SUMMARY
Tumor-associated myeloid cells modulate the tumor microenvironment and affect tumor progression. Type I interferon (IFN-I) has multiple effects on tumors and immune response, and ubiquitin-specific peptidase 18 (USP18) functions as a negative regulator of IFN-I signal transduction. This study aims to examine the function of IFN-I in myeloid cells during tumor progression. Here, we show that deletion of USP18 in myeloid cells suppresses tumor progression. Enhanced IFN-I signaling and blocked USP18 expression prompt downregulation of colony stimulating factor 1 receptor (CSF1R) and polarization of tumor-associated macrophages toward pro-inflammatory phenotypes. Further in vitro experiments reveal that downregulation of CSF1R is mediated by ubiquitin-proteasome degradation via E3 ligase neural precursor cell-expressed, developmentaly downregulated 4 (NEDD4) and the IFN-induced increase in ubiquitin E2 ubiquitin-conjugating enzyme H5. USP18 impairs ubiquitination and subsequent degradation of CSF1R by interrupting NEDD4 binding to CSF1R. These results reveal a previously unappreciated role of IFN-I in macrophage polarization by regulating CSF1R via USP18 and suggest targeting USP18 in myeloid-lineage cells as an effective strategy for IFN-based therapies.
Graphical Abstract
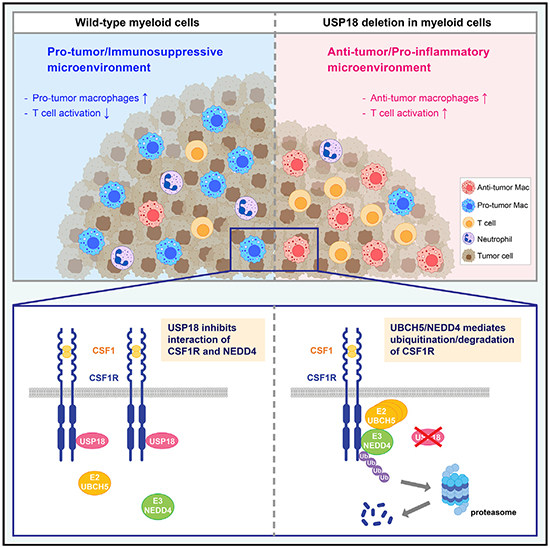
In brief
Miyauchi et al. report that deletion of USP18 in myeloid cells suppresses tumor progression by downregulation of CSF1R and increase in anti-tumor macrophages. USP18 impairs degradation of CSF1R by interrupting E3 ligase NEDD4 binding to CSF1R. These results reveal roles of IFN-I in macrophage polarization via the CSF1R-USP18 axis.
INTRODUCTION
Accumulating evidence reveals that the tumor microenvironment (TME) has a fundamental effect on tumor development, growth, and therapeutic outcome. The TME is a complex network containing cytokines, chemokines, extracellular matrix, endothelial cells, fibroblasts, and immune cells. The constant interaction and communication among these cells and tumor cells support cancer development. Therefore, it is important to understand the molecular basis of establishment of the pro-cancer TME and to identify therapeutic interventions to modulate the TME for suppressing cancer development. Myeloid cells, including macrophages, dendritic cells (DCs), and granulocytes, are important in initiating both innate and adaptive immune responses as well as in supporting and inhibiting neoplasms.1,2 The most frequently identified non-tumor cells in the TME are tumor-associated macrophages (TAMs). They are considered immunosuppressive cells in the TME and promote tumor growth and metastasis by stimulation of matrix remodeling, angiogenesis, and secretion of growth factors and cytokines. Clinical data indicate that TAMs are generally associated with high tumor grade and poor prognosis in many human cancer types, such as cancers of the breast, bladder, prostate, and head and neck; glioma; melanoma; and non-Hodgkin’s lymphoma.3–5 However, increased TAMs have also been reported with better prognosis in colorectal and gastric cancers, suggesting that TAMs may have diversified functions in the TME. In fact, macrophages are known to have high plasticity and present different phenotypes in response to a variety of stimulations and environments. In response to the TME, TAMs are known to be polarized to either a pro-inflammatory/anti-tumorigenic phenotype or an immunosuppressive/pro-tumorigenic phenotype. Immunosuppressive macrophages produce anti-inflammatory cytokines and growth factors, such as IL-10 and TGF-β, creating a tumor-promoting microenvironment, whereas anti-tumor macrophages produce pro-inflammatory cytokines, such as TNF-α and IL-6.6 Importantly, most TAMs may have a spectrum of mixed phenotypes between the extreme anti-tumor and the pro-tumor phenotypes, and upon different stimulations, they are able to gain or lose those phenotypes.5,7 Therefore, it is significant to investigate how to reprogram TAMs toward the anti-tumor phenotypes to suppress tumor growth.
Type I interferons (IFN-Is) have direct effects on tumor cells, inhibiting their proliferation and inducing apoptosis.8 IFN-Is also have effects on immune cells through various mechanisms.9 IFN-Is support cytotoxic T lymphocytes (CTLs) by enhancing cross-priming from dendritic cells, boosting immune effector functions, and promoting their survival.10,11 IFN-Is also stimulate macrophages, leading to the release of pro-inflammatory cytokines, such as IL-1β and IL-18.12 Moreover, IFN-Is can decrease the immunosuppressive function of regulatory T cells.13 Further investigations are still required to understand the precise role of IFN-Is in anti-tumor immunity. In the past decades, IFN-I has been used for cancer treatment via systemic administration; however, there are limitations for clinical use due to the limited efficacy and the adverse effects such as fatigue, anorexia, flu-like symptoms, and hepatotoxicity. Thus, approaches to enhance effects but lower toxicity via targeted therapy and specific delivery of IFN-I should be considered.
Ubiquitin-specific peptidase 18 (USP18, aka UBP43) was first reported as a ubiquitin-like modifier ISG15-specific deconjugating enzyme and is responsible for removing ISG15 from ISGylated proteins.14 In addition to increased levels of ISG15-modified proteins, USP18-deficient cells have much stronger IFN-I signaling.15 Further studies reveal that USP18 is a potent negative regulator of IFN-I signaling via disruption of STAT2 binding to IFN-I receptor subunit R2 and blocking of JAK kinase activation.16,17 Deletion of USP18 enhances and prolongs IFN-I signaling17,18 and expands the pool of IFN-inducible genes.19 In addition to regulation of IFN-I signaling and protein ISG15 conjugation, USP18 is reported to regulate type III IFN signal transduction20 and expression of EGFR, CCND1, and other regulators in cancer cells.21–23 Altogether, USP18 regulates IFN signaling and cancer-related target gene expression, suggesting a potential, promising benefit of USP18-related studies for development of targeted anti-cancer therapies.
Despite its importance in the anti-tumor immune response, IFN-I in myeloid cells, especially in TAMs, is underinvestigated. Furthermore, there has been no report about the role of USP18 in TAMs. The aim of the current study is to examine the function of USP18 and IFN-I in macrophages during tumor development by utilizing myeloid-lineage-specific USP18-knockout (KO) mice. Here, we report that deletion of USP18 in myeloid cells suppresses tumor growth and enhances activation of cytotoxic CD8+ cells. This anti-tumor effect is induced by an increase in anti-tumor macrophages caused by reduction of a well-known pro-tumor macrophage promoter, CSF1R. Mechanistically, deletion of USP18 enhances interaction of the ubiquitin E3 enzyme NEDD4 and CSF1R and increases expression of an IFN-I-induced ubiquitin E2 enzyme, ubiquitin-conjugating enzyme H5 (UBCH5), which consequently enhances ubiquitination and degradation of CSF1R protein through the proteasome. Our results indicate the potential of targeting USP18 in macrophages as an IFN-based immunotherapy.
RESULTS
Generation of myeloid-lineage-specific USP18 conditional knockout mice
To study USP18 in regulation of IFN signaling under different biological settings, we generated a conditional Usp18 gene-KO mouse model (Figure 1A). USP18 is a potent inhibitor of IFN-I signal transduction and subsequent IFN-stimulated gene (ISG) expression.19 To investigate IFN-mediated changes in the TME to promote tumor growth, we generated myeloid-specific USP18-deficient mice by crossing conditional Usp18-KO mice (Usp18f/f) with LysM-Cre transgenic mice that express Cre recombinase in myeloid cells.24 Cre-mediated deletion was observed in the peritoneal macrophages (PMs) from Usp18f/f LysM-Cre+/+ mice, but not in those from Usp18+/+ (wild type [WT]) or Usp18f/f LysM-Cre−/− mice (Figure 1B). No Usp18 gene deletion was observed in the tail DNA from the Usp18f/f LysM-Cre+/+ mice. Consistent with the results of genomic DNA, Usp18 mRNA in CD11b+ myeloid cells from bone marrow was decreased in Usp18f/f LysM-Cre+/+ mice, but not in CD3+ T cells (Figure 1C). As the downregulation of Usp18 mRNA expression in Usp18f/f LysM-Cre+/− was comparable to that in Usp18f/f LysM-Cre+/+ myeloid cells, we used both genotypes as myeloid-lineage-specific Usp18-KO mice (described as Usp18Δ/Δ mice hereafter) in further experiments. Deletion of USP18 and enhanced IFN-I response as indicated by increased ISG15 expression were also confirmed at the protein level in both bone-marrow-derived macrophages (BMDMs) and PMs (Figure 1D). Hematological analyses showed that deletion of Usp18 in myeloid cells did not affect normal hematopoiesis (Figure S1A). Furthermore, the deletion of Usp18 did not affect the cell viability of myeloid cells (Figure S1B).
Figure 1. Deletion of Usp18 in myeloid-lineage cells.
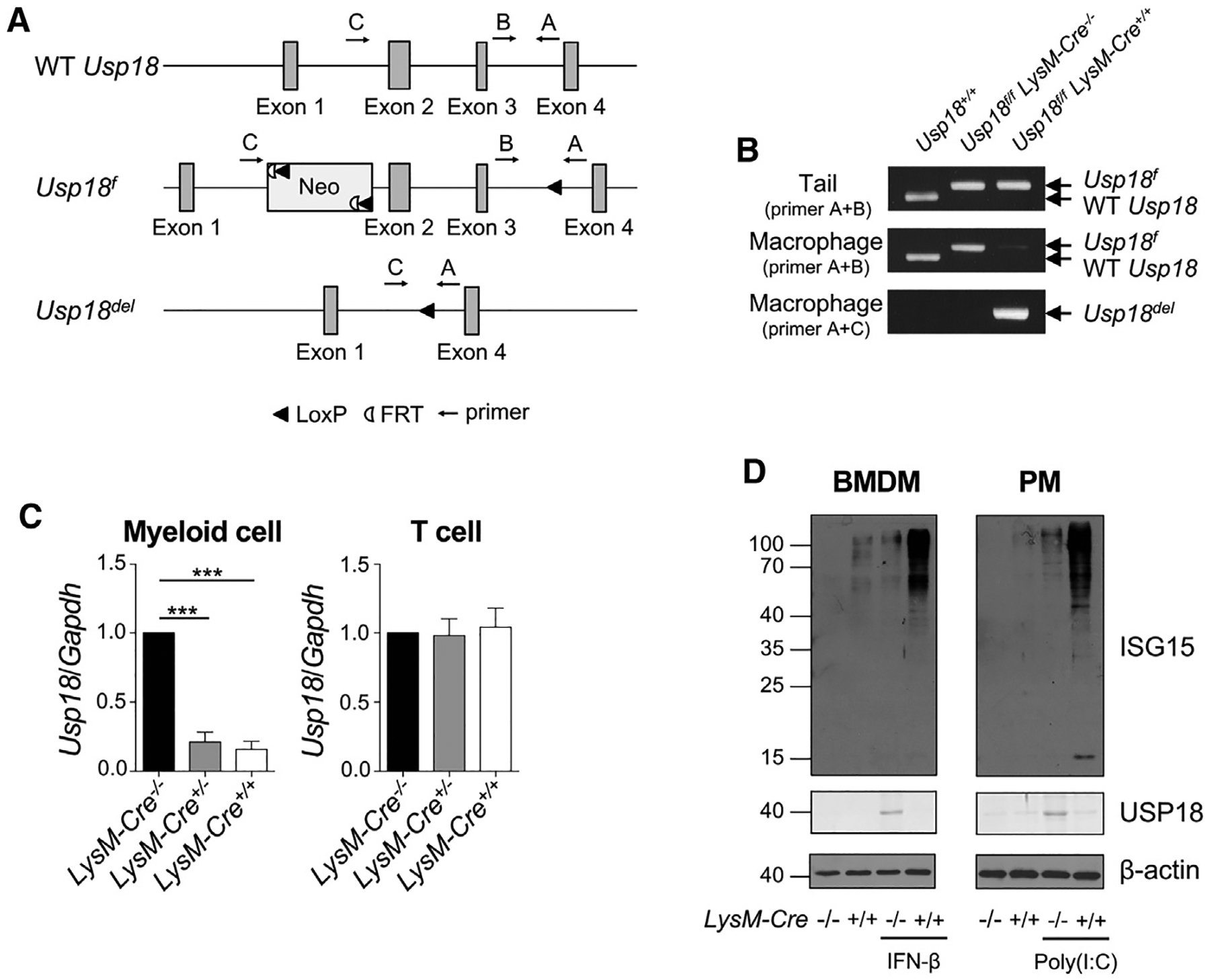
(A) The wild-type Usp18 locus (WT Usp18), the targeted Usp18 locus in the loxP-flanked Usp18 allele (Usp18f), and the Usp18 locus in myeloid-lineage cells from Usp18f/f LysM-Cre mice (Usp18del). The positions of PCR primers A, B, and C are shown. The loxP sites flanked 4.4 kb.
