Abstract
Primary/intrinsic and treatment-induced acquired resistance limit the initial response rate to and long-term efficacy of direct inhibitors of the KRASG12C mutant in cancer. To identify potential mechanisms of resistance, we applied a CRISPR/Cas9 loss-of-function screen and observed loss of multiple components of the Hippo tumor suppressor pathway, which acts to suppress YAP1/TAZ-regulated gene transcription. YAP1/TAZ activation impaired the anti-proliferative and pro-apoptotic effects of KRASG12C inhibitor (G12Ci) treatment in KRASG12C-mutant cancer cell lines. Conversely, genetic suppression of YAP1/WWTR1 (TAZ) enhanced G12Ci sensitivity. YAP1/TAZ activity overcame KRAS dependency through two distinct TEAD transcription factor-dependent mechanisms that phenocopy KRAS effector signaling. First, TEAD stimulated ERK-independent transcription of genes normally regulated by ERK (BIRC5, CDC20, ECT2, FOSL1 and MYC) to promote progression through the cell cycle. Second, TEAD caused activation of PI3K-AKT-mTOR signaling to overcome apoptosis. G12Ci treatment-induced acquired resistance was also caused by YAP1/TAZ-TEAD activation. Accordingly, concurrent treatment with pharmacologic inhibitors of TEAD synergistically enhanced KRASG12C inhibitor anti-tumor activity in vitro and prolonged tumor suppression in vivo. In summary, these observations reveal YAP1/TAZ-TEAD signaling as a crucial driver of primary and acquired resistance to KRAS inhibition and support the use of TEAD inhibitors to enhance the anti-tumor efficacy of KRAS-targeted therapies.
Keywords: ERK, Hippo pathway, KRAS, PI3K, TEAD, YAP
Introduction
Mutational activation of the KRAS oncogene occurs at the highest frequencies in lung (~30%), colorectal (~50%) and pancreatic cancers (~95%) (1,2), which comprise the top three leading causes of cancer-related deaths in the United States (3). Until recently, KRAS was considered an undruggable cancer target. However, the seminal discovery of a cryptic binding pocket during a screen for small molecules that selectively targeted one KRAS mutant, KRASG12C (4), set off intensive efforts to develop clinically active KRASG12C-selective inhibitors. In 2021, the first direct KRASG12C-selective inhibitor, sotorasib (AMG510), was approved (5), followed by the 2022 approval of adagrasib (MRTX849) (6), for the treatment of KRASG12C-mutant non-small cell lung cancer (NSCLC).
While the development of KRASG12C inhibitors marked a significant milestone in the field, their clinical efficacy as monotherapy has been severely compromised by primary/intrinsic and treatment-associated acquired resistance. Both initial and long-term responses to G12C-selective inhibitors have fallen short compared to other oncogene-targeted therapies in NSCLC (1,7,8), as indicated by an overall response rate of only 34–43% in NSCLC patients whose tumors harbor KRASG12C mutations, and by progression-free survival ranging from only 6.5 to 6.8 months (5,6,9). Therefore, overcoming both primary and acquired resistance to G12Ci is critical to enhancing the clinical efficacy of these inhibitors.
Retrospective analyses of clinical trial outcomes have provided very limited insight into the molecular basis of primary resistance. Other than mutational inactivation of KEAP1 associated with a reduced response, it remains to be elucidated why overall response rates to mutant-selective inhibitors are below 50% even in patients selected for treatment specifically on the basis of tumor expression of the targeted mutant (5,6,9). Genomic DNA analyses of relapsed patients have provided a more detailed molecular basis for acquired resistance (10–12). To date, gain- or loss-of-function mutations have been reported, involving signaling components upstream or downstream of KRAS, or at the level of KRAS or other RAS isoforms. Collectively, these present a general portrait where reactivation of RAS effector signaling provides a mechanistic basis for acquired resistance. However, potential resistance mechanisms have not yet been identified in approximately 50% of patients who develop acquired resistance to sotorasib or adagrasib monotherapy, indicating that the full complexity of mechanisms of acquired resistance also remains to be elucidated.
To uncover additional genetic modulators of G12Ci sensitivity, we applied a CRISPR-Cas9 loss-of-function screen and identified loss of multiple components of the Hippo tumor suppressor pathway, and therefore activation of the YAP1/TAZ transcriptional coactivators, as a mediator of G12C inhibitor resistance. We established that YAP1/TAZ activation can drive both primary and acquired resistance, through TEAD transcription factor-dependent signaling mechanisms that restore KRAS effector signaling functions in a KRAS-independent manner. We then demonstrated that pharmacologic inhibition of TEAD enhanced G12C inhibitor anti-tumor cell activity in vitro and in vivo. Our findings support TEAD inhibition as a therapeutic combination strategy to enhance the response rate and durability of direct KRAS inhibitor therapies.
Materials and Methods
CRISPR-Cas9 screen
The druggable genome CRISPR-Cas9 library targets 2,240 genes with five unique sgRNAs per gene and 50 sgRNA non-targeting controls, as described previously (13). MIA PaCa-2 cells were infected for 24 hours with lentiviral particles containing the library at an MOI of 0.2, followed by 24 hours in fresh media. Infected cells were selected in puromycin (2 μg/mL) for two days and genomic DNA (gDNA) was harvested (day −7) to determine library coverage. To ensure stable integration of the library at 1,000x coverage, cells were cultured in puromycin (1 μg/mL) for seven days and gDNA harvested (day 0, 9 days post infection). Cells were then grown as adherent cultures (2D) or non-adherent cultures in 0.5% methylcellulose (3D) and treated with either vehicle (DMSO) or G12Ci (MRTX1257, 10 nM) in triplicate. After two weeks of treatment, the concentration of MRTX1257 for both 2D and 3D cultures was increased to 20 nM. gDNA was harvested at four weeks of cumulative treatment using the DNeasy Blood and Tissue Kit (QIAGEN) and analyzed using next generation sequencing on a NextSeq 500 DNA sequencer (Illumina). Relative enrichment/depletion of sgRNAs for each gene was determined using MaGECK analysis (14). Sample quality was assessed using principal component analysis of normalized sgRNA counts. Experimental validation was performed by evaluating essential and non-essential genes (14) using receiver operator characteristic curves.
Cell lines and inhibitors
All cancer cell lines utilized in this study, in addition to relevant information regarding the culture media and purchase/procurement information, are listed in Supplementary Table S1. Briefly, cells were maintained in a humidified chamber (37°C and 5% CO2) and grown in DMEM (Gibco), RPMI1640 (Gibco), or McCoy’s 5a (Corning) medium supplemented with 10% fetal bovine serum (FBS, Gibco) and 0.1% penicillin/streptomycin for no more than 15 passages. Cells were routinely tested for Mycoplasma using MycoAlert (Lonza). Short tandem repeat (STR) profiling was performed on all cell lines. All pharmacological and other inhibitors utilized in this study and their sources are listed in Supplementary Table S2.
2D viability assays
All 2D viability assays were performed by plating cancer cells at 1,000–5,000 cells/well, depending on cell line, in a flat/clear-bottomed 96-well plate (Corning). Following genetic and/or pharmacological treatments, cells were cultured for five to seven days as indicated in the relevant figure legend. Pharmacological treatments were dispensed by a D300e Digital Dispenser (Tecan). Following treatment, viable cells were fluorescently labeled for 20 minutes using Calcein AM (500 nM, Invitrogen) and counted using the SpectraMax i3x Multi-Mode Detection Platform (Molecular Devices). Cell counts were normalized to day zero values determined from an independent culture plate.
Matrigel viability assays
Cells were plated in ice-cold medium composed of 10% Matrigel (Corning) at a density of 10,000 cells per well in low attachment, round-bottomed, white-welled 96-well plates. Cells were then cultured for two days before drug was dispensed using the D300e Digital Dispenser. Following five days of drug treatment, viability was determined using the CellTiter-Glo 3D Cell Viability Assay (Promega) according to the manufacturer’s recommendation, and read using the SpectraMax i3x.
Annexin-FITC/propidium iodide apoptosis assays
Measurements of treatment-induced apoptosis were performed using the TACS® Annexin V-FITC Kit (R&D Systems). Cells were plated at low density in 6-well plates and allowed to grow for five days under the indicated treatment. Detached cells were collected and placed in a collection tube. Attached cells were lifted using TrypLE™ (Gibco) for 15 minutes at 37°C. Collected cells were centrifuged at 500xg for five minutes, washed with 1x ice-cold phosphate-buffered saline (PBS), and incubated for 15 minutes at room temperature in 100 μL of staining solution (1:200 Annexin-FITC, 1x binding buffer (R&D Systems), 1:10 propidium iodide (PI) in H2O). Cells were diluted 1:4 in 1x binding buffer and 10–30,000 cellular events were analyzed via flow cytometry (Cytoflex S, Beckman Coulter). To differentiate cells from debris, FSC-A (x) and SSC-A (y) gating was performed using Cytobank (RRID:SCR_014043). Finally, apoptotic cells were defined as cells positively labelled for Annexin-FITC staining and further defined as either PI-positive or -negative.
Immunoblotting
Following treatment, cells were washed 2x with ice-cold PBS and lysed using RIPA buffer (50 mM TRIS-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease (Roche) and phosphatase inhibitors (Sigma). Cell lysates were scraped into 1.5 mL Eppendorf tubes and incubated on ice for 10–15 minutes with intermittent vortexing. Lysates were centrifuged 18,213xg at 4°C for 10 minutes to pellet cellular debris and organelles. Protein concentrations were normalized following quantification using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific). 4x Laemmli sample buffer (Bio-Rad) supplemented 1:10 with beta-mercaptoethanol (Sigma) was added to protein-normalized lysates. Samples then were heated at 95°C for 10 minutes. Standard immunoblotting techniques were followed. Buffers used were: running buffer (0.3% Trizma base, 1.44% glycine, 0.1% SDS in ddH2O), transfer buffer (3.03% Trizma base, 14.41% glycine, 20% methanol in ddH2O), and blocking buffer (5% milk in Tris-buffered saline with 0.05% Tween 20, TBS-T). Following transfer to PVDF membranes (Thermo Fisher), membranes were imaged using the ChemiDoc™ MP Imaging System (Bio-Rad) and analyzed using Image Lab (Bio-Rad) and FIJI (ImageJ, RRID:SCR_003070). Primary and secondary antibodies used in this study (Supplementary Table S3) were dissolved in five or three percent bovine serum albumin (BSA) in TBS-T, respectively.
RNA extraction, cDNA synthesis, and RT-qPCR
Cells were plated, treated, and collected as indicated. Upon collection, media was aspirated, and cells were washed 2x with ice cold PBS. RNA was either extracted immediately or stored (−80°C) for later analyses. RNA was extracted into molecular grade ddH2O using the RNAeasy Mini Kit (Qiagen) according to the manufacturer’s guidelines. RNA concentration and purity were determined using the NanoDrop 2000c (Thermo Fisher). cDNA was generated using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). PCR product (cDNA) was diluted 10x in molecular grade ddH2O. Real-time quantitative PCR was performed on the QuantStudio 6 Flex (Thermo Fisher) using the TaqMan PCR master mix (Applied Biosciences), FAM-labelled target probes, and the VIC/TAMRA-labelled endogenous control probe, β-actin. All probes utilized in this study are included in Supplementary Table S4. All samples were run in technical triplicates with delta-delta CT scores calculated for target and control probes in each sample. All samples were normalized internally to endogenous control and experimentally to vehicle controls (DMSO, NS).
siRNA transfections
Cells were reverse transfected with 10 nM per siRNA and Lipofectamine RNAiMAX (Thermo Fisher) according to the manufacturer’s instructions. siRNA and OptiMEM + RNAiMAX solutions were prepared separately, combined, and then allowed to equilibrate for at least 15 minutes at room temperature. Following incubation, the siRNA-OptiMEM-RNAiMAX solution was transferred to dishes, onto which cells were plated. Cells remained in knockdown solution until collection. For all siRNA constructs used, knockdown was confirmed by immunoblot, and for each gene target, two distinct siRNA constructs were utilized. Finally, for studies combining drug and siRNA treatment, cells were reverse transfected with siRNA 24 hours before the indicated drug treatments. All siRNA oligonucleotides utilized are listed in Supplementary Table S5.
Colony formation assays
Cells were plated in 12-well plates at 1,000–5,000 cells per well in 1 mL of medium. The following day (day 1), medium was replaced and 1 mL of medium containing drug or the equivalent volume of vehicle (DMSO) was added. Cells were allowed to grow for 10 days and medium was changed every three days. Following treatment, cells were washed with PBS, and stained with 0.05% crystal violet in 4% formaldehyde/PBS. Images were taken using the Typhoon™ FLA 9500 Biomolecular Imager (GE Healthcare), and percent cell coverage was determined using FIJI. All samples were normalized to their respective DMSO controls.
Luciferase reporter assay
Cells were plated at 2,000–5,000 cells per well in a flat, clear-bottom, white-walled, 96-well dish (Corning). The next day (day 1), medium was discarded, and fresh treatment (inhibitor) medium was added. On day 2, TransIT-2020 (Mirus Bio) was used according to the manufacturer’s guidelines to transfect 150 ng of 8xGTIIC-luciferase (15) (1:1 ratio of DNA:transfection reagent) per well. At forty-eight hours post transfection (day 4), viability was measured using Calcein AM (500 nM, 20 min). Immediately afterwards, 50 μL of 2x luciferin (Bright-Glo™ Luciferase Assay System, Promega) was added to each well, incubated for five minutes, and luminescence was read using the SpectraMax i3x. Within each well, luminescence values were normalized to cell count (viability) and vehicle control (DMSO).
