Abstract
The EGFR-C797S resistance mutation to third-generation drugs has been overcome by fourth-generation inhibitors, allosteric inhibitors, namely EAI045 and has reached phase 3 clinical trials, so the Allosteric Site is currently an attractive target for development. In this study, researchers are interested in knowing the activity of metabolite compounds from marine natural ingredients Clathria Sp. against the Allosteric Site of EGFR computationally. The methods used include molecular docking using Autodock4 software and Molecular Dynamics simulation performed using GROMACS software. The research began with the preparation of metabolite samples from Clathria Sp. through the KnapSack database site and the preparation of EGFR receptors that have been complexed with allosteric inhibitors, namely proteins with PDB code 5D41. Each compound was docked to the Allosteric Site of the natural ligand and then molecular dynamics simulations were performed on the compound with the best docking energy compared to the natural ligand. From the docking results, the Clathrin_A compound showed the lowest binding energy compared to other metabolites, and the value was close to the natural ligand. Then from the molecular dynamics results, the clathrin_A compound shows good stability and resembles the natural ligand, which is analyzed through RMSD, RMSF, SASA, Rg, and PCA, and shows the binding free energy from MMPBSA analysis which is close to the natural ligand. It can be concluded, Clathrin_A compound has potential as an allosteric inhibitor.
Keywords: Marine-natural products, EGFR, Allosteric inhibitor
Graphical abstract
Highlights
-
•
It was found compound from the Clathria Sp. based on molecular docking which has good affinity on EGFR allosteric site.
-
•
Compound from Clathria Sp. predicted have strong amino acid interaction.
-
•
The compounds that are screened have good complex stability during molecular dynamics simulation.
1. Introduction
The use of EGFR tyrosine kinase inhibitors (EGFR-TK) in treatment is currently facing various challenges due to multiple mutations (Sabbah et al., 2020; Dong et al., 2021). A significant obstacle is the emergence of a secondary deactivation mutation known as the T790M mutation in exon 20 (Leonetti et al., 2019; Kashima et al., 2020). This mutation is a major contributor to the failure of first-generation EGFR tyrosine kinase inhibitors like Erlotinib and Gefitinib, particularly in patients with NSCLC who initially responded to this therapy (Leonetti et al., 2019; Kashima et al., 2020). Second-generation EGFR inhibitors, such as Afatinib, target both mutants and the wild-type EGFR (WT), which can lead to side effects and dose-limiting toxicity (Harvey et al., 2020). To address these challenges, third-generation EGFR inhibitors have been developed and are currently in clinical and pre-clinical trials (Nagasaka et al., 2021; Cooper et al., 2022). These third-generation drugs can effectively inhibit positive EGFR-TK mutants with the T790M mutation by forming covalent bonds with amino acid C797 (Roskoski, 2019). Recent research has shown that the use of drugs like osimertinib and others from the third generation can overcome resistance caused by C797S mutations, preventing the formation of covalent bonds (Lu et al., 2018).
In 2016, reported the fourth-generation EGFR tyrosine kinase inhibitor, EGFR Allosteric Inhibitor 001 (EAI001) and EGFR Allosteric Inhibitor 045 (EAI045). These were the first allosteric inhibitors designed to target T790M and C797S mutations in EGFR (Jia et al., 2016; Wang et al., 2017). This represented a significant advancement as these drugs are the first of their kind in the EGFR tyrosine kinase inhibitor category that occupy an allosteric site on the EGFR and do not compete for binding.
The potential for natural products to serve as sources for drug candidate compounds is extensive, cost-effective, and readily accessible. In various studies we've encountered, numerous in silico investigations have been conducted on several natural compounds as potential inhibitors of the EGFR Allosteric Site (Saini et al., 2023). Hence, we are keen on investigating the capability of compounds found in natural substances to function as EGFR allosteric inhibitors. In this regard, we are particularly focusing on marine natural products derived from marine organisms, specifically Clathria Sp. Marine organisms have a long history of use in traditional medicine, yet research on their potential as pharmaceutical agents is still in its early stages (Negm et al., 2023), while the novelty of the marine natural products tends to be higher than terrestrial natural products (Negm et al., 2023). Clathria is a genus within the family Microconidia, comprising 544 species, and one of these species is known to be present in the Southeast Sulawesi Province of Indonesia (Sahidin et al., 2019). Based on that, this study aims to analyze the inhibitory potential of the compounds contained in the marine species, Clathria Sp., on the crystal structure of the EGFR that complexed with its allosteric inhibitor.
