J. Infect. Dis. 2020;221:S292–S307. doi: 10.1093/infdis/jiz286. [DOI] [PubMed] [Google Scholar]
- 7.Adelino T.É.R., Giovanetti M., Fonseca V., Xavier J., de Abreu Á.S., do Nascimento V.A., Demarchi L.H.F., Oliveira M.A.A., da Silva V.L., de Mello A.L.E.S., et al. Field and classroom initiatives for portable sequence-based monitoring of dengue virus in Brazil. Nat. Commun. 2021;12:2296. doi: 10.1038/s41467-021-22607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xavier J., Alcantara L.C.J., Fonseca V., Lima M., Castro E., Fritsch H., Oliveira C., Guimarães N., Adelino T., Evaristo M., et al. Increased interregional virus exchange and nucleotide diversity outline the expansion of chikungunya virus in Brazil. Nat. Commun. 2023;14:4413. doi: 10.1038/s41467-023-40099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giovanetti M., Slavov S.N., Fonseca V., Wilkinson E., Tegally H., Patané J.S.L., Viala V.L., San E.J., Rodrigues E.S., Santos E.V., et al. Genomic epidemiology of the SARS-CoV-2 epidemic in Brazil. Nat. Microbiol. 2022;7:1490–1500. doi: 10.1038/s41564-022-01191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel P., Landt O., Kaiser M., Faye O., Koppe T., Lass U., Sall A.A., Niedrig M. Development of one-step quantitative reverse transcription PCR for the rapid detection of flaviviruses. Virol. J. 2013;10:58. doi: 10.1186/1743-422X-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quick J., Grubaugh N.D., Pullan S.T., Claro I.M., Smith A.D., Gangavarapu K., Oliveira G., Robles-Sikisaka R., Rogers T.F., Beutler N.A., et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017;12:1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanciotti R.S., Kosoy O.L., Laven J.J., Velez J.O., Lambert A.J., Johnson A.J., Stanfield S.M., Duffy M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerging Infect. Dis. 2008;14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanciotti R.S., Kosoy O.L., Laven J.J., Panella A.J., Velez J.O., Lambert A.J., Campbell G.L. Chikungunya virus in US travelers returning from India, 2006. Emerging Infect. Dis. 2007;13:764–767. doi: 10.3201/eid1305.070015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson B.W., Russell B.J., Lanciotti R.S. Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J. Clin. Microbiol. 2005;43:4977–4983. doi: 10.1128/JCM.43.10.4977-4983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domingo C., Patel P., Yillah J., Weidmann M., Méndez J.A., Nakouné E.R., Niedrig M. Advanced yellow fever virus genome detection in point-of-care facilities and reference laboratories. J. Clin. Microbiol. 2012;50:4054–4060. doi: 10.1128/JCM.01799-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fonseca V., Libin P.J.K., Theys K., Faria N.R., Nunes M.R.T., Restovic M.I., Freire M., Giovanetti M., Cuypers L., Nowé A., et al. A computational method for the identification of Dengue, Zika and Chikungunya virus species and genotypes. PLoS Negl. Trop. Dis. 2019;13:e0007231. doi: 10.1371/journal.pntd.0007231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vilsker M., Moosa Y., Nooij S., Fonseca V., Ghysens Y., Dumon K., Pauwels R., Alcantara L.C., Vanden Eynden E., Vandamme A.-M., et al. Genome Detective: An automated system for virus identification from high-throughput sequencing data. Bioinformatics. 2019;35:871–873. doi: 10.1093/bioinformatics/bty695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M., Fu A., Hu B., Tong Y., Liu R., Liu Z., Gu J., Xiang B., Liu J., Jiang W., et al. Nanopore Targeted Sequencing for the Accurate and Comprehensive Detection of SARS-CoV-2 and Other Respiratory Viruses. Small. 2020;16:e2002169. doi: 10.1002/smll.202002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgueno A., Giovanetti M., Fonseca V., Morel M.N., Lima M., Castro E., Guimaraes N.R., Iani F.C.M., Bormida V., Cortinas M.N., et al. Genomic and eco-epidemiological investigations in Uruguay reveal local Chikungunya virus transmission dynamics during its expansion across the Americas in 2023. medRxiv. 2023 doi: 10.1101/2023.08.17.23294156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giovanetti M., Pereira L.A., Santiago G.A., Fonseca V., Mendoza M.P.G., de Oliveira C., de Moraes L., Xavier J., Tosta S., Fristch H., et al. Emergence of dengue virus serotype 2 cosmopolitan genotype, brazil. Emerging Infect. Dis. 2022;28:1725–1727. doi: 10.3201/eid2808.220550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grubaugh N.D., Ladner J.T., Lemey P., Pybus O.G., Rambaut A., Holmes E.C., Andersen K.G. Tracking virus outbreaks in the twenty-first century. Nat. Microbiol. 2019;4:10–19. doi: 10.1038/s41564-018-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alcantara L.C.J., Amenga-Etego L., Andersson R., Bhaumik M., Choi Y.K., Decaluwe H., Geoghegan J., Haagmans B.L., López S., Mukhtar M.M., et al. Methods for fighting emerging pathogens. Nat. Methods. 2022;19:395–397. doi: 10.1038/s41592-022-01441-2. [DOI] [PubMed] [Google Scholar]
- 24.De Paula S.O., de Melo Lima C., Torres M.P., Pereira M.R.G., Lopes da Fonseca B.A. One-Step RT-PCR protocols improve the rate of dengue diagnosis compared to Two-Step RT-PCR approaches. J. Clin. Virol. 2004;30:297–301. doi: 10.1016/j.jcv.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Naveca F.G., Claro I., Giovanetti M., de Jesus J.G., Xavier J., de Melo Iani F.C., do Nascimento V.A., de Souza V.C., Silveira P.P., Lourenço J., et al. Genomic, epidemiological and digital surveillance of Chikungunya virus in the Brazilian Amazon. PLoS Negl. Trop. Dis. 2019;13:e0007065. doi: 10.1371/journal.pntd.0007065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Centers for Disease Control and Prevention-CDC CDC DENV-1-4 rRT-PCR Multiplex and Trioplex rRT-PCR Assays—Dengue—CDC. [(accessed on 30 August 2023)]; Available online: https://www.cdc.gov/dengue/healthcare-providers/testing/molecular-tests/assays.html.
- 27.Chan K.R., Ismail A.A., Thergarajan G., Raju C.S., Yam H.C., Rishya M., Sekaran S.D. Serological cross-reactivity among common flaviviruses. Front. Cell. Infect. Microbiol. 2022;12:975398. doi: 10.3389/fcimb.2022.975398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardy J.L., Loman N.J. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 2018;19:9–20. doi: 10.1038/nrg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The new sequences generated in this study have been deposited in NCBI GenBank under the accession numbers listed in Supplementary Table S6. Input data used for the phylogenetic analyses are provided in the repository https://doi.org/10.6084/m9.figshare.24270724.v2.