Fluorescence, immunofluorescence
Cells were plated on glass-bottomed dishes (MatTek Corporation). For collection, wells were aspirated and washed 2x with PBS, then fixed for 20 minutes at room temperature using 4% paraformaldehyde in PBS. Following fixation, cells were washed and then permeabilized with 0.5% Triton/PBS for five minutes. Cells were then blocked for 30 minutes in 2% BSA in PBS at room temperature. YAP1/TAZ primary antibody (Santa Cruz Biotechnology, 1:50) was incubated at room temperature for one hour in 1% BSA in PBS. Cells were then washed and incubated for 45 minutes at room temperature in goat anti-mouse secondary antibody (Alexa Fluor™ 488, Thermo Fisher, 1:100) and Alexa Fluor™ 647 phalloidin (Thermo Fisher, 1:200) in 0.5% BSA/PBS. Finally, cells were washed 3x in PBS, with DAPI (Thermo Fisher, 1:10,000) included for the last five-minute wash. Cells were imaged on a Zeiss LSM 900 confocal laser scanning microscope with a PlanApo 63X/1.4 oil objective, and Z-stacks were collected for each image, or on an EVOS M7000 wide-field microscope with a 40× 0.65 NA objective. Images were processed using FIJI and a Z-projection image was generated.
Construction of plasmids and creation of stable cell lines
pLX304 Luciferase-V5 blast was a gift from Kevin Janes (Addgene plasmid # 98580; http://n2t.net/addgene:98580; RRID:Addgene_98580) (16). YAP1-V5 in pLX304 was a gift from William Hahn (Addgene plasmid # 42555; http://n2t.net/addgene:42555; RRID:Addgene_42555) (17). HA-TAZ (Addgene plasmid # 32839; http://n2t.net/addgene:32839; RRID:Addgene_32839) and HA-TAZS89A (Addgene plasmid # 32840; http://n2t.net/addgene:32840; RRID:Addgene_32840) were gifts from Kun-Liang Guan (18), and pInducer20 EGFP-TEADi was a gift from Ramiro Iglesias-Bartolome (Addgene plasmid # 140145; http://n2t.net/addgene:140145; RRID:Addgene_140145) (19). psPAX2 (Addgene plasmid # 12260; http://n2t.net/addgene:12260; RRID:Addgene_12260) and pMD2.G (Addgene plasmid # 12259; http://n2t.net/addgene:12259; RRID:Addgene_12259) were gifts from Didier Trono. pDONR223 was purchased from the Lineberger Tissue Culture Facility at UNC Chapel Hill. Supplementary Table S6 contains a full list of plasmids utilized or created during this study. Using Gateway cloning (BP reaction, Invitrogen), YAP1 was moved from the pLX304 to the pDONR223 backbone. Site-directed mutagenesis was performed using Phusion High Fidelity Polymerase (New England Biolabs (NEB)), and the primers listed in Supplementary Table S7, to create mutants of YAP1 and TAZ. Following sequence verification, AGE1-YAP1 FWD and HA-tagged-MLU1-YAP1 REV primers were used to amplify YAP1 cDNA, which was subsequently ligated into the AGE1- and MLU1-digested pInducer-EGFP backbone (20). WWTR1 cDNA (encodes TAZ) was amplified using HA-tagged-TAZ FWD and TAZ REV primers compatible with Gibson cloning. WWTR1 cDNA was again cloned into the digested pInducer-EGFP backbone, this time using the NEBuilder® HiFi DNA Assembly Master Mix (NEB). All plasmids were sequenced upstream, downstream, and within the insertion sites to ensure the entire cDNA insertion was correct.
Second-generation lentiviral packaging was utilized to create all stable cell lines. To create lentivirus, early passage HEK293T cells were plated at 106 cells/flask in a T25 flask with 5 mL DMEM + 10% FBS. Twenty-four hours post plating, medium was changed to 3 mL of DMEM + 10% FBS, 24 μL of Fugene 6 (used according to the manufacturer’s guidelines), 3 μg of psPAX2 (packaging plasmid), 1 μg of pMD2.G (envelope plasmid), 4 μg of transfer plasmid (containing ORF of interest), and 400 μL of OptiMEM. Following overnight incubation, the culture medium was changed to 5.5 mL of DMEM + 20% FBS and cells were allowed to incubate for an additional 48 hours. Following final incubation, viral supernatant was harvested and filtered using a 0.45-micron filter. Cleared viral supernatant was then either added to cells (discussed next) or stored at −80°C. To create stable cell lines, parental cells were plated in T25 flasks at approximately 75% confluency. The next day, viral supernatant and polybrene (8 μg/mL) was added to cells and allowed to incubate overnight. Following overnight incubation, virus was aspirated and fresh medium was added to the cells. The following morning, selection medium was added to the cells. Twenty-four hours later, cells were lifted and re-plated in selection medium until the “kill plate” viability reached zero.
Generation of pInducer20-EGFP-TEADi cell lines
Cells stably expressing the pInducer20-EGFP-TEADi construct (19) were created as described above. Twenty-four hours before collection, 1 μg/mL of doxycycline was added to cells to induce EGFP expression. The next day, 15–25 million cells were collected and spun down at 300xg for five minutes. Supernatant was discarded, and cells were resuspended in 3 mL of sorting medium (0.1% penicillin-streptomycin, 50 μg/mL gentamicin, 5 μg/mL Plasmocin®, 13% FBS, 1 μg/mL doxycycline, 1 μg/mL propidium iodide, and 0.1 mM DNAse in Hank’s balanced salt solution (HBSS)). Cells resuspended in sorting medium were passed through a 35-micron filter into polystyrene tubes (Corning) and kept on ice until sorting. During sorting, the top 10% of cells expressing EGFP were collected (~500K cells) into collection medium (same as sorting medium, but without doxycycline and propidium iodide).
Cancer Cell Line Encyclopedia (CCLE) and DepMap data analysis
All transcriptional (EH7525), mutational (EH7526), and Chronos dependency (EH7523) data were accessed from ExperimentHub (22Q1) using the depmap (21) package in R-studio. Mutation calls were used to filter for cell lines containing the G12C point mutation in KRAS.
To identify signaling pathways associated with primary G12Ci-resistance, KRASG12C-mutant cell lines were stratified according to their sensitivity to G12Ci. Differential expression was performed on raw counts (downloaded from Depmap.org) between G12Ci-sensitive and -resistant cell lines using EdgeR (22). Gene set enrichment analysis (GSEA, RRID:SCR_003199) was performed on differentially expressed genes using the C6 and Hallmark signatures (MSigDB) and select YAP1/TAZ signatures (Cluster 2 in Pham et al. (2021) YAP1/TAZ (23) and Wang et al. (2018) YAP1/TAZ (24)), using clusterProfiler (25). Individual GSEA plots were generated for indicated gene sets using gseaplot2 (enrichplot).
Single sample gene set enrichment (ssGSEA) was performed using the ssGSEA-gpmodule repository available on the GSEA-MSigDB github page. RNA expression data for KRASG12C-mutant cell lines was filtered for genes with transcripts per million (TPM) values above 0.25 before being evaluated using ssGSEA. RNA expression was rank normalized by ssGSEA, and a default weight of 0.75 was used with 1,000 permutations. Resulting net enrichment scores (NES) were used to compare to sum of YAP1 and WWTR1 Chronos gene dependencies reported for each cell line in DepMap data.
RNA sequencing and analyses
The RNA-Seq experiments for KRAS-mutant PDAC cell lines treated with shRNA KRAS knockdown or the ERK inhibitor (ERKi) SCH772984 are described elsewhere (26). HPAC cells were removed from the shRNA KRAS RNA Seq dataset due to the extremely poor KRAS knockdown efficiency observed in this cell line. For both data sets, TrimGalore (v0.4.5) (27) was used for quality control and adapter trimming via FastQC (28) and Cutadapt (29). We retained only high-quality paired end reads (> 35 base pairs for each read with >28 Sanger/Illumina 1.9 quality score). Read mapping to the human genome was conducted with STAR aligner (v2.7.8a, RRID:SCR_004463) (30) and Gencode (v30) (31) basic transcriptome (GRCh38.p12). The quantification and summarization of transcripts to gene level was accomplished with Salmon (v0.11.3) (32) and tximport (v1.22.0) (33), respectively. BiomaRt (v2.50.3, RRID:SCR_019214) (34) was used for annotations and data were managed and normalized via TMM with edgeR (v3.36.0) (22). Genes were filtered to include only protein coding, non-mitochondrial and non-Y chromosome genes prior to differential expression analyses with the glmQLFTest function (EdgeR). Multiple testing was addressed using FDR (false discovery rate) adjusted p-values via the Benjamini and Hochberg method.
Identification of KRAS-ERK and YAP1/TAZ-TEAD co-regulated genes
Transcriptional signatures driven by KRAS-ERK and YAP1/TAZ were compared using previously described (above) RNA-sequencing data following KRAS knockdown (shRNA) and 24 hours of ERKi treatment (26) in PDAC cell lines, and published YAP1/TAZ signatures (Cordenonsi YAP Conserved Signature (35), Pham et al. (2021) YAP1/TAZ (cluster 2) (23), and Wang et al. (2018) YAP1/TAZ (24)). Additionally, given the reported ability of both KRAS (36) and YAP1 (37) to regulate MYC expression, this gene was added manually. Chronos dependency scores (DepMap, EH7523, described above) were then used to stratify co-regulated genes according to dependency within a panel of KRASG12C-mutant cancer cell lines.
Generation of adagrasib-resistant lines
Adagrasib-resistant MIA PaCa-2 and H358 cells able to proliferate in 2 and 1 μM adagrasib, respectively, were generated over several months by sequential dose escalation, whereby high dose adagrasib was added until cells began to proliferate, at which point drug concentration was doubled or tripled until maximal doses were achieved. Age-matched control cells grown in equivalent concentrations of DMSO were passaged alongside resistant cell lines. Unless otherwise noted, parental and resistant cell lines were plated and grown in DMSO or adagrasib, respectively.
Mouse xenograft studies
All the procedures related to animal handling, care, and treatment were approved by the Institutional Animal Care and Use Committee (IACUC) of WuXi AppTec or Crown Bioscience, Inc., following the guidance of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Adagrasib (MRTX849) was formulated in vehicle 1 (10% Captisol® in 50 mM citrate buffer, pH 5.0), and TEAD inhibitor (VT104 or VT3989) was formulated in vehicle 2 (5% DMSO + 10% Solutol® + 85% D5W; D5W = 5% glucose). The formulated compounds were orally administered once a day, every day.
For the SW837 CDX model study, female NOD SCID mice were implanted subcutaneously at the right flank with SW837 tumor cells (5 × 106) in 0.2 mL of a 1:1 mixture of PBS and Matrigel. Tumor-bearing animals were randomized, and treatment was started when the average tumor size reached 123 mm3 (n = 8 per treatment group). For the CR6243 PDX study, female BALB/c nude mice were implanted subcutaneously at the right flank with primary human tumor xenograft model CR6243 tumor fragment (2–3 mm in diameter). Tumor-bearing animals were randomized, and treatment was started when the mean tumor size reached approximately 153 mm3 (n = 10 per treatment group). Tumor size and animal weights were monitored twice weekly. Tumor volume in mm3 was calculated using the formula: V = 0.5 a x b2, where a and b are the long and short diameters of the tumor, respectively.
Statistical analysis
Data are represented as mean or median ± SEM or SD (specified within each figure legend). Statistical significance was determined using Student’s t test, or one-way or two-way ANOVA. Tukey or Dunnett multiple comparisons testing following one-way or two-way ANOVA was utilized as appropriate and described in figure legends. All statistical analysis was performed using PRISM 9 (Graphpad Software, RRID:SCR_000306). Significance values were labelled in the figure or clearly stated in the figure legend.
Data availability
All plasmids generated from this study are to be deposited in Addgene. The shKRAS and ERKi RNA sequencing datasets analyzed in this study were obtained from the EMBL-EBI European Nucleotide Archive (ENA) database at http://www.ebi.ac.uk/ena/ under accession numbers PRJEB25797 and PRJEB25806, respectively. All CRISPR-Cas9 loss-of-function dependency data, and CCLE transcriptional and mutational data (22Q1) were obtained from the Broad Institute’s Cancer Dependency Map (DepMap) at https://depmap.org/portal/download/all/ or through the depmap (21) package in R-studio. All additional data generated to support the findings of this study are available from the corresponding author upon request.
Code availability
No unique code was generated from this study.