2. Materials and methods
2.1. Ligand preparation
The ligands chosen for this study were marine natural products sourced from sponges, specifically Clathria species. The chemical structures of these compounds were retrieved from the KnapSack database (http://www.knapsackfamily.com/KNApSAcK/(Afendi et al., 2012) and KnapSack 3D database (http://knapsack3d.sakura.ne.jp/) (Nakamura et al., 2013). Initially, the ligands were stored in. mol format and were subsequently converted to. pdb format using the Discovery Studio 2017 software (Ramadhan et al., 2021).
2.2. Protein preparation
The 3D structure of the EGR with Allosteric Site was acquired from the Protein Data Bank (PDB) (www.rcsb.org) using the PDB code 5D41 (with a resolution of 2.31 Å and obtained from X-RAY DIFFRACTION method). The native ligand was isolated using Discovery Studio 2017, and water molecules and unique ligands were removed. The protein was assigned partial charges using the Kollman charge force field and polar hydrogens were added. All protein preparation procedures were performed using the AutoDock4 software (Jia et al., 2016; Ramadhan et al., 2021; Hikmawati et al., 2022).
2.3. Molecular docking simulation
The allosteric inhibitor, which was native ligand, was subjected to validated the docking process. Native ligand preparation included adjusting the ligand torque to its default of each ligand using torsion tree tools and adding necessary charges in autodock software. The docking protocol was subsequently validated by re-docking the native ligands, and the best conformation of the docking result was compared to the initial conformation of the native ligand. The docking protocol was considered validated if the root mean square deviation (RMSD) value was less than 2.0 Å. The grid coordinates for the native ligand on the template protein during the docking process were set at X = −23.037, Y = 31.467, Z = 12.091, and the dimensions of the grid box used were X = 40 Å, Y = 40 Å, Z = 40 Å. These same coordinates were also employed for docking the metabolite compound (Jia et al., 2016; Bell and Zhang, 2019). The number of Genetic Algorithm runs in 100 conformation of each ligand. The 3D structure of the compounds contained in Clathria Sp. were docked to the active site, which the binding energy and amino acid interactions were analyzed. The docking results were analyzed using Discovery Studio 17 (Pitaloka et al., 2021).
2.4. Molecular dynamics and binding free energy calculation
The compound with the best binding energy was subjected to a 100 ns?Molecular Dynamics (MD) simulation with a 2 fs timestep, using Gromacs 2016.3 software and the AMBER99SB-ILDN force field (Abraham et al., 2015). Ligand topology and parameters were generated using ACPYPE (Sousa da Silva and Vranken, 2012). The Particle Mesh Ewald (PME) method was employed to calculate electrostatic forces over a distance (Essmann et al., 1995). Solvation was performed using the TIP3P water cube model (Mark and Nilsson, 2001), and system neutralization was achieved by adding Na+ and Cl-ions through autoionization. The simulation process involved several stages, starting with minimization, followed by heating to 310 K, temperature equilibration, pressure equilibration, and the actual MD simulation. Subsequently, the stability of the system was assessed through Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), Solvent Accessible Surface Area (SASA), Radius of Gyration (Rg), and Principal Component Analysis (PCA) (Lolok et al., 2021). Furthermore, the binding free energy was calculated using the Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) method. The g_mmpbsa package, integrated with Gromacs, was used for these calculations. The binding free energy (ΔGbind) of the complex was determined as the difference between the free energies of the complex (ΔGcomplex) and the unbound receptor (ΔGrec) and ligand (ΔGligand), theoretically described by the equations: ΔGbind = Gcomplex - Grec - Gligand; ΔGbind = ΔEMM + ΔGsol - TΔS. The ΔEMM component includes ΔEbond, ΔEangle, ΔEtorsion, ΔEvdw, and ΔEELE (Ren et al., 2020). The polar desolvation energy was calculated using the Poisson-Boltzmann equation with a 0.5 Å grid size, and a solvent dielectric constant of 80 was used to represent water. The nonpolar contribution was determined based on the solvent-accessible surface area with a solvent radius of 1.4 Å. The binding free energy of the complexes was computed from the MD simulation outputs, considering 500 snapshots from 1 to 100 ns of the simulation trajectories (Pitaloka et al., 2021).