(B) PCR analysis of genomic DNA from tails (top) and peritoneal macrophages (middle and bottom) with the primers indicated in (A). Primers A and B were used in the top and middle images, and primers A and C were used in the bottom image. The PCR product obtained with primers A and B from the wild-type Usp18 locus is 385 bp (WT Usp18) and that from the Usp18f locus is 456 bp (Usp18f). In Usp18f/f LysM-Cre+/+ mice, a band of 509 bp obtained with primers A and C indicates Cre-mediated deletion of Usp18 (Usp18del).
(C) Quantitative PCR analysis of mRNA from myeloid cells and T cells in Usp18f/f LysM-Cre−/−, Usp18f/f LysM-Cre+/−, and Usp18f/f LysM-Cre+/+ mice. Myeloid cells from bone marrow and T cells from spleen were isolated. The values of Usp18f/f LysM-Cre−/− were considered as 1. Mean ± SEM, n = 3 in each group. One-way ANOVA multiple comparison test with post hoc Tukey test, ***p < 0.001. (D) Western blot of bone-marrow-derived macrophages (BMDM) with or without IFN-β treatment (100 U/mL, 24 h) and peritoneal macrophages (PM) from poly(I:C)-injected or uninjected mice (5 μg/g body weight, 48 h). Cells were harvested from Usp18f/f LysM-Cre−/− or Usp18f/f LysM-Cre+/+ mice. Cell lysates were analyzed with the indicated antibodies. See also Figure S1.
Myeloid-lineage-specific deletion of USP18 suppresses tumor progression
To elucidate the role of USP18 in myeloid cells on tumor progression, B16F10 melanoma, EL4 lymphoma, or LLC lung carcinoma cells were subcutaneously injected into Usp18f/f and Usp18Δ/Δ mice. Delayed tumor growth was observed in all of these tumor models in Usp18Δ/Δ mice (Figure 2A). In addition to an inhibitory effect on IFN-I stimulation, USP18 deconjugates ubiquitin-like modifier ISG15 from ISGylated proteins.14,17 Therefore, we next explored whether USP18-regulated protein ISGylation in the TME contributed to this tumor-suppressive phenotype. We generated myeloid-specific Usp18-KO mice that have an additional deletion of Isg15 (Usp18Δ/Δ Isg15−/−) and repeated tumor growth studies using B16F10 and EL4 tumor cells. More ISG15-modified proteins were accumulated substantially in USP18-deficient myeloid cells (Usp18Δ/Δ) compared with control (Usp18f/f). In contrast, Usp18Δ/Δ Isg15−/− mice did not have protein ISGylation nor free ISG15 (Figure 2B). Compared with Usp18f/f mice, tumor growth was significantly reduced in both Usp18Δ/Δ mice and Usp18Δ/Δ Isg15−/− mice. We did not observe significant differences between Usp18Δ/Δ mice and Usp18Δ/Δ Isg15−/− mice (Figure 2C). These results suggest that the function of USP18 in the regulation of protein ISGylation in myeloid cells is not involved in the enhanced anti-tumor activity observed in Usp18Δ/Δ mice. To further analyze the function of USP18 in regulation of IFN response, we examined tumor growth with additional IFN stimulation by poly(I:C) treatment to stimulate endogenous IFN production. Poly(I:C) treatment did not affect tumor growth in Usp18f/f mice; however, the anti-tumor effect was further enhanced by poly(I:C) treatment in Usp18Δ/Δ mice (Figure 2D). Moreover, an IFN-I receptor (IFNAR1)-blocking antibody abrogated the anti-tumor effect observed in Usp18Δ/Δ mice (Figure 2E). These results indicate that this anti-tumor effect is likely mediated by the enhanced IFN-I response and not by protein ISGylation in myeloid cells. In addition, we confirmed that there were no effects caused by Cre recombinase expression on tumor progression by comparing tumor growth between Usp18+/+ LysM-Cre−/− mice and Usp18+/+ LysM-Cre+/+ mice (Figure 2F). Together, these results indicate that depletion of USP18 in myeloid cells enhances anti-tumor immune responses, which is not due to the increased protein modification by ISG15 and is likely related to the increased IFN-I response in the TME.
Figure 2. Suppressed tumor growth in Usp18Δ/Δ mice.
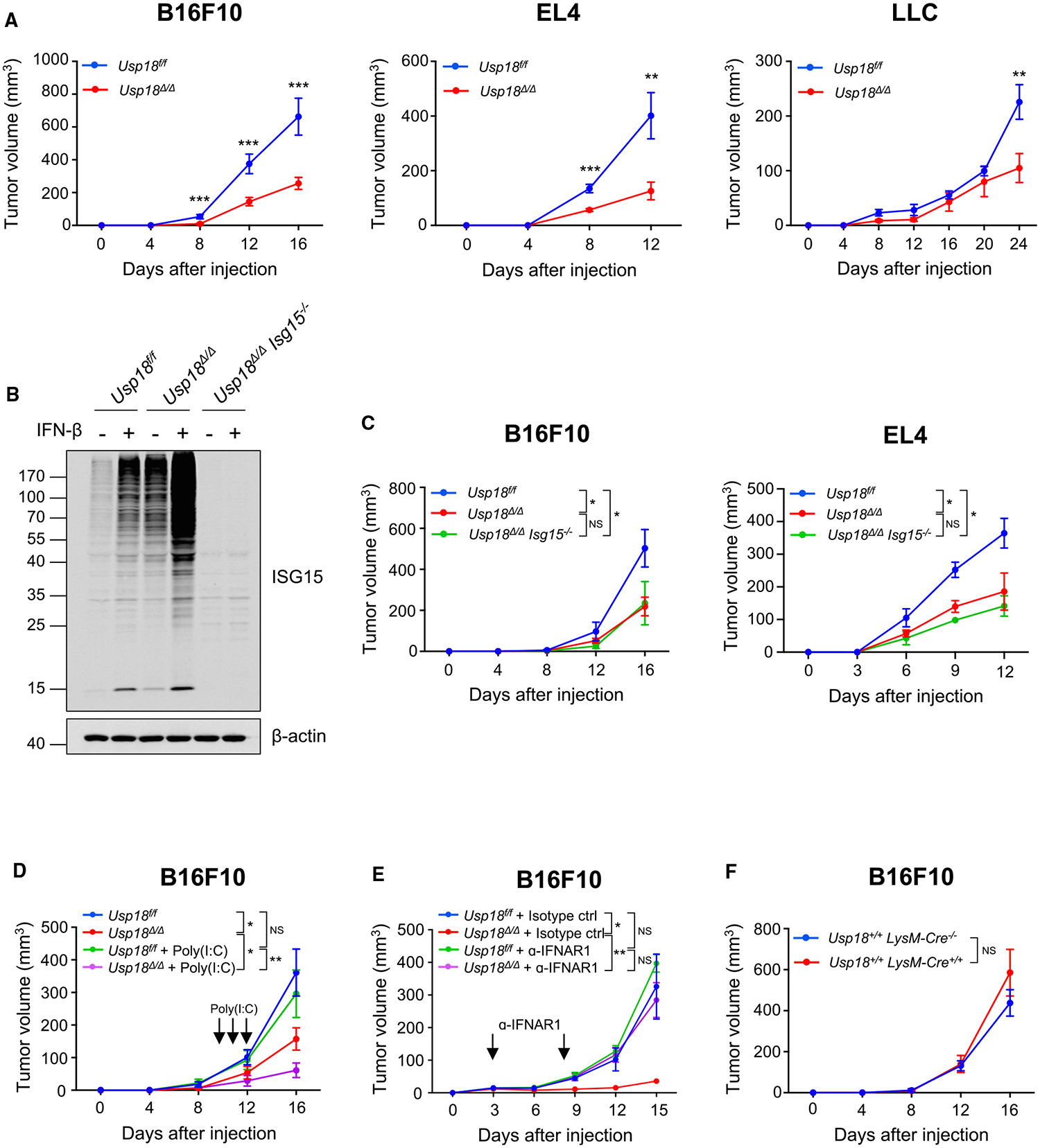
(A) B16F10 melanoma (1 × 105), EL4 thymoma (5 × 106), and LLC (5 × 105) cells were subcutaneously injected into Usp18f/f or Usp18Δ/Δ mice (B16F10: Usp18f/f, n = 12; Usp18Δ/Δ, n = 17; EL4: Usp18f/f, n = 7; Usp18Δ/Δ, n = 9; LLC: Usp18f/f, n = 18; Usp18Δ/Δ, n = 10).
(B) Western blot of bone-marrow-derived macrophages from Usp18f/f, Usp18Δ/Δ, and Usp18Δ/Δ Isg15−/− mice with or without IFN-β treatment (100 U/mL, 24 h). Cell lysates were analyzed with the indicated antibodies.
(C) B16F10 melanoma (1 × 105) or EL4 thymoma (5 × 106) cells were subcutaneously injected into Usp18f/f, Usp18Δ/Δ, or Usp18Δ/Δ Isg15−/− mice (B16F10: Usp18f/f, n = 6; Usp18Δ/Δ, n = 8; Usp18Δ/Δ Isg15−/−, n = 5; EL4: Usp18f/f, n = 7; Usp18Δ/Δ, n = 8; Usp18Δ/Δ Isg15−/−, n = 4).
(D) B16F10 melanoma cells (1 × 105) were subcutaneously injected into Usp18f/f or Usp18Δ/Δ mice with or without poly(I:C) treatment. Poly(I:C) (5 μg/g body weight) was intraperitoneally injected on days 10, 11, and 12 (Usp18f/f, n = 5; Usp18Δ/Δ, n = 9; Usp18f/f + poly(I:C), n = 5; Usp18Δ/Δ + poly(I:C), n = 11).
(E) B16F10 melanoma cells (1 × 105) were subcutaneously injected into Usp18f/f or Usp18Δ/Δ mice treated with isotype control or anti-IFNAR1 antibodies. The antibodies (200 μg) were intravenously injected on days 3 and 8 (Usp18f/f + isotype control, n = 6; Usp18Δ/Δ + isotype control, n = 6; Usp18f/f + α-IFNAR1, n = 6; Usp18Δ/Δ + α-IFNAR1, n = 5).
(F) B16F10 melanoma cells (1 × 105) were subcutaneously injected into Usp18+/+ LysM-Cre−/− or Usp18+/+ LysM-Cre+/+ mice (Usp18+/+ LysM-Cre−/−, n = 6; Usp18+/+ LysM-Cre+/+, n = 5). Mean ± SEM. Two-tailed unpaired t test and ordinary one-way ANOVA multiple comparison test with post hoc Tukey test were conducted for comparisons of two groups and more than two groups, respectively. NS, not significant; *p < 0.05, **p < 0.01, ***p < 0.001.
