Abstract
Objectives
The aim of this study was to investigate the clinical and molecular characteristics of Klebsiella pneumoniae infection from a tertiary general hospital in Wuhan, China.
Methods
From December 2019 to August 2022, 311 non-duplicate isolates of K. pneumoniae were collected from a tertiary hospital in Wuhan. These comprised 140 carbapenem-resistant K. pneumoniae (CRKP) isolates and 171 carbapenem-susceptible K. pneumoniae (CSKP) isolates. The clinical characteristics of patients with K. pneumoniae infection were retrospectively collected. Polymerase chain reaction (PCR) assays were used to identify the main carbapenem resistance genes, virulence genes and multi-locus sequence typing (MLST) profiles of the isolates, and the Galleria mellonella infection model was used to determine their virulence phenotypes.
Results
Independent risk factors for CRKP infection were hypertension, neurological disorders, being admitted to the intensive care unit (ICU) and prior use of antibiotics. Patient with CRKP infection had higher mortality than those with CSKP infection (23.6% vs 14.0%, P < 0.05). One hundred and two sequence types (STs) were identified among the K. pneumoniae isolates, and the most prevalent ST type was ST11 (112/311, 36.0%). All of the ST11 isolates were CRKP. Among the 112 ST11 isolates, 105 (93.8%) harboured the carbapenem resistance gene blaKPC-2 (ST11-KPC-2), and of these isolates, 78 (74.3%, 78/105) contained all of the four virulence genes, namely rmpA, rmpA2, iroN and iucA, suggesting that these genes were widespread among the isolates responsible for K. pneumoniae infections.
Conclusion
In this study, ST11-KPC-2 was responsible for most of the K. pneumoniae infection cases. Carbapenem resistance rather than the co-occurrence of the virulence genes rmpA, rmpA2, iroN and iucA was associated with K. pneumoniae infection-related mortality during hospitalisation. Furthermore, a high proportion of ST11-KPC-2 isolates carried all of the four virulence genes.
Keywords: CRKP, CSKP, ST11, Virulence genes, BlaKPC-2, BlaNDM like, BlaOXA-48 like
Introduction
Klebsiella pneumoniae is a gram-negative, nonmotile, encapsulated opportunistic pathogen that generally colonises human mucous membranes, including the nasopharynx and gastrointestinal tract and is ubiquitously found in soils and the healthcare environment such as on the surfaces of medical devices [1, 2]. In immunocompromised patients, infants and older adults, it can cause a variety of infectious diseases, including lower respiratory tract infections, urinary tract infections, bacteraemia and sepsis, and can even lead to infection-related death [3–5]. Furthermore, infection caused by drug-resistant K. pneumoniae, especially by carbapenem-resistant K. pneumoniae (CRKP), are known to be associated with longer hospital stays, higher medical costs and a higher risk of mortalities than those caused by carbapenem-susceptible K. pneumoniae [6].
Carbapenem is regarded as an effective antimicrobial agent for treating serious K. pneumoniae infections [7]. However, carbapenem resistance has been found in K. pneumoniae isolates in many regions worldwide and has been recognised as a major public health concern due to its rapid transmission [8]. In China, the average resistance rates of K. pneumoniae to imipenem and meropenem were 22.6% and 24.2%, respectively, in 2022, which were significantly higher than the rates of 2.9% and 3.0% reported in 2005 (http://www.chinets.com) [9]. K. pneumoniae carbapenemase (KPC) and New Delhi metallo-β-lactamase (NDM) encoded by blaKPC and blaNDM, respectively, are the commonest factors responsible for carbapenem resistance in K. pneumoniae in most countries worldwide, whereas oxacillinase-48 (OXA-48) is most common in Africa [10–12]. Moreover, co-occurrence of blaKPC-2 and blaNDM in K. pneumoniae has been reported in the USA, China and Brazil [13, 14]. A promising new β-lactamase inhibitor combination for treating infections caused by KPC- and NDM-co-producing K. pneumoniae is aztreonam-avibactam [15, 16].
Hypervirulent CRKP (hv-CRKP) is a growing public concern due to its high drug resistance and virulence, posing a serious threat to human health [17]. In 2018, in a hospital outbreak of ST11 hv-CRKP in Zhejiang province, China, the fatality rate was 100%. This was found to be attributable to the presence of pLVPK, a virulence plasmid (~ 170 kb long) containing two capsular polysaccharide regulatory genes (rmpA and rmpA2) and multiple siderophore gene clusters (iucABCD/iutA/iroBCDN), which imparted concurrent hypervirulence, multidrug resistance and high transmissibility. It was also shown that the occurrence of the virulence genes rmpA, rmpA2, iroN and iucA was correlated with the presence of the virulence plasmid pLVPK [18–20]. Furthermore, an increasing occurrence of hv-CRKP is also being observed in other sequence types (STs), including ST15 and ST147 [21–24]. Therefore, this study aimed to find the risk factors for and mortality associated with CRKP infection by comparing the clinical characteristics of CRKP infection cases with those of CSKP infection cases and to identify the molecular characteristics, namely, the presence of carbapenem resistance genes (blaKPC-2, blaNDM like and blaOXA-48 like) and four virulence genes (rmpA, rmpA2, iroN and iucA), that are correlated with the resistance and virulence of CRKP isolates.
Methods
Collection of bacterial isolates and medical records
The study was carried out in the 3300-bed Zhongnan Hospital of Wuhan University, a grade A tertiary general hospital in Hubei province. Three hundred and eleven non-duplicate isolates of K. pneumoniae were randomly collected from patients with K. pneumoniae infection admitted to this hospital from December 2019 to August 2022. These isolates were obtained from diverse clinical specimens, including sputum (n = 119, 38.3%), urine (n = 81, 26%), bronchoscopic lavage fluid (n = 32, 10.3%), blood (n = 27, 8.7%), drainage fluid (n = 18, 5.8%) and others fluids (ascites, bile, cerebrospinal fluid, hydrothorax secretion, perianal swab, pus, wound secretion and vaginal secretion) (n = 34, 10.9%).
