Version Changes
Revised. Amendments from Version 1
The revised article shows significant improvements. First, the introduction now includes the latest reference articles. Second, the explanation of criteria for selecting harmful nsSNPs improves the transparency of the methodology. Figure 1 has been carefully modified after re-analysis, making it a clearer visual representation. Lastly, a detailed explanation for Figure 5 enhances reader comprehension.
Abstract
Introduction
The sirtuin (Silent mating type information regulation 2 homolog)1(SIRT1) protein plays a vital role in many disorders such as diabetes, cancer, obesity, inflammation, and neurodegenerative and cardiovascular diseases. The objective of this in silico analysis of SIRT1's functional single nucleotide polymorphisms (SNPs) was to gain valuable insight into the harmful effects of non-synonymous SNPs (nsSNPs) on the protein. The objective of the study was to use bioinformatics methods to investigate the genetic variations and modifications that may have an impact on the SIRT1 gene's expression and function.
Methods
nsSNPs of SIRT1 protein were collected from the dbSNP site, from its three (3) different protein accession IDs. These were then fed to various bioinformatic tools such as SIFT, Provean, and I- Mutant to find the most deleterious ones. Functional and structural effects were examined using the HOPE server and I-Tasser. Gene interactions were predicted by STRING software. The SIFT, Provean, and I-Mutant tools detected the most deleterious three nsSNPs (rs769519031, rs778184510, and rs199983221).
Results
Out of 252 nsSNPs, SIFT analysis showed that 94 were deleterious, Provean listed 67 dangerous, and I-Mutant found 58 nsSNPs resulting in lowered stability of proteins. HOPE modelling of rs199983221 and rs769519031 suggested reduced hydrophobicity due to Ile 4Thr and Ile223Ser resulting in decreased hydrophobic interactions. In contrast, on modelling rs778184510, the mutant protein had a higher hydrophobicity than the wild type.
Conclusions
Our study reports that three nsSNPs (D357A, I223S, I4T) are the most damaging mutations of the SIRT1 gene. Mutations may result in altered protein structure and functions. Such altered protein may be the basis for various disorders. Our findings may be a crucial guide in establishing the pathogenesis of various disorders.
Keywords: SIRT1, nsSNP, bioinformatics, protein modelling
Introduction
In silico analysis of SIRT1 Gene
Sirtuins are nicotinamide adenine dinucleotide (NAD+)-dependent deacetylases that regulate transcriptional activity intracellularly. They are present in a wide range of tissues, such as the adipose, kidney, brain, liver, and muscle tissues. 1 , 2 SIRT1 (Silent mating-type information regulation 2 homolog 1) is known to regulate a variety of cellular processes, including lipid and glucose metabolism, stress tolerance, autophagy, circadian rhythms, and mitochondrial biogenesis, according to several studies. 3 – 5 SIRT1 gene expression modulates its downstream pathways in diabetes, cancer, obesity, inflammation, and neurodegenerative and cardiovascular diseases by focusing on numerous cellular proteins, including nuclear factor-κB (NF-κB), endothelial nitric oxide synthase (eNOS), forkhead transcriptional factors (FoxOs), AMP-activated protein kinase (AMPK), protein tyrosine phosphatase (PTP). However, considerable evidence suggests that in a variety of malignant cell types, SIRT1 is upregulated and that SIRT1 antagonists prevented the development of cancer cells. 6 – 8
Single nucleotide polymorphisms (SNPs) are variations in the DNA sequence that result from the alterations in a single nucleotide (A, T, C, or G). Around 90% of human genetic variation is made up of SNPs. The three-billion-base human genome has SNPs at every 100–300 bases, with varying densities between regions. 9 The genome's coding and noncoding sections can both present SNPs. SNPs can have a wide spectrum of effects on how cells behave, from having no effect to causing disease or altering the reaction to a drug. Since they are responsible for about half of the genetic differences associated with human hereditary diseases, non-synonymous SNPs (nsSNPs) that result in an amino acid residue substitution in the protein product are also of high relevance. 10 There may be effects on transcription factor binding, splicing, or gene expression from coding synonymous SNPs (sSNPs) and SNPs that aren't in the gene promoter or coding regions. 11 , 12
Human reactions to viruses, medications, vaccinations, and other agents are significantly influenced by SNPs. SNPs are therefore useful in biomedical research, the creation of pharmaceutical products, the improvement of medical diagnostics, and the application of personalised medicine. 13 SNPs are responsible for specific phenotypes and therefore it is very important to identify them. This is a difficult task as it necessitates repeatedly evaluating thousands of SNPs in candidate genes. Selecting a group of SNPs for a study to determine the role of an SNP in a disease is a challenging endeavour; in these situations, a bioinformatics tool may be very helpful to distinguish between neutral and functional SNPs. They might also show the structural underpinnings of the mutations. These bioinformatics applications are used to assess the SNPs’ functional significance.
To find the SIRT1 protein's most dangerous nsSNPs, we applied bioinformatics techniques. We hypothesised that SIRT1 protein would be harmful because of nsSNPs on the gene. This is the first study of its kind for the SIRT1 gene to include both protein structure prediction and mutation analysis.
Methods
Multiple steps were used to complete the study. The figure below shows the equipment used to complete the task ( Figure 1).
Figure 1. Sorting of nsSNPs of SIRT1.
Extraction of nsSNPs
The NCBI SNP database was accessed. Information on the entire SIRT1 gene, including its nsSNPs, was obtained. As the query sequence, filtered nsSNPs from the dbSNP database were examined. The NCBI Protein accession IDs NP_ 036370.2, NP_001135970.1, and NP_002294.2 for the SIRT1 gene were used.
Identification of damaging nsSNPs
SIFT, Provean, and I-Mutant software were used to identify the impact of spotted nsSNPs on the SIRT1 gene.
SIFT (Sorting Intolerant from Tolerant) server
The SIFT server is a web-based bioinformatics tool that forecasts the detrimental effects of nucleotide substitution and frame shift (insertion/deletion) on protein function based on the degree of amino acid residue maintenance in sequence alignments obtained from highly associated sequences, with the primary assumption that mutations in evolutionarily conserved regions primarily affect its function. 14 The distinct input data order for the SIFT server includes protein sequence, chromosome location, and dbSNP reference number. SNPs and Indels were separated from the overall number in order to use this tool, and they were provided with the chromosome positions for frame shift indels and the residue number (rs) ID numbers for missense, nonsense, and stop gain SNPs. Each residue was given a value from 0 to 1 by the SIFT server, with scores below 0.05 indicating detrimental amino acid changes and scores above 0.05 indicating tolerance. 15 The website hosts SIFT version 5.2.2.
Provean
A protein's biological activity may be impacted by an amino acid substitution or indel, according to predictions made by the programme Protein Variation Effect Analyzer (PROVEAN). When filtering sequence variants, PROVEAN is useful for locating nonsynonymous or indel variations that are anticipated to be functionally significant. 16 The tool takes as input a protein sequence and several amino acid combinations, runs a BLAST search to find related sequences (supporting sequences), and outputs PROVEAN scores. The interpretation was done using the score thresholds. The default threshold is -2.5, meaning that variants with a score of -2.5 or less are deemed harmful, whereas variants with a score of -2.5 or more are considered neutral. http://provean.jcvi.org/index.php can be visited to access Provean.
I-Mutant 2.0
I-Mutant 2.0 was used in the investigation to analyse the stability of the targeted SIRT1 protein. This website server estimates any mutation-related changes to protein stability. 17 By adjusting the pH to 7 and the temperature to 25°C, this technique was used to analyse the SIRT1 protein sequences. It gives the opportunity to forecast how the protein's stability will be altered in response to single-site changes in the protein's structure or sequence. The design of the I-Mutant outcome is as follows: Free energy change value; Delta Delta G (DDG)= 0 is neutral, DDG > 0 is an increase in stability, and DDG <0 is a reduction in stability (I-Mutant website)
Examining the functional and structural effect of nsSNPs
To understand the effect of nsSNPs on the SIRT1 protein structure, the study used Polyphen, HOPE and I-Tasser software. The three most deleterious and damaging nsSNPs of the SIRT1 gene from each of its isoforms were chosen and processed to examine their structural and functional effects on SIRT1 protein.
Polyphen
Using simple physical and comparative considerations, polymorphism phenotyping v2(PolyPhen) is a method that estimates the potential effects of an amino acid substitution on the structure and functionality of a human protein. 18
HOPE modelling
The HOPE server analyses mutations automatically and can show the structural repercussions of a mutation. In addition to predictions from DAS services, sequence annotations from the UniProt database and calculations on the 3D coordinates of the protein using WHAT IF Web services are just a few of the data sources that HOPE uses to compile its information. 19
Iterative Threading ASSEmbly Refinement ( I-Tasser)
I-Tasser is a software package for protein structure and function modelling. The Template modelling score was used to compare the wild and mutant models. The estimated values of root mean square deviation (RMSD) and melting temperature (TM) allowed for the precise determination of similarity score. According to statistics, a TM-score of 0.17 or less indicates that two randomly chosen structures from the Protein DataBank library are comparable, while a score of 0.5 or more indicates that two structures have a similar topology. Studies have demonstrated a clear correlation between a high level of RMSD value and a high amount of change between wild-type and mutant. 20 , 21 The harmful mutations were then introduced into I-Tasser by adhering to their values. 22 – 24 Chimera 1.11 was used to study the molecular characteristics and interactive visualisation of the final protein structure. 25
Gene-gene interactions of the SIRT1 gene
Managing protein interactions is essential for maintaining the system's homeostasis. STRING's task is to display the total score of interaction genes. In this stage, SIRT1 served as the input target gene, and analysis was completed.