3. Results and discussion
3.1. Molecular docking simulation
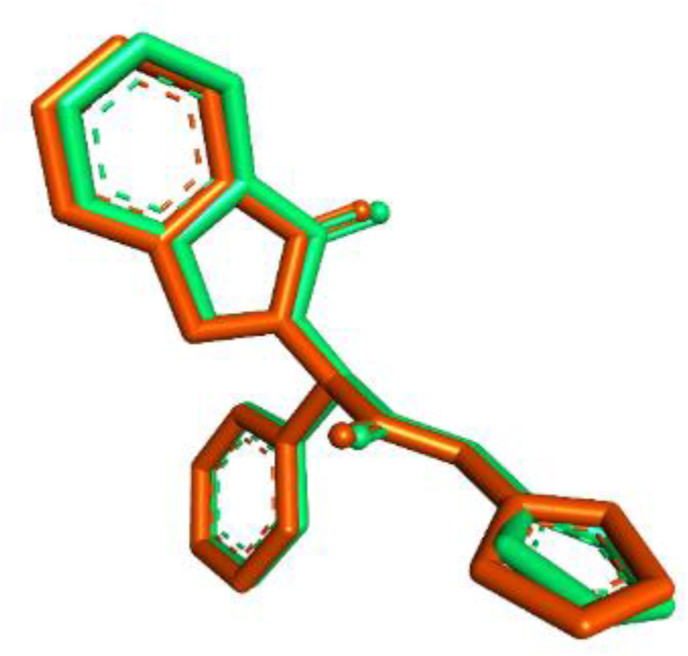
The process of molecular docking commenced with the re-docking of the native ligand (EAI) into the binding pocket of the 5D41 protein to validate the docking methodology. The RMSD value measured from all heavy atoms and obtained 1.134 Å (as depicted in Fig. 1), indicating a satisfactory outcome (RMSD below 2.0 Å showed good validation results) and its confirm the docking method was valid. The binding energy of the re-docked EAI was determined to be −11.09 kcal/mol.
Fig. 1.
Overlapped ligand structures. Green is native ligand conformation before docking process and Red is native ligand conformation after docking process.
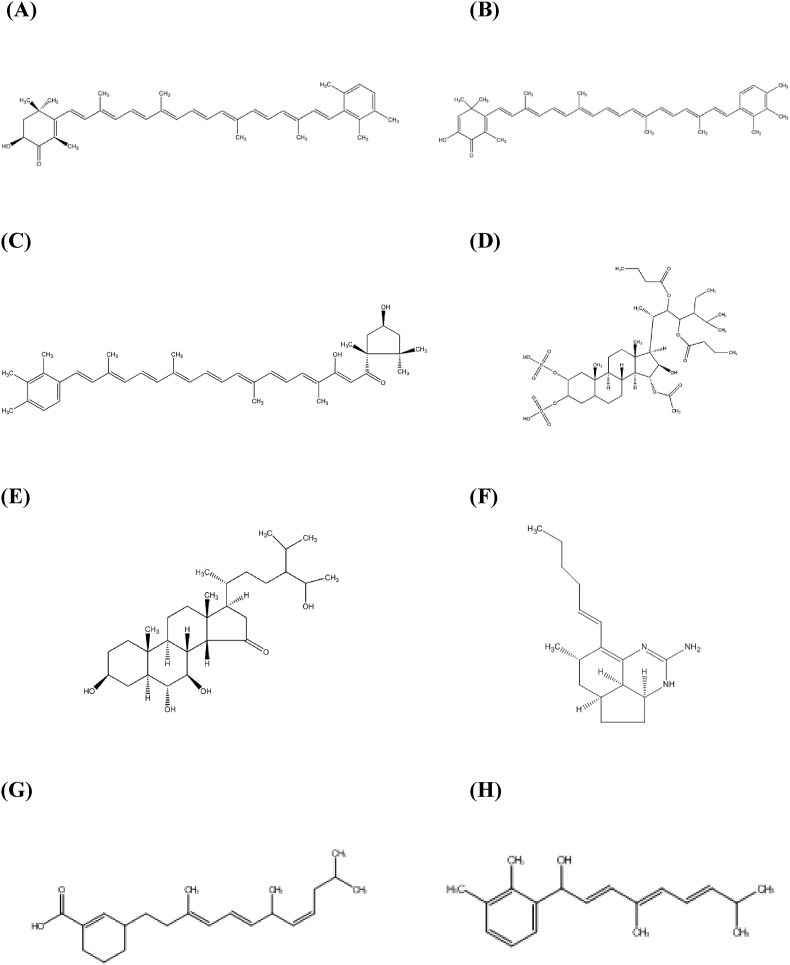
The ligands tested consisted of eight bioactive compounds contained in Clathria Sp., including Isoclathriaxanthin, Tedanin, Trikentriorhodin, Clathsterol, Clathriol, Mirabilin G, Clathrin A, Clathrin B (Fig. 2). The binding energy of each ligand can be seen in Table 1. From the results of binding energy analysis, each metabolite shows diverse binding energy. Clathsterol, Isoclathriaxanthin, Tedanin, and Trikentriorhodin ligands gave positive values, indicating they have no binding and inhibition ability towards the receptor. The very large positive values shown by these ligands are due to the very bulky structure with very long hydrocarbon chains of these ligands, causing a size mismatch on the active side of the receptor.
Fig. 2.
2D structure of ligands isoclathriaxanthin (A), tedanin (B), trikentriorhodin (C), clathsterol (D), clathriol (E), mirabilin g (F), clathrin a (G), clathrin B (H).
Table 1.