Patients with K. pneumoniae infection were included in the study and classified as follows: K. pneumoniae infection cases were classified as bacteraemia (n = 27, 8.7%) when a blood culture was positive for a K. pneumoniae strain and the patient had clinical signs of systemic inflammatory response syndrome; K. pneumoniae infections were classified as nonbacteraemia through nonblood cultures, typically for cases such as pneumonia (n = 151, 48.5%), urinary tract infection (n = 81, 26%), liver abscess (n = 9, 2.8%), abdominal infection (n = 16, 5.1%), cholecystitis (n = 8, 2.6%) and others (n = 19, 6.1%). Electronic medical records of the patients were retrospectively collected, and the following data were extracted: gender; age; smoking status; comorbidities such as diabetes, hypertension, liver abscess, respiratory failure, cancer (including haematological malignancies) and neurological disorders, use of antibiotics in the 3 months before the current hospitalisation and during the current hospitalisation until a positive K. pneumoniae culture was obtained; initial severity (in the ICU); immunosuppressive conditions containing immunosuppressives therapy, the carry of human immunodeficiency virus (HIV) and high dosage administration of corticoids; the coexistence of carbapenem-resistant Acinetobacter baumannii in the clinical specimen and mortality outcome of this hospitalisation. This study was approved by the Ethics Committee of Zhongnan Hospital, Wuhan University.
K. pneumoniae isolate identification and antimicrobial susceptibility testing (AST)
All K. pneumoniae isolates were identified using matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry (Vitek MS, bioMérieux). The AST for the K. pneumoniae isolates was carried out using a VITEK 2 Compact System and the VITEK 2 AST-GN16 Test Kit (bioMerieux Inc., Durham, NC, USA), which contains the following antibiotics: amikacin (AN), amoxicillin–clavulanicn acid (AMC), ampicillin (AM), aztreonam (ATM), cefazolin (CZ), cefepime (FEP), cefoxitin (FOX), ceftriaxone (CRO), ciprofloxacin (CIP), gentamicin (GM), imipenem (IPM), levofloxacin (LEV), piperacillin–tazobactam (TZP), tobramycin (TM) and trimethoprim–sulfamethoxazole (SXT). In addition, the Kirby–Bauer method was used to test the ceftazidime–avibactam (CZA) susceptibility of the CRKP isolates according to the manufacturer’s instructions. Antibiotic susceptibility was defined according to the antibiotic breakpoints established by the Clinical & Laboratory Standards Institute. Following the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines, the minimum inhibitory concentrations (MICs) of polymyxin B for CRKP strains were determined by the broth microdilution method, and the breakpoints for colistin were as reported by EUCAST (breakpoint tables for the interpretation of MICs and zone diameters, Version 11.0).
Detection of antimicrobial resistance and virulence genes
Genomic DNA was extracted from the K. pneumoniae isolates using a DNA extraction kit (Aidlab, Beijing) according to the manufacturer’s instructions. All primers used in the present study are shown in Table 1. The presence of three carbapenemase genes, blaKPC-2, blaNDM like and blaOXA-48 like, in the CRKP isolates was tested using a polymerase chain reaction (PCR) assay. The final volume of the PCR reaction mixture was 25 µL, which included 12.5 µL of 2 × buffer (Aidlab, Beijing), 9.5 µL ddH2O, 1 µL forward primer (10 µM), 1 µL reverse primer (10 µM) and 1 µL genomic DNA. The PCR amplification conditions were as follows: denaturation at 95 °C for 3 min, followed by 42 cycles of 95 °C for 30 s, 57 °C for 20 s and 72 °C for 20 s and final extension at 72 °C for 5 min. The PCR products were then subjected to 1.5–2.0% agarose gel electrophoresis for detection. The primers of the virulence genes rmpA, rmpA2, iroN and iucA were those used in a previous study [20]. The annealing temperature and elongation time were determined according to the primer and product length.
Table 1.
Primers used to screen for the presence of blaKPC-2, blaNDM like and blaOXA-48 like in K. pneumoniae
| Target genes | Primer name | Primer sequence | Product size | Reference |
|---|---|---|---|---|
| blaKPC-2 | blaKPC-2-F | ATGCGCTCTATCGGCGATAC | 91 bp | This study |
| blaKPC-2-R | ATGAGGTATCGCGCGCATC | |||
| blaNDM like | blaNDM like-F | GGGCCGTATGAGTGATTGCG | 213 bp | This study |
| blaNDM like-R | ATATCACCGTTGGGATCGAC | |||
| blaOXA-48 like | blaOXA-48 like -F | TTAAGTGGGATGGACAGACG | 179 bp | This study |
| blaOXA-48 like-R | ATTGCCCGAAATGTCCTCAT |
Multi-locus sequence typing (MLST)
Seven house-keeping genes (infB, pgi, mdh, phoE, gapA, tonB and rpoB) were amplified from the genomic DNA of the K. pneumoniae isolates for MLST as described previously [25]. The amplified products were purified and subjected to Sanger’s dideoxy DNA sequencing (Tianyi Huiyuan, China). The resulting sequences were compared with the existing K. pneumoniae gene sequences available on the Pasteur Institute MLST website (http://bigsdb.pasteur.fr/klebsiella/). Each clinical isolate was then classified by the ST.