Deletion of USP18 promotes macrophage polarization toward anti-tumor phenotypes
To examine the effects of myeloid-specific deletion of USP18 on the TME, single-cell RNA sequencing (RNA-seq) of tumor-infiltrating immune cells was performed. Tumor-infiltrating immune cells (CD45+ cells) from B16F10 tumors in Usp18f/f and Usp18Δ/Δ mice were sorted and analyzed. Both datasets were integrated, and non-linear dimensionality reduction was performed, shown in uniform manifold approximation and projection (UMAP) (Figure 3A). Each cluster was defined by using known markers25–28 (Figure S2A). Initial clustering analysis showed that there were fewer B cells (cluster 0) and neutrophils (cluster 6) in Usp18Δ/Δ mice (Figure S2B). To increase resolution and define the myeloid-lineage populations more accurately, a subpopulation of monocytes, macrophages, dendritic cells, and neutrophils was further investigated by subclustering analysis (Figure 3B). A combination of automated and manual annotation methods was used for cell-type annotation. First, cells were annotated using SingleR28 with the ImmGen reference dataset.29 Then, if needed, the annotations were refined manually based on gene expression and TAM classification from a published article30 (Figures S2C and S2D). When Usp18Δ/Δ mice were compared with Usp18f/f mice, we observed an increase in clusters 7 and 12 (clusters of macrophages) and a decrease in clusters 3 and 17 (clusters of neutrophils) (Figure 3C). It is known that TAMs polarize and differentiate into functionally distinct subsets that change the TME. To further validate our cluster annotations and investigate how USP18-deletion-dependent alterations in macrophage clusters may correlate with TAM polarization and affect the TME, we performed pathway analysis on macrophage-annotated clusters using gene set enrichment analysis (GSEA).31,32 The top hit pathways in cluster 12 were IFN-α and IFN-γ responses (Figure 3D). Indeed, type I IFN-inducible genes were highly expressed in this cluster (Figure S3A). Accordingly, cluster 12 was annotated as IFN-primed TAMs with additional support from a recent review article on TAM classification.30 Similarly, other macrophage clusters were annotated based on enriched functional terms, highly expressed genes, and the recent TAM classification. Clusters 1, 2, 6, and 14 were annotated as lipid-associated TAMs, pro-angiogenic TAMs, immune-regulatory TAMs, and proliferating TAMs, respectively (Figure S3B). Interestingly, cluster 7 cells did not highly express a defined set of marker genes. Metascape analysis33 of reported cluster 7 markers reported several top hit pathways, including “positive regulation of inflammatory response” (Figure 3E); however, the functional associations were not as clear compared with the other TAM clusters. Therefore, we performed a subclustering analysis on cluster 7 for further characterization. The majority of the cells in cluster 7 were from Usp18Δ/Δ, and interestingly, cells from Usp18f/f and Usp18Δ/Δ conditions distributed into almost distinct clusters (Figure S3C). Pathway enrichment analysis on differentially expressed genes (DEGs) between Usp18f/f and Usp18Δ/Δ cells in cluster 7 revealed that the top hit pathways were pro-inflammatory responses, including IFN-α and IFN-γ responses (Figure 3F). Based on the results from Metascape and the subclustering analysis, cluster 7 was annotated as pro-inflammatory TAMs.
Figure 3. Increased anti-tumor macrophages in Usp18Δ/Δ mice.
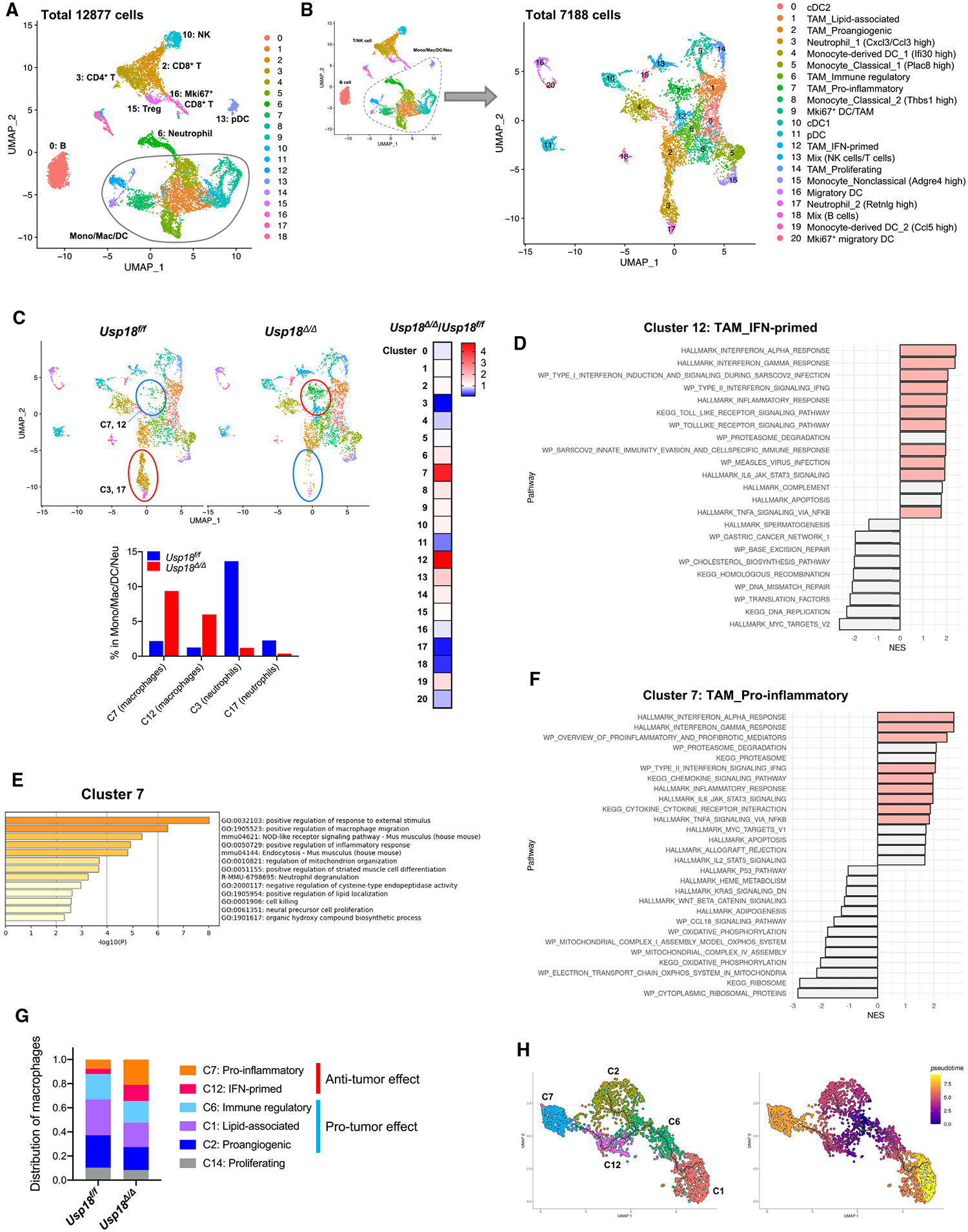
B16F10 melanomas grown in Usp18f/f and Usp18Δ/Δ mice were harvested, and CD45+ cells isolated from the tumors were analyzed by single-cell RNA-seq.
(A) UMAP plot of intratumoral CD45+ cells from the merged sample.
(B) UMAP plot of the monocytes/macrophages/dendritic cells/neutrophils subpopulation (Mono/Mac/DC/Neu).
(C) UMAP plot of the myeloid population split by Usp18f/f and Usp18Δ/Δ mice. Heatmap shows the changes in all the clusters. The bar graph shows the percentage of cells in the clusters with changes after the deletion of Usp18.
(D) Pathway enrichment analysis of DEGs in cluster 12 by GSEA. The colored pathways indicate relevance to the annotated function of the cluster.
(E) Pathway enrichment analysis of DEGs in cluster 7 by Metascape.
(F) Pathway enrichment analysis of DEGs of Usp18Δ/Δ in cluster 7. The colored pathways indicate relevance to the annotated function of the cluster.
(G) Distribution of clusters in the macrophage population.
(H) Trajectory inference analysis of TAM clusters (clusters 1, 2, 6, 7, and 12) analyzed by Monocle 3. See also Figures S2 and S3.
It has been reported that pro-inflammatory macrophages (cluster 7) and IFN-primed macrophages (cluster 12) have anti-tumor effects, whereas immune-regulatory macrophages (cluster 6), lipid-associated macrophages (cluster 1), and pro-angiogenic macrophages (cluster 2) have pro-tumor effects.25,30,34–40 The effect of proliferating macrophages (cluster 14) in mice is controversial, and further investigation is required. Thus, in the macrophage fraction of Usp18Δ/Δ mice, we observe an increase in the macrophages with anti-tumor functions and a decrease in the macrophages with pro-tumor functions (Figure 3G).
To further analyze the polarization and differentiation of TAM subsets, we performed trajectory inference analysis using Monocle 3.41–43 TAMs frequently have immunosuppressive/pro-tumor phenotypes in the TME. Indeed, the higher infiltration/presence of TAMs in the tumor is associated with worse clinical outcomes in many cancer types.3–5 Using immune-regulatory TAMs (cluster 6) as a root, we identified three branching trajectories associated with distinct TAM clusters (Figure 3H). As the tumor progresses, immune-regulatory TAMs (cluster 6) differentiate into two types of pro-tumor TAMs along two branches: lipid-associated TAMs (cluster 1) and pro-angiogenic TAMs (cluster 2). Cells were also differentiated into anti-tumor TAMs along a third branch: IFN-primed TAMs (cluster 12) and pro-inflammatory TAMs (cluster 7). The clusters on this trajectory were increased by the deletion of Usp18, suggesting that the deletion of Usp18 may induce the reprogramming of TAMs toward an anti-tumor phenotype. Taken together, single-cell RNA-seq analysis results revealed that USP18 deletion in myeloid cells enhances macrophage polarization toward anti-tumor/pro-inflammatory phenotypes, which likely contributes to the anti-tumor microenvironment.
Deletion of USP18 downregulates CSF1R expression and promotes activation of CD8+ T cells
We next performed immune phenotyping in B16F10 tumor and tumor-draining lymph nodes (TDLNs) by flow cytometry (Figures S4A and S4B). Among myeloid subsets, TAMs (CD11b+F4/80+Gr-1−) and neutrophils (CD11b+Ly6G+) were decreased in Usp18Δ/Δ mice (Figures 4A and S4C), which was consistent with the results from single-cell RNA-seq analysis (Figure 3C). We also observed that deletion of USP18 enhanced anti-tumor macrophage polarization in single-cell RNA-seq analysis (Figure 3G). One of the key molecules that regulates macrophage differentiation and polarization is colony-stimulating factor 1 (CSF1). CSF1 and other cytokines and chemokines recruit circulating monocytes and myeloid-derived suppressor cells (MDSCs) to tumor.44 Furthermore, CSF1 promotes macrophage survival and polarization signals that induce immunosuppressive macrophages.45–48 We, therefore, hypothesized that CSF1 signaling is involved in the phenotypes that we have observed and examined whether USP18 affects CSF1R expression by using flow cytometry. Compared with control (Usp18f/f), a significant reduction in CSF1R on TAMs was observed in Usp18Δ/Δ mice (Figure 4B). Furthermore, to analyze the role of USP18 in CSF1R-mediated regulation of macrophage polarization, BMDMs from Usp18+/+ and Usp18Δ/Δ mice were treated with the CSF1R inhibitor PLX3397 prior to IFN-I treatment. Then, macrophage polarization was analyzed by flow cytometry using CD206, a marker of pro-tumor/immunosuppressive macrophages.49 PLX3397 abrogated USP18 deficiency-induced reduction in pro-tumor/immunosuppressive macrophages (Figure 4C), supporting that USP18-mediated downregulation of CSF1R expression contributes to macrophage polarization.
Figure 4. Decreased CSF1R expression in tumor-associated macrophages and increased activation of CD8+ T cells in Usp18Δ/Δ mice.