Results and discussion
Extraction of nsSNPs
The number of SNPs of the SIRT1 gene obtained from the NCBI database were 15,865. Three isoforms for SIRT1 were found ( isoform a, isoform b and isoform c). Isoform a (NP_036370.2) comprised 597 nsSNPs, isoform b (NP_001135970.1) comprised a total of 330 nsSNPs and isoform c (NP_001300978.1) 331 nsSNPs. The diagrammatical depiction is shown in Figure 1. The nsSNPs listed in all the isoforms are listed and the duplicates were deleted; the total number of nsSNPs included in all the isoforms was 252 ( Table 1).
Table 1. SIFT, I-Mutant, and Provean analyses for the nsSNPs of the SIRT1 Gene.
| Coordinates | Known protein ID | Substitution | dbSNP ID | Prediction | SIFT score | DDG (Kcal/mol/stability) | Provean score | prediction |
|---|---|---|---|---|---|---|---|---|
| 10,69647216,1,C/T | Q96EB6 | H158Y | rs767540399 | TOLERATED | 1 | 0.06/increased | -1.115 | Neutral |
| 10,69648720,1,A/G | Q96EB6 | I210V | rs369379188 | TOLERATED | 0.82 | -0.61/decreased | 0.333 | Neutral |
| 10,69644769,1,C/A | Q96EB6 | A97E | rs550317521 | TOLERATED | 0.76 | -0.21/decreased | -0.457 | Neutral |
| 10,69647188,1,C/A | Q96EB6 | F148L | rs773494159 | TOLERATED | 0.75 | -1.27/decreased | 0.215 | Neutral |
| 10,69644762,1,A/T | Q96EB6 | T95S | rs767360333 | TOLERATED | 0.75 | -0.03/decreased | -0.254 | Neutral |
| 10,69647286,1,G/A | Q96EB6 | R181Q | rs547102478 | TOLERATED | 0.72 | -1.13/decreased | 0.115 | Neutral |
| 10,69648649,1,C/G | Q96EB6 | T186S | rs201841214 | TOLERATED | 0.64 | -0.59/decreased | 0.061 | Neutral |
| 10,69644828,1,G/A | Q96EB6 | A117T | rs756786838 | TOLERATED | 0.59 | -0.77/decreased | 0.599 | Neutal |
| 10,69644675,1,G/C | Q96EB6 | G66R | rs531048058 | TOLERATED | 0.59 | -0.20/decreased | 0.25 | Neutral |
| 10,69644864,1,G/A | Q96EB6 | G129S | rs746381892 | TOLERATED | 0.53 | -0.58/decreased | -0.191 | Neutral |
| 10,69648723,1,C/A | Q96EB6 | P211T | rs201007799 | TOLERATED | 0.51 | -0.92/decreased | -0.014 | Neutral |
| 10,69647222,1,T/A | Q96EB6 | C160S | rs763883280 | TOLERATED | 0.49 | -0.82/decreased | 0.042 | Neutral |
| 10,69644778,1,A/G | Q96EB6 | E100G | rs199624804 | TOLERATED | 0.49 | -0.73/decreased | -1.003 | Neutral |
| 10,69648724,1,C/T | Q96EB6 | P211L | rs201668639 | TOLERATED | 0.48 | -0.43/decreased | 1.077 | Neutral |
| 10,69647273,1,A/G | Q96EB6 | T177A | rs565217830 | TOLERATED | 0.47 | -1.23/decreased | -0.59 | Neutral |
| 10,69644820,1,C/A | Q96EB6 | P114Q | rs182199697 | TOLERATED | 0.45 | -1.37/decreased | -0.658 | Neutral |
| 10,69648744,1,A/G | Q96EB6 | M218V | rs745979871 | TOLERATED | 0.4 | -0.63/decreased | -1.14 | Neutral |
| 10,69644688,1,C/T | Q96EB6 | A70V | rs751564985 | TOLERATED | 0.37 | -0.05/decreased | 0.69 | Neutral |
| 10,69647183,1,C/G | Q96EB6 | L147V | rs748384143 | TOLERATED | 0.34 | -1.7/decreased | -0.174 | Neutral |
| 10,69648722,1,A/G | Q96EB6 | I210M | rs201862792 | TOLERATED | 0.32 | -1.39/decreased | -0.025 | Neutral |
| 10,69647233,1,T/A | Q96EB6 | D163E | rs761508157 | TOLERATED | 0.31 | -0.06/decreased | -1.813 | Neutral |
| 10,69644519,1,T/C | Q96EB6 | S14P | rs201230502 | TOLERATED | 0.27 | 0.06/increased | -0.631 | Neutral |
| 10,69647181,1,T/C | Q96EB6 | L146P | rs772106776 | TOLERATED | 0.26 | -1.89/decreased | -0.95 | Neutral |
| 10,69644589,1,C/T | Q96EB6 | P37L | rs548590752 | TOLERATED | 0.26 | -0.27/decreased | 0.112 | Neutral |
| 10,69648765,1,A/G | Q96EB6 | I225V | rs559057403 | TOLERATED | 0.22 | -1.10/decreased | -0.362 | Neutral |
| 10,69648669,1,A/G | Q96EB6 | M193V | rs762526864 | TOLERATED | 0.22 | -0.49/decreased | -0.716 | Neutral |
| 10,69647253,1,A/G | Q96EB6 | H170R | rs144124002 | TOLERATED | 0.21 | -0.19/decreased | -1.457 | Neutral |
| 10,69644799,1,T/C | Q96EB6 | L107P | rs587776957 | TOLERATED | 0.21 | -1.31/decreased | -1.226 | Neutral |
| 10,69647175,1,A/G | Q96EB6 | D144G | rs142378619 | TOLERATED | 0.2 | -0.61/decreased | 1.231 | Neutral |
| 10,69647219,1,T/C | Q96EB6 | S159P | rs200660028 | TOLERATED | 0.2 | -0.40/decreased | -1.69 | Neutral |
| 10,69647276,1,C/G | Q96EB6 | P178A | rs752035789 | TOLERATED | 0.17 | E | -2.017 | Neutral |
| 10,69648861,1,A/G | Q96EB6 | I257V | rs765326045 | TOLERATED | 0.14 | -0.79/decreased | -0.642 | Neutral |
| 10,69648670,1,T/C | Q96EB6 | M193T | rs200994303 | TOLERATED | 0.12 | -0.81/decreased | -1.398 | Neutral |
| 10,69648666,1,C/G | Q96EB6 | L192V | rs772852024 | TOLERATED | 0.11 | -1.28/decreased | -0.843 | Neutral |
| 10,69647289,1,T/G | Q96EB6 | I182R | rs749341245 | TOLERATED | 0.09 | -1.63/decreased | -0.979 | Neutral |
| 10,69644622,1,C/T | Q96EB6 | P48L | rs568432780 | TOLERATED | 0.08 | -0.20/decreased | -0.384 | Neutral |
| 10,69648819,1,A/G | Q96EB6 | I243V | rs200711525 | TOLERATED | 0.06 | -1.08/decreased | -0.89 | Neutral |
| 10,69644886,1,C/G | Q96EB6 | A136G | rs775978922 | DAMAGING | 0.05 | -1.07/decreased | -0.329 | Neutral |
| 10,69648718,1,C/G | Q96EB6 | T209R | rs756483371 | DAMAGING | 0.05 | -0.16/decreased | -2.574 | Deleterious |
| 10,69648745,1,T/C | Q96EB6 | M218T | rs201583175 | DAMAGING | 0.04 | -0.70/decreased | -1.991 | Neutral |
| 10,69647279,1,A/G | Q96EB6 | R179G | rs200805107 | DAMAGING | 0.04 | -1.66/decreased | -1.894 | Neutral |
| 10,69647220,1,C/A | Q96EB6 | S159Y | rs760482544 | DAMAGING | 0.04 | -0.23/decreased | -2.272 | Neutral |
| 10,69647264,1,A/C | Q96EB6 | S174R | rs758805613 | DAMAGING | 0.04 | -0.27/decreased | -2.166 | Neutral |
| 10,69651254,1,C/G | Q96EB6 | A295G | rs368002483 | DAMAGING | 0.01 | -1.33/decreased | -3.508 | Deleterious |
| 10,69647193,1,A/T | Q96EB6 | D150V | rs776925394 | DAMAGING | 0.01 | -0.13/decreased | -2.979 | Deleterious |
| 10,69651200,1,A/G | Q96EB6 | D277G | rs776933716 | DAMAGING | 0.01 | -1.50/decreased | -4.86 | Deleterious |
| 10,69651236,1,A/G | Q96EB6 | D289G | rs375090685 | DAMAGING | 0.01 | -0.90/decreased | -5.977 | Deleterious |
| 10,69644516,1,G/T | Q96EB6 | G13C | rs200005116 | DAMAGING *Warning! Low confidence. | 0.01 | -0.64/decreased | -0.496 | Neutral |
| 10,69648820,1,T/C | Q96EB6 | I243T | rs776084517 | DAMAGING | 0.01 | -1.91/decreased | -2.831 | Deleterious |
| 10,69648712,1,C/T | Q96EB6 | P207L | rs375988661 | DAMAGING | 0.01 | -0.48/decreased | -6.177 | Deleterious |
| 10,69648786,1,C/A | Q96EB6 | P232T | rs750787952 | DAMAGING | 0.01 | -0.98/decreased | -4.805 | Deleterious |
| 10,69648748,1,C/G | Q96EB6 | T219R | rs199716245 | DAMAGING | 0.01 | -0.24/decreased | -4.312 | Deleterious |