A Galleria mellonella model of K. pneumoniae infection
To test the virulence of the K. pneumoniae isolates, G. mellonella was purchased from Aidlab (China) and stored in the dark at 4 °C until testing. Overnight cultures of K. pneumoniae strains were washed with phosphate-buffered saline (PBS) and further adjusted with PBS to concentrations of 1 × 106 CFU/mL. Each bacterial isolate was injected into 10 G. mellonella, and the survival rates of the G. mellonella were recorded. The experiment was repeated three times.
Statistical analysis
SPSS 27.0 software was used for data processing. The risk factors for CRKP infection and infection-related mortality were analysed by univariate logistic regression analysis. The variables with P < 0.05 in the univariate analysis were included in the multivariate logistic regression model, and backward stepwise regression analysis was performed to identify independent risk factors. A chi-square test was used to analyse the co-occurrence of the four virulence genes in the CRKP and CSKP isolates. P < 0.05 was considered to indicate a statistically significant difference.
Results
Risk factors for CRKP infection
Factors associated with CRKP infection included prior use of two or more antibiotics (including carbapenems), prior use of carbapenems, hypertension, cancer, neurological disorders, ICU admission, and the coexistence of carbapenem-resistant A. baumannii in clinical specimens (P < 0.05) (Table 2). Among them, hypertension (odds ratio [OR], 1.824; 95% confidence interval [CI], 1.053–3.161; P = 0.032), neurological disorders (OR, 2.890; 95% CI, 1.568–5.326; P < 0.001), ICU admission (OR, 2.099; 95% CI, 1.148–3.836; P = 0.016), prior use of two or more antibiotics (OR, 4.124; 95% CI, 2.142–7.939; P < 0.001) and prior use of carbapenems (OR, 2.613; 95% CI, 1.314–5.196; P = 0.006) were independent risk factors for CRKP infection (Table 2).
Table 2.
Clinical characteristics of patients infected with K. pneumoniae
| Risk factors | Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| CRKP (n = 140) | CSKP (n = 171) | P | OR (95% CI) | P | |
| Age (years) | 63.6 18.7 | 63.7 16.3 | 0.949 | ||
| Male, n (%) | 91 (65.0%) | 120 (70.2%) | 0.333 | ||
| Smoking | 30 (21.4%) | 41 (24.0%) | 0.596 | ||
| Comorbidities, n (%) | |||||
| Diabetes | 45 (32.1%) | 51 (29.8%) | 0.661 | ||
| Hypertension | 73 (52.1%) | 70 (40.9%) | 0.049 | 1.824 (1.053–3.161) | 0.032 |
| Liver abscess | 4 (2.9%) | 12 (7.0%) | 0.099 | ||
| Respiratory failure | 31 (22.1%) | 25 (14.6%) | 0.086 | ||
| Cancer | 19 (13.6%) | 40(23.4%) | 0.028 | 1.163 (0.545–2.480) | 0.696 |
| Neurological disorders | 78 (55.7%) | 46 (26.9%) | < 0.001 | 2.890 (1.568–5.326) | < 0.001 |
| Initial severity, n (%) | |||||
| ICU | 97 (69.3%) | 63 (36.8%) | < 0.001 | 2.099 (1.148–3.836) | 0.016 |
| Immunosuppressive conditions, n (%) | 36 (25.9%) | 51 (29.8%) | 0.446 | ||
| Antibiotic use before K. pneumoniae infection, n (%) | |||||
| Two or more antibiotics | 113 (80.7%) | 56(32.7%) | < 0.001 | 4.124 (2.142–7.939) | < 0.001 |
| Carbapenem antibiotic | 71 (50.7%) | 28 (16.4%) | < 0.001 | 2.613 (1.314–5.196) | 0.006 |
| The coexistence of carbapenem-resistant A. baumannii in clinical specimens, n (%) | 28 (20.0%) | 16 (9.4%) | 0.007 | 0.695 (0.313–1.541) | 0.370 |
|
Outcomes Death, n (%) |
33 (23.6%) | 24 (14.0%) | 0.031 | ||
OR: odds ratio; CI: confidence interval; ICU: intensive care unit. The variables with P < 0.05 screened in univariate analysis were included in multivariate logistic regression analysis
Clinical cohort study of mortality: survival versus death in patients with K. pneumoniae infection
Among the 311 patients with K. pneumoniae infection, 57 (18.3%) patients died. Univariate analysis showed that respiratory failure, ICU admission, and prior use of carbapenems were factors related to death of in these patients. Multivariate analysis confirmed that respiratory failure (OR, 5.909; 95% CI, 2.911–11.996; P < 0.001) and ICU admission (OR, 12.285; 95% CI, 4.180–36.109; P < 0.001) were independent risk factors for death in these patients (Table 3).
Table 3.