Binding energy and inhibition Constant of ligands obtained from docking results.
| Ligands | Binding energy (kcal/mol) | Inhibition Constant (Ki) |
|---|---|---|
| Native Ligand (57N) | −11.09 | 7.4 nM |
| Trikentriorhodin | 864,000 | – |
| Tedanin | 1,400,000 | – |
| Mirabilin G | −8.38 | 714.12 nM |
| Isoclathriaxanthin | 1,390,000.0 | – |
| Clathsterol | 96.39 | – |
| Clathriol | −8.46 | 634.14 nM |
| Clathrin B | −8.3 | 821.06 nM |
| Clathrin A | −9.43 | 122.05 nM |
Furthermore, the ligands Clathrin A, Clathrin B, Clathriol, and Mirabilin G showed negative values, indicating their ability to inhibit EGFR through their Allosteric Site. Overall, Clathrin A ligand gave the most satisfactory result with a docking score of - 9.43 kcal/mol, which is close to the docking score of EAI natural ligand which is - 11.09 kcal/mol.
3.2. Ligand-receptor interaction
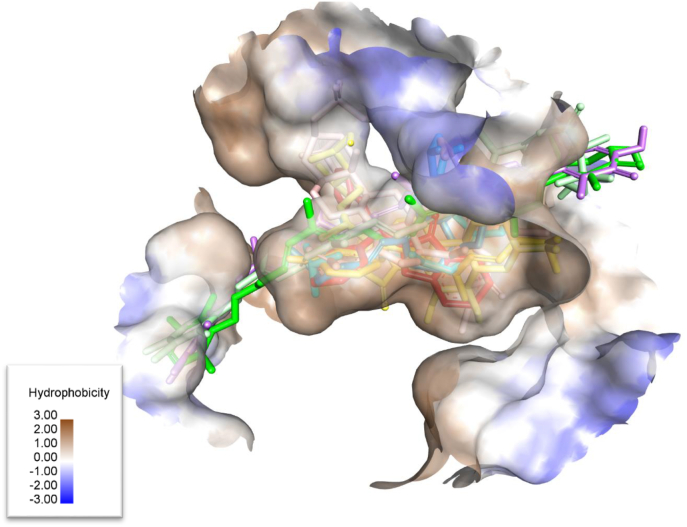
Next, we observed the molecular interactions to evaluate the binding of each compound. Observed from the visualization of 3D ligand-ligand binding (Fig. 3), the active site where the natural ligand and other compounds bind, is dominant in the hydrophobic region of the receptor. Furthermore, observed from Table 2, each compound shows a similar number of hydrophobic interactions, in the range of 7–10 hydrophobic interacting residues. Then, in terms of total interactions, compounds with a total of a dozen residues have a negative binding energy similar to the native ligand. However, compounds with total interactions above 20, show positive binding energy. This is due to the large structure of these compounds which causes steric collisions on the small Allosteric Site of EGFR.