Results
CRISPR-Cas9 screen identifies loss of Hippo pathway components as mediators of G12Ci resistance
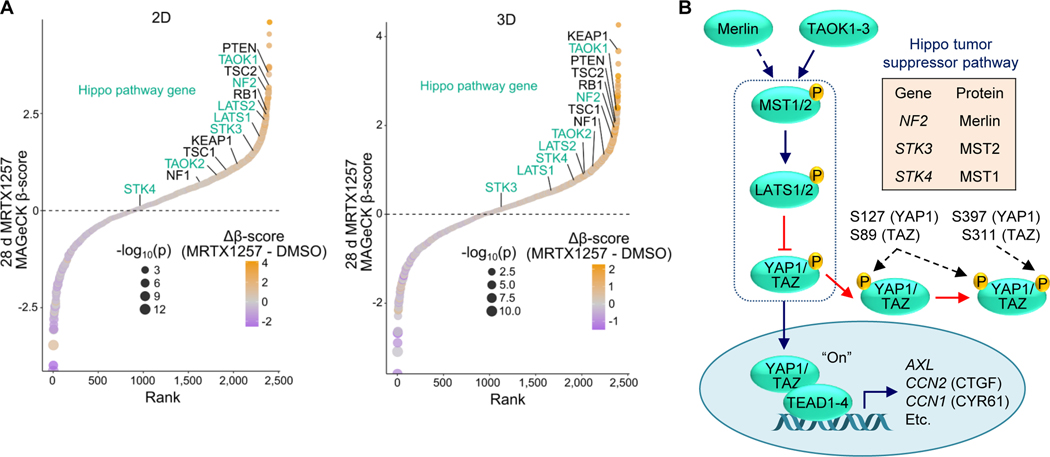
To identify genes that modulate sensitivity to direct pharmacologic inhibition of KRASG12C, we performed a CRISPR-Cas9 loss-of-function screen targeting major cancer signaling networks (13). We treated the KRASG12C-mutant pancreatic ductal adenocarcinoma (PDAC) cell line, MIA PaCa-2, with the KRASG12C-specific covalent inhibitor MRTX1257 (herein designated G12Ci), an analogue of the approved G12C-specific inhibitor adagrasib/MRTX849 (38). Recent studies determined that mutant KRAS-dependent cancer cell growth is better modelled by anchorage-independent three-dimensional (3D) growth conditions (39). Additionally, 3D cell culture CRISPR-Cas9 screens more closely model genetic dependencies found in vivo compared to 2D (40). Therefore, we performed our screen in both 2D anchorage-dependent and 3D anchorage-independent growth conditions. Following four-week inhibitor treatment, samples were sequenced and the relative enrichment and depletion of sgRNAs were determined (Supplementary Fig. 1A-C).
We found that genetic dependencies in 2D and 3D conditions without inhibitor treatment were highly similar (Supplementary Fig. 1D). Likewise, G12Ci-selective genetic dependencies were highly correlated in 2D and 3D conditions (Supplementary Fig. 1E). Among the genes with enriched sgRNAs, we found tumor suppressors such as PTEN, NF1, KEAP1 and RB1 that were recently reported to be lost in initially G12Ci-responsive patients who later relapsed due to acquired resistance (10–12) (Fig. 1A). In addition to these previously identified tumor suppressors, we identified the loss of nearly every component of the core Hippo tumor suppressor pathway, namely NF2, LATS1/2, TAOK1/2 and STK3/4 (Fig. 1A and B).
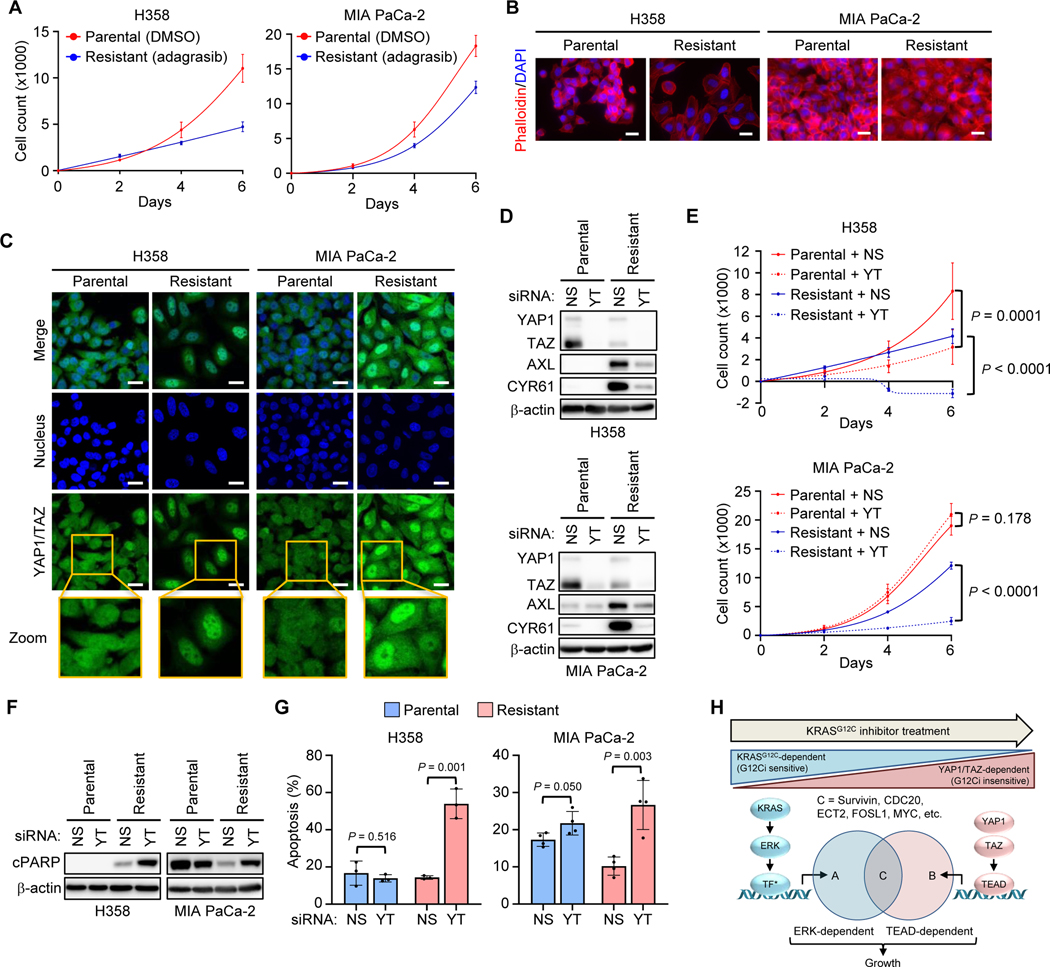
Figure 1.
CRISPR-Cas9 screen identifies loss of Hippo pathway components as mediators of G12Ci resistance. A, CRISPR/Cas9 loss-of-function screen performed in MIA PaCa-2 cells treated with the G12Ci, MRTX1257, and plated in 2D (left) or 3D (0.5% methylcellulose, right) for 28 days. Tumor suppressor genes previously identified in patients or preclinically are in black, while components of the Hippo pathway are in turquoise. Genes are ranked by relative enrichment/depletion (β-scores) in the G12Ci condition. Relative enrichment/depletion in G12Ci relative to DMSO condition (Δβ-scores) are also shown. B, Schematic of the Hippo tumor suppressor pathway, with key inhibitory LATS1/2-mediated phosphorylation sites on YAP1/TAZ denoted.
YAP1 and TAZ exhibit equivalent functions in driving resistance to pharmacologic inhibition of KRASG12C
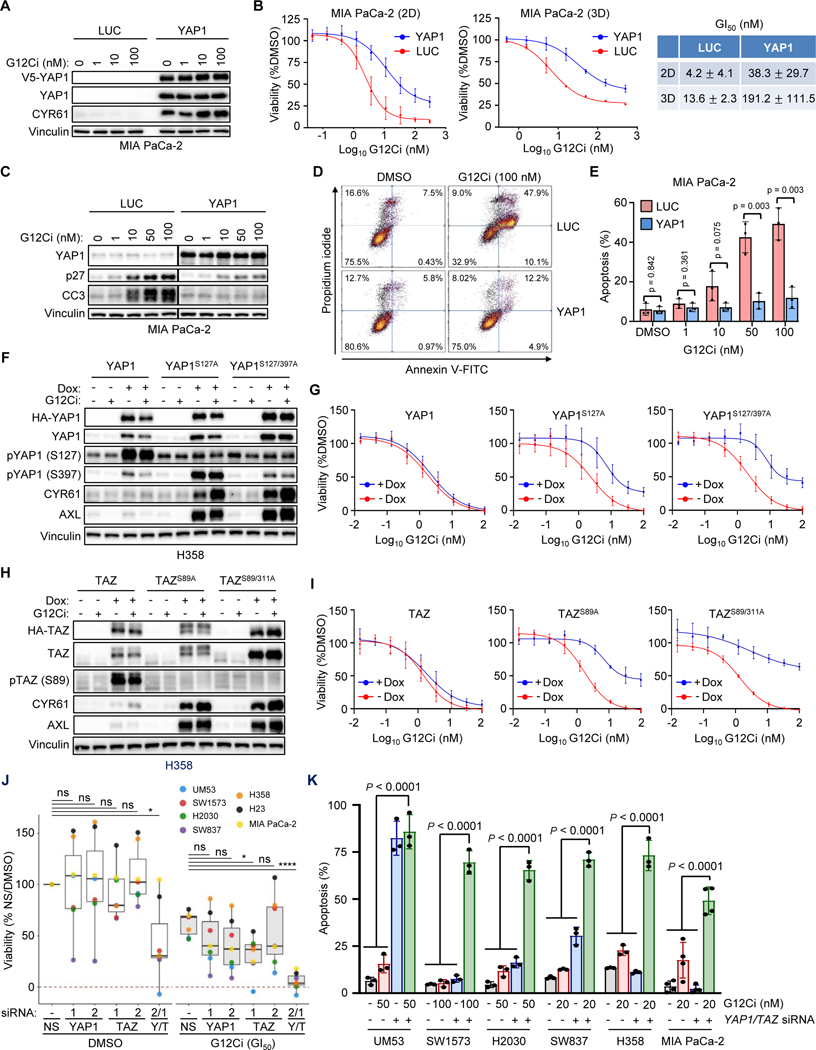
The Hippo signaling cascade negatively regulates the highly related transcriptional co-activator paralogs, YAP1 and TAZ (41) (Fig. 1B). Previous studies found that YAP1 overexpression or activation overcame cancer cell dependency on mutant KRAS (42,43). Therefore, we hypothesized that YAP1 activation can similarly drive resistance to treatment with pharmacologic inhibitors of KRASG12C. To assess YAP1-driven resistance to G12Ci treatment, we established MIA PaCa-2 cell lines that stably overexpressed either V5 epitope-tagged YAP1 wild-type (WT) or the luciferase (LUC) negative control (Fig. 2A). Supporting increased YAP1 activity, we also observed elevated expression of the canonical YAP1 gene target protein, CYR61 (44).
Figure 2.
YAP1 and TAZ exhibit equivalent functions in driving resistance to pharmacologic inhibition of KRASG12C. A, Immunoblot of ectopic V5 epitope-tagged YAP1WT- and LUC-overexpressing MIA PaCa-2 cells following 24 hours of G12Ci treatment at indicated concentrations. B, Cell viability assay following five days of G12Ci treatment of cells as in A, grown in 2D (plastic, left) and 3D (Matrigel, right) conditions. Data represent the mean ± SD for 3+ (2D) and 2 (3D) independent biological replicates. G12Ci concentrations that reduce viability by 50% (GI50) are shown (right) as mean ± SD. C, Immunoblot of cells as in A. LUC and YAP1 immunoblots were run and imaged together, so all conditions can be directly compared. D, FACS gating strategy for 5-day apoptosis assay following G12Ci treatment (100 nM) of cells as in A. E, Quantification of 5-day apoptosis assay of cells as in A across indicated G12Ci concentrations. Mean ± SD is shown for n = three biological replicates; two-tailed unpaired t-test. F-I, YAP1/TAZ overexpression was induced by Dox (1 μg/mL) 24 hours prior to the indicated treatment. F, Immunoblot of H358 cells overexpressing YAP1 constructs treated for 24 hours with DMSO or G12Ci (20 nM). G, Five-day viability assays following G12Ci treatment of cells as in F. Each data point represents the mean ± SD for three or more biological replicates. H, Immunoblot of H358 cells overexpressing designated TAZ constructs treated for 24 hours with DMSO or G12Ci (20 nM). I, Five-day viability assay following G12Ci treatment of cells as in H. Data points represent the mean ± SD for three biological replicates. J, Five-day viability assay following individual or concurrent YAP1/TAZ knockdown (Y2, T1) in the presence of DMSO or ~GI50 doses of G12Ci (H358: 1 nM; MIA PaCa-2: 5 nM; SW837: 10 nM; UM53, H2030, H23: 50 nM; SW1573: 100 nM). Knockdown was performed 24 hours prior to addition of DMSO or G12Ci. Viability is normalized to DMSO/NS-treated control cells. Data points represent the median viability across three or more biological replicates for each indicated cell line and condition. Statistics represent one-way ANOVA with Dunnett post hoc multiple comparisons testing: *, P < 0.05; ****, P < 0.0001; ns, not significant. K, Five-day apoptosis assay (annexin V-FITC/propidium iodide) in indicated cell lines following treatment with G12Ci, siYAP1/TAZ (Y2, T1), or the combination. Knockdown with non-targeting control (NS, “-“) or YAP1/TAZ (YT, “+”) was performed 24 hours prior to addition of DMSO or G12Ci. Statistics represent one-way ANOVA with Tukey post hoc multiple comparisons testing.
We determined that ectopic YAP1 overexpression attenuated the G12Ci-mediated reduction in cell viability under both 2D and 3D growth conditions, consistent with YAP1-driven resistance to G12Ci (Fig. 2B). We next investigated whether YAP1 overexpression altered G12Ci-induced changes in cell cycle regulation and apoptosis. While G12Ci treatment induced accumulation of cyclin-dependent kinase inhibitor p27 KIP1, a driver of G1 cell cycle arrest, YAP1-overexpressing cells reduced this accumulation (Fig. 2C). Similarly, accumulation of the apoptosis marker cleaved caspase 3 (CC3) was reduced in YAP1-overexpressing cells. YAP1 overexpression also almost completely prevented G12Ci-induced apoptosis, as measured by annexin V-FITC/propidium iodide staining (Fig. 2D and E). Taken together, we found that overexpression of YAP1 attenuated G12Ci-mediated growth suppression in MIA PaCa-2 cells, in part through reduced cell cycle arrest and apoptosis.