| 10,69651253,1,G/A | Q96EB6 | A295T | rs751811485 | DAMAGING | 0 | -0.81/decreased | -3.491 | Deleterious |
| 10,69648827,1,T/A | Q96EB6 | D245E | rs201479376 | DAMAGING | 0 | -0.28/decreased | -3.358 | Deleterious |
| 10,69648825,1,G/C | Q96EB6 | D245H | rs761327480 | DAMAGING | 0 | -0.41/decreased | -5.393 | Deleterious |
| 10,69644488,1,C/A | Q96EB6 | D3E | rs35671182 | DAMAGING *Warning! Low confidence. | 0 | 0.59/increase | -0.464 | Neutral |
| 10,69648760,1,T/G | Q96EB6 | I223S | rs769519031 | DAMAGING | 0 | -2.25/decreased | -4.47 | Deleterious |
| 10,69648811,1,T/C | Q96EB6 | I240T | rs199618656 | DAMAGING | 0 | -1.77/decreased | -3.43 | Deleterious |
| 10,69648867,1,C/G | Q96EB6 | L259V | rs750671807 | DAMAGING | 0 | -1.35/decreased | -2.769 | Deleterious |
| 10,69651218,1,T/G | Q96EB6 | L283R | rs773632625 | DAMAGING | 0 | -2/decreased | -5.537 | Deleterious |
| 10,69651215,1,G/A | Q96EB6 | R282H | rs762393274 | DAMAGING | 0 | -1.60/decreased | -4.481 | Deleterious |
| 10,69651193,1,T/A | Q96EB6 | S275T | rs779685735 | DAMAGING | 0 | -0.67/decreased | -2.769 | Deleterious |
| 10,69651285,1,T/G | NP_001135970 | D10E | rs780983084 | DAMAGING | 0.01 | -0.52/decreased | -3.322 | Deleterious |
| 10,69651266,1,T/C | NP_001135970 | I4T | rs199983221 | DAMAGING | 0 | -2.2/decreased | -4.39 | Deleterious |
| 10,69651258,1,G/T | NP_001135970 | M1I | rs753708973 | DAMAGING | 0 | -0.17/decreased | -3.705 | Deleterious |
| 10,69651292,1,C/G | NP_001135970 | P13A | rs200902165 | DAMAGING | 0 | -1.26/decreased | -7.539 | Deleterious |
| 10,69672378,1,G/A | B0QZ35 | C199Y | rs150099719 | TOLERATED | 1 | -0.64/decreased | -1.163 | Neutral |
| 10,69676275,1,T/A | B0QZ35 | D420E | rs775109357 | TOLERATED | 1 | -0.31/decreased | -0.41 | Neutral |
| 10,69676297,1,A/G | B0QZ35 | I428V | rs35224060 | TOLERATED | 1 | 0.33/increased | 0.108 | Neutral |
| 10,69669103,1,C/G | B0QZ35 | Q118E | rs746762337 | TOLERATED | 1 | -1.24/decreased | 0.07 | Neutral |
| 10,69666602,1,C/T | B0QZ35 | S30L | s200610338 | TOLERATED | 1 | E | 3.993 | Neutral |
| 10,69667859,1,G/A | B0QZ35 | V80I | rs750895479 | TOLERATED | 1 | 0.37/increased | 0.385 | Neutral |
| 10,69672512,1,C/T | B0QZ35 | P244S | rs201723648 | TOLERATED | 0.94 | E | 0.648 | Neutral |
| 10,69672620,1,C/A | B0QZ35 | Q280K | rs769257926 | TOLERATED | 0.92 | -0.94/decreased | -0.787 | Neutral |
| 10,69672599,1,G/A | B0QZ35 | E273K | rs779678154 | TOLERATED | 0.85 | -1.3/decreased | -1.264 | Neutral |
| 10,69669016,1,G/A | B0QZ35 | V89I | rs201152568 | TOLERATED | 0.85 | -0.27/decreased | 0.201 | Neutral |
| 10,69672597,1,T/C | B0QZ35 | M272T | rs757828571 | TOLERATED | 0.81 | -0.33//decreased | -0.187 | Neutral |
| 10,69676028,1,A/G | B0QZ35 | Q338R | rs766102594 | TOLERATED | 0.81 | -1.7/decreased | -1.453 | Neutral |
| 10,69672692,1,A/G | B0QZ35 | T304A | rs779413432 | TOLERATED | 0.8 | -0.83/decreased | -0.746 | Neutral |
| 10,69672720,1,T/C | B0QZ35 | V313A | rs781614748 | TOLERATED | 0.76 | -1.24/decreased | -0.056 | Neutral |
| 10,69672590,1,G/A | B0QZ35 | G270S | rs144625497 | TOLERATED | 0.73 | -0.58/decreased | 0.377 | Neutral |
| 10,69676289,1,A/G | B0QZ35 | N425S | rs761406151 | TOLERATED | 0.72 | -0.12/decreased | -0.158 | Neutral |
| 10,69672755,1,G/C | B0QZ35 | V325L | rs567829185 | TOLERATED | 0.71 | -0.86/decreased | -0.246 | Neutral |
| 10,69672683,1,G/A | B0QZ35 | G301S | rs200296961 | TOLERATED | 0.67 | -0.77/decreased | 0.011 | Neutral |
| 10,69672680,1,G/C | B0QZ35 | V300L | rs750092788 | TOLERATED | 0.62 | -0.18/decreased | -0.652 | Neutral |
| 10,69672383,1,C/A | B0QZ35 | P201T | rs116499760 | TOLERATED | 0.59 | -0.28/decreased | -0.581 | Neutral |
| 10,69672605,1,A/C | B0QZ35 | K275Q | rs747447296 | TOLERATED | 0.58 | -0.69/decreased | -1.15 | Neutral |
| 10,69672684,1,G/A | B0QZ35 | G301D | rs533321736 | TOLERATED | 0.57 | -0.55/decreased | 0.908 | Neutral |
| 10,69672243,1,A/G | B0QZ35 | H154R | rs146837595 | TOLERATED | 0.57 | -0.17/decreased | -1.947 | Neutral |
| 10,69672479,1,G/A | B0QZ35 | E233K | rs116040871 | TOLERATED | 0.54 | -1.18/decreased | -0.433 | Neutral |
| 10,69672675,1,A/G | B0QZ35 | K298R | rs765178020 | TOLERATED | 0.52 | -0.11/decreased | -0.612 | Neutral |
| 10,69672597,1,T/A | B0QZ35 | M272K | rs757828571 | TOLERATED | 0.52 | -0.31/decreased | -1.385 | Neutral |
| 10,69672539,1,T/G | B0QZ35 | L253V | rs377735046 | TOLERATED | 0.51 | -0.27/decreased | 0.246 | Neutral |
| 10,69672495,1,C/G | B0QZ35 | P238R | rs768902657 | TOLERATED | 0.51 | 0 | -2.575 | Deleterious |
| 10,69672426,1,A/G | B0QZ35 | Q215R | rs201058854 | TOLERATED | 0.49 | -1.37/decreased | -1.134 | Neutral |
| 10,69672630,1,G/C | B0QZ35 | R283T | rs773365044 | TOLERATED | 0.49 | -0.44/decreased | 0.351 | Neutral |
| 10,69676289,1,A/C | B0QZ35 | N425T | rs761406151 | TOLERATED | 0.48 | -0.03/decreased | -0.034 | Neutral |
| 10,69672438,1,C/T | B0QZ35 | A219V | rs61754500 | TOLERATED | 0.44 | -0.82/decreased | -0.93 | Neutral |
| 10,69672644,1,A/G | B0QZ35 | I288V | rs370128548 | TOLERATED | 0.42 | -0.19/decreased | 0.248 | Neutral |
| 10,69669104,1,A/G | B0QZ35 | Q118R | rs371326132 | TOLERATED | 0.42 | -1.07/decreased | -1.537 | Neutral |
| 10,69672357,1,G/A | B0QZ35 | G192D | rs369274325 | TOLERATED | 0.41 | -0.2/decreased | -2.13 | Neutral |
| 10,69672380,1,A/G | B0QZ35 | N200D | rs201090108 | TOLERATED | 0.41 | -0.01/decreased | -1.016 | Neutral |
| 10,69672657,1,T/C | B0QZ35 | M292T | rs139635382 | TOLERATED | 0.39 | 0.26/increased | -0.466 | Neutral |
| 10,69676240,1,G/A | B0QZ35 | A409T | rs114182972 | TOLERATED | 0.38 | -0.23/decreased | -0.877 | Neutral |
| 10,69672593,1,T/G | B0QZ35 | C271G | rs745527350 | TOLERATED | 0.38 | -2.81/decreased | -1.457 | Neutral |
| 10,69672335,1,G/C | B0QZ35 | E185Q | rs771126114 | TOLERATED | 0.38 | -0.9/decreased | -0.676 | Neutral |
| 10,69676099,1,G/A | B0QZ35 | V362I | rs201730062 | TOLERATED | 0.38 | -0.66/decreased | -0.29 | Neutral |
| 10,69676168,1,C/T | B0QZ35 | P385S | rs757740493 | TOLERATED | 0.37 | -0.82/decreased | -0.649 | Neutral |
| 10,69672564,1,A/G | B0QZ35 | D261G | s756116247 | TOLERATED | 0.36 | E | -0.904 | Neutral |
| 10,69672567,1,A/G | B0QZ35 | D262G | rs763773932 | TOLERATED | 0.36 | E | -1.552 | Neutral |
| 10,69669136,1,G/A | B0QZ35 | E129K | rs763496433 | TOLERATED | 0.35 | -0.86/decreased | -1.352 | Neutral |