Analysis of associated risk factors for death associated with K. pneumoniae infection
| Risk factors | Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Death (n = 57) | Survival (n = 254) | P | OR (95% CI) | P | |
| Age (years) | 65.9 ± 18.8 | 63.1 ± 17.1 | 0.265 | ||
| Male, n (%) | 37 (64.9%) | 174 (68.5%) | 0.601 | ||
| Smoking | 9 (15.8%) | 62 (24.4%) | 0.162 | ||
| Accompanying disease, n (%) | |||||
| Diabetes | 18 (31.6%) | 78 (30.7%) | 0.898 | ||
| Hypertension | 27 (47.4%) | 116 (45.7%) | 0.817 | ||
| Liver abscess | 1 (1.8%) | 15 (5.9%) | 0.201 | ||
| Respiratory failure | 30 (52.6%) | 26 (10.2%) | < 0.001 | 5.909 (2.911–11.996) | < 0.001 |
| Cancer | 8 (14.0%) | 51 (20.1%) | 0.294 | ||
| Neurological disorders | 25 (43.9%) | 99 (39.0%) | 0.498 | ||
| Initial severity, n (%) | |||||
| ICU | 53 (93.0%) | 107 (42.1%) | < 0.001 | 12.285 (4.180–36.109) | < 0.001 |
| Immunosuppressive conditions, n (%) | 19 (33.3%) | 68 (26.9%) | 0.329 | ||
| Antibiotic use before K. pneumoniae infection, n (%) | |||||
| Two or more antibiotics | 37 (64.9%) | 132 (52.0%) | 0.077 | ||
| Carbapenem antibiotic | 26 (45.6%) | 73 (28.7%) | 0.013 | 1.124 (0.564–2.239) | 0.739 |
| The coexistence of carbapenem-resistant A. baumannii in clinical specimens (%) | 12 (21.1%) | 32 (12.6%) | 0.099 | ||
| Co-carry rmpA, rmpA2, iroN and iucA, n (%) | 25 (43.9%) | 108 (42.5%) | 0.854 | ||
AST of the K. pneumoniae isolates
All of the K. pneumoniae isolates were resistant to AM. In total, 140 (45.0%) isolates were categorised as CRKP based on their IPM susceptibility. Compared with the CSKP isolates, the CRKP isolates had higher resistance rates to all antibiotics except ampicillin, to which all isolates were resistant. In addition, the CRKP isolates showed resistance rates of 1.4% and 6.4% for polymyxin B and ceftazidime–avibactam, respectively (Table 4). The detailed results of antibiotic susceptibility testing of CSKP and CRKP isolates are presented in Table 4.
Table 4.
Antimicrobial resistance profiles of K. pneumoniae
| All K. pneumoniae (n = 311) | CRKP (n = 140) | CSKP (n = 171) |
P (comparison between CRKP and CSKP) | |
|---|---|---|---|---|
| Antibiotics | Resistance | Resistance | Resistance | |
| IPM, n (%) | 140 (45.0) | 140 (100.0) | 0 (0.0) | / |
| AM | 311 (100.0) | 140 (100.0) | 171 (100.0) | / |
| AN | 115 (37.0) | 114 (81.4) | 1 (0.6) | < 0.001 |
| ATM | 173 (55.6) | 134 (95.7) | 39(22.8) | < 0.001 |
| AMC | 155 (49.8) | 135 (96.4) | 20 (11.7) | < 0.001 |
| CZ | 218 (70.1) | 137 (97.9) | 81(47.4) | < 0.001 |
| CRO | 184 (59.2) | 135 (96.4) | 49 (28.7) | < 0.001 |
| CIP | 193 (62.1) | 136 (97.1) | 57 (33.3) | < 0.001 |
| FOX | 154(50.0) | 133 (95.0) | 21 (12.3) | < 0.001 |
| FEP | 181(58.2) | 134 (95.7) | 47 (27.5) | < 0.001 |
| GM | 140(45.0) | 122 (87.1) | 18 (10.5) | < 0.001 |
| LEV | 179(57.6) | 136 (97.1) | 43 (25.1) | < 0.001 |
| SXT | 154(49.5) | 113 (80.7) | 41 (24.0) | < 0.001 |
| TZP | 141(45.3) | 135 (96.4) | 6 (3.5) | < 0.001 |
| TM | 128(41.2) | 119 (85.0) | 9 (5.3) | < 0.001 |
| PB | / | 2 (1.4) | / | / |
| CZA | / | 9 (6.4) | / | / |
AM: ampicillin; AN: amikacin; ATM: aztreonam; AMC: amoxicillin-clavulanicn acid; cefazolin; ceftriaxone; CIP: ciprofloxacin; FOX: cefoxitin; FEP: cefepime; GM: gentamicin; LEV: levofloxacin; SXT: trimethoprim/sulfamethoxazole; TZP: piperacillin/tazobactam; tobramycin; PB: polymyxin B; CZA: ceftazidime-avibactam
MLST of the K. pneumoniae isolates
All isolates were successfully typed by MLST and assigned to 102 STs. The most prevalent ST was ST11 (112, 36.0%), followed by ST23 (15, 4.8%), ST37 (9, 2.9%) and ST15 (9, 2.9%). Notably, all of the 112 ST11 isolates were CRKP. Fourteen STs were found among the 140 CRKP isolates. The detailed results of MLST for CRKP isolates are provided in Fig. 1. Among the CSKP isolates, 95 STs were found, with ST23 (13, 7.6%) being the most common ST, followed by ST68 (8, 4.7%), ST37 (6, 3.5%), ST25 (5, 2.9%), ST15 (4, 2.3%), ST412 (4, 2.3%), ST36 (4, 2.3%) and ST307 (4, 2.3%).
Fig. 1.