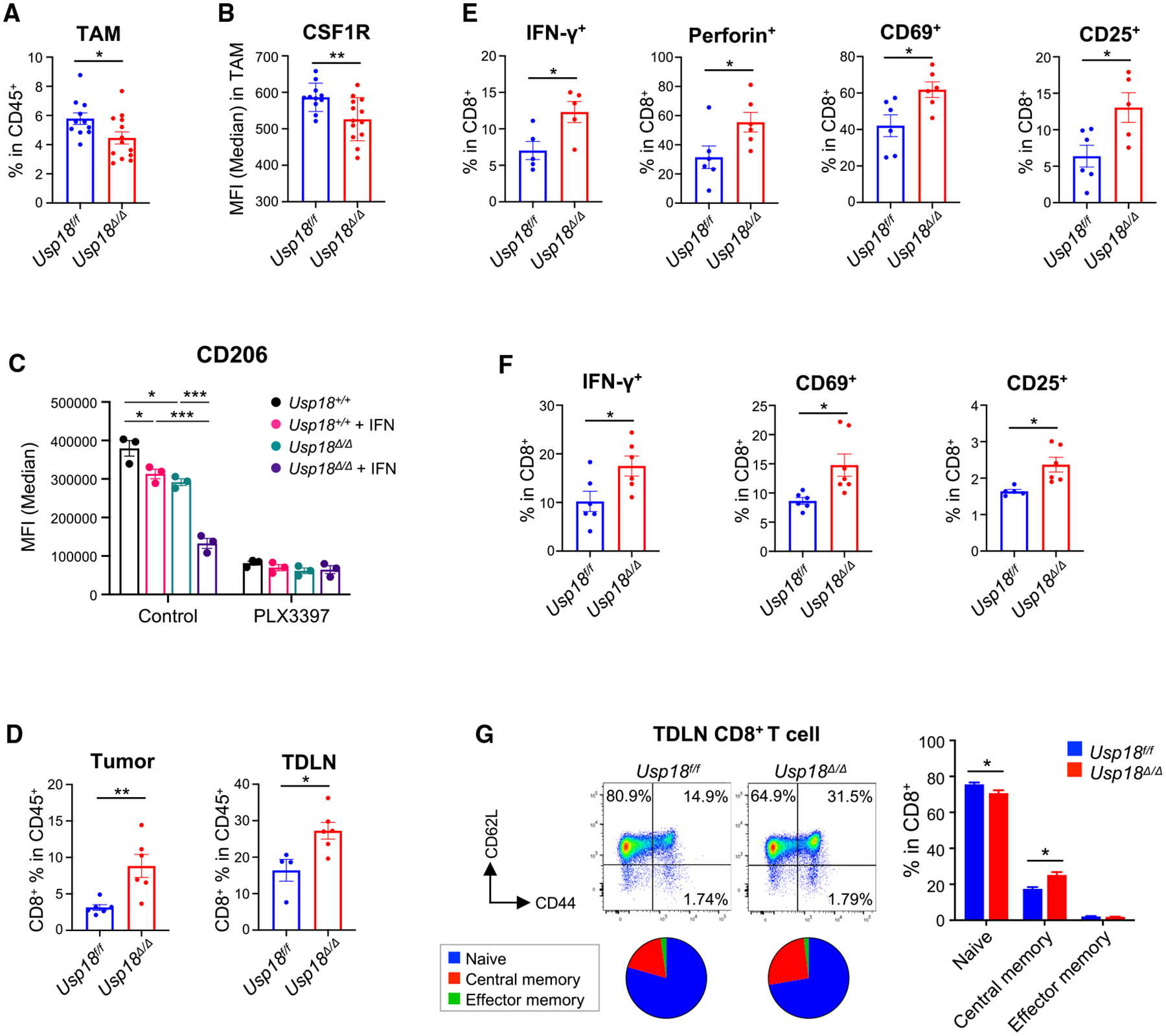
(A, B, and D‒G) B16F10 melanomas in Usp18f/f and Usp18Δ/Δ mice were harvested on day 16, and single-cell suspensions from tumors and tumor-draining lymph nodes (TDLNs) were analyzed by flow cytometry.
(A and B) Percentage of TAMs in CD45+ cells (A) and MFI of CSF1R in TAMs (B) from tumor samples. Usp18f/f, n = 11; Usp18Δ/Δ, n = 13. Mean ± SEM.
(C) BMDMs from Usp18+/+ or Usp18Δ/Δ mice were treated with IFN-β (100 U/mL) for 24 h. As indicated, cells were treated with CSF1R inhibitor PLX3397 (50 μM) for 2 h prior to IFN-β treatment. BMDMs were also stimulated with IL-4 (40 ng/mL), IL-13 (20 ng/mL), and CSF1 (20 ng/mL) for 24 h to polarize toward an immunosuppressive phenotype and then analyzed by flow cytometry (n = 3). Mean ± SEM.
(D‒F) Percentage of CD8+ T cells in CD45+ cells from tumor and TDLN samples (D), and cytokines and activation markers of CD8+ T cells from tumor (E) and TDLN
(F) samples. Usp18f/f, n = 6; Usp18Δ/Δ, n = 6. Mean ± SEM.
(G) Memory subsets of CD8+ T cells in TDLNs (naive, CD62L+CD44−; central memory, CD62L+CD44+; effector memory, CD62L−CD44+). Usp18f/f, n = 11; Usp18Δ/Δ, n = 13. Mean ± SEM. Two-tailed unpaired t test and ordinary one-way ANOVA multiple comparison test with post hoc Tukey test were conducted for comparisons of two groups and more than two groups, respectively. *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S4.
In addition to the changes in TAM population, there were significant changes in T cell population. Usp18Δ/Δ mice showed an increased frequency of CD8+ T cells in tumor and TDLN (Figure 4D). Furthermore, intracellular cytokines and lymphocyte activation markers were significantly increased in CD8+ T cells from the tumor (Figure 4E). Similar results were also observed in CD8+ T cells in TDLNs (Figure 4F). For induction of antigen-specific anti-tumor immune response, development of memory T cell subsets in lymph nodes is a crucial step. Interestingly, we observed enhanced memory CD8+ T cell formation in Usp18Δ/Δ mice (Figure 4G). These results suggest that USP18 deletion leads to CSF1R downregulation in TAMs and a decrease in pro-tumor/immunosuppressive macrophages, contributing to enhanced anti-tumor macrophage polarization, consequently promoting a Th1-dominant TME and enhancing CD8+ T cell-mediated anti-tumor immunity, in agreement with previous reports.6,50–52
IFN-I downregulates CSF1R protein expression, but not mRNA expression, in USP18-deficient macrophages
To elucidate further details of the downregulation of CSF1R in USP18-deficient macrophages, we investigated the effects of IFN-I on CSF1R expression in vitro. BMDMs from Usp18f/f or Usp18Δ/Δ mice were utilized for the analyses. The dose-dependent IFN-I-induced downregulation of total CSF1R protein expression was detected by western blotting (Figure 5A). Flow cytometric analysis showed that cell-surface expression of CSF1R was also downregulated (Figure 5B). These results support the observation in the TME (Figure 4B). Although downregulation of cell-surface CSF1R expression was also observed in BMDMs from Usp18f/f mice upon prolonged IFN treatment, the decrease was much greater in BMDMs from Usp18Δ/Δ mice (Figure 5B). To check whether USP18 regulates CSF1R at the transcript level, we performed RT-qPCR analysis. There was no significant decrease in Csf1r mRNA (Figure 5C), indicating that the downregulation of CSF1R happened mainly at the post-transcriptional level. Consistent with this, there was no difference in Csf1r expression in single-cell RNA-seq analysis (Figure S2E). Furthermore, USP18 and ISG15 double-KO BMDMs also showed CSF1R downregulation to the same degree (Figure 5D), supporting the idea that USP18-mediated regulation of protein ISGylation is not involved in the reduction of CSF1R protein.
Figure 5. IFN-I-induced downregulation of CSF1R in a dose-dependent manner.
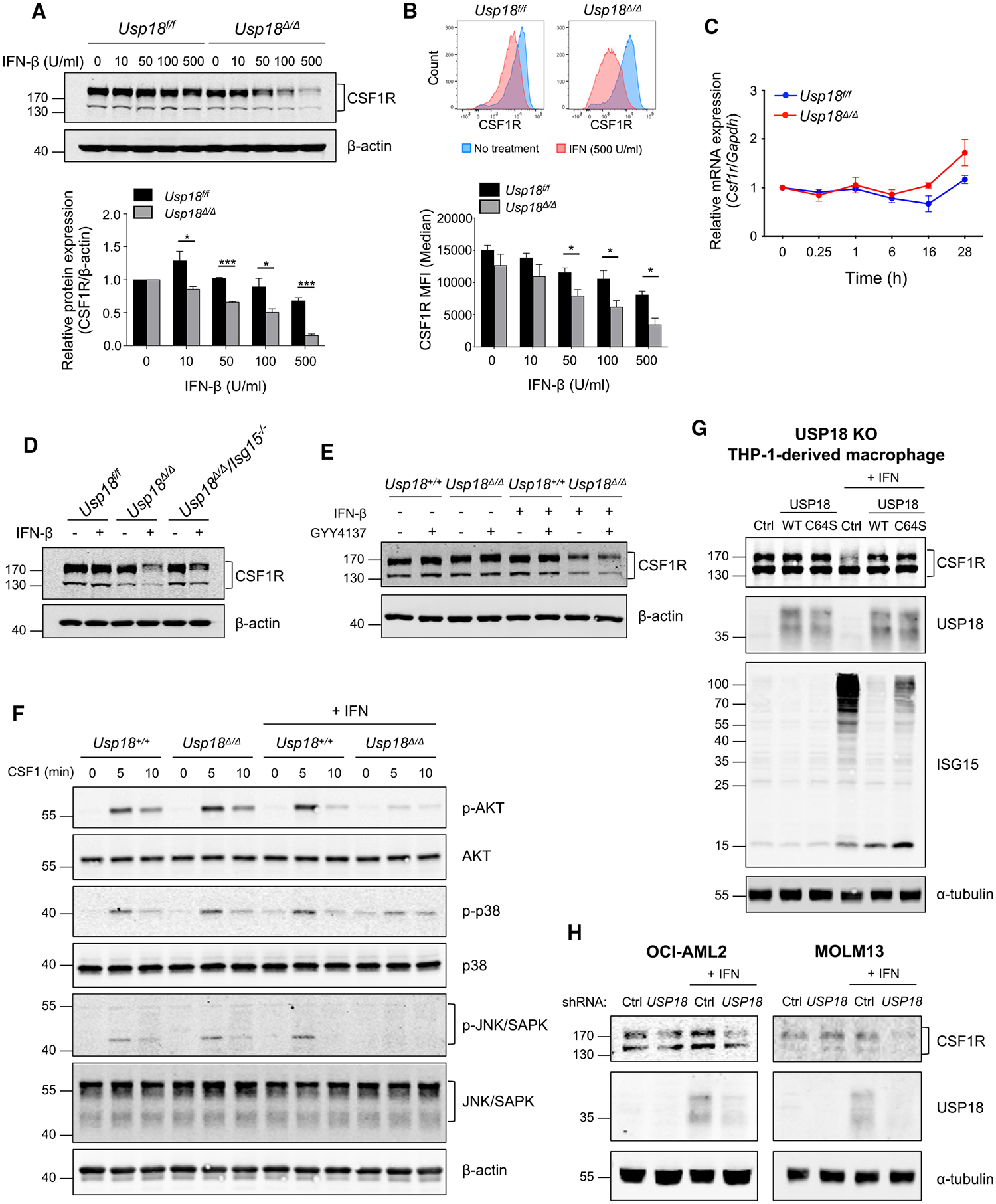
(A) Western blot of bone-marrow-derived macrophages (BMDMs) from Usp18f/f and Usp18Δ/Δ mice with or without IFN-β treatment for 24 h at the indicated concentrations. Cell lysates were analyzed with the indicated antibodies (n = 3). Mean ± SEM, two-tailed unpaired t test, *p < 0.05, ***p < 0.001.
(B) CSF1R cell-surface expression on BMDMs from Usp18f/f and Usp18Δ/Δ mice with or without IFN-β treatment for 24 h at the indicated concentrations was analyzed by flow cytometry (n = 3). Mean ± SEM, two-tailed unpaired t test, *p < 0.05.
(C) Csf1r mRNA expression in BMDMs from Usp18f/f and Usp18Δ/Δ mice with or without IFN-β treatment (100 U/mL for the indicated times) was analyzed by RT-qPCR (n = 3).
(D) Western blot of BMDMs from Usp18f/f, Usp18Δ/Δ, and Usp18Δ/Δ Isg15−/− mice with or without IFN-β treatment (100 U/mL for 24 h).
(E) BMDMs from Usp18f/f UBC-Cre-ERT2 mice were treated with β-estradiol (2 μM) for 48 h. Cells were then treated with IFN-β (100 U/mL) for 24 h with or without NF-κB inhibitor GYY4137 (100 μM) for 2 h prior to IFN-β treatment. Cell lysates were analyzed with the indicated antibodies by western blotting.