| 10,69672456,1,C/T | B0QZ35 | P225L | rs770755510 | TOLERATED | 0.35 | 0.36/increased | -1.813 | Neutral |
| 10,69667871,1,A/G | B0QZ35 | I84V | rs17855431 | TOLERATED | 0.34 | -0.28/decreased | -0.432 | Neutral |
| 10,69672377,1,T/C | B0QZ35 | C199R | rs202116390 | TOLERATED | 0.33 | -0.43/decreased | -3.055 | Deleterious |
| 10,69672632,1,A/G | B0QZ35 | N284D | rs763220339 | TOLERATED | 0.33 | -1.22/decreased | -0.461 | Neutral |
| 10,69667878,1,A/G | B0QZ35 | N86S | rs17855432 | TOLERATED | 0.33 | -0.7/decreased | -0.927 | Neutral |
| 10,69667859,1,G/C | B0QZ35 | V80L | rs750895479 | TOLERATED | 0.33 | 0.19/increased | -0.68 | Neutral |
| 10,69672552,1,C/G | B0QZ35 | A257G | rs762955289 | TOLERATED | 0.32 | -3/decreased | -0.793 | Neutral |
| 10,69676118,1,G/C | B0QZ35 | C368S | rs114575266 | TOLERATED | 0.32 | -0.76/decreased | -2.036 | Neutral |
| 10,69676243,1,G/C | B0QZ35 | G410R | rs771866041 | TOLERATED | 0.32 | -0.14/decreased | -0.936 | Neutral |
| 10,69672621,1,A/C | B0QZ35 | Q280P | rs201096600 | TOLERATED | 0.32 | -0.63/decreased | -1.255 | Neutral |
| 10,69672456,1,C/A | B0QZ35 | P225Q | rs770755510 | TOLERATED | 0.31 | E | -1.222 | Neutral |
| 10,69676231,1,C/A | B0QZ35 | P406T | rs770824011 | TOLERATED | 0.31 | -0.66/decreased | -1.383 | Neutral |
| 10,69672684,1,G/T | B0QZ35 | G301V | rs533321736 | TOLERATED | 0.3 | -0.13/decreased | -0.94 | Neutral |
| 10,69676190,1,T/C | B0QZ35 | I392T | rs140030776 | TOLERATED | 0.3 | -0.26/decreased | -0.248 | Neutral |
| 10,69672490,1,T/G | B0QZ35 | S236R | rs192990424 | TOLERATED | 0.3 | -0.99/decreased | -1.914 | Neutral |
| 10,69672629,1,A/G | B0QZ35 | R283G | rs202044007 | TOLERATED | 0.29 | -1.1/decreased | -1.032 | Neutral |
| 10,69669023,1,G/A | B0QZ35 | R91Q | rs766737138 | TOLERATED | 0.29 | -0.85/decreased | -1.062 | Neutral |
| 10,69676297,1,A/C | B0QZ35 | I428L | rs35224060 | TOLERATED | 0.28 | 0.72/increased | -0.316 | Neutral |
| 10,69672622,1,A/C | B0QZ35 | Q280H | rs777147954 | TOLERATED | 0.28 | -1.18/decreased | -1.047 | Neutral |
| 10,69669058,1,A/G | B0QZ35 | I103V | rs757140711 | TOLERATED | 0.27 | -0.32/decreased | -0.427 | Neutral |
| 10,69667830,1,T/G | B0QZ35 | I70S | rs763566753 | TOLERATED | 0.27 | E | 0.432 | Neutral |
| 10,69672662,1,A/G | B0QZ35 | N294D | rs753828684 | TOLERATED | 0.27 | -0.4/decreased | -0.577 | Neutral |
| 10,69676282,1,G/T | B0QZ35 | A423S | rs776051187 | TOLERATED | 0.26 | -0.93/decreased | -0.122 | Neutral |
| 10,69672488,1,A/G | B0QZ35 | S236G | rs199497583 | TOLERATED | 0.26 | -2.12/decreased | -1.407 | Neutral |
| 10,69672369,1,A/G | B0QZ35 | K196R | rs549636735 | TOLERATED | 0.25 | -0.15/decreased | -0.838 | Neutral |
| 10,69672430,1,A/T | B0QZ35 | K216N | rs200415719 | TOLERATED | 0.25 | -0.97/decreased | -0.117 | Neutral |
| 10,69676231,1,C/T | B0QZ35 | P406S | rs770824011 | TOLERATED | 0.25 | -0.84/decreased | -1.273 | Neutral |
| 10,69672504,1,C/G | B0QZ35 | T241S | rs777098039 | TOLERATED | 0.25 | -0.74/decreased | -0.77 | Neutral |
| 10,69676286,1,T/C | B0QZ35 | I424T | rs202021325 | TOLERATED | 0.23 | E | -0.394 | Neutral |
| 10,69676298,1,T/C | B0QZ35 | I428T | rs758781892 | TOLERATED | 0.23 | 0.03/increased | -0.426 | Neutral |
| 10,69676169,1,C/A | B0QZ35 | P385H | rs765609303 | TOLERATED | 0.23 | -0.92/decreased | -1.139 | Neutral |
| 10,69666569,1,A/G | B0QZ35 | Q19R | rs532201569 | TOLERATED | 0.22 | -0.1/decreased | -1.062 | Neutral |
| 10,69676276,1,C/G | B0QZ35 | Q421E | rs201854199 | TOLERATED | 0.22 | -0.33/decreased | -0.47 | Neutral |
| 10,69676238,1,G/C | B0QZ35 | R408T | rs774376548 | TOLERATED | 0.22 | -0.42/decreased | -0.622 | Neutral |
| 10,69676156,1,A/G | B0QZ35 | S381G | rs114572830 | TOLERATED | 0.22 | -1.11/decreased | -1.35 | Neutral |
| 10,69672755,1,G/A | B0QZ35 | V325M | rs567829185 | TOLERATED | 0.21 | -1.01/decreased | -0.598 | Neutral |
| 10,69669028,1,C/T | B0QZ35 | P93S | rs752020872 | TOLERATED | 0.2 | -1.86/decreased | -5.682 | Deleterious |
| 10,69666581,1,G/C | B0QZ35 | C23S | rs200987359 | TOLERATED | 0.19 | -1.24/decreased | -6.059 | Deleterious |
| 10,69672453,1,T/G | B0QZ35 | L224W | rs748917510 | TOLERATED | 0.19 | 0.09/increased | -1.771 | Neutral |
| 10,69676205,1,A/G | B0QZ35 | N397S | rs116459300 | TOLERATED | 0.19 | -0.44//decreased | -1.479 | Neutral |
| 10,69672369,1,A/C | B0QZ35 | K196T | rs549636735 | TOLERATED | 0.18 | -0.26/decreased | -2.109 | Neutral |
| 10,69676340,1,A/G | B0QZ35 | N442S | rs756220511 | TOLERATED | 0.18 | -1.64/decreased | -0.75 | Neutral |
| 10,69676075,1,G/T | B0QZ35 | V354L | rs199502996 | TOLERATED | 0.18 | -0.53/decreased | -1.596 | Neutral |
| 10,69667850,1,T/G | B0QZ35 | C77G | rs779440453 | TOLERATED | 0.17 | -2.97/decreased | -3.709 | Deleterious |
| 10,69669041,1,C/G | B0QZ35 | A97G | rs755416990 | TOLERATED | 0.16 | -2.16/decreased | -0.743 | Neutral |
| 10,69669041,1,C/T | B0QZ35 | A97V | rs755416990 | TOLERATED | 0.16 | E | -0.179 | Neutral |
| 10,69676087,1,T/C | B0QZ35 | S358P | rs754313614 | TOLERATED | 0.15 | 0.18/increased | -0.896 | Neutral |
| 10,69669139,1,G/A | B0QZ35 | V130I | rs745364339 | TOLERATED | 0.15 | -0.14/decreased | -0.482 | Neutral |
| 10,69676075,1,G/A | B0QZ35 | V354I | rs199502996 | TOLERATED | 0.15 | -0.27/decreased | -0.369 | Neutral |
| 10,69676303,1,G/A | B0QZ35 | V430M | rs766908589 | TOLERATED | 0.15 | 0.45/increased | -0.04 | Neutral |
| 10,69676169,1,C/G | B0QZ35 | P385R | rs765609303 | TOLERATED | 0.14 | -0.37/decreased | -1.564 | Neutral |
| 10,69666562,1,C/G | B0QZ35 | Q17E | rs772390347 | TOLERATED | 0.14 | -1.38/decreased | -1.579 | Neutral |
| 10,69672655,1,G/C | B0QZ35 | Q291H | rs759555466 | TOLERATED | 0.14 | -1.17/decreased | -1.134 | Neutral |
| 10,69669105,1,G/C | B0QZ35 | Q118H | rs776212608 | TOLERATED | 0.13 | -1.92/decreased | -1.877 | Neutral |
| 10,69672447,1,C/T | B0QZ35 | S222L | rs199649997 | TOLERATED | 0.13 | -0.25/decreased | -1.509 | Neutral |
| 10,69667865,1,G/A | B0QZ35 | G82R | rs758727461 | TOLERATED | 0.12 | -0.26/decreased | -0.355 | Neutral |
| 10,69669131,1,A/G | B0QZ35 | K127R | rs772640272 | TOLERATED | 0.12 | -1.33/decreased | -2.53 | Deleterious |
| 10,69666607,1,A/G | B0QZ35 | K32E | rs751410492 | TOLERATED | 0.12 | 0.31/increased | -1.657 | Neutral |
| 10,69669097,1,C/T | B0QZ35 | P116S | rs775426483 | TOLERATED | 0.12 | -1.84/decreased | -6.457 | Deleterious |
| 10,69672627,1,C/G | B0QZ35 | S282C | rs748473217 | TOLERATED | 0.11 | -0.44/decreased | -1.282 | Neutral |