We next extended our analysis to a second cancer type, utilizing the non-small cell lung cancer (NSCLC) cell line H358, which is both KRASG12C-mutant and G12Ci-sensitive. In contrast to MIA PaCa-2 cells, stable overexpression of V5-YAP1 WT did not significantly increase expression of the YAP1 target CYR61 or attenuate G12Ci-induced growth inhibition, and caused only limited reduction in apoptosis (Supplementary Fig. 2A-C). Addressing a potential basis for this, we found high levels of YAP1 phosphorylation at two LATS-mediated phosphorylation sites (S127 and S397) that drive YAP1 inactivation (Supplementary Fig. 2A). LATS phosphorylation at S127 promotes YAP1 cytoplasmic sequestration and at S397 creates a phosphodegron that signals for β-TrCP-driven E3 ligase-mediated degradation (44).
We speculated that, despite high YAP1 overexpression, Hippo pathway activity may inactivate YAP1 in these cells, providing a possible basis for the absence of increased YAP1 transcriptional activity. To evaluate this possibility, we utilized two activated YAP1 mutants: a single (YAP1S127A) and a double (YAP1S127/397A) phosphodeficient mutant, each of which is refractory to upstream Hippo-mediated pathway phosphorylation and inactivation (44). We established H358 cell lines stably infected with doxycycline (Dox)-inducible lentivirus expression vectors encoding HA epitope-tagged WT YAP1 or the constitutively activated YAP1 mutants. Dox transiently (24 hours) induced elevated levels of WT and phosphodeficient YAP1, the latter of which were not phosphorylated at their corresponding mutated (formerly serine) residues (Fig. 2F). As expected, because the YAP1 mutants could not be phosphorylated and thereby inactivated, cell lines expressing YAP1S127A and YAP1S127/397A, but not YAP1 WT, exhibited high expression of CYR61 and of a second YAP1 transcriptional target, AXL, and significantly attenuated G12Ci-mediated growth inhibition (Fig. 2G; Supplementary Fig. 2D). Additionally, activated YAP1 reduced G12Ci-induced p27 accumulation and apoptosis (Supplementary Fig. 2E and F). We conclude that loss of Hippo pathway regulation together with YAP1 overexpression are both required for YAP1-mediated resistance to G12Ci in H358 cells.
Although YAP1 and TAZ are commonly considered functionally redundant, the two paralogs interact with both shared and specific transcription factors, control nonidentical transcriptional programs, and exhibit distinct regulation and biological activities (45). While YAP1 has been described as a bypass mechanism to overcome KRAS addiction (42,43), a role for TAZ in this context has not been described. We therefore determined if TAZ activation can also drive resistance to G12Ci. We established H358 cells stably infected with expression vectors encoding Dox-inducible WT TAZ and TAZ mutants with amino acid substitutions at residues analogous to the Hippo-insensitive YAP1 mutants, TAZS89A and TAZS89/311A (Fig. 2H). Transient (24 hours) Dox-induced expression of mutant but not WT TAZ resulted in high expression of AXL and CYR61, indicating TAZ activation. Whereas expression of TAZ WT did not alter G12Ci sensitivity, TAZS89A caused partial resistance and TAZS89/311A drove near-complete resistance to G12Ci (Fig. 2I; Supplementary Fig. 2G). Thus, activation of either YAP1 or TAZ alone can drive resistance to pharmacologic inhibition of KRASG12C.
To further determine if YAP1 and TAZ share potentially redundant roles in driving resistance to G12C inhibition, we next examined the consequences of suppression of YAP1 and/or TAZ protein expression on G12Ci sensitivity. We observed that siRNA-mediated knockdown of YAP1 or WWTR1 (TAZ) alone in a panel of seven KRASG12C-mutant cell lines partially reduced CYR61 expression, ranging from 11–90%, with maximal inhibition observed upon concurrent knockdown of YAP1 and TAZ (Supplementary Fig. 2H). Interestingly, we observed cell line variation in the relative importance of YAP1 and TAZ to stimulate CYR61 expression, in that YAP1 was more significant in SW1573 and SW837 cells and TAZ was the predominant driver in UM53 cells.
We next evaluated if YAP1 and/or TAZ knockdown decreased cell viability and induced apoptosis alone or in combination with G12Ci. We found that siRNA suppression of YAP1 or TAZ alone had variable effects on cell viability across cell lines, with maximal inhibition observed upon concurrent YAP1 and TAZ knockdown (Fig. 2J). Strikingly, concurrent YAP1 and TAZ knockdown sensitized all cell lines to G12Ci treatment, even in cell lines such as MIA PaCa-2 and H23, where YAP1/TAZ knockdown had no effect on cell viability under basal conditions. The ability of YAP1/TAZ knockdown to enhance G12Ci efficacy was observed along the entire dose-response curve for both G12Ci/MRTX1257 and adagrasib/MRTX849 (Supplementary Fig. 2I). When we extended our analysis to measure apoptosis, we found that G12Ci or YAP1/TAZ knockdown alone induced varying levels, whereas the combination resulted in significant apoptosis in all cell lines (Fig. 2K). We conclude that YAP1 and TAZ can serve overlapping and distinct roles in driving resistance to G12Ci.
YAP1 and TAZ drive resistance to G12Ci independent of ERK
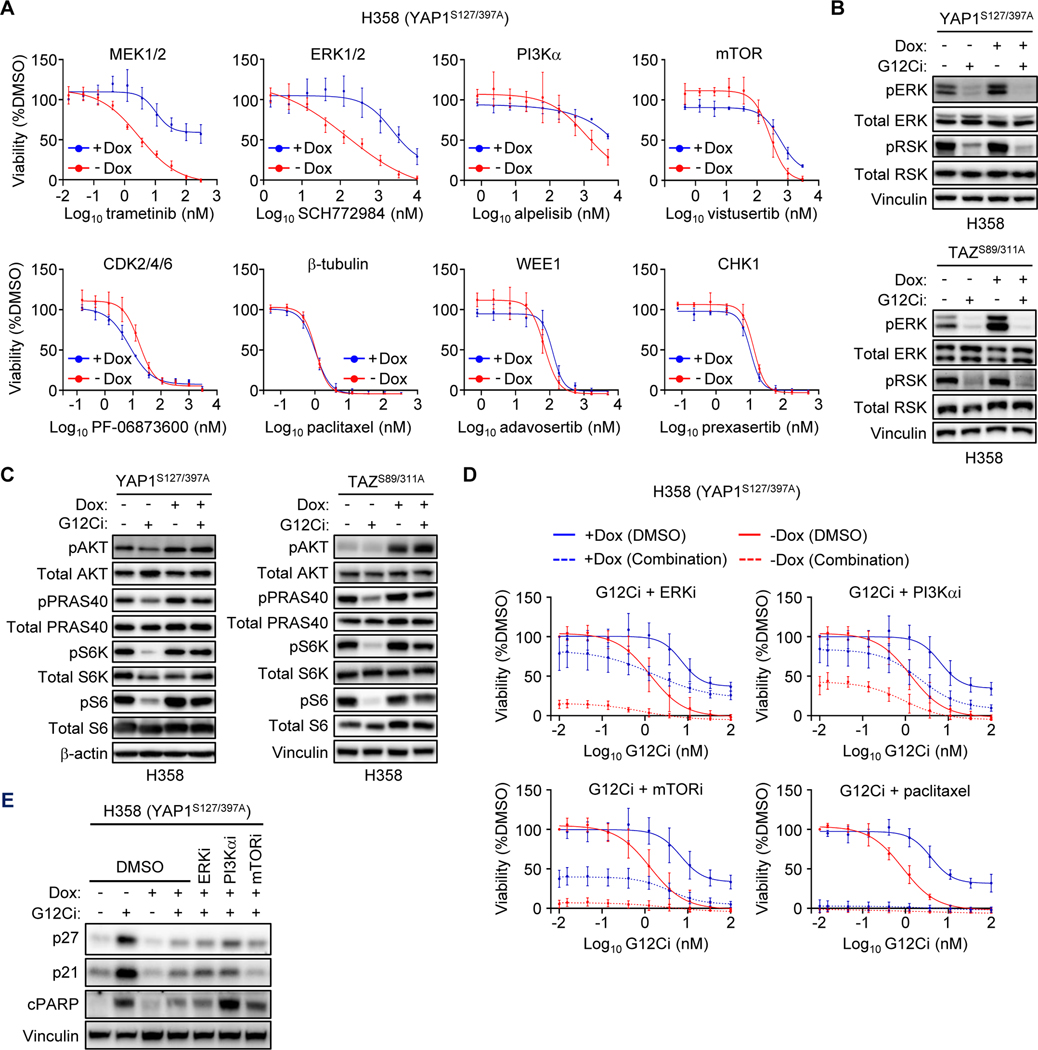
We next investigated the mechanism by which YAP1/TAZ activity drives resistance to G12Ci. The Hippo pathway has been described to modulate resistance to a diversity of mechanistically distinct anti-cancer drugs (46,47). Therefore, we evaluated whether YAP1/TAZ activation in KRASG12C-mutant cancer cells drove resistance to additional chemotherapeutic drugs that target cellular components related to or distinct from KRAS. We determined the sensitivity of H358 cells expressing Hippo-independent YAP1S127/397A to inhibitors of RAS effector signaling pathways (ERK, PI3Kα, or mTORC1/2), cell cycle progression (CDK2/4/6), DNA damage checkpoint (WEE1 and CHK1) and mitosis (β-tubulin) (Fig. 3A). We found that active YAP1 reduced H358 sensitivity to inhibitors of MEK and ERK, but not to inhibitors of tubulin, cell cycle, DNA damage, or the PI3K-mTOR pathway. Similar results were also seen in MIA PaCa-2 cells ectopically expressing V5-YAP1 WT (Supplementary Fig. 3A), consistent with their increased sensitivity to WT YAP1 compared to H358 cells. H358 cells expressing Dox-inducible activated TAZS89A/S311A also showed reduced sensitivity to ERKi (Supplementary Fig. 3B). Collectively, these data suggest that YAP1/TAZ-driven resistance is selective and mechanistically related to the KRAS-ERK MAPK signaling pathway.
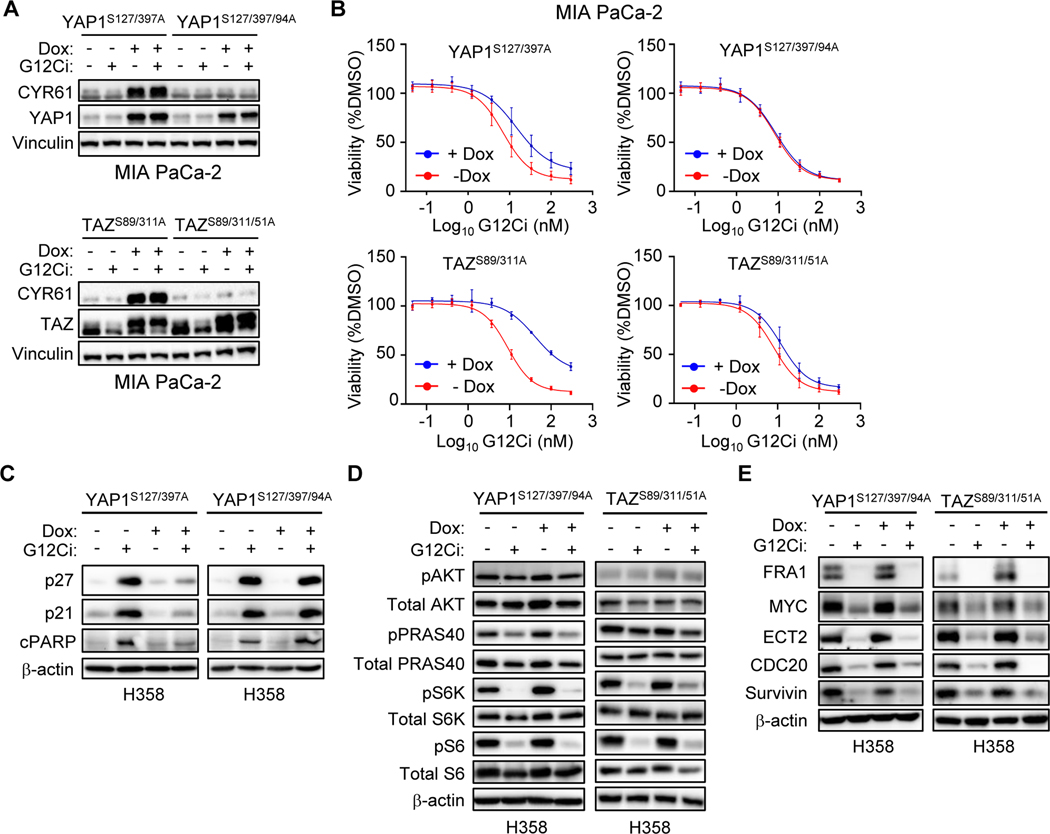
Figure 3.
YAP1 and TAZ drive resistance to G12Ci independent of ERK. A-E, Expression of activated YAP1 and TAZ in H358 cells was induced by Dox (1 μg/mL) 24 hours prior to the indicated treatments. A, Five-day viability assay in YAP1S127/397A overexpressing H358 cells treated with the indicated inhibitors. Each data point represents the mean ± SD for three biological replicates. B, Immunoblot to detect ERK inhibition in YAP1S127/397A and TAZS89/311A overexpressing H358 cells following 24 hours of treatment with DMSO or G12Ci (20 nM). C, Immunoblot to detect inhibition of PI3K signaling as in B. D, Five-day viability assay of YAP1S127/397A overexpressing H358 cells treated with increasing doses of G12Ci alone or in combination with ERKi (SCH772984, 1 μM), PI3Kαi (alpelisib, 1 μM), mTORC1/2i (vistusertib, 1 μM), or paclitaxel (10 nM). Data points represent the mean ± SD for three or more biological replicates. Some combinations displayed in different panels were performed alongside the shared G12Ci treatment alone (−/+ Dox). E, Immunoblots in cells overexpressing YAP1S127/397A and treated for 24 hours with DMSO or G12Ci (20 nM) alone or in combination with inhibitors as in D. Blots are from the same lysates as Supplementary Fig. S3C.