(F) BMDMs from Usp18f/f and Usp18Δ/Δ mice were treated with IFN-β (100 U/mL for 24 h) and with CSF1 (50 ng/mL) for the indicated times after CSF1 starvation for 4 h. Cell lysates were analyzed with the indicated antibodies by western blotting.
(G) USP18-KO THP-1-derived macrophages with the expression of empty vector control, wild-type USP18, and USP18 C64S were treated with IFN-I (1,000 U/mL for 24 h) and analyzed by western blotting.
(H) CSF1R in OCL-AML2 and MOLM13 cells infected with control or USP18 short hairpin RNA (shRNA) with or without IFN-I (1,000 U/mL for 24 h) was analyzed by western blotting.
We previously reported that the deletion of USP18 activates the expression of NF-κB-regulated genes.19 As NF-κB is one of the major transcription factors regulating the expression of pro-inflammatory genes, we investigated whether the polarization toward the pro-inflammatory phenotype observed in Usp18Δ/Δ macrophages caused by CSF1R downregulation relies on the NF-κB pathway. The NF-κB inhibitor GYY4137 did not affect CSF1R downregulation in Usp18Δ/Δ BMDMs (Figure 5E), suggesting that NF-κB pathways were not involved in the IFN-mediated downregulation of CSF1R. The downregulation of CSF1R expression also led to the suppression of PI3K and MAPK signaling, the major downstream pathways of CSF1R, in Usp18Δ/Δ macrophages (Figure 5F). To investigate if IFN-I-induced downregulation of CSF1R can also be observed in a human system, USP18-KO THP-1-derived macrophages were analyzed. CSF1R expression was downregulated after IFN-I treatment in USP18-KO THP-1 (Figure 5G). As USP18 also functions as a deconjugating enzyme, it may mediate the deubiquitination of CSF1R, which inhibits its protein degradation. To examine if the deconjugating activity of USP18 is involved in the regulation of CSF1R expression, either WT USP18 or an enzymatically inactive USP18 C64S mutant was expressed in the USP18-KO THP-1 cells. The CSF1R protein level was restored with the reexpression of either WT USP18 or USP18 C64S in the USP18-KO THP-1 cells, supporting the idea that the enzymatic activity of USP18 is not responsible for the downregulation of CSF1R (Figure 5G). We also confirmed that the IFN-I-mediated downregulation of CSF1R was observed in other human myeloid cell lines, OCI-AML2 and MOLM13 (Figure 5H). Together, these data suggest that the deletion of USP18 mediates the downregulation of CSF1R at the post-transcriptional level through enhanced IFN-I response.
NEDD4 interacts with USP18 and mediates ubiquitin-dependent proteasomal degradation of CSF1R
To clarify further mechanisms of downregulation of CSF1R, a protein stability cycloheximide (CHX) chase assay was performed. It revealed that degradation of CSF1R was faster in BMDMs from Usp18Δ/Δ mice than in control cells, indicating that USP18 deletion enhanced the process of CSF1R degradation (Figure 6A). Protein ubiquitination plays a major role in degradation of cellular proteins. One ubiquitin E3 ligase, neural precursor cell-expressed, developmentally downregulated 4 (NEDD4), was previously detected as a USP18-interacting protein in our yeast two-hybrid study. NEDD4 is widely expressed in mammalian tissues, and more than 50 proteins are reported as targets of NEDD4, including IGF-1R, PTEN, and EGFR.53 We hypothesized that NEDD4 also mediates CSF1R degradation and that USP18 modulates the process. We first used co-immunoprecipitation to analyze the interaction of USP18 and NEDD4. The interaction between exogenously expressed USP18 and NEDD4 was observed (Figure 6B). In addition, we detected the interaction between endogenous USP18 and NEDD4 in THP-1-derived macrophages (Figure 6C). Furthermore, our reciprocal co-immunoprecipitation results demonstrate the interaction of NEDD4 and CSF1R (Figure 6D), suggesting that CSF1R is a ubiquitination substrate of NEDD4.
Figure 6. NEDD4-mediated ubiquitin-dependent proteasomal degradation of CSF1R.
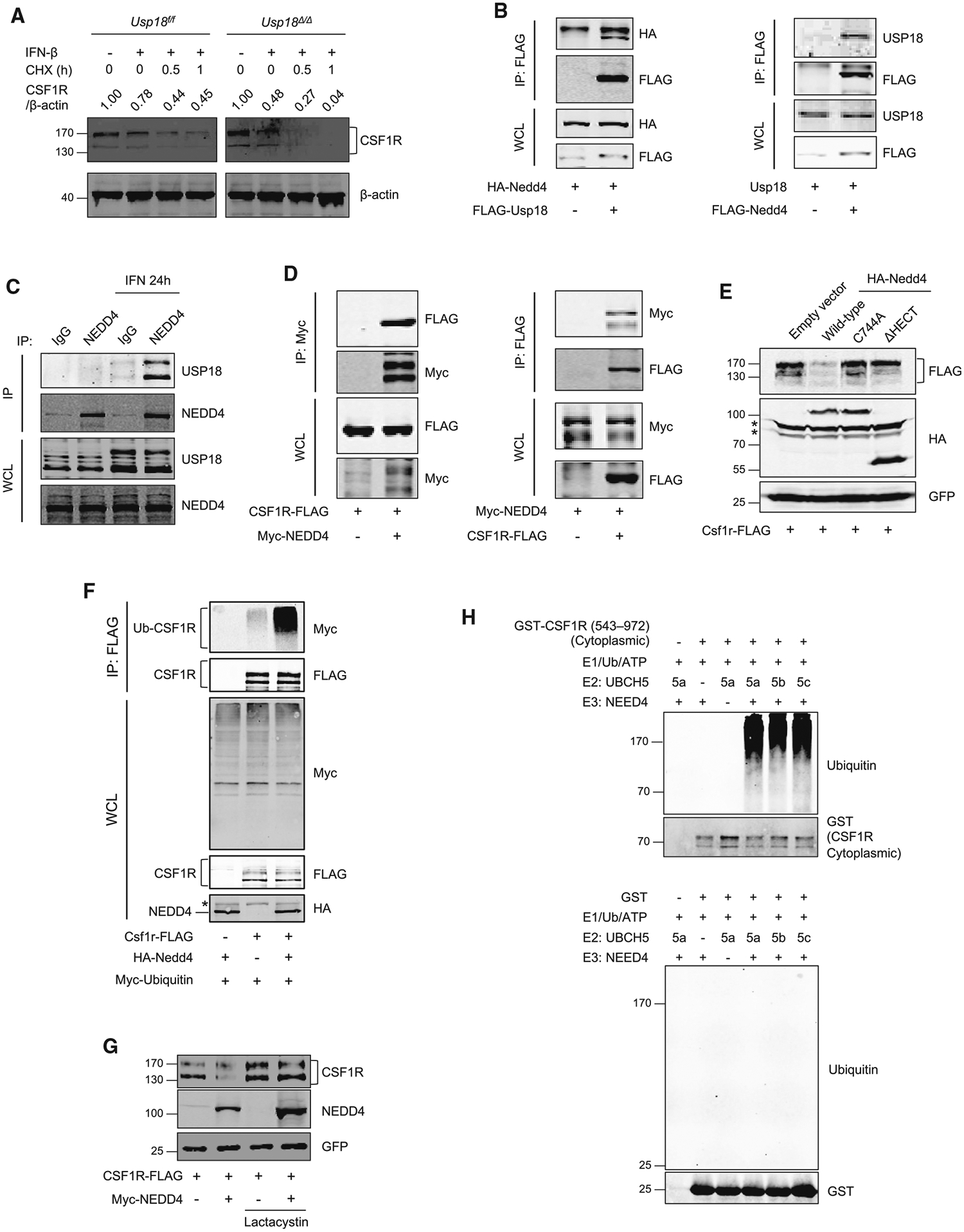
(A) Bone-marrow-derived macrophages (BMDMs) from Usp18f/f and Usp18Δ/Δ mice and wild type were treated with CHX (50 μg/mL) for the indicated times, and CSF1R expression was analyzed by western blotting.
(B) HEK293T cells were transfected for 24 h as indicated. Protein interaction was analyzed by co-immunoprecipitation.
(C) Interaction of endogenous USP18 and NEDD4 in THP-1-derived macrophages with or without IFN-I (1,000 U/mL for 24 h) was analyzed by co-immunoprecipitation by using anti-NEDD4 antibody.
(D) HEK293T cells were transfected as indicated. Twenty-four hours after transfection, protein interaction was analyzed by co-immunoprecipitation.
(E) HEK293T cells were transfected with FLAG-CSF1R and either wile-type or mutated HA-NEDD4 for 36 h. GFP was transfected as a control. Asterisks (*) indicate non-specific signals.
(F) HEK293T cells were transfected as indicated with lactacystin for 24 h. Ubiquitination of mouse CSF1R was analyzed after immunoprecipitation. Asterisk (*) indicates a non-specific signal.
(G) HEK293T cells were transfected as indicated with or without lactacystin for 24 h. Human CSF1R expression was analyzed by western blotting. GFP was transfected as a control.
(H) In vitro ubiquitination assay of CSF1R substrate protein with E2 enzymes UBCH5a/5b/5c and E3 enzyme NEDD4. Reaction mixtures as indicated were incubated at 30°C for 1 h. The eluate was analyzed by western blotting.
To examine whether NEDD4 E3 ligase mediates degradation of CSF1R, WT and mutant NEDD4 without enzyme activity were expressed with CSF1R. Since NEDD4 is an E3 in the HECT-domain ubiquitin ligase family, we generated the HECT-domain-deletion (ΔHECT) mutant and C744A-enzymatic-active-site mutant. CSF1R was downregulated by only WT NEDD4 and not the two mutants of NEDD4 (Figure 6E). A proteasome inhibitor, lactacystin, was used to further examine the involvement of the ubiquitin-proteasome pathway in the NEDD4-mediated degradation of CSF1R. Enhanced ubiquitination of CSF1R (Figure 6F) and restoration of CSF1R protein level (Figure 6G) were observed with lactacystin treatment. Furthermore, in vitro ubiquitination assays showed that CSF1R but not GST protein was able to be ubiquitinated by E3 ligase NEDD4 (Figure 6H). These data demonstrate that NEDD4 mediates the degradation of CSF1R via the ubiquitin-proteasome pathway.
USP18 impairs degradation of CSF1R by inhibiting CSF1R-NEDD4 interaction and regulating ubiquitin E2 UBCH5 expression
To reveal how USP18 modulates the NEDD4-mediated degradation of CSF1R, the interaction of CSF1R and NEDD4 was analyzed in the presence of USP18. USP18 inhibited the interaction of CSF1R and NEDD4 in a dose-dependent manner (Figure 7A). We then analyzed USP18 in NEDD4-mediated ubiquitination in the presence of the proteasome inhibitor lactacystin. NEDD4-mediated ubiquitination of CSF1R (Figure 7B, lanes 3 and 4) was impaired in the presence of USP18 (Figure 7B, lanes 5 and 6). These data indicate that USP18 inhibits the interaction of CSF1R and NEDD4, which diminishes the NEDD4-mediated ubiquitination of CSF1R and results in the inhibition of CSF1R degradation. To confirm these findings, NEDD4 was knocked down in BMDMs and THP-1-derived macrophages. Knockdown of NEDD4 showed restoration of CSF1R expression in USP18-KO (Usp18Δ/Δ) BMDMs (Figure 7C). Also, IFN-induced downregulation of CSF1R in USP18-KO cells was restored by the knockdown of NEDD4 (Figure 7D). Together, these results support the idea that NEDD4 is a modulator of CSF1R downregulation. Interestingly, although NEDD4-mediated CSF1R degradation was enhanced by IFN-I treatment, NEDD4 itself is not induced by IFN-I, suggesting that other regulators in the ubiquitin-proteasome pathway may be responsible for IFN-I-mediated CSF1R degradation. We analyzed an IFN-I-inducible E2 ubiquitin-conjugating enzyme, UBCH5.54 Our RNA-seq analysis of THP-1 cells showed that the expression of UBE2D3 (UBCH5) was significantly upregulated in parental and USP18-KO cells upon IFN-I treatment (Figure 7E). Furthermore, UBCH5 protein was induced and CSF1R was downregulated in USP18-KO THP-1-derived macrophages after IFN-I treatment (Figure 7F). Importantly, the downregulation of CSF1R was restored by knocking down UBCH5 expression. Together, these data show that UBCH5 expression is enhanced upon IFN-I stimulation and that UBCH5 mediates IFN-I-dependent degradation of CSF1R.