Fig. 3.
3D interaction of amino acids on the Allosteric Site of EGFR with (a) Native Ligand (red), (b) Trichentriorhodin (light green) (c) Tedanin (purple), (d) Mirabilin G (blue), (e) Isoclathriaxanthin (dark green), (f) Clathsterol (salem pink), (g) Clathriol (yellow), (h) Clathrin B (mustard), (i) Clathrin A (aqua blue).
Table 2.
Recap of ligand-receptor amino acid interactions.
| Ligands | Hydrogen Bond | Total Residue with Hydophobic Interaction | Total Residue Interaction |
|---|---|---|---|
| Native Ligand (57N) | ASP A:855 | 10 | 14 |
| Trikentriorhodin | – | 9 | 27 |
| Tedanin | – | 9 | 27 |
| Mirabilin G | – | 5 | 16 |
| Isoclathriaxanthin | – | 7 | 25 |
| Clathsterol | LYS A:745, ILE A:774, TYR A:727 | 7 | 33 |
| Clathriol | ASP A:855, ILE A:759 | 8 | 21 |
| Clathrin B | – | 7 | 16 |
| Clathrin A | – | 8 | 17 |
3.3. Molecular dynamics analysis
Identification of the affinity of Clathrin A to the allosteric active site of EGFR was followed by molecular dynamics simulations. Molecular dynamics was successfully performed for 100 ns of both complexes, native-receptor and clathrin A-receptor. The stability of the system during simulation was measured through RMSD, RMSF, and SASA (Fig. 4). The stability of the clathrin A complex at the beginning of the simulation up to 80 ns is slightly higher than the native ligand at 0.25 A - 0.4 Å, while the native ligand is in the range of 0.15–0.3 Å. However, both complexes show similarities in the fluctuations measured through the RMSD graph at the last 20 ns of simulation. Furthermore, the calculated average RMSD fluctuations for each system were 0.29 Å (clathrin A-receptor) and 0.23 Å (native-receptor). The amino acid fluctuations of the two complex systems calculated by RMSF showed the same pattern in all regions.
Fig. 4.
Graph of RMSD (A), RMSF (B), and SASA value of acarbose-amylase complex (red) and ursolic acid-amylase complex (black).
SASA is performed to predict the extent of protein conformational changes during the simulation, which can be accessed by water molecules. SASA was analyzed during 100 ns of MD trajectory simulation, which is shown in the SASA analysis shows the value of the clathrin A-receptor complex compared to the native-receptor complex shows similar fluctuations. From the calculation of the average value, the SASA value for the clathrin A-receptor complex is 160.81 nm2 and the native-receptor complex is 157.57 nm2. The SASA analysis also showed very similar results between the two complexes. Based on these analyses, it positively indicates that clathrin A exhibits an amino acid binding similar to EGFR allosteric inhibitors and clathrin A might work as a potential inhibitor similar to existing EGFR Allosteric Inhibitors.
3.4. Binding free energy calculation
The binding free energies were calculated from the molecular dynamics pathways of both complexes using the MM-PBSA method in the time range of 0–100 ns (see Table 3). The analysis results indicated that in both complex systems, the polar solvation energy is opposite to the binding energy due to its negative value, while the van der Waals, electrostatic, and SASA energies favor the formation of a bond. The total binding free energy of the clathrin A-receptor shows a figure almost equivalent to that of the native-clathrin complex (Total binding free energy of the clathrin A-receptor complex is −116.565 kJ/mol; native-receptor is −133.246 kJ/mol). This occurs because the Van de waals energy (KJ/mol) and Electrostatic energy (KJ/mol) in the native-receptor complex are more negative than the clathrin A-receptor. However, the values between the two complexes are not too far apart.