Furthermore, our finding that YAP1/TAZ activation drove selective resistance to inhibitors of KRAS-ERK signaling is consistent with a mechanism where YAP1/TAZ activation causes RAS-independent activation of ERK. However, in H358 cells expressing activated YAP1S127/397A or TAZS89/311A, we found that G12Ci still suppressed pERK and phosphorylation of the ERK substrate RSK (pRSK) (Fig. 3B). These data show that activated YAP1/TAZ drives resistance to G12Ci by a mechanism independent of ERK reactivation.
The PI3K-AKT-mTOR signaling network comprises a second effector pathway important in maintaining KRAS-dependent cancer growth (48,49). Additionally, PI3K signaling can drive resistance to KRAS-ERK inhibitors (1,2). We therefore investigated whether YAP1/TAZ activation can render G12Ci refractory to inhibition of PI3K effector signaling. G12Ci treatment reduced the levels of phosphorylated and activated substrates of PI3K (pAKT), AKT (pPRAS40), mTORC1 (pS6K) and S6K (pS6) in control (-Dox) but not Dox-stimulated H358 cells expressing activated YAP1S127/397A or TAZS89/311A (Fig. 3C). YAP1 has been shown to activate PI3K-AKT-mTOR signaling through a variety of mechanisms (50–52).
Addressing the significance of PI3K signaling in resistance, we determined that concurrent inhibition of PI3K or mTORC1/2 partially (40–50%) reversed the G12Ci resistance caused by activated YAP1S127/397A (Fig. 3D). Interestingly, concurrent PI3K and to a lesser degree mTOR, but not ERK inhibition, restored G12Ci apoptosis but not cell cycle inhibitory activity (Fig. 3E; Supplementary Fig. 3C). Thus, YAP1/TAZ activation of PI3K-AKT-mTOR signaling appears to drive KRAS-independence in part through promotion of cell survival. We conclude that YAP1 can drive KRASG12C-independent activation of PI3K-mTOR but not ERK to cause partial resistance to G12Ci.
YAP1/TAZ gene signature is associated with primary resistance to G12Ci
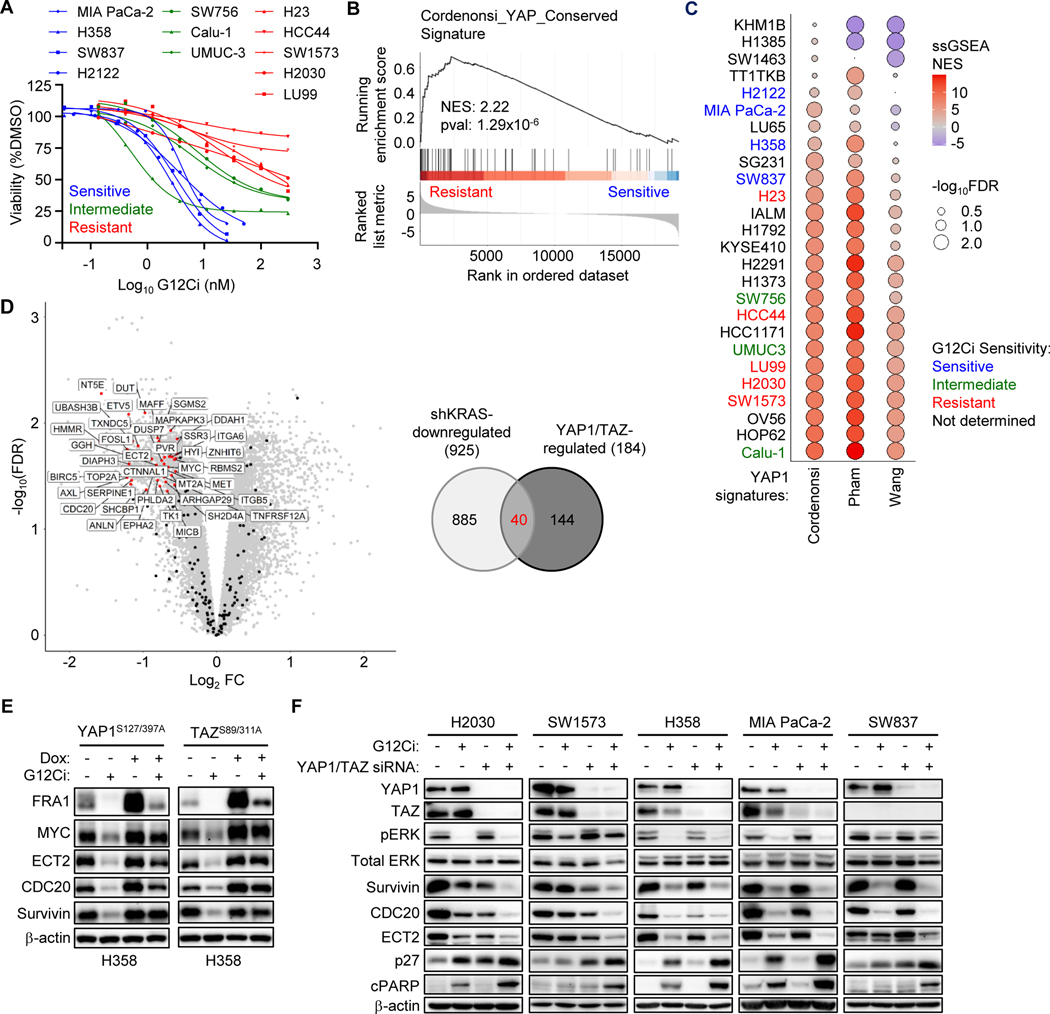
Our analyses above showed that YAP1/TAZ selectively drove resistance to inhibitors of KRAS-ERK signaling by a mechanism independent of ERK. That is, rather than reactivating ERK, YAP1/TAZ can substitute for ERK signaling. Since the major outputs of both YAP1/TAZ and ERK signaling networks involve regulation of gene transcription, we next assessed the possibility that YAP1/TAZ and ERK may regulate overlapping gene transcription signatures to mediate resistance to G12Ci. To evaluate this possibility, we first determined if a gene transcription signature was associated with G12Ci resistance. In agreement with previous studies (38,39,53,54), we observed a broad range of G12Ci sensitivities in a panel of KRASG12C-mutant cell lines (Fig. 4A). We then evaluated available RNA-Seq datasets from the Cancer Cell Line Encyclopedia to assess differential gene expression between G12Ci-resistant and G12Ci-sensitive cell lines.
Figure 4.
YAP1/TAZ gene signature is associated with primary resistance to G12Ci. A, Five-day viability assay following treatment of 12 KRASG12C-mutant cell lines with increasing concentrations of G12Ci. Each data point represents the mean for three or more biological replicates. Treatment response was stratified into three categories: sensitive (blue), intermediate (green), and resistant (red). Many curves were generated from the G12Ci-alone (-TEADi) condition in Supplementary Fig. S7D-E. B, Cordenonsi YAP Conserved Signature (MSigDB) enrichment plot of differentially regulated genes between four G12Ci-sensitive and five resistant cell lines. C, Single sample GSEA (ssGSEA) net enrichment scores (NES) for three YAP1/TAZ signatures across 26 KRASG12C-mutant cancer cell lines. Cell line labels are colored according to their observed G12Ci sensitivity. D, Volcano plot (left) showing differentially expressed genes following KRAS knockdown (shRNA) in a panel of human PDAC cell lines (26). Genes with a LogFC < −0.5 and an FDR < 0.05 are broadly defined as “shKRAS-downregulated” (n = 925). Genes within published YAP1/TAZ gene signatures (those used in C), along with MYC, which was added manually given the reported ability of both YAP1/TAZ (37) and KRAS (36) to regulate its expression, are overlaid (n = 184 genes). Venn diagram (right) showing the overlap between KRAS-, YAP1/TAZ-, and potentially co-regulated genes. Forty co-regulated genes are labeled. E, Immunoblot of YAP1S127/397A and TAZS89/311A overexpressing H358 cells following 24 hours of treatment with DMSO or G12Ci (20 nM). Dox (1 μg/mL) was added 24 hours before G12Ci. F, Immunoblot of cell lines following 24 hours of treatment with G12Ci (H2030, SW1573: 200 nM; H358, MIA PaCa-2, SW837: 20 nM) with or without YAP1/TAZ knockdown. Reverse knockdown with non-targeting control (siNS, “-“) or YAP1/TAZ (siYT, “+”) was performed 24 hours prior to addition of DMSO or G12Ci.
We applied gene set enrichment analysis (GSEA) to differentially expressed genes (Supplementary Fig. 4A). Consistent with a previous GSEA of sotorasib-sensitive and -resistant KRASG12C-mutant cell lines (54), we also found that the Molecular Signatures Database (MSigDB) Singh KRAS Dependency Signature (55) and the Hallmark Epithelial Mesenchymal Transition (56) gene sets were enriched within G12Ci-sensitive and -resistant cell lines, respectively (Supplementary Fig. 4A and B). We additionally identified the MSigDB Cordenonsi YAP Conserved Signature (35) as the most enriched of the 189 MSigDB C6 oncogenic signatures within resistant cell lines (Fig. 4B; Supplementary Fig. 4A). Furthermore, resistant cell lines were similarly enriched for two additional published YAP1/TAZ signatures (23,24) (Supplementary Fig. 4C).
We then performed single sample GSEA on the transcript abundances (TPM) for each individual cell line, utilizing the previously described Cordenonsi (35), Wang (24) and Pham (23) YAP1/TAZ gene signatures. Despite low agreement among the three YAP1/TAZ gene signatures, with only seven genes shared among all three (Supplementary Fig. 4D), we found the highest enrichment in G12Ci-resistant cell lines and the lowest enrichment in G12Ci-sensitive cell lines (Fig. 4C). Taken together, we found that YAP1/TAZ transcriptional signatures are enriched in cell lines exhibiting primary resistance to G12Ci. We therefore also hypothesized that cell lines enriched for YAP1/TAZ transcriptional signatures should be particularly sensitive to their loss. Indeed, we found that previously described YAP1/TAZ transcriptional signatures were significantly correlated with YAP1/TAZ genetic dependency (DepMap) across KRASG12C-mutant cell lines (Supplementary Fig. 4E).
We had hypothesized that YAP1/TAZ-driven resistance towards KRAS and ERK-MAPK pathway inhibitors is supported by the maintenance of essential ERK-regulated transcriptional targets. To address this, we next compared all genes identified in the three YAP1 gene signatures to genes downregulated following shRNA-mediated KRAS knockdown in a panel of KRAS-mutant PDAC cell lines (26) and identified a 40-gene set overlap of KRAS- and YAP1/TAZ-upregulated genes (Fig. 4D). The KRAS/YAP1 overlapping gene set was also found in our previously determined ERK-dependent gene signature established in KRAS-mutant PDAC cell lines (26) (Supplementary Fig. 4F).
We then used the Cancer Dependency Map (DepMap) CRISPR dataset and identified 10 of the co-regulated genes as critical for growth in 21 KRASG12C-mutant cell lines (Supplementary Fig. 4F-H). From this set of genes, we then focused on five (MYC, FOSL1, BIRC5, CDC20, and ECT2) that have been shown previously to support mutant KRAS-dependent cancer growth (36,57–62). We utilized H358 cells with Dox-inducible expression of activated YAP1 and TAZ mutants to evaluate KRAS and YAP1/TAZ co-regulation of these five genes. In the absence of Dox, G12Ci treatment reduced the levels of all five proteins encoded by these genes, supporting their regulation by KRAS. Dox induction of YAP1 or TAZ further increased the levels of all five proteins, and G12Ci caused only partial reduction (Fig. 4E). Additional concurrent inhibition of ERK, PI3Kα or mTOR did not further diminish YAP1-driven gene expression, indicating that YAP1 regulation of expression of these genes is independent of activation of ERK and of PI3K-AKT-mTOR signaling (Supplementary Figs. 3C and 4I).
Given our previous results demonstrating the ability of YAP1/TAZ to rescue G12Ci by maintaining expression of KRAS- and ERK-regulated essential genes, we reasoned that, conversely, YAP1/TAZ suppression would lead to synergistic loss of expression of these genes upon G12Ci treatment. Indeed, we found that co-regulated essential genes were decreased upon either G12Ci or siRNA knockdown of YAP1/TAZ, with the combination leading to maximal reduction (Fig. 4F). The combination also led to increased expression of p27 and cPARP, indicators of cell cycle arrest and apoptosis, respectively. Our results support a model where YAP1/TAZ activation reduces KRAS-dependency by driving expression of normally ERK-regulated proteins that are essential to support cancer growth.
YAP1/TAZ activation drives treatment-induced acquired resistance to G12Ci
Although a subset of patients treated with KRASG12C inhibitors respond initially with tumor regression, relapse due to treatment-induced acquired resistance is a continuing challenge (10–12). To investigate the potential role of YAP1/TAZ activation in acquired resistance to G12C-specific inhibitors, we established mass populations of drug-resistant H358 and MIA PaCa-2 cell lines by sequential dose escalation of adagrasib over several months, resulting in the outgrowth of resistant cells that proliferated in high concentrations of adagrasib (1 and 2 μM, respectively) (Fig. 5A). Age-matched control cells grown in equivalent concentrations of DMSO were consistently passaged alongside adagrasib-resistant cell lines.
Figure 5.