Figure 7. Inhibitory effect of USP18 on NEDD4-mediated ubiquitination of CSF1R.
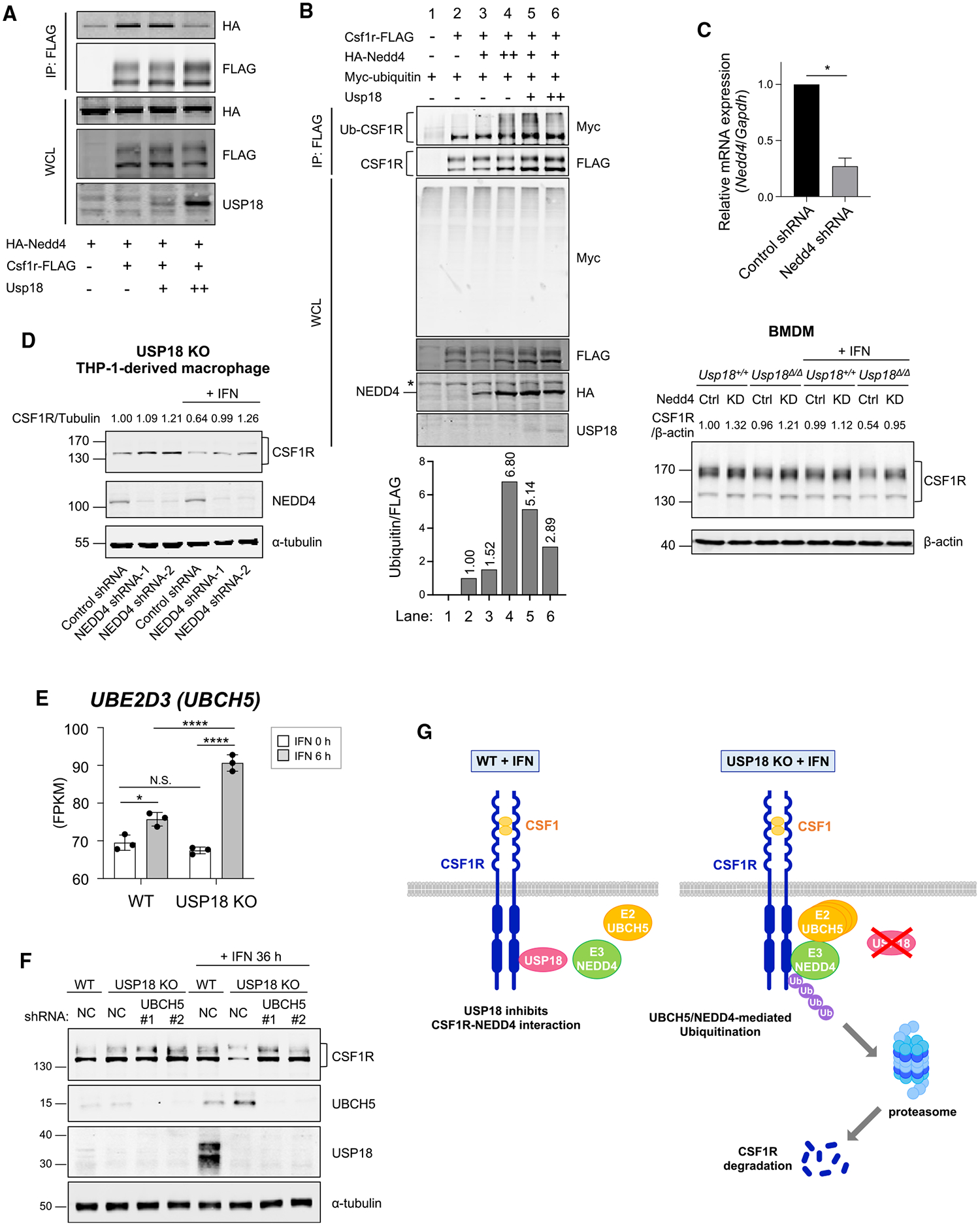
(A) Interaction of CSF1R and NEDD4 in the presence of USP18 was analyzed. HEK293T cells were transfected as indicated for 24 h. Co-immunoprecipitation followed by western blotting was performed.
(B) HEK293T cells were transfected as indicated for 24 h. All samples were treated with lactacystin (10 μM) for 2 h. Co-immunoprecipitation followed by western blotting was performed. Asterisk (*) indicates a non-specific signal.
(C) Nedd4-knockdown or control bone-marrow-derived macrophages (BMDMs) from Usp18f/f UBC-Cre-ERT2 mice were treated with β-estradiol (2 μM) for 48 h. Knockdown of Nedd4 was confirmed by qPCR (top). CSF1R expression in the BMDMs with or without IFN-β treatment (100 U/mL for 24 h) was analyzed by western blotting (bottom). Mean ± SEM, two-tailed unpaired t test, *p < 0.05.
(D) NEDD4 was knocked down by shRNA in USP18-knockout THP-1-derived macrophages. The expression of CSF1R was analyzed by western blotting.
(E) UBE2D3 expression in WT or USP18-KO THP-1 cells with or without IFN-I treatment analyzed by bulk RNA-seq (n = 3). Mean ± SD. One-way ANOVA multiple comparison test with post hoc Tukey test; N.S., not significant; *p < 0.05, ****p < 0.0001.
(F) UBCH5C and CSF1R expression in THP-1-derived macrophages with knockdown of UBCH5C was analyzed by western blotting.
(G) Scheme of the inhibitory effect of USP18 on NEDD4-dependent downregulation of CSF1R.
Figure 7G is the scheme of the mechanism we propose based on the current studies. IFN-Is are produced in the TME, which induces the expression of USP18 and the ubiquitin E2 enzyme UBCH5. In WT macrophages, USP18 inhibits the interaction of CSF1R and the ubiquitin E3 NEDD4, which suppresses ubiquitination and subsequent degradation of CSF1R. On the other hand, in USP18-KO macrophages, NEDD4 binds to CSF1R, which enhances UBCH5 and NEDD4-mediated ubiquitination and subsequent degradation of CSF1R, leading to inhibition of polarization toward immunosuppressive macrophages.
DISCUSSION
In this study, we sought to reveal the functions of IFN-I in myeloid cells during tumor development. We utilized a myeloid-lineage-specific USP18-deletion mouse model, which showed that deletion of USP18 delayed tumor growth. Our single-cell transcriptional characterization of tumor-infiltrated immune cells uncovered differences after USP18 depletion, including an increase in anti-tumor macrophages. CSF1R expression, a key signal for the polarization toward an immunosuppressive/pro-tumorigenic phenotype, in macrophages was downregulated by USP18 KO in vivo and in vitro. We found that NEDD4 is a ubiquitin E3 ligase for CSF1R, and USP18 inhibits its degradation by interfering with the interaction of CSF1R and NEDD4. Furthermore, depletion of the negative regulator of IFN-I signaling, USP18, enhanced expression of ubiquitin E2 UBCH5. Together, our data demonstrate that suppression of USP18 promotes UBCH5 and NEDD4-mediated proteasome degradation of CSF1R, leading to an increase in anti-tumor macrophages in the TME. This is, at least in part, a mechanism of enhanced anti-tumor activity observed in USP18-KO mice.
A previous report demonstrated that deletion of adenosine deaminase acting on RNA 1 (ADAR1), a suppressor of IFN-I response, decreased immunosuppressive macrophages and myeloid-derived suppressor cells and enhanced anti-tumor immunity, which indicates the importance of IFN-I in TME.26 Also, it was reported that IFN-Is inhibited the generation of TAMs by using an IFN-α/β receptor-KO mouse.55 These authors also showed that IFN-Is significantly inhibited the generation of bone-marrow-cell-derived macrophages in response to CSF1 in vitro. This may be explained by our finding that IFN-I mediated downregulation of CSF1R (Figure 5). We previously reported that deletion of USP18 activates expression of NF-κB-regulated genes.19 In this study, we observed that IFN-I-mediated CSF1R downregulation in USP18-KO macrophages was not regulated by NF-κB (Figure 5E). However, NF-κB is known to be one of the key transcription factors related to polarization toward anti-tumor/pro-inflammatory macrophages.56 NF-κB regulates the expression of a large number of inflammatory genes, including TNF-α, IL-1β, cyclooxygenase-2 (COX-2), IL-6, and IL-12p40.57 Therefore, it is likely that anti-tumor/pro-inflammatory macrophage polarization observed in USP18-KO macrophages also relies on the NF-κB pathway in addition to the CSF1R-mediated regulation.
TAMs are good targets for cancer therapy. In addition to their immunosuppressive functions in the TME, TAMs are also known to mediate resistance to standard therapies, including chemo-therapy and radiation therapy.48,58–60 As CSF1 is required for macrophage differentiation, different types of CSF1R blockades have been approved for clinical use, including monoclonal antibodies against CSF1R and tyrosine kinase inhibitors. In our current study, deletion of USP18 in macrophages downregulated CSF1R expression on TAMs and reduced the frequency of immunosuppressive TAMs in the TME. More importantly, we demonstrated that deletion of USP18 created an anti-tumor microenvironment by repolarization of TAMs toward anti-tumor macrophages (Figures 3 and 4). The above therapeutic applications support the clinical impact of our current studies. Accumulating evidence suggests the importance of macrophage polarization in tumor progression. For example, Pyonteck et al. reported that CSF1R blockade reduces immunosuppressive macrophage polarization, which improved disease outcomes in their glioma models.45 Other reports also demonstrated the impact of TAM polarization on tumor progression,61 supporting the idea that reprogramming of TAMs toward the anti-tumor phenotype induced by USP18 deletion has potential for clinical use.
Tong et al. reported that IFN-I downregulates CSF1R expression via miR-155.62 IFN-I inhibits differentiation of Ly6C+ monocytes to TAMs by inhibiting upregulation of CSF1R in monocytes during differentiation to macrophages. Consistent with our finding shown in Figure 5C, mRNA expression of Csf1r was not reduced after IFN treatment in the report. They found that the inhibitory effect of IFN on CSF1R occurs at the level of mRNA translation mediated by IFN-induced miR-155. In our current study, we revealed another mechanism of CSF1R regulation mediated by USP18 and NEDD4.
Reduction of CSF1R on cell membranes has been reported by shedding with TNF-α-converting enzyme TACE63 and γ-secretase64 and by CSF1 or Toll-like receptor (TLR) agonist-stimulated internalization and lysozyme degradation.65,66 However, such changes happened within 30 min after stimulation. In contrast, IFN-I-dependent downregulation of CSF1R takes much longer, supporting the idea that IFN-I induces a mechanism of degradation different from the one induced by CSF1 or TLR agonists. Also, proto-oncoprotein c-Cbl is reported as an E3 ligase for CSF1R.67–69 This c-Cbl-induced ubiquitination of CSF1R leads to internalization and endocytosis of the receptor, followed by receptor degradation in lysosomes.70 Therefore, this degradation process is completely separate from our findings. Here, we identified NEDD4 as a ubiquitin E3 ligase for CSF1R and demonstrated that UBCH5 served as its E2 enzyme. The UBCH5 gene is a known IFN-I-inducible gene, whereas the NEDD4 gene is not.54 Therefore, under the circumstances of enhanced IFN-I signaling caused by USP18 deletion, IFN-I-inducible E2 enzyme UBCH5 likely enhances the activity of E3 ligase NEDD4, leading to degradation of CSF1R via the ubiquitin-proteasome system.
Since NEDD4 is frequently overexpressed in cancers, including prostate, bladder, and colon cancers,71 NEDD4 was originally thought to be an oncogene. However, recently, NEDD4 is thought to have dual roles as an oncogene and a tumor suppressor in cancers by mediating the ubiquitination of substrates that have a variety of functions.72 Furthermore, accumulating evidence suggests that NEDD4 also has important roles in the immune system. NEDD4 enhances T cell activation and proliferation by promoting ubiquitin-mediated degradation of Cbl-b, which negatively regulates T cell activation.73–75 With regard to B cells, NEDD4 promotes the activation of the CD40-Akt pathway by ubiquitination of TRAF3, inducing immunoglobulin class switching, which is essential for humoral immunity.76 It has also been reported that NEDD4 plays an important role in macrophages during the innate immune response and inflammation. NEDD4 regulates TNF-α expression from macrophages by mediating ubiquitination of p38α77 and enhances killing of intracellular bacterial pathogens by promoting autophagy.78 Furthermore, Nuro-Gyina et al. showed that NEDD4 is essential for anti-fungal innate immune response by using Nedd4f/f LysM-Cre mice.79 However, our current study shows the function of NEDD4 in TAMs.