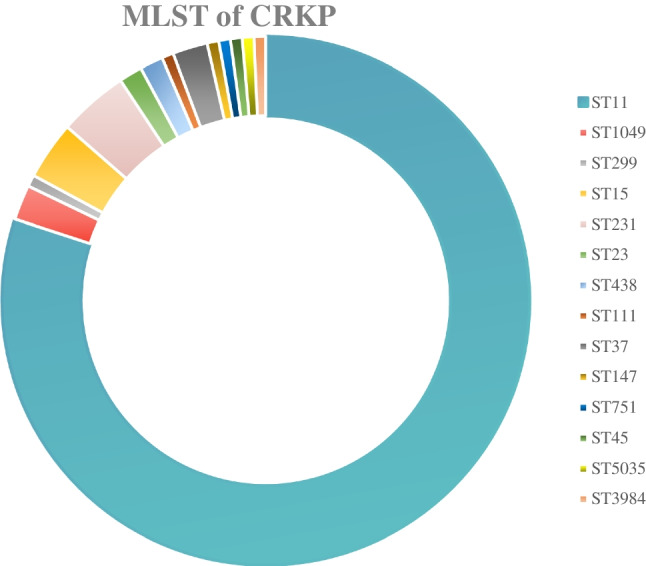
Classification characteristics of MLST of CRKP
Virulence gene and resistance gene profiles
Three hundred and eleven K. pneumoniae isolates were tested for the presence of the virulence genes rmpA, rmpA2, iroN and iucA. Overall, the positive rates of rmpA, rmpA2, iroN and iucA were 58.2% (181), 48.6% (151), 51.1% (159) and 46.6% (145), respectively. Furthermore, 133 (42.8%) of the total isolates contained all of the four virulence genes, 53 (17.0%) isolates contained one to three of the virulence genes, whereas 125 (40.2%) isolates carried none of the virulence genes. Among the 140 CRKP isolates, the positive rates of rmpA, rmpA2, iroN and iucA were 67.1% (94), 62.9% (88), 60.0% (84) and 59.3% (83), respectively; 83 (59.3%) of these isolates contained all of the four virulence genes, 13 (0.9%) isolates contained one to three of the virulence genes, while 44 (31.4%) isolates carried none of the virulence genes. Among the 171 CSKP isolates, the positive rates of rmpA, rmpA2, iroN and iucA were 50.9% (87), 36.8% (63), 43.9% (75) and 36.3% (62) respectively; 50 (29.2%) of these isolates contained all of the four virulence genes, 40 (23.4%) isolates contained one to three of the virulence genes, while 81 (47.4%) isolates carried none of the virulence genes. As shown in Table 5, CRKP was more likely than CSKP to carry all of the four virulence genes.
Table 5.
Analysis of K. pneumoniae carrying all of the four virulence genes
| Virulence genes | CRKP (n = 140) | CSKP (n = 171) | P |
|---|---|---|---|
| Co-carry rmpA, rmpA2, iroN and iucA, n (%) | 83 (59.3%) | 50 (29.2%) | < 0.001 |
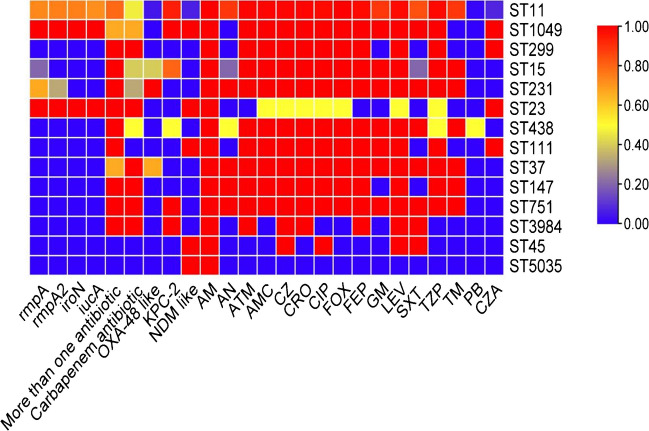
Three common carbapenemase-encoding genes—blaKPC-2, blaNDM like and blaOXA-48 like—were detected using a PCR assay. Among the 140 CRKP isolates, the positive rates of blaKPC-2, blaNDM like and blaOXA-48 like were 82.1%, 10.7% and 10.7%, respectively. The distribution of resistance genes in different STs is shown in Fig. 2. Of the 112 ST11 isolates, 105 (93.8%) harboured blaKPC-2; moreover, 78 (74.3%) of these 105 ST11-KPC-2 isolates contained all of the four virulence genes rmpA, rmpA2, iroN and iucA. The proportional relationships between STs and prior antibiotic use, resistance genes, virulence genes, and resistance ratios to different antibiotics are shown in Fig. 3.
Fig. 2.
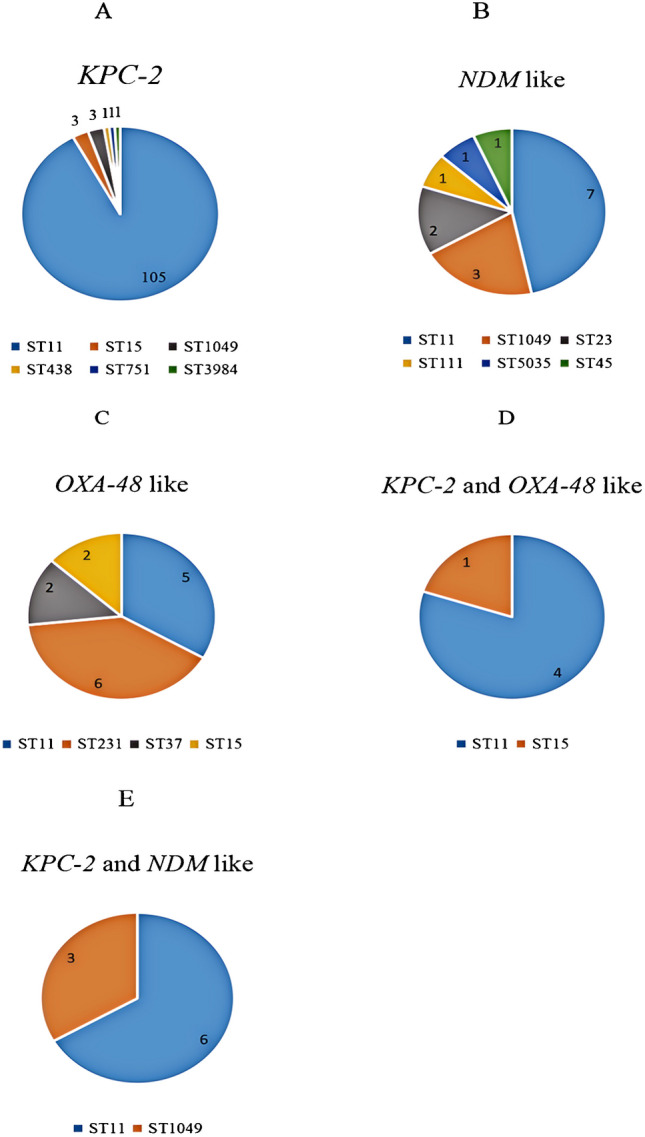
Genotyping characteristics of various drug resistance genes of CRKP
Fig. 3.