YAP1/TAZ activation drives treatment-induced acquired resistance to G12Ci. A-G, Parental and resistant cell lines were grown in DMSO or adagrasib (H358: 1 μM; MIA PaCa-2: 2 μM), respectively. A, Six-day viability assay of parental and adagrasib-resistant H358 (left) and MIA PaCa-2 (right) cell lines. Each data point represents the mean ± SD for three biological replicates. B, Representative widefield fluorescence images of merged DAPI (blue) and phalloidin (red) staining in H358 (left) and MIA PaCa-2 (right) parental and adagrasib-resistant cell lines. Scale bars = 25 μm. C, Representative confocal immunofluorescence images of individual and merged staining for YAP1/TAZ (green) and DAPI (blue) in cell lines as in B. Scale bars = 25 μm. D, Immunoblot following 72-hour knockdown of YAP1/TAZ (YT) or non-targeting control (NS) in H358 (top) and MIA PaCa-2 (bottom) parental and adagrasib-resistant cell lines. E, Six-day viability assay in cell lines as in D. Each data point represents the mean ± SD for three biological replicates; two-way ANOVA with Tukey post hoc multiple comparisons testing. F, Immunoblot following 72-hour knockdown in cell lines as in D. G, Five-day apoptosis assay (annexin V-FITC/propidium iodide) in cell lines as in D. N = three biological replicates with the mean ± SD shown; two-tailed unpaired t test. H, Schematic representing convergence of KRAS-ERK and YAP1/TAZ signaling.
Adagrasib retained the ability to bind KRASG12C in drug-resistant H358 and MIA PaCa-2 cells maintained continuously in high adagrasib, as indicated by the presence of the covalently modified form of KRAS that displays altered mobility (Supplementary Fig. 5A). pERK levels were reduced (H358) or elevated (MIA PaCa-2) compared to those in parental cells. Notably, adagrasib-resistant cell lines were morphologically distinct from their parental counterparts, exhibiting a flattened and less refractile appearance (Supplementary Fig. 5B). We applied phalloidin staining to assess whether F-actin cytoskeletal changes were associated with the altered morphology, and found striking actin reorganization in resistant cells (Fig. 5B).
Actin reorganization can cause LATS-dependent or -independent activation of YAP1/TAZ (41,63–65). We evaluated YAP1/TAZ subcellular distribution and found increased nuclear accumulation, suggesting increased activity, in resistant cells compared to parental cell lines (Fig. 5C). Increased YAP1/TAZ activity was supported by a substantial increase in AXL and CYR61 expression, which was abrogated by YAP1/TAZ knockdown (Fig. 5D).
We next determined if the resistant lines exhibited greater growth dependence on YAP1/TAZ compared to parental lines. Concurrent knockdown of YAP1 and TAZ completely reduced growth of resistant H358 cells but only partially reduced growth in parental H358 cells; likewise, it caused near-complete reduction of growth in resistant MIA PaCa-2 cells but did not affect the growth of parental MIA PaCa-2 cells (Fig. 5E; Supplementary Fig. 5C). The increased growth dependence of resistant cell lines was associated with increased apoptosis, as indicated by suppression of YAP1/TAZ increasing cPARP in the resistant but not parental cell lines (Fig. 5F). Similarly, we observed a nearly three-fold increase in apoptosis as determined by annexin V and propidium iodide staining upon YAP1/TAZ knockdown in the resistant but not parental counterparts (Fig. 5G). Taken together, these data support a model where acquired resistance to G12C-specific inhibitors is associated with YAP1/TAZ-dependent growth that is independent of KRASG12C (Fig. 5H). We found that activated YAP1/TAZ expression supports the growth of G12Ci-treated cells by maintaining expression of KRAS- and ERK-regulated genes (Fig. 4E). Consistently, adagrasib-resistant cells maintain expression of these genes in a YAP1/TAZ-dependent manner (Supplementary Fig. 5D).
YAP1 and TAZ require TEAD to drive resistance to KRASG12C inhibition
YAP1/TAZ proteins lack a DNA-binding domain, and therefore require association with transcription factors to drive gene transcription. The most significant and best-studied YAP1/TAZ transcription factor partners are members of the TEAD family (TEAD1–4). However, YAP1/TAZ have also been shown to associate with other transcription factors (41,44,63). Previous studies reached conflicting conclusions. One study determined that Yap1 supported KrasG12D-independent mouse PDAC growth through a TEAD-dependent mechanism (42). In contrast, a second study determined that YAP-mediated KRASG13D-independence in human colon cancer cells involved a TEAD-independent function through the FOS transcription factor (43).
To determine the role of TEAD in supporting YAP1- and TAZ-driven resistance to G12Ci, we applied the same strategy taken in the previous studies (42,43). YAP1 and TAZ association with TEAD proteins requires a conserved serine in their conserved amino-terminal TEAD-binding domains, and consequently can be abolished by mutation of those serines (66,67). Therefore, to assess the TEAD-dependence of our Hippo-independent YAP1/TAZ mutants, we introduced those mutations, S94A and S51A, into the TEAD binding domains of activated YAP1S127/397A and TAZS89/311A, respectively. We found that the TEAD binding-deficient mutants failed to induce expression of CYR61 or AXL in MIA PaCa-2 and H358 cells (Fig. 6A; Supplementary Fig. 6A) and did not alter sensitivity to treatment with G12Ci or ERKi (Fig. 6B; Supplementary Fig. 6B and 6C) or adagrasib (Supplementary Fig. 6D and 6E). Consistent with this, loss of TEAD association also prevented activated YAP1 from protecting against G12Ci induction of cell cycle inhibition or apoptosis (Fig. 6C). We conclude that both YAP1 and TAZ drive resistance to pharmacologic inhibition of KRASG12C through a TEAD-dependent mechanism.
Figure 6.
YAP1 and TAZ require TEAD to drive resistance to KRASG12C inhibition. A-E, Mutant YAP1 and TAZ overexpression was induced by Dox (1 μg/mL) 24 hours prior to indicated treatments; YAP1S127/397A and TAZS89/311A (activated), YAP1S127/397/94A and TAZS89/311/51A (activated, TEAD binding-deficient). A, Immunoblot of MIA PaCa-2 cells overexpressing the indicated YAP1 (top) and TAZ (bottom) mutants following 24-hour treatment with DMSO or G12Ci (20 nM). B, Five-day viability assay following G12Ci treatment of cells as in A. Data points represent the mean ± SD for three or more biological replicates. C-E, Immunoblot of H358 cells overexpressing indicated YAP1 and TAZ mutants following 24-hour treatment with DMSO or G12Ci (20 nM). Immunoblots detect markers of cell viability (C), PI3K signaling (D), or protein abundance of KRAS-ERK and YAP1/TAZ co-regulated genes (E).
We also found that activated YAP1 and TAZ stimulation of PI3K-AKT-mTORC1 signaling was TEAD-dependent (Fig. 3C and 6D). Furthermore, YAP1 and TAZ induction of the ERK-regulated proteins FRA1, ECT2, Survivin, MYC and CDC20 was lost when TEAD association was impaired (Fig. 4E and 6E). Collectively, our data support a model where YAP1/TAZ-driven resistance to G12Ci is dependent on TEAD-mediated gene transcription and PI3K-AKT-mTOR activation.
TEAD inhibition enhances G12Ci initial and long-term efficacy
Our analyses with TEAD binding-deficient mutants of activated YAP1/TAZ implicated a requirement for TEAD to enable YAP1/TAZ-driven resistance to G12Ci, suggesting that blocking TEAD function may be an effective strategy to block this resistance. To further address the role of TEAD in YAP1/TAZ-driven G12Ci resistance, we utilized a TEAD dominant negative, comprised of a tandem array of TEAD binding domains from VGLL4, YAP1 and TAZ (termed TEADi; herein referred to as TEAD-DN) (19). TEAD-DN binds to TEAD and prevents its interaction with YAP1 and TAZ.
For these analyses we utilized two KRASG12C-mutant cell lines (UM53 and H2030) that displayed high enrichment in YAP1/TAZ gene signatures and significant growth inhibition upon siRNA-mediated YAP1/TAZ knockdown (Fig. 2J). Dox induction of GFP-tagged TEAD-DN expression resulted in suppression of the YAP1/TAZ targets AXL and CYR61 (Fig. 7A). TEAD-DN also reduced protein levels from the KRAS-ERK- and YAP1/TAZ-co-regulated genes (MYC, FOSL1, CDC20, ECT2, and BIRC5), which were further suppressed with G12Ci. Similar to YAP1/TAZ knockdown, TEAD-DN potently inhibited proliferation (Supplementary Fig. 7A) and sensitized cells to G12Ci to cause complete growth inhibition (Fig. 7B). We conclude that blocking YAP1/TAZ interaction with TEAD impaired endogenous YAP1/TAZ-mediated gene transcription, causing inhibition of growth and increased sensitivity to G12Ci.
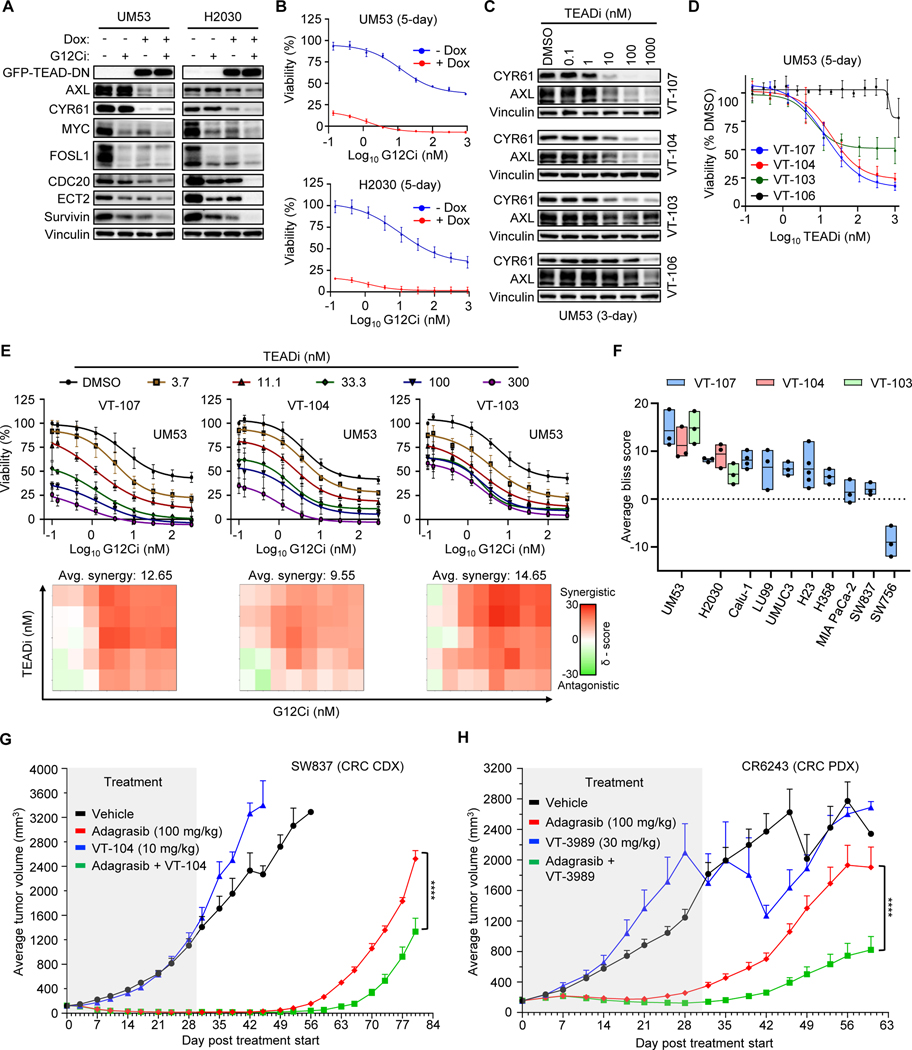
Figure 7.
TEAD inhibition enhances initial and long-term efficacy of KRASG12C-selective inhibitors. A, Immunoblot of UM53 and H2030 cells expressing dox-inducible TEAD dominant-negative peptide (TEAD-DN) (19) and treated with DMSO or G12Ci (200 nM) for 24 hours. Dox (1 μg/mL) was added 24 hours before drug treatment. B, Five-day viability assay following G12Ci treatment of cell lines as in A. Each data point represents the mean ± SD for three biological replicates. C, Immunoblot following 72-hour treatment with pan-TEAD inhibitors, VT-104 and VT-107; TEAD1-selective inhibitor, VT-103; and an inactive VT-107 enantiomer, VT-106, at the indicated concentrations. D, Five-day viability assay performed in cells as in C. Each data point represents the mean ± SD for three biological replicates. E, Top – five-day viability assay in UM53 cells following increasing doses of both G12Ci and the indicated TEADi. Each data point represents the mean ± SD for three biological replicates. Bottom - Excess over Bliss synergy values calculated from data represented above using SynergyFinder. Tiles in the 2D contour represents the synergy score at each combination. Representative plots are shown. F, Average synergy scores taken from combination studies in E and Supplementary Fig. S7D-E. Boxplots represent the mean and range for average synergy scores taken from three or more independent biological replicates. G, Adagrasib (100 mg/kg), VT-104 (10 mg/kg) or the combination was administered daily by oral gavage to NOD-SCID mice bearing the SW837 subcutaneous cell line-derived xenografts (n = 8 per treatment). Treatment (grey tile) was stopped at day 30. H, Adagrasib (100 mg/kg), VT-3989 (30 mg/kg) or the combination was administered daily via oral gavage to BALB/c mice bearing the CR6243 subcutaneous patient-derived xenografts (n = 10 per treatment). Treatment (grey tile) was stopped at day 31. (G, H), Data are shown as tumor volume mean ± SEM. Statistical significance between adagrasib and combination treatment groups was determined by two-way ANOVA; ****, P < 0.0001.