Given that USP18 inhibits NEDD4-mediated ubiquitination and subsequent degradation of CSF1R, we hypothesized that USP18 may also regulate NEDD4-mediated ubiquitination of other target proteins. It has been reported that RAP2A is one of the targets of the NEDD4-mediated ubiquitination.80 To investigate if the interaction of USP18 and NEDD4 affects the ubiquitin conjugation to RAP2A, NEDD4, RAP2A, ubiquitin, and USP18 were co-expressed in HEK293T cells. We confirmed that NEDD4 enhanced ubiquitination of RAP2A; however, it was not affected in the presence of USP18, suggesting that not all of the NEDD4-mediated ubiquitination was inhibited by USP18. This could be due to the difference in other ubiquitination components, such as E2 enzymes. RAP2A is just one example among more than 20 of the reported substrates of NEDD4, and there may be other targets of NEDD4-mediated ubiquitination that are regulated by USP18. If so, USP18 could be involved in other diseases and be a potential therapeutic target to enhance the function of NEDD4. Further investigation is warranted.
Our findings suggest that targeting USP18 in macrophages has the potential to reprogram TAM to enhance anti-tumor activity in different types of cancers. This would be a good therapeutic strategy for the following reasons. In most tumors, myeloid cells are the most abundant cell types among tumor-infiltrating immune cells. Although T cell-based anti-tumor therapies, such as checkpoint blockade immunotherapy, are currently used, one of the limitations of T cell-targeting therapy is the low number of tumor-infiltrating T cells. As myeloid cells usually account for 30%–50% of infiltrating immune cells in tumors, it would be feasible and more efficient to target them to regulate T cells as presented in our current studies. In addition, circulating monocytes are one of the main sources of macrophages in tumors,81 which can be targeted by USP18 depletion as well. Importantly, deletion of USP18 in myeloid-lineage cells did not show any negative effects on normal hematopoiesis (Figure S1). Given these advantages, targeting USP18 in myeloid cells is potentially a promising therapeutic strategy across different types of cancers and warrants further investigation.
Limitations of the study
In the current study, the tumor-infiltrating immune cells were analyzed only by single-cell RNA-seq. There was no information on protein expression, which requires investigation by cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) or other, similar methods to support our conclusion. Also, cellular neighborhoods in the TME remain to be investigated by digital-spatial analysis. Moreover, additional functional analysis of the different macrophage populations will further improve the annotation of the macrophage clusters and help us to understand the more precise mechanism of USP18-mediated reprogramming of macrophages.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dong-Er Zhang (d7zhang@health.ucsd.edu).
Materials availability
The following plasmids were generated in this study: Plasmid: pCX4-bsr-mouse Usp18, pCX4-bsr -FLAG-mouse Usp18, pCX4-bsr-human USP18 (sgRNA resistant), pCX4-bsr-human USP18 C64S (sgRNA resistant), pCAG-mouse Csf1r-FLAG, pCAG-human CSF1R-FLAG, pcDNA-Myc-human NEDD4, pcDNA-HA-mouse Nedd4, pcDNA-HA-mouse Nedd4 (C744A mutant), pcDNA-HA-mouse Nedd4 (HECT domain deletion mutant), pcDNA-FLAG-mouse Nedd4, pcDNA-Myc-Ubiquitin, pSUPER.retro.puro-human USP18 shRNA, pLKO.1 Negative control (GFP).
All materials generated in this study are available from the lead contact with a completed materials transfer agreement.
Data and code availability
Single-cell and bulk RNAseq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| InVivoMAb anti-mouse IFNAR-1 (Clone MAR1-5A3) | Bio X Cell | Cat #: BE0241; RRID: AB_2687723 |
| InVivoMAb mouse IgG1 isotype control (Clone MOPC-21) | Bio X Cell | Cat #: BE0083; RRID: AB_1107784 |
| Rat monoclonal anti-mouse CD16/CD32 (Clone: 93) | Thermo Fisher Scientific (eBioscience) | Cat #: 14-0161-82; RRID: AB_467133 |
| Mouse monoclonal anti-mouse CD45.2 (clone 104) Brilliant Violet 605 | BioLegend | Cat #: 109841; RRID: AB_2563485 |
| Mouse monoclonal anti-mouse CD45.2 (clone 104) PerCP-Cyanine5.5 | BioLegend | Cat #: 109828; RRID: AB_893350 |
| Mouse monoclonal anti-mouse CD45.2 (clone 104) Alexa Fluor 700 | BioLegend | Cat #: 109822; RRID: AB_493731 |
| Rat monoclonal anti-mouse/human CD11b (clone M1/70) Alexa Fluor 700 | BD Biosciences | Cat #: 557960; RRID: AB_396960 |
| Rat monoclonal anti-mouse/human CD11b (clone M1/70) FITC | BD Biosciences | Cat #: 553310; RRID: AB_394774 |
| Rat monoclonal anti-mouse/human CD11b (clone M1/70) PE-Cyanine7 | BioLegend | Cat #: 101216; RRID: AB_312799 |
| Armenian Hamster monoclonal anti-mouse CD11c (clone N418) Pacific Blue | BioLegend | Cat #: 117322; RRID: AB_755988 |
| Rat monoclonal anti-mouse F4/80 (clone BM8) Brilliant Violet 605 | BioLegend | Cat #: 123133; RRID: AB_2562305 |
| Rat monoclonal anti-mouse F4/80 (clone BM8) APC | BioLegend | Cat #: 123116; RRID: AB_893481 |
| Rat monoclonal anti-mouse Gr-1 (Ly6G/Ly6C) (clone RB6-8C5) PE-Cyanine7 | BD Biosciences | Cat #: 565033; RRID: AB_2739049 |
| Rat monoclonal anti-mouse CD115 (CSF-1R) (clone AFS98) APC | BioLegend | Cat #: 135510; RRID: AB_2085221 |
| Rat monoclonal anti-mouse CD115 (CSF-1R) (clone AFS98) PE | BioLegend | Cat #: 135506; RRID: AB_1937253 |
| Rat monoclonal anti-mouse CD3 (clone 17A2) Brilliant Violet 510 | BioLegend | Cat #: 100234; RRID: AB_2562555 |
| Armenian Hamster monoclonal anti-mouse CD3e (clone 145-2C11) Brilliant Violet 650 | BD Biosciences | Cat #: 564378; RRID: AB_2738779 |
| Rat monoclonal anti-mouse CD4 (clone RM4-5) Pacific Blue | BD Biosciences | Cat #: 558107; RRID: AB_397030 |
| Rat monoclonal anti-mouse CD4 (clone RM4-5) Brilliant Violet 570 | BioLegend | Cat #: 100542; RRID: AB_2563051 |
| Rat monoclonal anti-mouse CD8a (clone 5H10) Pacific Orange | Thermo Fisher Scientific (Invitrogen) | Cat #: MCD0830; RRID: AB_10376311 |
| Rat monoclonal anti-mouse CD8a (clone 53-6.7) PE-Cyanine5 | BioLegend | Cat #: 100710; RRID: AB_312749 |
| Rat monoclonal anti-mouse Ly6G (clone 1A8) APC-Cyanine7 | BioLegend | Cat #: 127623; RRID: AB_10645331 |
| Rat monoclonal anti-mouse Ly6C (clone HK1.4) PE-Dazzle594 | BioLegend | Cat #: 128043; RRID: AB_2566576 |
| Mouse monoclonal anti-mouse NK-1.1 (clone PK136) PE | BioLegend | Cat #: 108708; RRID: AB_313395 |
| Rat monoclonal anti-mouse/human CD45R/B220 (clone RA3-6B2) Alexa Fluor 700 | BioLegend | Cat #: 103232; RRID: AB_493717 |
| Rat monoclonal anti-mouse CD25 (clone PC61) PE-Cyanine7 | BioLegend | Cat #: 102016; RRID: AB_312865 |
| Rat monoclonal anti-mouse Foxp3 (clone MF23) Alexa Fluor 647 | BD Biosciences | Cat #: 560401; RRID: AB_1645201 |
| Rat monoclonal anti-mouse Foxp3 (clone FJK-16s) APC | Thermo Fisher Scientific (eBioscience) | Cat #: 17-5773-82; RRID: AB_469457 |
| Rat monoclonal anti-mouse/human CD44 (clone IM7) FITC | BioLegend | Cat #: 103006; RRID: AB_312957 |
| Rat monoclonal anti-mouse CD62L (clone MEL-14) PerCP-Cyanine5.5 | BioLegend | Cat #: 104432; RRID: AB_2285839 |
| Armenian Hamster monoclonal anti-mouse CD69 (clone H1.2F3) PE-Dazzle594 | BioLegend | Cat #: 104536; RRID: AB_2565583 |
| Rat monoclonal anti-mouse IFN-γ (clone XMG1.2) PE | BioLegend | Cat #: 505807; RRID: AB_315401 |
| Rat monoclonal anti-mouse Perforin (clone S16009A) PE-Dazzle594 | BioLegend | Cat #: 154315; RRID: AB_2922482 |
| Rat monoclonal anti-mouse CD206 (MMR) (clone C068C2) PE | BioLegend | Cat #: 141705; RRID: AB_10896421 |
| Rabbit polyclonal anti-CSF-1R/M-CSF-R | Cell Signaling Technology | Cat #: 3152; RRID: AB_2085233 |
| Rabbit monoclonal anti-CSF-1R/M-CSF-R (clone D3O9X) | Cell Signaling Technology | Cat #: 67455; RRID: AB_2799725 |
| Rabbit polyclonal anti-NEDD4 | Cell Signaling Technology | Cat #: 2740; RRID: AB_2149312 |
| Rabbit monoclonal anti-UbcH5C (clone D60E2) | Cell Signaling Technology | Cat #: 4330; RRID: AB_10544697 |
| Rabbit polyclonal anti-Akt | Cell Signaling Technology | Cat #: 9272; RRID: AB_329827 |
| Rabbit monoclonal anti-Phospho-Akt (Ser473) (clone D9E) | Cell Signaling Technology | Cat #: 4060; RRID: AB_2315049 |
| Rabbit polyclonal anti-p38 MAPK | Cell Signaling Technology | Cat #: 9212; RRID: AB_330713 |
| Rabbit monoclonal anti-Phospho-p38 (Thr180/Tyr182) (clone 12F8) | Cell Signaling Technology | Cat #: 4631; RRID: AB_331765 |
| Rabbit monoclonal anti-SAPK/JNK (clone 56G8) | Cell Signaling Technology | Cat #: 9258; RRID: AB_2141027 |
| Rabbit polyclonal anti-Phospho-SAPK/JNK (Thr183/Tyr185) | Cell Signaling Technology | Cat #: 9251; RRID: AB_331659 |
| Mouse monoclonal anti-β-actin (clone AC-15) | Sigma-Aldrich | Cat #: A1978; RRID: AB_476692 |
| Mouse monoclonal anti-α-tubulin (clone DM1A) | Sigma-Aldrich | Cat #: T6199; RRID: AB_477583 |
| Mouse monoclonal anti-FLAG M2 | Sigma-Aldrich | Cat #: F3165; RRID: AB_259529 |
| Mouse monoclonal anti-Myc (clone 9E10) | Santa Cruz Biotechnology | Cat #: sc-40; RRID: AB_627268 |
| Rat monoclonal anti-HA (clone 3F10) | Roche | Cat #: 11867423001; RRID: AB_390918 |
| Rabbit polyclonal anti-GFP | Thermo Fisher Scientific (Invitrogen) | Cat#: A-11122; RRID: AB_221569 |
| Anti-ISG15 | Malakhov et al.14 | N/A |
| Anti-USP18 | Malkahova et al.15 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| BD DIFCO™ Thioglycollate Medium, Brewer Modified | Becton Dickinson | Cat#: 211716 |