| 10,69672524,1,G/A | B0QZ35 | V248M | rs758444346 | TOLERATED | 0.11 | -0.69/decreased | -0.538 | Neutral |
| 10,69669050,1,C/A | B0QZ35 | P100Q | rs748394475 | TOLERATED | 0.1 | -0.85/decreased | -1.547 | Neutral |
| 10,69672276,1,C/G | B0QZ35 | P165R | rs777705431 | TOLERATED | 0.1 | -0.76/decreased | -5.303 | Deleterious |
| 10,69669139,1,G/C | B0QZ35 | V130L | rs745364339 | TOLERATED | 0.1 | -0.48/decreased | -1.206 | Neutral |
| 10,69666595,1,G/A | B0QZ35 | A28T | rs201340003 | TOLERATED | 0.09 | -1.02/decreased | -1.057 | Neutral |
| 10,69676223,1,C/G | B0QZ35 | P403R | rs749218988 | TOLERATED | 0.08 | -0.77/decreased | -0.966 | Neutral |
| 10,69672519,1,C/G | B0QZ35 | S246C | rs373331174 | TOLERATED | 0.08 | -1.16/decreased | -0.839 | Neutral |
| 10,69676033,1,C/G | B0QZ35 | L340V | rs751212678 | TOLERATED | 0.07 | -0.67/decreased | -0.868 | Neutral |
| 10,69676204,1,A/C | B0QZ35 | N397H | rs747251569 | TOLERATED | 0.07 | -0.59/decreased | -1.742 | Neutral |
| 10,69669029,1,C/T | B0QZ35 | P93L | rs757269120 | TOLERATED | 0.07 | -1.34/decreased | -7.329 | Deleterious |
| 10,69672780,1,G/A | B0QZ35 | R333Q | rs771135356 | TOLERATED | 0.07 | -1.35/decreased | -2.059 | Neutral |
| 10,69669115,1,G/A | B0QZ35 | A122T | rs200690371 | TOLERATED | 0.06 | -0.03/decreased | -1.862 | Neutral |
| 10,69667848,1,A/C | B0QZ35 | D76A | rs756786753 | TOLERATED | 0.06 | -0.91/decreased | -3.303 | Deleterious |
| 10,69676215,1,A/C | B0QZ35 | E400D | rs201327262 | TOLERATED | 0.06 | -1.36/decreased | -0.816 | Neutral |
| 10,69676215,1,A/T | B0QZ35 | E400D | rs201327262 | TOLERATED | 0.06 | -1.36/decreased | -0.816 | Neutral |
| 10,69672530,1,G/T | B0QZ35 | V250F | rs773456695 | TOLERATED | 0.06 | -0.26/decreased | -0.799 | Neutral |
| 10,69667869,1,A/C | B0QZ35 | D83A | rs17855430:C | DAMAGING | 0.05 | -1.35/decreased | -5.798 | Deleterious |
| 10,69672285,1,A/G | B0QZ35 | H168R | rs779026786 | DAMAGING | 0.05 | -1.31/decreased | -7.23 | Deleterious |
| 10,69672324,1,T/A | B0QZ35 | V181D | rs1063111 | DAMAGING | 0.05 | -1.59/decreased | -6.395 | Deleterious |
| 10,69666665,1,C/T | B0QZ35 | A51V | rs141528984 | DAMAGING | 0.04 | 0.69/increased | -3.419 | Deleterious |
| 10,69676219,1,G/A | B0QZ35 | E402K | rs778261267 | DAMAGING *Warning! Low confidence. | 0.04 | -0.42/decreased | -1.024 | Neutral |
| 10,69672282,1,C/T | B0QZ35 | P167L | rs201948258 | DAMAGING | 0.04 | -0.72/decreased | -5.575 | Deleterious |
| 10,69666675,1,A/C | B0QZ35 | Q54H | s116374368 | DAMAGING | 0.04 | -1.4/decreased | -2.377 | Neutral |
| 10,69666675,1,A/T | B0QZ35 | Q54H | rs116374368 | DAMAGING | 0.04 | -1.4/decreased | -2.377 | Neutral |
| 10,69672374,1,T/C | B0QZ35 | C198R | rs772656917 | DAMAGING | 0.03 | -1.56/decreased | -8.169 | Deleterious |
| 10,69676279,1,G/A | B0QZ35 | E422K | rs201112743 | DAMAGING *Warning! Low confidence. | 0.03 | -0.95/decreased | -0.509 | Neutral |
| 10,69672290,1,C/T | B0QZ35 | H170Y | rs751498023 | DAMAGING | 0.03 | 0.1/decreased | -2.752 | Deleterious |
| 10,69667835,1,A/G | B0QZ35 | K72E | rs753398498 | DAMAGING | 0.03 | -0.42/decreased | -2.77 | Deleterious |
| 10,69676156,1,A/C | B0QZ35 | S381R | rs114572830 | DAMAGING *Warning! Low confidence. | 0.03 | -0.49/decreased | -1.398 | Neutral |
| 10,69669116,1,C/G | B0QZ35 | A122G | rs201635394 | DAMAGING | 0.02 | -1.07/decreased | -2.995 | Deleterious |
| 10,69667871,1,A/C | B0QZ35 | I84L | rs17855431 | DAMAGING | 0.02 | -0.09/decreased | -1.848 | Neutral |
| 10,69669050,1,C/T | B0QZ35 | P100L | rs748394475 | DAMAGING | 0.02 | -0.33/decreased | -3.331 | Deleterious |
| 10,69676105,1,T/G | B0QZ35 | S364A | rs772469360 | DAMAGING | 0.02 | -1.07/decreased | -1.339 | Neutral |
| 10,69676123,1,A/G | B0QZ35 | S370G | rs377449611 | DAMAGING | 0.02 | -1.98/decreased | -2.004 | Neutral |
| 10,69676129,1,A/G | B0QZ35 | S372G | rs759347614 | DAMAGING | 0.02 | E | -2.414 | Neutral |
| 10,69672405,1,C/T | B0QZ35 | T208I | rs750553699 | DAMAGING | 0.02 | 1.34/increased | -3.254 | Deleterious |
| 10,69669115,1,G/C | B0QZ35 | A122P | rs200690371 | DAMAGING | 0.01 | -0.12/decreased | -3.21 | Deleterious |
| 10,69666664,1,G/T | B0QZ35 | A51S | rs777323664 | DAMAGING | 0.01 | -0.35/decreased | -2.737 | Deleterious |
| 10,69667850,1,T/C | B0QZ35 | C77R | rs779440453 | DAMAGING | 0.01 | -1.55/decreased | -4.545 | Deleterious |
| 10,69676085,1,A/C | B0QZ35 | D357A | rs778184510 | DAMAGING | 0.01 | -2.58/decreased | -4.873 | Deleterious |
| 10,69676217,1,A/C | B0QZ35 | D401A | rs151026272 | DAMAGING *Warning! Low confidence. | 0.01 | -0.19/decreased | -1.941 | Neutral |
| 10,69667869,1,A/G | B0QZ35 | D83G | rs17855430 | DAMAGING | 0.01 | -2.13/decreased | -5.487 | Deleterious |
| 10,69669090,1,A/C | B0QZ35 | E113D | rs771950281 | DAMAGING | 0.01 | -0.78/decreased | -2.83 | Deleterious |
| 10,69669073,1,A/G | B0QZ35 | I108V | rs149206117 | DAMAGING | 0.01 | -0.86/decreased | -0.932 | Neutral |
| 10,69666592,1,A/G | B0QZ35 | I27V | rs552023236 | DAMAGING | 0.01 | -0.06/decreased | -0.823 | Neutral |
| 10,69672761,1,A/G | B0QZ35 | K327E | rs199770148 | DAMAGING | 0.01 | -0.33/decreased | -2.073 | Neutral |
| 10,69669148,1,C/T | B0QZ35 | L133F | rs755327185 | DAMAGING | 0.01 | 0.05/increased | -3.628 | Deleterious |
| 10,69672308,1,C/T | B0QZ35 | L176F | rs754951946 | DAMAGING | 0.01 | 0.36/increased | -3.79 | Deleterious |
| 10,69676160,1,T/C | B0QZ35 | L382S | rs754362414 | DAMAGING *Warning! Low confidence. | 0.01 | E | -2.203 | Neutral |
| 10,69676172,1,T/G | B0QZ35 | M386R | rs750707792 | DAMAGING *Warning! Low confidence. | 0.01 | -0.04/decreased | -1.467 | Neutral |
| 10,69676205,1,A/T | B0QZ35 | N397I | rs116459300 | DAMAGING *Warning! Low confidence. | 0.01 | 0.29/increased | -2.884 | Deleterious |
| 10,69669097,1,C/A | B0QZ35 | P116T | rs775426483 | DAMAGING | 0.01 | -1.89/decreased | -6.707 | Deleterious |
| 10,69672239,1,C/G | B0QZ35 | P153A | rs201348222 | DAMAGING | 0.01 | -1.64/decreased | -7.352 | Deleterious |
| 10,69672239,1,C/A | B0QZ35 | P153T | rs201348222 | DAMAGING | 0.01 | -1.39/decreased | -7.319 | Deleterious |
| 10,69672270,1,G/A | B0QZ35 | R163K | rs764179598 | DAMAGING | 0.01 | -0.45/decreased | -2.743 | Deleterious |
| 10,69676088,1,C/T | B0QZ35 | S358F | rs757624637 | DAMAGING | 0.01 | 0.21/increased | -3.193 | Deleterious |
| 10,69676105,1,T/C | B0QZ35 | S364P | rs772469360 | DAMAGING | 0.01 | -0.48/decreased | -1.779 | Neutral |