These observations support TEAD as a target for blocking YAP1/TAZ-driven resistance to G12Ci. Potent and specific small molecule inhibitors targeting the TEAD family of transcription factors that prevent YAP1/TAZ interaction are under clinical evaluation in NF2-deficient mesothelioma (68). To assess the efficacy of pharmacologic inhibition of TEAD to overcome G12Ci resistance, we treated UM53 cells with two distinct pan-TEAD inhibitors (VT-107 and VT-104), a TEAD1-specific inhibitor (VT-103), and an inactive enantiomer of VT-107 (VT-106) (69). To monitor target inhibition, we used a TEAD-binding transcriptional reporter (8xGTIIC) comprised of eight repeating TEAD transcriptional elements upstream of luciferase (15). We found strong suppression of luciferase at low nanomolar concentrations of all three TEAD inhibitors but not the inactive enantiomer (Supplementary Fig. 7B). This dose-dependent inhibition corresponded to suppression of YAP1/TAZ target genes (CCN1, CCN2, and ANKRD1) (Supplementary Fig. 7C) and encoded proteins (AXL and CYR61) (Fig. 7C), and to growth suppression (Fig. 7D). These results demonstrate the potency and specificity of these TEAD inhibitors and validate TEAD as a target for pharmacologic inhibition of YAP1/TAZ transcriptional activity.
We next determined if concurrent TEAD inhibition would increase G12Ci sensitivity. We found that both pan-TEAD and TEAD1-selective inhibitors sensitized UM53 and H2030 cells to G12Ci in a dose-dependent manner (Fig. 7E; Supplementary Fig. 7D). Performing Bliss synergy analysis, we found the combination of TEADi and G12Ci to be synergistic at almost all concentrations in both cell lines. In a panel of KRASG12C-mutant PDAC, NSCLC, CRC, and bladder cancer cell lines, we found highly consistent results across cancer types (Fig. 7F; Supplementary Fig. 7E). Pan-TEAD inhibitor treatment potentiated the cytotoxic effects of G12Ci treatment in UM53 and H2030 cells (Supplementary Fig. 7F).
We also evaluated whether TEADi would impair acquired resistance to pharmacological inhibition of KRASG12C. We found that adagrasib-resistant H358 and MIA PaCa-2 cell lines were preferentially sensitive to pan-TEAD inhibition relative to their parental counterparts (Supplementary Fig. 7G). To determine whether TEADi could delay the acquisition of acquired resistance, we performed 10-day clonogenic growth assays. We found that most cell lines were able to reestablish growth by ten days of G12Ci treatment (Supplementary Fig. 7H). Similarly, while we observed significant growth suppression after 5-day treatment with the pan-TEADi VT-104, no growth inhibitory activity was seen after ten days. However, the combination of G12Ci and TEADi caused near-complete growth inhibition across cell lines in that timeframe. These results suggest that TEADi has limited long-term efficacy as a single agent but is sufficient to enhance the long-term efficacy of G12Ci.
Finally, tumor-bearing mice carrying SW837 cell line-derived xenografts (CDX) were treated daily with VT-104 and adagrasib for 30 days, then monitored for an additional 50 days. Treatment with adagrasib alone caused complete tumor regression, as we have shown previously for this cell line (38), but tumor regrowth began by 19 days after the end of treatment (Fig. 7G; Supplementary Fig. 7I). Whereas the pan-TEADi VT-104 alone did not inhibit tumor growth, when given in combination with adagrasib, it delayed tumor regrowth by nearly nine days. We extended these observations to a patient-derived xenograft (PDX; CR6243) model of CRC and a TEADi currently under clinical evaluation, VT3989 (70). Despite strong initial tumor inhibition by adagrasib, tumor growth resumed while mice were still on adagrasib treatment and continued after treatment was stopped (Fig. 7H; Supplementary Fig. 7J). VT3989 alone did not cause significant inhibition of tumor growth, but when given in combination with adagrasib significantly delayed tumor regrowth (Fig. 7H). The combinations of TEADi and KRASG12C inhibition did not display significant toxicity as monitored by body weight (Supplementary Fig. 7K and L). Together with our cell culture analyses, these results support concurrent TEAD inhibition as a therapeutic approach to improve the long-term efficacy of pharmacological inhibition of KRASG12C.
Discussion
The limited objective response rate (<50%) and duration of response (~6 months) reflect the limited clinical efficacy of single agent treatment with KRASG12C inhibitors (5,6). Analyses of patient genetic information have begun to elucidate a genetic basis for both primary/intrinsic and acquired mechanisms of resistance (9–12). Genomic DNA analyses of tumors from patients who relapsed upon treatment with KRASG12C-selective inhibitors have identified mutational activation or inactivation of signaling components, either upstream or downstream, or at the level of RAS. However, approximately 50% of patients who relapse on G12C inhibitor treatment do not exhibit alterations at the genomic DNA level, indicating that more mechanisms remain to be discovered. In this study, we applied an unbiased genetic screen and identified the loss of components of the Hippo tumor suppressor signaling network, which inhibits YAP1 and TAZ regulation of gene transcription, as drivers of resistance to G12C inhibitors. We verified that gain- or loss-of-function of YAP1/TAZ caused resistance or enhanced sensitivity, respectively, to G12C inhibitors. We identified a TEAD transcription factor-dependent mechanism whereby YAP1/TAZ activation drives RAS-independent signaling activities that phenocopy activation of two key KRAS effector signaling networks (Supplementary Fig. 7M). Finally, we showed that concurrent pharmacologic inhibition of TEAD enhanced KRASG12C inhibitor anti-tumor cell activity in mouse models of KRASG12C-driven human cancers. With TEAD inhibitors now entering clinical evaluation (68,70), our study supports the use of TEAD inhibitors in combination drug strategies to prolong the efficacy of direct KRAS inhibitors.
Our identification of Hippo pathway inactivation as a driver of resistance to pharmacologic inhibition of KRASG12C is consistent with previous studies that identified YAP1 overexpression or activation as a mechanism by which KRAS-mutant cancers can escape their addiction to and dependency on continued aberrant KRAS function (42,43). We additionally demonstrated that activation of the related YAP1 paralog, TAZ, exhibits both overlapping and distinct functions in driving resistance to G12C-selective inhibitors. With additional direct KRAS inhibitors targeting other KRAS mutations under preclinical and clinical evaluation (1), we suspect that YAP1/TAZ activation will emerge as a driver of resistance to all KRAS inhibitors. Finally, a recent study evaluating a molecular basis for acquired resistance in a lung cancer patient treated with sotorasib did not identify any DNA mutations, and instead, found upregulation of YAP1-associated gene transcription (71), further supporting the clinical relevance of YAP1 and TAZ in both primary and acquired resistance in patients treated with KRAS inhibitors.
YAP1/TAZ engage a spectrum of transcription factors to regulate gene transcription. Previous studies have reached conflicting conclusions regarding the importance of specific transcription factors in overcoming addiction to mutant KRAS, identifying both TEAD-dependent and -independent mechanisms (42,43). Here we validated the importance of the TEAD family of transcription factors in YAP1/TAZ-induced resistance to pharmacological inhibition of KRASG12C. We further defined a molecular basis for TEAD function, where TEAD stimulated signaling activities that phenocopied the two key KRAS effector signaling networks that support KRAS-dependent cancer growth (Supplementary Fig. 7M). Previous studies proposed that YAP1 and KRAS regulate distinct but overlapping gene signatures to support cancer growth (43). Our comparison of published YAP1 and/or TAZ gene signatures with our KRAS- and ERK-regulated gene signatures (26) identified co-regulated genes that are well-established dependencies for the growth of KRAS-mutant cancers. Previous analyses of mouse models of KRAS-driven cancers showed that these genes are essential for the progression and maintenance of tumorigenic growth (36,57–62). Given that published YAP1 or YAP1/TAZ gene signatures show limited overlap, likely reflecting tissue type and genetic context differences, establishing the TEAD- and KRAS-dependent gene transcriptomes in the same cancer setting will be needed to establish a comprehensive molecular portrait of how TEAD-regulated gene transcription drives resistance to KRAS inhibitors. This information may then identify molecular markers to better define the subset of patients who will respond to KRAS-targeted therapies and define combination approaches to prolong drug efficacy.
In addition to stimulating an ERK-independent gene transcription program that partially substitutes for the loss of KRAS-ERK signaling, we also determined that YAP1/TAZ activates TEAD-dependent, KRAS-independent activation of PI3K-AKT-mTOR signaling. Whereas TEAD regulation of normally ERK-regulated genes drives cell cycle progression, we found that PI3K-AKT-mTOR activation offsets the apoptotic consequences of KRAS inhibition. The importance of ERK MAPK and PI3K signaling in KRAS-mutant cancer growth is well demonstrated by the synergistic action of concurrent pharmacologic inhibition of these effector pathways to cause tumor regression (2).
Upon validating TEAD as a key driver of YAP1/TAZ-stimulated drug resistance, we then demonstrated that pharmacologic inhibition of TEAD synergistically enhances adagrasib anti-tumor activity in vivo. While TEAD inhibition alone did not exhibit significant anti-tumor activity in KRASG12C-mutant xenograft tumors, concurrent treatment prolonged the response to adagrasib treatment. Currently, TEAD inhibitors are under clinical evaluation for NF2-deficient cancers (68). Our findings support the use of TEAD inhibitors, in combination with KRAS inhibitors, for KRAS-mutant cancers.
During the course of preparing our study for publication, four independent publications described the utility of small molecule TEAD inhibitors to overcome resistance to KRASG12C inhibitors (72–75). Very similar to our study, Mukhopadhyay et al. applied a genome-wide CRISPR/Cas9 screen and identified components of the YAP1/TAZ-TEAD pathway as genes that are synthetically lethal with adagrasib treatment in KRAS/STK11-mutant NSCLC lines (75). Together, these studies provide independent validation of our major observation, that concurrent TEAD inhibition can enhance G12Ci anti-tumor activity. In addition to complementing and extending these findings, our study provides significant mechanistic and translational insight. In agreement with Adachi et al. (72), we conclude that loss of Hippo pathway signaling occurs following G12Ci long-term treatment. We extend these findings by providing rigorous mechanistic evidence for the required loss of Hippo pathway activity in promoting YAP1/TAZ-driven KRAS resistance. Furthermore, we provide evidence for redundant and complementary roles of TAZ in modulating G12Ci sensitivity. Additionally, while these studies correctly speculated that TEAD family transcription factors are the principal YAP1/TAZ transcriptional binding partner, we provide experimental evidence for the necessity of TEAD in YAP1/TAZ-driven G12Ci resistance. Finally, as summarized above, our study provides extended insight into the mechanistic basis of YAP1/TAZ-driven G12Ci resistance. We found that YAP1/TAZ is sufficient to sustain PI3K-AKT-mTOR signaling and ERK-MAPK transcriptional activity, independent of ERK-MAPK pathway reactivation. The latter finding differs from the conclusion that YAP1 causes ERK reactivation through the RAS-related GTPase MRAS (72), where we found no evidence for ERK reactivation in YAP1/TAZ-driven G12Ci resistance. Finally, whereas previous studies focused primarily on lung and colorectal cancer, we have extended these findings into KRASG12C-mutant pancreatic cancer cell lines. In summary, TEAD inhibition has emerged as a powerful means by which to impair YAP1/TAZ activity and thereby enhance initial and long-term efficacy of targeting KRAS.
Supplementary Material
Significance:
YAP1/TAZ-TEAD activation compensates for loss of KRAS effector signaling, establishing a mechanistic basis for concurrent inhibition of TEAD to enhance the efficacy of KRASG12C-selective inhibitor treatment of KRASG12C-mutant cancers.
Acknowledgements
Support was provided by grants to A.D. Cox and/or C.J. Der from the National Cancer Institute (NCI; R01CA42978, P50CA196510, P50CA257911, U01CA199235, P01CA203657 and R35CA232113), to C.J. Der from the Pancreatic Cancer Action Network/AACR (15-90-25-DER) and the Pancreatic Cancer Action Network (22-WG-DERB), and the Department of Defense (W81XWH2110692). A.C. Edwards was supported by NIGMS T32GM119999 and NCI F31CA275260. C.A.S. was supported by NCI T32CA009156 and F32CA232529. J.E. Klomp was supported by NCI T32CA009156, F32 CA239328, and K99 CA276700, and American Cancer Society (ACS) PF-20-069. A.M. Waters was supported by ACS PF-18-061.
C.J. Der is a consultant/advisory board member for Cullgen, Deciphera Pharmaceuticals, Mirati Therapeutics, Reactive Biosciences, Revere Pharmaceuticals, Revolution Medicines, Sanofi and SHY Therapeutics. C.J. Der has received research funding support from Deciphera Pharmaceuticals, Mirati Therapeutics, Reactive Biosciences, Revolution Medicines, and SpringWorks Therapeutics. A.D. Cox has consulted for Eli Lilly and Mirati Therapeutics. N. Sudhakar, J. Hallin and P. Olson are employees and shareholders of Mirati Therapeutics, Inc. J.G. Christensen is the EVP and Chief Scientific Officer of, and a shareholder in, Mirati Therapeutics, Inc. T. T. Tang and L. Post report employment with Vivace Therapeutics and hold equity interest in Vivace Therapeutics.
Footnotes
Conflict of Interest
The other authors declare no competing interests.