Heatmap of virulence gene carrier rates, multiple antibiotic use rates, carbapenem antibiotic use rates and antimicrobial resistance rates in different MLST CRKP strains
Comparison of the G. mellonella virulence test
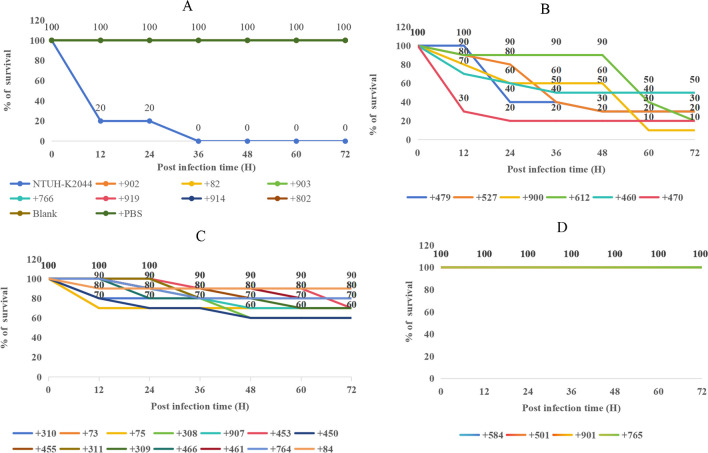
Patients with CRKP infection had a higher mortality rate than those with CSKP infection (23.6% vs 14.0%, P < 0.05) (Table 2); however, co-carrying rmpA, rmpA2, iroN and iucA was not associated with mortality in patients with K. pneumoniae infection (43.9% vs 42.5%, P = 0.854) (Table 3). To further investigate this phenomenon, 24 isolates carrying all of the four virulence genes were randomly selected and subjected to the G. mellonella virulence test. It was found that six of these isolates exhibited high virulence with a 72-h G. mellonella survival rate < 50%, 14 isolates showed low virulence with a 72-h G. mellonella survival rate > 50%, and four isolates were found to be nonvirulent with a 72-h G. mellonella survival rate of 100%. The hypervirulent positive-control K. pneumoniae strain NTUH-K2044 was found to have a 36-h G. mellonella survival rate of 0%, whereas seven K. pneumoniae isolates without these four virulence genes and the negative control strain had a 72-h G. mellonella survival rate of 100% (Fig. 4).
Fig. 4.
Experiments with different K. pneumoniae infecting Galleria mellonella. A Survival rates of NTUH-K2044 and 7 strains K. pneumoniae without these four virulence genes, + PBS empty control and blank control. Survival rates of 24 strains containing these four virulence genes; B 6 strains were high virulent, with a survival rate of 50% or less within 72 h; C 14 strains showed low virulence with a 72-h survival rate greater than 50%; D 4 strains were nonvirulent with 100% survival at 72 h. NTUH-K2044 carried these virulence genes: iroB, iroC, iroD, iroN, iucA, iucB, iucC, iucD, iutA, rmpA and rmpA2
Discussion
To improve the efficacy of CRKP infection treatment, we investigated the risk factors for CRKP infection and found ICU admission and prior antibiotic use to be associated with CRKP infection, consistent with previous studies [26]. Hypertension has been reported to be associated with bloodstream infection of CRKP [27], and our study showed that it was also one of the risk factors for CRKP infection. K. pneumoniae and A. baumannii have a mutually beneficial relationship, protecting and feeding each other to promote their survival [28]. Plasmids carrying resistance genes can also be transferred between them [29]. Accordingly, we found that the coexistence of carbapenem-resistant A. baumannii was associated with CRKP infection. Our results found that patients with neurological disorders were more susceptible to CRKP infection than those without neurological disorders, which may be related to the former patients’ need for absolute bed rest. We also found that patients with cancer were more susceptible to CSKP infection than those without cancer, but the reason was not clear and needs to be explored in a further study.
Our results demonstrated that different STs tended to carry different resistance genes. As in many other studies in China, a high clonal diversity of K. pneumoniae was observed in our study, with ST11 being the predominant ST [8, 30–32]. Another important phenomenon observed was that all of the ST11 isolates were CRKP with a blaKPC-2 positive rate of 93.8%, and ST11 isolates harbouring blaKPC-2 (ST11-KPC-2) accounted for 80.0% of the CRKP isolates and 36.0% of the total K. pneumoniae isolates. This suggests that ST11-KPC-2 was responsible for most of the K. pneumoniae infection cases. Thus, there is an urgent need to prevent and control the further spread of ST11-KPC-2 in hospitals across China. Furthermore, this study was the first to identify ST438, ST751 and ST3984 carrying blaKPC-2; ST111 and ST5035 carrying blaNDM like; and ST1049 carrying both blaKPC-2 and blaNDM like.