| 10,69676115,1,C/T | B0QZ35 | S367F | rs768864413 | DAMAGING | 0.01 | -0.32/decreased | -2.558 | Deleterious |
| 10,69676115,1,C/A | B0QZ35 | S367Y | rs768864413 | DAMAGING | 0.01 | -0.47/decreased | -2.064 | Neutral |
| 10,69676129,1,A/T | B0QZ35 | S372C | rs759347614 | DAMAGING | 0.01 | -0.82/decreased | -2.959 | Deleterious |
| 10,69666661,1,G/C | B0QZ35 | V50L | rs200058231 | DAMAGING | 0.01 | -0.01/decreased | -2.384 | Neutral |
| 10,69672320,1,G/A | B0QZ35 | D180N | rs145326137 | DAMAGING | 0 | -0.7/decreased | -4.738 | Deleterious |
| 10,69666668,1,G/A | B0QZ35 | G52E | rs757804740 | DAMAGING | 0 | -1.9/decreased | -6.479 | Deleterious |
| 10,69669152,1,T/C | B0QZ35 | I134T | rs768051584 | DAMAGING | 0 | -0.23/decreased | -4.51 | Deleterious |
| 10,69672238,1,A/G | B0QZ35 | I152M | rs200021101 | DAMAGING | 0 | -1.03/decreased | -2.226 | Neutral |
| 10,69672258,1,T/A | B0QZ35 | I159K | rs140677498 | DAMAGING | 0 | -0.37/decreased | -6.333 | Deleterious |
| 10,69672327,1,T/C | B0QZ35 | I182T | rs1063112 | DAMAGING | 0 | -0.4/decreased | -4.671 | Deleterious |
| 10,69669173,1,A/G | B0QZ35 | K141R | rs756329197 | DAMAGING | 0 | -1.54/decreased | -2.846 | Deleterious |
| 10,69669145,1,C/T | B0QZ35 | L132F | rs199593180 | DAMAGING | 0 | 0.67/increased | -3.795 | Deleterious |
| 10,69669146,1,T/A | B0QZ35 | L132H | rs766945174 | DAMAGING | 0 | -0.73/decreased | -6.641 | Deleterious |
| 10,69669145,1,C/G | B0QZ35 | L132V | rs199593180 | DAMAGING | 0 | 0.23/increased | -2.846 | Deleterious |
| 10,69672262,1,A/T | B0QZ35 | L160F | rs761122062 | DAMAGING | 0 | -1.43/decreased | -3.79 | Deleterious |
| 10,69666629,1,A/C | B0QZ35 | N39T | rs767148239 | DAMAGING | 0 | -0.32/decreased | -5.535 | Deleterious |
| 10,69669196,1,C/T | B0QZ35 | P149S | rs267602551 | DAMAGING | 0 | -0.6/decreased | -7.579 | Deleterious |
| 10,69672417,1,C/T | B0QZ35 | P212L | rs201863201 | DAMAGING | 0 | E | -3.214 | Deleterious |
| 10,69672779,1,C/T | B0QZ35 | R333W | rs201647881 | DAMAGING | 0 | -0.75/decreased | -4.292 | Deleterious |
| 10,69666625,1,C/T | B0QZ35 | R38C | rs201583982 | DAMAGING | 0 | -0.52/decreased | -7.38 | Deleterious |
| 10,69666626,1,G/A | B0QZ35 | R38H | rs147909071 | DAMAGING | 0 | -0.72/decreased | -4.613 | Deleterious |
| 10,69669164,1,C/T | B0QZ35 | S138F | rs752958196 | DAMAGING | 0 | -1.61/decreased | -5.693 | Deleterious |
| 10,69676106,1,C/T | B0QZ35 | S364F | rs780449017 | DAMAGING | 0 | -0.1/decreased | -2.214 | Neutral |
| 10,69666661,1,G/T | B0QZ35 | V50F | rs200058231 | DAMAGING | 0 | 0.23/increased | -4.016 | Deleterious |
Identification of damaging nsSNPs
The following bioinformatics tools have provided the supplied data to further detect the influence of 252 nsSNPs on the structure and function of the SIRT1 gene. Because the resulting values were lower than the Tolerance Index (0.05), the SIFT software revealed 94 nsSNPs to be intolerant ( Table 1).
Protein stability changed depending on which amino acid was substituted and 216 nsSNPs demonstrated a decline in stability based on DDG value received from I-Mutant server ( Table 1).
PROVEAN identified 77 nsSNPs as having a negative impact since the final score of the variations was lower than the specified value of threshold (-2.5).
In the SIRT1 gene, three isoforms were identified, resulting in a total of 252 identified Single Nucleotide Polymorphisms (SNPs). Among them, SIFT software identified 94 non-synonymous SNPs (nsSNPs) as intolerant (using a cutoff value of <0.05). Subsequently, these 94 intolerant nsSNPs underwent Provean analysis, revealing 67 nsSNPs as harmful (with a cutoff value of <-2.5). The selected 67 nsSNPs then underwent I-Mutant analysis, which indicated 58 nsSNPs with decreased stability (using a cutoff value of 0). From this subset, we selected three nsSNPs from each isoform, rs778184510, rs769519031, rs199983221respectively, based on decreasing protein stability.
Structural and functional effect of nsSNPs
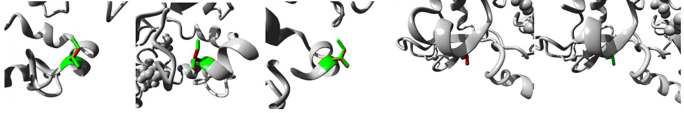
I-Mutant predicted the three (3) nsSNPs which played a role in decreasing SIRT1 stability (from each isoform - rs778184510, rs769519031, rs199983221), and they were selected for finding the impact of substitution of amino acid on structure and function of human protein (using Polyphen) and for the comparison of protein model (using I-Tasser). To generate the SIRT1 protein structure, SIRT1 protein sequences, single amino acid from the wild type, and mutations were uploaded to I-Tasser, which is one of the most accurate and sophisticated technique for predicting protein structure ( Figure 2). Then, using this technique, five models for each SIRT1 mutation and protein were produced.
Figure 2. Predicted protein structure models by I Tasser of SIRT1 Gene for each SNPs rs778184510, rs769519031, rs199983221 respectively.
When the native structure is known, TM score and RMSD can be used to compare the structural similarity of two structures. 26 The proposed TM score is supposed to solve the RMSD issue, which is prone to local errors. A local error (such as a mismatched tail) will raise the RMSD score even if the overall topology is good, since the RMSD measures the average distance between all residue pairs between two structures. The TM-score is insensitive to the local modelling error, nevertheless, because the short distance is weighted more severely than the long distance. A model with a proper topology is indicated by a TM-score and IT greater than 0.5, while a random similarity is indicated by a TM-score & IT less than 0.17 ( Table 2). These cut-offs are independent of the length of the protein.
Table 2. Results of Polyphen and I-Tasser analyses of three nsSNPs.
| Protein ID | Residual change | Polyphen | I-Tasser | ||||
|---|---|---|---|---|---|---|---|
| Score | Sensitivity | specificity | TM Score | RMSD values | C Score | ||
| rs778184510 | D357A | 0.983/Damaging | 0.74 | 0.96 | 0.29±0.09 | 17.0±2.8Ǻ | -3.92 |
| rs769519031 | I223S | 0.997/Damaging | 0.27 | 0.98 | 0.42±0.14 | 14.6±3.7Ǻ | -2.55 |
| rs199983221 | I4T | 1.00/Damaging | 0.00 | 1.00 | 0.48±0.15 | 11.7±4.5Ǻ | 1.93 |
By calculating a confidence score, or C-score, I-Tasser evaluates the accuracy of anticipated models. The convergence parameters from simulations of the structure assembly and the significance of threading template alignments are used to make this determination. A model with a high level of confidence also has a higher C-score. The C-score typically ranges from (-5,2).