References
- 1.Punekar SR, Velcheti V, Neel BG, Wong K-K. The current state of the art and future trends in RAS-targeted cancer therapies. Nat Rev Clin Oncol 2022;19:637–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore AR, Rosenberg SC, McCormick F, Malek S. RAS-targeted therapies: is the undruggable drugged? Nat Rev Drug Discov 2020;19:533–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17–48. [DOI] [PubMed] [Google Scholar]
- 4.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013;503:548–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N Engl J Med 2021;384:2371–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jänne PA, Riely GJ, Gadgeel SM, Heist RS, Ou S-HI, Pacheco JM, et al. Adagrasib in Non-Small-Cell Lung Cancer Harboring a KRASG12C Mutation. N Engl J Med 2022;387:120–31. [DOI] [PubMed] [Google Scholar]
- 7.Soria J-C, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113–25. [DOI] [PubMed] [Google Scholar]
- 8.Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim D-W, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:829–38. [DOI] [PubMed] [Google Scholar]
- 9.Negrao MV, Araujo HA, Lamberti G, Cooper AJ, Akhave NS, Zhou T, et al. Comutations and KRASG12C Inhibitor Efficacy in Advanced NSCLC. Cancer Discov 2023;13:1556–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, Murciano-Goroff YR, Xue JY, Ang A, Lucas J, Mai TT, et al. Diverse alterations associated with resistance to KRAS(G12C) inhibition. Nature 2021;1–5. [DOI] [PMC free article] [PubMed]
- 11.Awad MM, Liu S, Rybkin II, Arbour KC, Dilly J, Zhu VW, et al. Acquired Resistance to KRASG12C Inhibition in Cancer. N Engl J Med 2021;384:2382–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka N, Lin JJ, Li C, Ryan MB, Zhang J, Kiedrowski LA, et al. Clinical acquired resistance to KRASG12C inhibition through a novel KRAS switch-II pocket mutation and polyclonal alterations converging on RAS-MAPK reactivation. Cancer Discov 2021;11(8):1913–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin KH, Rutter JC, Xie A, Pardieu B, Winn ET, Bello RD, et al. Using antagonistic pleiotropy to design a chemotherapy-induced evolutionary trap to target drug resistance in cancer. Nat Genet 2020;52:408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang B, Wang M, Zhang W, Xiao T, Chen C-H, Wu A, et al. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat Protoc 2019;14:756–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in mechanotransduction. Nature 2011;474:179–83. [DOI] [PubMed] [Google Scholar]
- 16.Bajikar SS, Wang C-C, Borten MA, Pereira EJ, Atkins KA, Janes KA. Tumor-Suppressor Inactivation of GDF11 Occurs by Precursor Sequestration in Triple-Negative Breast Cancer. Dev Cell 2017;43:418–435.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, et al. β-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 2012;151:1457–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei Q-Y, Zhang H, Zhao B, Zha Z-Y, Bai F, Pei X-H, et al. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol 2008;28:2426–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Y, Park J, Feng A, Awasthi P, Wang Z, Chen Q, et al. YAP1/TAZ-TEAD transcriptional networks maintain skin homeostasis by regulating cell proliferation and limiting KLF4 activity. Nat Commun 2020;11:1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carver J, Dexheimer TS, Hsu D, Weng M-T, Smith JL, Guha R, et al. A High-Throughput Assay for Small Molecule Destabilizers of the KRAS Oncoprotein. PLOS ONE 2014;9:e103836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Killian T, Gatto L. Exploiting the DepMap cancer dependency data using the depmap R package [Internet]. F1000Research 2021. Available from: https://f1000research.com/articles/10-416.
- 22.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pham TH, Hagenbeek TJ, Lee H-J, Li J, Rose CM, Lin E, et al. Machine-Learning and Chemicogenomics Approach Defines and Predicts Cross-Talk of Hippo and MAPK Pathways. Cancer Discov 2021;11:778–93. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Xu X, Maglic D, Dill MT, Mojumdar K, Ng PK-S, et al. Comprehensive Molecular Characterization of the Hippo Signaling Pathway in Cancer. Cell Rep 2018;25:1304–1317.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bryant KL, Stalnecker CA, Zeitouni D, Klomp JE, Peng S, Tikunov AP, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med 2019;25:628–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krueger F. Trim Galore! [Internet]. Babraham Bioinformatics 2012. Available from: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- 28.Andrews S. FastQC [Internet]. Babraham Bioinformatics 2010. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 29.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j 2011;17:10. [Google Scholar]
- 30.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 2019;47:D766–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017;14:417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 2016;4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 2005;21:3439–40. [DOI] [PubMed] [Google Scholar]
- 35.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 2011;147:759–72. [DOI] [PubMed] [Google Scholar]
- 36.Vaseva AV, Blake DR, Gilbert TSK, Ng S, Hostetter G, Azam SH, et al. KRAS Suppression-Induced Degradation of MYC Is Antagonized by a MEK5-ERK5 Compensatory Mechanism. Cancer Cell 2018;34:807–822.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami S, Nemazanyy I, White SM, Chen H, Nguyen CDK, Graham GT, et al. A Yap-Myc-Sox2-p53 Regulatory Network Dictates Metabolic Homeostasis and Differentiation in Kras-Driven Pancreatic Ductal Adenocarcinomas. Dev Cell 2019;51:113–128.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRASG12C Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov 2020;10:54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janes MR, Zhang J, Li L-S, Hansen R, Peters U, Guo X, et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018;172:578–589.e17. [DOI] [PubMed] [Google Scholar]
- 40.Han K, Pierce SE, Li A, Spees K, Anderson GR, Seoane JA, et al. CRISPR screens in cancer spheroids identify 3D growth specific vulnerabilities. Nature 2020;580:136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer Cell 2016;29:783–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 2014;158:185–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 2014;158:171–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piccolo S, Dupont S, Cordenonsi M. The Biology of YAP/TAZ: Hippo Signaling and Beyond. Physiol Rev 2014;94:1287–312. [DOI] [PubMed] [Google Scholar]
- 45.Reggiani F, Gobbi G, Ciarrocchi A, Sancisi V. YAP and TAZ Are Not Identical Twins. Trends Biochem Sci 2021;46:154–68. [DOI] [PubMed] [Google Scholar]
- 46.Zeng R, Dong J. The Hippo Signaling Pathway in Drug Resistance in Cancer. Cancers (Basel) 2021;13:318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen C, Yi C. YAP/TAZ Signaling and Resistance to Cancer Therapy. Trends Cancer 2019;5:283–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castellano E, Downward J. RAS Interaction with PI3K: More Than Just Another Effector Pathway. Genes Cancer 2011;2:261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasan N, Cantley LC. At a crossroads: how to translate the roles of PI3K in oncogenic and metabolic signalling into improvements in cancer therapy. Nat Rev Clin Oncol 2022;19:471–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tumaneng K, Schlegelmilch K, Russell RC, Yimlamai D, Basnet H, Mahadevan N, et al. YAP mediates crosstalk between the Hippo and PI(3)K–TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol 2012;14:1322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White SM, Avantaggiati ML, Nemazanyy I, Di Poto C, Yang Y, Pende M, et al. YAP/TAZ Inhibition Induces Metabolic and Signaling Rewiring Resulting in Targetable Vulnerabilities in NF2-Deficient Tumor Cells. Dev Cell 2019;49:425–443.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park Y-Y, Sohn BH, Johnson RL, Kang M-H, Kim SB, Shim J-J, et al. YAP1 and TAZ Activates mTORC1 Pathway by Regulating Amino Acid Transporters in hepatocellular carcinoma. Hepatology 2016;63:159–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Misale S, Fatherree JP, Cortez E, Li C, Bilton S, Timonina D, et al. KRAS G12C NSCLC Models Are Sensitive to Direct Targeting of KRAS in Combination with PI3K Inhibition. Clin Cancer Res 2019;25:796–807. [DOI] [PubMed] [Google Scholar]
- 54.Adachi Y, Ito K, Hayashi Y, Kimura R, Tan TZ, Yamaguchi R, et al. Epithelial-to-Mesenchymal Transition is a Cause of Both Intrinsic and Acquired Resistance to KRAS G12C Inhibitor in KRAS G12C–Mutant Non–Small Cell Lung Cancer. Clin Cancer Res 2020;26:5962–73. [DOI] [PubMed] [Google Scholar]
- 55.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 2009;15:489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang W-H, Nguyen T-TT, Hsu C-H, Bryant KL, Kim HJ, Ying H, et al. KRAS-dependent cancer cells promote survival by producing exosomes enriched in Survivin. Cancer Lett 2021;517:66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cook DR, Kang M, Martin TD, Galanko JA, Loeza GH, Trembath DG, et al. Aberrant Expression and Subcellular Localization of ECT2 Drives Colorectal Cancer Progression and Growth. Cancer Res 2022;82:90–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vallejo A, Perurena N, Guruceaga E, Mazur PK, Martinez-Canarias S, Zandueta C, et al. An integrative approach unveils FOSL1 as an oncogene vulnerability in KRAS-driven lung and pancreatic cancer. Nat Commun 2017;8:14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Volonte D, Sedorovitz M, Galbiati F. Impaired Cdc20 signaling promotes senescence in normal cells and apoptosis in non–small cell lung cancer cells. J Biol Chem 2022;298:102405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarthy AV, Morgan-Lappe SE, Zakula D, Vernetti L, Schurdak M, Packer JCL, et al. Survivin depletion preferentially reduces the survival of activated K-Ras-transformed cells. Mol Cancer Ther 2007;6:269–76. [DOI] [PubMed] [Google Scholar]
- 62.Justilien V, Ali SA, Jamieson L, Yin N, Cox AD, Der CJ, et al. Ect2-Dependent rRNA Synthesis Is Required for KRAS-TRP53-Driven Lung Adenocarcinoma. Cancer Cell 2017;31:256–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma S, Meng Z, Chen R, Guan K-L. The Hippo Pathway: Biology and Pathophysiology. Annu Rev Biochem 2019;88:577–604. [DOI] [PubMed] [Google Scholar]
- 64.Piccolo S, Panciera T, Contessotto P, Cordenonsi M. YAP/TAZ as master regulators in cancer: modulation, function and therapeutic approaches. Nat Cancer 2023;4:9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Franklin JM, Wu Z, Guan K-L. Insights into recent findings and clinical application of YAP and TAZ in cancer. Nat Rev Cancer 2023;23:512–25. [DOI] [PubMed] [Google Scholar]
- 66.Zhao B, Ye X, Yu J, Li L, Li W, Li S, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 2008;22:1962–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang H, Liu C-Y, Zha Z-Y, Zhao B, Yao J, Zhao S, et al. TEAD Transcription Factors Mediate the Function of TAZ in Cell Growth and Epithelial-Mesenchymal Transition. J Biol Chem 2009;284:13355–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pobbati AV, Kumar R, Rubin BP, Hong W. Therapeutic targeting of TEAD transcription factors in cancer. Trends Biochem Sci 2023;48:450–62. [DOI] [PubMed] [Google Scholar]
- 69.Tang TT, Konradi AW, Feng Y, Peng X, Ma M, Li J, et al. Small Molecule Inhibitors of TEAD Auto-palmitoylation Selectively Inhibit Proliferation and Tumor Growth of NF2-deficient Mesothelioma. Mol Cancer Ther 2021;20:986–98. [DOI] [PubMed] [Google Scholar]
- 70.YAP TA, Kwiatkowski DJ, Desai J, Dagogo-Jack I, Millward M, Kindler HL, et al. First-in-class, first-in-human phase 1 trial of VT3989, an inhibitor of yes-associated protein (YAP)/transcriptional enhancer activator domain (TEAD), in patients (pts) with advanced solid tumors enriched for malignant mesothelioma and other tumors with neurofibromatosis 2 (NF2) mutations [abstract]. In: Proceedings of the 114th Annual Meeting of the American Association for Cancer Research; 2023 April 14–19; Orlando, FL. Philadelphia (PA): AACR; 2023. Abstract nr. CT-006. [Google Scholar]
- 71.Tsai YS, Woodcock MG, Azam SH, Thorne LB, Kanchi KL, Parker JS, et al. Rapid idiosyncratic mechanisms of clinical resistance to KRAS G12C inhibition. J Clin Invest 2022;132:e155523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adachi Y, Kimura R, Hirade K, Yanase S, Nishioka Y, Kasuga N, et al. Scribble mis-localization induces adaptive resistance to KRAS G12C inhibitors through feedback activation of MAPK signaling mediated by YAP-induced MRAS. Nat Cancer 2023;1–15. [DOI] [PubMed]
- 73.Hagenbeek TJ, Zbieg JR, Hafner M, Mroue R, Lacap JA, Sodir NM, et al. An allosteric pan-TEAD inhibitor blocks oncogenic YAP/TAZ signaling and overcomes KRAS G12C inhibitor resistance. Nat Cancer 2023;4:812–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tammaccaro SL, Prigent P, Le Bail J-C, Dos-Santos O, Dassencourt L, Eskandar M, et al. TEAD Inhibitors Sensitize KRASG12C Inhibitors via Dual Cell Cycle Arrest in KRASG12C-Mutant NSCLC. Pharmaceuticals (Basel) 2023;16:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mukhopadhyay S, Huang H-Y, Lin Z, Ranieri M, Li S, Sahu S, et al. Genome-Wide CRISPR Screens Identify Multiple Synthetic Lethal Targets That Enhance KRASG12C Inhibitor Efficacy. Cancer Res 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All plasmids generated from this study are to be deposited in Addgene. The shKRAS and ERKi RNA sequencing datasets analyzed in this study were obtained from the EMBL-EBI European Nucleotide Archive (ENA) database at http://www.ebi.ac.uk/ena/ under accession numbers PRJEB25797 and PRJEB25806, respectively. All CRISPR-Cas9 loss-of-function dependency data, and CCLE transcriptional and mutational data (22Q1) were obtained from the Broad Institute’s Cancer Dependency Map (DepMap) at https://depmap.org/portal/download/all/ or through the depmap (21) package in R-studio. All additional data generated to support the findings of this study are available from the corresponding author upon request.