Infection cases attributable to hv-CRKP isolates are increasing by the year in hospitals, and plasmid recombination and fusion events have been found to be the two important evolutionary pathways responsible for the emergence of hv-CRKP isolates [22, 33]. The virulence plasmid pLVPK containing two capsular polysaccharide regulator genes (rmpA and rmpA2) and several siderophore gene clusters (e.g. iucABCD/iutA/iroBCDN clusters) is thought to contribute to the hypermucoviscous phenotype of K. pneumoniae [20, 18, 19]. Therefore, we further investigated the presence of four representative virulence genes carried by pLVPK in our isolates. We found that 133 (42.8%) of all K. pneumoniae isolates and 83 (59.3%) of the CRKP isolates carried all of the four virulence genes rmpA, rmpA2, iroN and iucA, and 78 of those 83 (94.0%) CRKP strains belonged to ST11-KPC-2. Such high proportions of K. pneumoniae, CRKP and ST11-KPC-2 isolates carrying all of the four virulence genes have not been previously reported. A study reported that among 1052 CRKP strains isolated during 2015–2017 from 56 centres across China, only 72 (6.8%) strains carried all of the four virulence genes [34]. In recent years, the highest prevalence of a single virulence gene in CRKP isolates was approximately 20% in China and Vietnam, followed by 10% in Qatar, and less than 10% in the USA and the UK [21, 35–37]. Thus, the mechanism underlying the simultaneous presence of all four of these virulence genes at high rates in K. pneumoniae, CRKP and ST11-KPC-2 isolates is currently unknown and needs to be investigated in the future.
In K. pneumoniae, the occurrence of pLVPK or pLVPK-like virulent plasmids was reported to have a strong correlation with highly hypervirulent phenotypes. Several experiments have confirmed the hypervirulent phenotype of pLVPK-positive isolates, including the string test, serum killing assay and the G. mellonella infection model [20, 22]. Highly hypervirulent phenotypes of K. pneumoniae have been reported to be associated with high mortality rates [38–41]. However, in the present study, carbapenem resistance rather than the co-occurrence of the virulence genes rmpA, rmpA2, iroN and iucA in the isolates was associated with patient death during hospitalisation. The possible reasons for the inconsistency include low expression levels of virulence genes under host immune pressure, prior use of two or more antibiotics or carbapenems, serious underlying diseases and a small sample size in our study. Supporting our speculation, only a small proportion of the K. pneumoniae isolates containing these four virulence genes was found to exhibit high virulence in the G. mellonella infection model.
Given our study’s limitations of being a single-centre study and having a small sample size, a multicentre study with a large sample size is warranted to confirm our results.
In conclusion, the independent risk factors for CRKP infection in our hospital were diverse. ST11-KPC-2, which carried all of the four virulence genes, was responsible for most of the K. pneumoniae infection cases. Furthermore, patient mortality was associated with carbapenem resistance rather than with the co-occurrence of all of the four virulence genes.
Author contribution
Yating Xiang and Yirong Li designed research; Hongpan Tian and Qingsong Chen performed research; Jihong Gu, Hongmao Liu and Cuixiang Wang developed software necessary to perform and record experiments.
Funding
This work was supported by the National Key Research and Development Program of China (2021YFC2701803).
Data availability
Not applicable.
Code availability
Not applicable.
Declarations
Ethics approval
Ethical approval for this study was obtained from the Ethics Committee of Zhongnan Hospital of Wuhan University.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Martin RM, Bachman MA (2018) Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol 8:4 [DOI] [PMC free article] [PubMed]
- 2.Ray CG, Ryan KJ. Sherris medical microbiology: an introduction to infectious diseases. NY: McGraw-Hill; 2004. [Google Scholar]
- 3.Bengoechea JA, Sa PJ. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev. 2019;43:123–144. doi: 10.1093/femsre/fuy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munoz-Price LS, Poirel L, Bonomo RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu C, Liu L, Jin MM, et al. Molecular epidemiology and risk factors of carbapenem-resistant Klebsiella Pneumoniae bloodstream infections in Wuhan. China Curr Med Sci. 2022;42:68–76. doi: 10.1007/s11596-021-2480-5. [DOI] [PubMed] [Google Scholar]
- 6.Tanwar J, Das S, Fatima Z, et al. Multidrug resistance: an emerging crisis. Interdiscip Perspect Infect Dis. 2014;541340:7. doi: 10.1155/2014/541340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu LF, Sun XX, Ma XL. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob. 2017;16(1):18. doi: 10.1186/s12941-017-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X, Sun Q, Li J, et al. Molecular epidemiology of carbapenem-resistant hypervirulent Klebsiella pneumoniae in China. Emerg Microbes Infect. 2022;11:841–849. doi: 10.1080/22221751.2022.2049458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tao G, Tan H, Ma J, et al. Resistance phenotype and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae isolated from Nanjing Children’s Hospital in Jiangsu Province. China Infect Drug Resist. 2022;15:5435–5447. doi: 10.2147/IDR.S377068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammer CC, Brainard J, Hunter PR. Risk factors and risk factor cascades for communicable disease outbreaks in complex humanitarian emergencies: a qualitative systematic review. BMJ Glob Health. 2018;3:e000647. doi: 10.1136/bmjgh-2017-000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Duin D, Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence. 2017;8:460–469. doi: 10.1080/21505594.2016.1222343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu L, Wang S, Zhang Z, et al. Whole genome sequence of bla NDM and bla KPC co-producing Klebsiella pneumoniae isolate KSH203 with capsular serotype K25 belonging to ST11 from China. J Glob Antimicrob Resist. 2020;20:272–274. doi: 10.1016/j.jgar.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Hu R, Li Q, Zhang F, et al. Characterisation of bla(NDM-5) and bla(KPC-2) co-occurrence in K64-ST11 carbapenem-resistant Klebsiella pneumoniae. J Glob Antimicrob Resist. 2021;27:63–66. doi: 10.1016/j.jgar.2021.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Bano J, Gutierrez-Gutierrez B, Machuca I, et al. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin Microbiol Rev. 2018;2:31. doi: 10.1128/CMR.00079-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isler B, Aslan AT, Akova M, et al. Treatment strategies for OXA-48-like and NDM producing Klebsiella pneumoniae infections. Expert Rev Anti Infect Ther. 2022;11(20):1389–1400. doi: 10.1080/14787210.2022.2128764. [DOI] [PubMed] [Google Scholar]