Table 3.
Binding free energy calculation using MMPBSA methods.
| Binding free energy calculations | Ligand complex in EGFR Allosteric Site |
|
|---|---|---|
| Native ligand | Mirabilin_G | |
| Van de waals energy (KJ/mol) | −221.199±11.680 | −188.038±14.341 |
| Electrostatic energy (KJ/mol) | −84.840±8.093 | −54.922±8.773 |
| Polar solvation energy (KJ/mol) | 191.053±11.444 | 146.510±15.903 |
| SASA energy (KJ/mol) | −18.261±0.724 | −20.116±0.793 |
| Total binding energy (KJ/mol) | −133.246±13.335 | −116.565±17.907 |
4. Conclusions
Based on molecular docking and dynamics analysis, clathrin_A compound of Clathria Sp. has the potential to be developed as an anticancer compound targeting the allosteric site of EGFR.
Disclosure statement
The authors declare no competing financial interests.
CRediT authorship contribution statement
Nurisyah: Conceptualization, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, Supervision, Project administration, Funding acquisition. Dwi Syah Fitra Ramadhan: Conceptualization, Methodology, Study design, Software, Validation, Writing – original draft, Writing – review & editing, Visualization. Ratnasari Dewi: Resources, Data curation. Asyhari asikin: Software, Validation. Dwi Rachmawaty Daswi: Formal analysis, Writing – review & editing. Adriyani adam: Methodology, Study design, Investigation, Resources. Chaerunnimah: Resources, Data curation. Sunarto: Writing – review & editing, Project administration. Rafika: Writing – review & editing, Project administration. Artati: Writing – review & editing, Project administration. Taufik Muhammad Fakih: Writing – review & editing, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are grateful for the support of the Poltekkes Kemenkes Makassar so the research can be carried out.
Handling Editor: Dr A Wlodawer
Data availability
No data was used for the research described in the article.
References
- Afendi F.M., et al. KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 2012;53(2):e1. doi: 10.1093/pcp/pcr165. [DOI] [PubMed] [Google Scholar]
- Bell E.W., Zhang Y. DockRMSD: an open-source tool for atom mapping and RMSD calculation of symmetric molecules through graph isomorphism. J. Cheminf. 2019;11(1):40. doi: 10.1186/s13321-019-0362-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A.J., Sequist L.V., Lin J.J. Third-generation EGFR and ALK inhibitors: mechanisms of resistance and management. Nat. Rev. Clin. Oncol. 2022;19(8):499–514. doi: 10.1038/s41571-022-00639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R.-F., et al. EGFR mutation mediates resistance to EGFR tyrosine kinase inhibitors in NSCLC: from molecular mechanisms to clinical research. Pharmacol. Res. 2021;167 doi: 10.1016/j.phrs.2021.105583. [DOI] [PubMed] [Google Scholar]
- Harvey R.D., et al. Afatinib for the treatment of EGFR mutation-positive NSCLC: a review of clinical findings. J. Oncol. Pharm. Pract. : official publication of the International Society of Oncology Pharmacy Practitioners. 2020;26(6):1461–1474. doi: 10.1177/1078155220931926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikmawati D., et al. Pharmacophore-guided virtual screening and dynamic simulation of Kallikrein-5 inhibitor: Discovery of potential molecules for rosacea therapy. Inform. Med. Unlocked. 2022;28 doi: 10.1016/j.imu.2022.100844. [DOI] [Google Scholar]
- Jia Y., et al. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature. 2016;534(7605):129–132. doi: 10.1038/nature17960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima K., et al. CH7233163 overcomes osimertinib-resistant EGFR-del19/t790m/C797S mutation. Mol. Cancer Therapeut. 2020;19(11):2288–2297. doi: 10.1158/1535-7163.MCT-20-0229. [DOI] [PubMed] [Google Scholar]