HOPE modelling for rs199983221
Isoleucine turned into threonine in position 4. The mutant residue was more compact than the wild-type residue. The mutant residue was also less hydrophobic than the wild-type residue. The mutation caused the hydrophobic contacts in the protein's core to disappear.
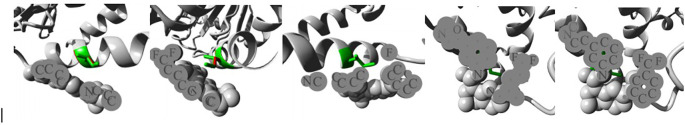
An overview of the protein is also displayed in the ribbon presentation ( Figure 3a). Additionally, there are five detailed pictures of the mutation site ( Figure 3b).
Figure 3a. Overview of the protein in ribbon presentation (I4T).
Figure 3b. Close-up of the mutation.
The protein is coloured grey, the side chains of both the wild-type and the mutant residue are shown and coloured green and red respectively. (I4T).
HOPE modelling of rs769519031
In this case, isoleucine turns into serine at position 223. The mutant residue was more compact than the wild-type residue. The mutant residue was less hydrophobic than the wild-type residue. This could lead to a lack of interactions with the other genes The mutation may cause the proteins' surface-bound hydrophobic interactions with other molecules to disappear. 19
An overview of the protein is also displayed in the ribbon presentation ( Figure 4a). There are also five enlargements of the mutation location ( Figure 4b).
Figure 4a. Overview of the protein in ribbon presentation (I223S).
Figure 4b. Close-up of the mutation.
The protein is coloured grey, the side chains of both the wild-type and the mutant residue are shown and coloured green and red respectively (I223S).
HOPE modelling of rs778184510
The mutant residue was smaller than the wild-type residue in this instance because alanine has replaced aspartic acid at position 357. In contrast to the wild-type residue charge, which was negative, the mutant residue charge was neutral. The mutant residue was more hydrophobic than the wild-type residue. The wild-type residue is expected to be located in its preferred secondary structure turn, according to the Reprof programme. The local conformation would only be slightly unstable since the mutant residue prefers a different secondary structure. The mutation places a more hydrophobic residue here. Hydrogen bonds may break as a result of this, and it may also prevent correct folding.
Gene interactions
STRING revealed the physical interactions between SIRT1 and other genes in the gene's interactions. In its pathways, it interacted with NFKB1, NFKB1A, DDX5, AURKA, BARD1, RPA1, UBEBA, ARNTL,CLOCK,CRY1, PPARGC1A, FOXO1, FOXO3, RELA, MYOD1, SUV39H1, MDM2, EP300, PPARG and TP53 ( Figure 5). The query proteins and the initial line of SIRT1's interaction are represented by coloured nodes on the picture. White nodes are the second interactional shell. Protein-protein interactions are represented by edges. The edges of the known interactions are blue and pink. Others illustrate the predicted interplay between proteins. In evidence mode, an edge may be drawn with up to 7 differently colored lines - these lines represent the existence of the seven types of evidence used in predicting the associations. Red lines indicate the presence of fusion evidence, green lines represent neighbourhood evidence, blue lines suggest cooccurrence evidence, purple lines correspond to experimental evidence, yellow lines denote text mining evidence, light blue lines signify database evidence, and black lines represent co-expression evidence. TP53 is more connected to and interdependent with SIRT1 than any other interaction on the list.
Figure 5. Gene interactions of SIRT1.
Numerous studies have been done in the past to determine the connection between the SIRT1 gene's polymorphism and a number of conditions, such as cancer, inflammation, obesity, diabetes, and cardiovascular and neurological illnesses. The most harmful nsSNPs in the SIRT1 gene that may be crucial in the development of certain disorders have been explored in this work.
The SIRT1 gene has 252 nsSNPs, according to our findings. The present study's SIFT findings revealed that the SIRT1 protein contains 94 harmful nsSNPs, 66 of which are detrimental as indicated by PROVEAN.
Provean scores were -4.873, -4.47, -4.39 for rs778184510, rs769519031 and rs199983221 respectively which were higher compared to other SNP’s and were chosen from each isoform of SIRT1 protein. These three nsSNPs, which cause high risk of altering normal functioning of SIRT1 gene, were selected for further evaluation based on the I-Mutant value, from each isoform of the SIRT1 protein.
D357A, I223S, and I4T’s respective Polyphen2 scores, which range from 0 to 1, were 0.983, 0.997, and 1.00, respectively; all three were classified as having probable damage by Polyphen2. Protein structure and functional activity depend on protein stability. 27 Thus, I-Mutant, which was used to assess the stability of protein, demonstrated the protein stability for D357A, I223S, and I4T as -2.58, -2.25 and -2.2, respectively, as the lowest values. Thus, these three SNPs affect the function and structure of SIRT1 protein.
By determining the RMSD values and TM scores for each mutant model, we expanded our analysis. While RMSD aids in calculating the average distance between the carbon backbones of wild and mutant models, the TM score is utilised to assess the topological similarity between wild- and mutant-type models. 20 , 21 The mutant model D357A demonstrated a greater RMSD value, which had a greater deviation from the wild type compared to the other two mutant models. To further establish the detrimental impacts of these nsSNPs, the SIRT1 protein structure was determined using I-Tasser, and the protein's FASTA sequence served as the sole input. Using I-Tasser, the prototypes are acquired, and the protein simulation is carried out. Following the introduction of the mutant models to the HOPE server, the server generated the effects of mutations on the contacts and the structural placement.
Mutations can affect a protein's stability, structure, and ultimately, function. Mutations are components of the "raw material" of evolution. The majority of, if not all, protein mutations are eliminated by negative, purifying selection, which lowers the probability of subsequent adaptations.
Because of this, under the influence of positive selection, only a small portion of all potential mutations will be resolved to take on a new function. Randomness or "neutral drift" might theoretically cause neutral mutations to randomly correct in small populations. At the level of the organism, the consequences of mutations on fitness are complicated and seldom ever correlate to the characteristics of a single gene or protein. Several levels of redundancy, resilience, and backup decrease the impact of numerous mutations. Understanding and predicting the impact of mutations at the organismal level present important problems for evolutionary biology. 28 , 29
The stability of the proteins is influenced by the quantity of functional protein present. According to previous research, stability and folding effects are responsible for 80% of the negative consequences of pathogenic mutations. 30 Protein dysfunctionalization is mostly caused by mutations that reduce the amount of soluble, functional proteins over a specified threshold (or DDG value). 30 Experimental studies on a variety of proteins indicated that between 33 and 40% of the time, a detrimental mutation is likely to occur. 29 As mutation rates increase, protein fitness therefore substantially decreases. When five mutations are introduced into a protein, its fitness is decreased by 20%.
Protein evolution rates, and maybe even the rates at which entire organisms evolve, seem to be primarily (though surely not solely) influenced by stability, 31 , 32 particularly but not entirely in connection with the acquisition of new functionalities. Stability appears to be the primary (though surely not the only) driver of how rapidly proteins change, despite the fact that a protein's starting stability might mitigate some of the destabilising effects of mutations.
For a small number of proteins, experimental datasets are frequently made accessible, and they generally focus on changes in mutation thermodynamic stability (DDG values). Recent developments in computing have made it possible for researchers to predict the DDG values of certain protein mutations. Some prediction methods strongly rely on sequence, whereas others mostly rely on three-dimensional structures. 33 , 34
New protein functions cannot be developed because of the destabilising impact of mutations. Neutral or non-adaptive mutational drifts have been found to be less disruptive and to occur frequently at buried residues as compared to new function or adaptive mutations. 35
The mutant study shows the decreased thermodynamic stability of the proteins, regardless of whether SIFT and Provean examinations of SNPs in the leptin and leptin receptor genes suggest that they are detrimental or tolerated. This might have an impact on how leptin and leptin receptor proteins function. This conclusion supports previous studies linking leptin, leptin gene polymorphisms, and the incidence of depression in obese individuals.
Despite several studies relating SNPs in different genes to a number of disorders, computational analysis of the functional effects of SNPs in SIRT1 is still lacking. To determine whether an amino acid change will have an impact on protein function, the SIFT technique examines sequence homology across related genes and domains across evolution. The physical-chemical properties of the residues of amino acids are also considered. According to estimates, SIFT has error rates of 31% and 20% for false negatives and positives, respectively. When amino acid changes are used as the test set, SIFT is roughly 80% effective in benchmarking trials and is thought to significantly reduce the residual activity of the variant protein.
However, utilizing SIFT and Provean, it is now feasible to analyse gene polymorphisms and forecast how a mutation will alter a protein's functionality. Since most disease mutations have an effect on protein stability, I-Mutant assessed the stability of the mutant proteins.