- 17.Han YL, Wen XH, Zhao W, et al. Epidemiological characteristics and molecular evolution mechanisms of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Front Microbiol. 2022;13:1003783. doi: 10.3389/fmicb.2022.1003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alcantar-Curiel MD, Giron JA. Klebsiella pneumoniae and the pyogenic liver abscess: implications and association of the presence of rpmA genes and expression of hypermucoviscosity. Virulence. 2015;6:407–409. doi: 10.1080/21505594.2015.1030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YT, Chang HY, Lai YC, et al. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004;337:189–198. doi: 10.1016/j.gene.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;1(18):37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 21.Tsui CK-M, Abid FB, Ismail KA, et al. Genomic epidemiology of carbapenem-resistant Klebsiella in Qatar: emergence and dissemination of hypervirulent Klebsiella pneumoniae sequence type 383 strains. Antimicrob Agents Chemother. 2023;13:e0003023. doi: 10.1128/aac.00030-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Dong N, Chan EW-C, et al. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 2021;29(1):65–83. doi: 10.1016/j.tim.2020.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Shankar C, Veeraraghavan B, Nabarro LEB, et al. Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol. 2018;18:6. doi: 10.1186/s12866-017-1148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eger E, Heiden SE, Becker K, et al. Hypervirulent Klebsiella pneumoniae sequence type 420 with a chromosomally inserted virulence plasmid. Int J Mol Sci. 2021;22:9196. doi: 10.3390/ijms22179196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diancourt L, Passet V, Verhoef J, et al. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiao Y, Qin Y, Liu J, et al. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection/colonization and predictors of mortality: a retrospective. Pathog Glob Health. 2015;109:68–74. doi: 10.1179/2047773215Y.0000000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Chen Y, Liu P, et al. Risk factors and mortality for elderly patients with bloodstream infection of carbapenem resistance Klebsiella pneumoniae: a 10-year longitudinal study. BMC Geriatr. 2022;573:22. doi: 10.1186/s12877-022-03275-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semenec L, Cain AK, Dawson CJ, et al. Cross-protection and cross-feeding between Klebsiella pneumoniae and Acinetobacter baumannii promotes their co-existence. Nat Commun. 2023;702:14. doi: 10.1038/s41467-023-36252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lv D, Zuo Y, Wang Y, et al. Predictors of occurrence and 30-day mortality for co-infection of carbapenem-resistant Klebsiella pneumoniae and carbapenem-resistant Acinetobacter baumannii. Front Cell Infect Microbiol. 2022;12:919414. doi: 10.3389/fcimb.2022.919414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi: 10.1016/j.ebiom.2017.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen CM, Guo MK, Ke SC, et al. Emergence and nosocomial spread of ST11 carbapenem-resistant Klebsiella pneumoniae co-producing OXA-48 and KPC-2 in a regional hospital in Taiwan. J Med Microbiol. 2018;67(7):957–964. doi: 10.1099/jmm.0.000771. [DOI] [PubMed] [Google Scholar]
- 32.Huang YH, Chou SH, Liang SW, et al. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother. 2018;73(8):2039–2046. doi: 10.1093/jac/dky164. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Zeng J, Liu W, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect. 2015;71(5):553–560. doi: 10.1016/j.jinf.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Jin L, Ouyang PE, et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother. 2020;75:327–336. doi: 10.1093/jac/dkz446. [DOI] [PubMed] [Google Scholar]
- 35.Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Micro-biol Rev. 2019;32:e00001–19. doi: 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turton JF, Payne Z, Coward A, et al. Virulence genes in isolates of Klebsiell apneumoniae from the UK during 2016, including among carbapenemase gene-positive hypervirulent K1 ST23 and “non-hypervirulent” types ST147, ST15 and ST383. J Med Microbiol. 2018;67:118–128. doi: 10.1099/jmm.0.000653. [DOI] [PubMed] [Google Scholar]
- 37.Chou A, Nuila RE, Franco LM, et al. Prevalence of hypervirulent Klebsiella pneumoniae-associated genes rmpA and magA in two tertiary hospitals in Houston, TX, USA. J Med Microbiol. 2016;65:1047–1048. doi: 10.1099/jmm.0.000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W, Sun G, Yu Y, et al. Increasing occurrence of anti-microbial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2014;58(2):225–232. doi: 10.1093/cid/cit675. [DOI] [PubMed] [Google Scholar]
- 39.Zhan L, Wang S, Guo Y, et al. Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a Tertiary Hospital in China. Front Cell Infect Microbiol. 2017;7:182. doi: 10.3389/fcimb.2017.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu H, Li D, Zhou H, et al. Bacteremiaandotherbodysite infection caused by hypervirulent and classic Klebsiella pneumoniae. Microb Pathog. 2017;104:254–262. doi: 10.1016/j.micpath.2017.01.049. [DOI] [PubMed] [Google Scholar]
- 41.Shankar C, Nabarro LE, DevangaRagupathi NK, et al. Draft genome sequences of three hypervirulent carbapenem-resistant Klebsiella pneumoniae isolates from bacteremia. Genome Announc. 2016;4(6):e01081–16. doi: 10.1128/genomeA.01081-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.