- Leonetti A., et al. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer. 2019;121(9):725–737. doi: 10.1038/s41416-019-0573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolok N., et al. Molecular docking and molecular dynamics studies of bioactive compounds contained in noni fruit (Morinda citrifolia L .) against human pancreatic α -amylase. J. Biomol. Struct. Dyn. 2021;0(0):1–8. doi: 10.1080/07391102.2021.1894981. [DOI] [PubMed] [Google Scholar]
- Lu X., et al. Targeting EGFR(L858R/T790M) and EGFR(L858R/T790M/C797S) resistance mutations in NSCLC: current developments in medicinal chemistry. Med. Res. Rev. 2018;38(5):1550–1581. doi: 10.1002/med.21488. [DOI] [PubMed] [Google Scholar]
- Nagasaka M., et al. Beyond osimertinib: the development of third-generation EGFR tyrosine kinase inhibitors for advanced EGFR+ NSCLC. J. Thorac. Oncol. : official publication of the International Association for the Study of Lung Cancer. 2021;16(5):740–763. doi: 10.1016/j.jtho.2020.11.028. [DOI] [PubMed] [Google Scholar]
- Nakamura K., et al. KNApSAcK-3D: a three-dimensional structure database of plant metabolites. Plant Cell Physiol. 2013;54(2):e4. doi: 10.1093/pcp/pcs186. [DOI] [PubMed] [Google Scholar]
- Negm W.A., Ezzat S.M., Zayed A. Marine organisms as potential sources of natural products for the prevention and treatment of malaria. RSC Adv. 2023;13(7):4436–4475. doi: 10.1039/d2ra07977a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitaloka D.A., et al. Docking-based virtual screening and molecular dynamics simulations of quercetin analogs as enoyl-acyl carrier protein reductase (InhA) inhibitors of Mycobacterium tuberculosis. Sci. Pharm. 2021 doi: 10.3390/scipharm89020020. [DOI] [Google Scholar]
- Ramadhan D.S.F., et al. In silico analysis of marine natural product from sponge (Clathria Sp.) for their activity as inhibitor of SARS-CoV-2 Main Protease. J. Biomol. Struct. Dynam. 2021:1–7. doi: 10.1080/07391102.2021.1959405. [DOI] [PubMed] [Google Scholar]
- Ren J., et al. Assessing the performance of the g_mmpbsa tools to simulate the inhibition of oseltamivir to influenza virus neuraminidase by molecular mechanics Poisson–Boltzmann surface area methods. J. Chin. Chem. Soc. 2020 doi: 10.1002/jccs.201900148. [Preprint] [DOI] [Google Scholar]
- Roskoski R.J. Small molecule inhibitors targeting the EGFR/ErbB family of protein-tyrosine kinases in human cancers. Pharmacol. Res. 2019;139:395–411. doi: 10.1016/j.phrs.2018.11.014. [DOI] [PubMed] [Google Scholar]
- Sabbah D.A., Hajjo R., Sweidan K. Review on epidermal growth factor receptor (EGFR) structure, signaling pathways, interactions, and recent updates of EGFR inhibitors. Curr. Top. Med. Chem. 2020;20(10):815–834. doi: 10.2174/1568026620666200303123102. [DOI] [PubMed] [Google Scholar]
- Sahidin I., et al. Structural relationship among steroids from Sulawesi Tenggara's sponge Clathria sp. and their radical scavenger activity. IOP Conf. Ser. Earth Environ. Sci. 2019;370(1) doi: 10.1088/1755-1315/370/1/012027. [DOI] [Google Scholar]
- Saini R., et al. Discovery of the allosteric inhibitor from actinomyces metabolites to target EGFRCSTMLR mutant protein: molecular modeling and free energy approach. Sci. Rep. 2023;13(1):8885. doi: 10.1038/s41598-023-33065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Song Y., Liu D. EAI045: the fourth-generation EGFR inhibitor overcoming T790M and C797S resistance. Cancer Lett. 2017;385:51–54. doi: 10.1016/j.canlet.2016.11.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.