To find, characterize, validate, and predict the functional consequences of harmful non-synonymous SNPs (nsSNPs) in the interleukin-8 gene, Dakal et al. carried out a comparable. 36
It may also be deduced that all three of the SIRT1 gene's most harmful nsSNPs eventually interfere with and disrupt the normal function of other expressive genes. Based on their interaction patterns and their correlation profiles with numerous diseases and their pathways, SIRT1 is involved in pathways with genes such as NFKB1, NFKB1A, DDX5, AURKA, BARD1, RPA1, UBEBA, ARNTL, CLOCK, CRY1, PPARGC1A, FOXO1, FOXO3, RELA, MYOD1, SUV39H1, MDM2, EP300, PPARG and TP53, which, therefore indicate its importance. 37
Conclusions
The SIRT1 protein plays a crucial role in various disorders, and its structural confirmation is essential for its proper functioning. Through our in-silico analysis of functional SNPs, we have gained significant insight into the potential detrimental effects of ns-SNPs on SIRT1 protein structure and functionality. Our findings highlight the three ns-SNPs (D357A, I223S, and I4T) could be the most harmful mutations, and these results may serve as a valuable reference point for future research on diagnostic and therapeutic approaches related to SIRT1-associated disorders. Large-scale experimental mutational validation will be necessary to validate these findings and advance our understanding of the role of SIRT1 in disease.
Author contributions
Desy TM and Usha Adiga designed the research, performed softwares and wrote the manuscript; Tirthal Rai and Sachidananda Adiga, Vijith Shetty revised the manuscript and involved in data analysis. All authors read and approved the fnal manuscript.
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 2; peer review: 2 approved]
Data availability
Underlying data
Biostudies: Identification Of The SIRT1 Gene's Most Harmful Non-Synonymous SNPs And Their Effects On Functional And Structural Features- An Insilico Analysis; Accession number: S-BSST944. https://identifiers.org/biostudies: S-BSST944
This project contains the following underlying data
-
-
Table 1: SIFT, I mutant analysis, Provean for the nsSNPs of SIRT1 Gene
-
-
Figure 1: Sorting of nsSNPs of SIRT1
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
References
- 1. Ashok K, Mona D-G: Sirtuins as NAD+-dependent deacetylases and their potential in medical therapy. Medical Epigenetics. 2nd ed. 2021; pp.633–664. 10.1016/B978-0-12-823928-5.00028-1 [DOI] [Google Scholar]
- 2. Yang T, Fu M, Pestell R, et al. : SIRT1 and endocrine signaling. Trends Endocrinol. Metab. 2006 Jul 1;17(5):186–191. 10.1016/j.tem.2006.04.002 [DOI] [PubMed] [Google Scholar]
- 3. Yeon-Hwa L, Su-Jung K, Young-Joon S: Role of Post-translational Modification of Silent Mating Type Information Regulator 2 Homolog 1 in Cancer and Other Disorders. J. Cancer Prev. 2022;27:157–169. 10.15430/JCP.2022.27.3.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yanting G, Dechun J, Pengcheng X, et al. : Protective Effect of Silent Mating Type Information Regulation 2 Homolog 1 on TGF- β1 Pathway via mTOR in Diabetic Nephropathy. J. Biosci. Med. 2023;11:194–207. 10.4236/jbm.2023.112015 [DOI] [Google Scholar]
- 5. Ding RB, Bao J, Deng CX: Emerging roles of SIRT1 in fatty liver diseases. Int. J. Biol. Sci. 2017;13(7):852–867. 10.7150/ijbs.19370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patricia S, Pardo, Aladin M, et al. : SIRT1 Regulation in Ageing and Obesity. Mech. Ageing Dev. 2020;188:111249. 10.1016/J.MAD.2020.111249 [DOI] [PubMed] [Google Scholar]
- 7. Kosgei VJ, Coelho D, Guéant-Rodriguez R-M, et al. : Sirt1-PPARS Cross-Talk in Complex Metabolic Diseases and Inherited Disorders of the One Carbon Metabolism. Cells. 2020;9. 10.3390/CELLS9081882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chalkiadaki A, Guarente L: The multifaceted functions of sirtuins in cancer. Nat. Rev. Cancer. 2015 Oct;15(10):608–624. 10.1038/nrc3985 [DOI] [PubMed] [Google Scholar]
- 9. Todd L: Single Nucleotide Polymorphisms (SNPs). 2022. 10.1016/b978-0-12-822563-9.00037-8 [DOI] [Google Scholar]
- 10. Metin Y, Metin Y, Pemra O: In Silico Tools and Approaches for the Prediction of Functional and Structural Effects of Single-Nucleotide Polymorphisms on Proteins: An Expert Review. Omics. 2021;25(1):23–37. 10.1089/OMI.2020.0141 [DOI] [PubMed] [Google Scholar]
- 11. Arina O, Degtyareva EV, Antontseva T, et al. : Regulatory SNPs: Altered Transcription Factor Binding Sites Implicated in Complex Traits and Diseases. Int. J. Mol. Sci. 2021;22. 10.3390/IJMS22126454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robert F, Pelletier J: Exploring the impact of single-nucleotide polymorphisms on translation. Front. Genet. 2018 Oct 30;9(9):507. 10.3389/fgene.2018.00507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jain KK, Jain KK: Molecular Diagnostics in Personalized Medicine. Textbook of Personalized Medicine. 2015; pp.35–89. 10.1007/978-3-030-62080-6_2 [DOI] [Google Scholar]
- 14. Sim NL, Kumar P, Hu J, et al. : SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012 Jul 1;40(W1):W452–W457. 10.1093/nar/gks539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kumar P, Henikoff S, Ng PC: Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009 Jul;4(7):1073–1081. 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 16. Choi Y, Chan AP: PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015 Aug 15;31(16):2745–2747. 10.1093/bioinformatics/btv195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Capriotti E, Fariselli P, Casadio R: I-Mutant2. 0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005 Jul 1;33(suppl_2):W306–W310. 10.1093/nar/gki375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adzhubei IA, Schmidt S, Peshkin L, et al. : A method and server for predicting damaging missense mutations. Nat. Methods. 2010 Apr;7(4):248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Venselaar H, Te Beek TA, Kuipers RK, et al. : Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC bioinformatics. 2010 Dec;11(1):1. 10.1186/1471-2105-11-548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carugo O, Pongor S: A normalized root-mean-spuare distance for comparing protein three-dimensional structures. Protein Sci. 2001 Jul;10(7):1470–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y, Skolnick J: TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005 Jan 1;33(7):2302–2309. 10.1093/nar/gki524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y: I-TASSER server for protein 3D structure prediction. BMC bioinformatics. 2008 Dec;9(1):1–8. 10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roy A, Kucukural A, Zhang Y: I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010 Apr;5(4):725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roy A, Kucukural A, Zhang Y: I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010 Apr;5(4):725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pettersen EF, Goddard TD, Huang CC, et al. : UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004 Oct;25(13):1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 26. Zhang Y, Skolnick J: Scoring function for automated assessment of protein structure template quality. Proteins: Structure, Function, and Bioinformatics. 2004 Dec 1;57(4):702–710. 10.1002/prot.20264 [DOI] [PubMed] [Google Scholar]
- 27. Deller MC, Kong L, Rupp B: Protein stability: a crystallographer's perspective. Acta Crystallographica Section F: Structural Biology Communications. 2016 Feb 1;72(2):72–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pál C, Papp B, Lercher MJ: An integrated view of protein evolution. Nat. Rev. Genet. 2006;7(5):337–348. 10.1038/nrg1838 [DOI] [PubMed] [Google Scholar]
- 29. Camps M, Herman A, Loh ER, et al. : Genetic constraints on protein evolution. Crit. Rev. Biochem. Mol. Biol. 2007;42(5):313–326. 10.1080/10409230701597642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yue P, Li Z, Moult J: Loss of protein structure stability as a major causative factor in monogenic disease. J. Mol. Biol. 2005;353(2):459–473. 10.1016/j.jmb.2005.08.020 [DOI] [PubMed] [Google Scholar]
- 31. Bloom JD, Silberg JJ, Wilke CO, et al. : Thermodynamic prediction of protein neutrality. Proc. Natl. Acad. Sci. 2005;102(3):606–611. 10.1073/pnas.0406744102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zeldovich KB, Chen P, Shakhnovich EI: Protein stability imposes limits on organism complexity and speed of molecular evolution. Proc. Natl. Acad. Sci. 2007;104(41):16152–16157. 10.1073/pnas.0705366104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang LT, Gromiha MM, Ho SY: iPTREE-STAB: interpretable decision tree based method for predicting protein stability changes upon mutations. Bioinformatics. 2007;23(10):1292–1293. 10.1093/bioinformatics/btm100 [DOI] [PubMed] [Google Scholar]
- 34. Parthiban V, Gromiha MM, Schomburg D: CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006;34(suppl_2):W239–W242. 10.1093/nar/gkl190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vendruscolo M, Tartaglia GG: Towards quantitative predictions in cell biology using chemical properties of proteins. Mol. BioSyst. 2008;4(12):1170–1175. 10.1039/b805710a [DOI] [PubMed] [Google Scholar]
- 36. Tokuriki N, Stricher F, Serrano L, et al. : How protein stability and new functions trade off. PLoS Comput. Biol. 2008;4(2):e1000002. 10.1371/journal.pcbi.1000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rahman S, Islam R: Mammalian Sirt1: insights on its biological functions. Cell Commun. Signal. 2011 Dec;9(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]