| Recombinant Murine M-CSF | Peprotech | Cat#: 315-02 |
| Recombinant Murine IL-4 | Peprotech | Cat#: 214-14 |
| Recombinant Murine IL-13 | Peprotech | Cat#: 210-13 |
| PMA (Phorbol 12-Myristate 13-Acetate) | Sigma-Aldrich | Cat#: P1585 |
| Mouse IFN-Beta, Mammalian | PBL Assay Science | Cat#: 12405 |
| Universal Type I IFN Protein | R&D Systems | Cat#: 11200-2 |
| Cycloheximide | Sigma-Aldrich | Cat#: 01810 |
| Lactacystin | Sigma-Aldrich | Cat #: L6785 |
| β-Estradiol | Sigma-Aldrich | Cat #: E2758 |
| Polyinosinic-polycytidylic acid sodium salt | Sigma-Aldrich | Cat#: P0913 |
| Pexidartinib (PLX3397) | Selleckchem | Cat#: S7818 |
| GYY4137, H2S donor | abcam | Cat#: ab142145 |
| Percoll™ | cytiva | Cat #: 17089101 |
| Propidium Iodide | Roche | Cat#: 11348639001 |
| eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set | Thermo Fisher Scientific (Invitrogen) | Cat #: 00–5523-00 |
| Allele-In-One Mouse Tail Direct Lysis Buffer | Allele Biotechnology | Cat #: ABP-PP-MT01500 |
| TRIzol™ Reagent | Thermo Fisher Scientific (Invitrogen) | Cat#: 15596018 |
| First Strand cDNA Synthesis Kit | MCLAB | Cat #: FSCS-200 |
| KAPA SYBR FAST qPCR Kit Master Mix (2X) Universal | KAPA Biosystems | Cat #: KK4602 |
| cOmplete™, EDTA-free Protease Inhibitor Cocktail | Sigma-Aldrich (Roche) | Cat#: 11873580001 |
| PhosSTOP™ | Sigma-Aldrich (Roche) | Cat #: 4906837001 |
| Protein G Plus/Protein A Agarose Suspension | EMD Millipore | Cat #: IP05 |
| Anti-FLAG M2 Affinity Gel | Sigma-Aldrich | Cat #: A2220 |
| PEI (Polyethylenimine, branched) | Sigma-Aldrich | Cat #: 408727 |
| Hexadimethrine bromide | Sigma-Aldrich | Cat #: H9268 |
| Glutathione Sepharose 4B | cytiva | Cat #: 17075601 |
| PreScission Protease | cytiva | Cat #: 27084301 |
| Critical commercial assays | ||
| CD11b MicroBeads, human and mouse | Miltenyi Biotec | Cat #: 130-049-601 |
| Dynabeads™ Untouched™ Mouse T Cells Kit | Invitrogen | Cat#: 11413D |
| Chromium Single Cell 3’Reagent Kit v3 | 10x Genomics | Cat#: PN-1000075 |
| Zombie Aqua™ Fixable Viability Kit | BioLegend | Cat#: 423101 |
| Zombie NIR™ Fixable Viability Kit | BioLegend | Cat#: 423106 |
| Ubiquitinylation kit | Enzo Life Sciences | Cat #: BML-UW9920-0001 |
| Deposited data | ||
| Single-cell RNAseq data | This paper | GEO: GSE173705 |
| Bulk RNAseq data | Arimoto et al.19 | GEO: GSE165428 |
| Experimental models: Cell lines | ||
| Mouse: B16F10 | Dr. Michiko Fukuda (Sanford Burnham Prebys Institute) | RRID: CVCL_0159 |
| Mouse: EL4 | American Type Culture Collection (ATCC) | Cat #: TIB-39; RRID: CVCL_0255 |
| Mouse: Lewis Lung Carcinoma (LLC) | National Cancer Institute | RRID: CVCL_4358 |
| Human: THP-1 | ATCC | Cat #: TIB-202; RRID: CVCL_0006 |
| Human: THP-1 USP18 knockout | Arimoto et al.19 | N/A |
| Human: MOLM13 | Dr. Lee Grimes (Cincinnati Children’s Hospital) | RRID:CVCL_2119 |
| Human: OCI-AML2 | Dr. Suming Huang (University of Florida) | RRID:CVCL_1619 |
| Human: HEK293T | ATCC | Cat #: CRL-3216; RRID: CVCL_0063 |
| Experimental models: Organisms/strains | ||
| Mouse: B6.129P2-Lyz2tm1(cre)Ifo/J | The Jackson Laboratory | Strain #: 004781; RRID: IMSR_JAX:004781 |
| Mouse: B6.Cg-Ndor1Tg(UBC-cre/ERT2)1Ejb/1J | The Jackson Laboratory | Strain #: 008085; RRID: IMSR_JAX:008085 |
| Mouse: Usp18f/f | Arimoto et al.19 | N/A |
| Mouse: Isg15 knockout | Osiak et al.82 | N/A |
| Oligonucleotides | ||
| Primer: mouse Gapdh Forward: TATGTCGTGGAGTCTACTGG |
This paper | N/A |
| Primer: mouse Gapdh Reverse: GAGTTGTCATATTTCTCGTG |
This paper | N/A |
| Primer: mouse Usp18 Forward: CAGCCCTCATGGTCTGGTTG |
This paper | N/A |
| Primer: mouse Usp18 Reverse: GCACTCCGAGGCACTGTTAT |
This paper | N/A |
| Primer: mouse Csf1r Forward: AATGCTAACGCCACCGAGA |
This paper | N/A |
| Primer: mouse Csf1r Reverse: CATGGAAAGTTCGGACACAGG |
This paper | N/A |
| Primer: mouse Nedd4 Forward: ACCAGCGTGCAGACAAAAAC |
This paper | N/A |
| Primer: mouse Nedd4 Reverse: AAAAGAATGCGGTGTCGCTG |
This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pCL-10A1 | Novus | Cat #: NBP2-29542 |
| Plasmid: pCL-Eco | Novus | Cat #: NBP2-29540 |
| Plasmid: psPAX2 | Dr. Didier Trono (Addgene) | RRID: Addgene_12260 |
| Plasmid: pMD2.G | Dr. Didier Trono (Addgene) | RRID: Addgene_12259 |
| Plasmid: pCX4-bsr | Dr. Tsuyoshi Akagi (KAN Research Institute) | N/A |
| Plasmid: pcDNA™3.1 (+) Mammalian Expression Vector | Thermo Fisher Scientific (Invitrogen) | Cat #: V79020 |
| Plasmid: pCX4-bsr-mouse Usp18 | This paper | N/A |
| Plasmid: pCX4-bsr -FLAG-mouse Usp18 | This paper | N/A |
| Plasmid: pCX4-bsr-human USP18 (sgRNA resistant) | This paper | N/A |
| Plasmid: pCX4-bsr-human USP18 C64S (sgRNA resistant) |
This paper | N/A |
| Plasmid: pCAG-mouse Csf1r-FLAG | This paper | N/A |
| Plasmid: pCAG-human CSF1R-FLAG | This paper | N/A |
| Plasmid: pcDNA-Myc-human NEDD4 | This paper | N/A |
| Plasmid: pcDNA -HA-mouse Nedd4 | This paper | N/A |
| Plasmid: pcDNA -HA-mouse Nedd4 (C744A mutant) | This paper | N/A |
| Plasmid: pcDNA -HA-mouse Nedd4 (HECT domain deletion mutant) | This paper | N/A |
| Plasmid: pcDNA-FLAG-mouse Nedd4 | This paper | N/A |
| Plasmid: pcDNA-Myc-Ubiquitin | This paper | N/A |
| Plasmid: pEGFP-N1 | Clontech | Cat #: 6085-1 |
| Plasmid: pSUPER.retro.puro | OligoEngine | Cat #: VEC-PRT-0002 |
| Plasmid: pSUPER.retro.puro-human USP18 shRNA | This paper | N/A |
| Plasmid: pLKO.1 Negative control (GFP) | This paper | N/A |
| Plasmid: pLKO.1-human UBCH5C shRNA | La Jolla Institute for Immunology | Cat #: TRCN0000038791 |
| Plasmid: pLKO.1-human UBCH5C shRNA | La Jolla Institute for Immunology | Cat #: TRCN0000038793 |
| Plasmid: pLKO.1-mouse Nedd4 shRNA | La Jolla Institute for Immunology | Cat #: TRCN0000092436 |
| Plasmid: pLKO.1-human NEDD4 shRNA | La Jolla Institute for Immunology | Cat #: TRCN0000007550 |
| Plasmid: pLKO.1-human NEDD4 shRNA | La Jolla Institute for Immunology | Cat #: TRCN0000007551 |
| Plasmid: pGEX-6P-1 | cytiva | Cat #: 28954648 |
| Plasmid: pGEX-6P-1-human CSF1R (543 – 972) | This paper | N/A |
| Plasmid: pGEX-6P-1-human NEDD4 | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad | N/A |
| FlowJo 10 | FlowJo, LLC | N/A |
| LI-COR Image Studio™ Software | LI-COR | N/A |
| 10x Genomics Cell Ranger (v3.0.1) | Zheng et al.83 | N/A |
| Seurat (v4.3.0) | Hao et al.84 | https://satijalab.org/seurat/ |
| STAR | Dobin et al.85 | N/A |
| DESeq2 | Love et al.86 | N/A |
| SingleR | Aran et al.28 | N/A |
| ImmGen (Immunological Genome Project) | Heng et al.29 | N/A |
| Gene Set Enrichment Analysis (GSEA) | Mootha et al.31
Subramanian et al.32 |
https://www.gsea-msigdb.org |
| Metascape | Zhou et al.33 | https://metascape.org |
| Monocle 3 | Cao et al.41
Qiu et al.42 Trapnell et al.43 |
N/A |
| Other | ||
| scil Vet abc Plus hematology analyzer | scil animal care company | N/A |
| MycoAlert PLUS Mycoplasma Detection Kit | Lonza | Cat#: LT07-710 |
| BD FACS Canto II Flow Cytometer | BD Biosciences | N/A |
| BD LSR II Flow Cytometer | BD Biosciences | N/A |
| BD FACS Aria II Cell Sorter | BD Biosciences | N/A |
| NovoCyte Advanteon Flow Cytometer | Agilent Technologies | N/A |
| CFX96 Thermal cycler | Bio-Rad Laboratories | N/A |
| Odyssey Imaging System | LI-COR Biotechnology | N/A |
| Countess II FL Automated Cell Counter | Thermo Fisher Scientific (Invitrogen) | N/A |
| Chromium controller | 10x Genomics | N/A |
EXPERIMENTAL MODELS AND STUDY PARTICIPANT DETAILS
Mouse studies
All animal studies were approved by the Institutional Animal Care and Use Committee of the University of California, San Diego (S07271). All mice were housed and bred in a specific pathogen-free vivarium at UCSD Moores Cancer Center accredited by the American Association for the Accreditation of Laboratory Animal Care. Usp18f/f mice were generated by Ingenious Targeting Laboratory as previously described.19 LysM-Cre and UBC-Cre-ERT2 mice were purchased from The Jackson Laboratory. Isg15 knockout mice were from Dr. Klaus-Peter Knobeloch.82 Six to ten-week-old female and male mice were used for experiments. Littermates of the same sex were randomly assigned to experimental groups. Hematological parameters in peripheral blood were analyzed by using Vet abc Plus (scil animal care company).
For the establishment of tumor models, 1 × 105 B16F10 melanoma, 5 × 106 EL4 lymphoma, or 5 × 105 LLC lung carcinoma were subcutaneously injected into the right flank. Tumor diameter was measured every 3–4 days with an electronic caliper and reported as a volume using the formula; tumor volume = (length × width × height)/2. For Poly(I:C) treatment, 5 μg/g body weight of Polyinosinic-polycytidylic acid sodium salt (Sigma-Aldrich) was intraperitoneally injected starting on Day 10 daily for 3 days. For IFNAR1 blocking experiment, α-IFNAR1 or isotype control antibodies (Bio X cell) were intravenously injected on Day 3 and 8 (200 μg/dose).
Cell lines and primary cell culture
B16F10 from Dr. Michiko Fukuda (Sanford Burnham Prebys Institute), EL4 from American Type Culture Collection (ATCC), and Lewis Lung Carcinoma (LLC) from National Cancer Institute were grown in DMEM supplemented with 10% FBS, 1% L-Glutamine, and 1% Penicillin/Streptomycin. THP-1 from ATCC, MOLM13 from Dr. Lee Grimes (Cincinnati Children’s Hospital), and OCI-AML2 from Dr. Suming Huang (University of Florida) were grown in RPMI 1640 supplemented with 10% FBS, 1% L-Glutamine, and 1% Penicillin/Streptomycin. HEK293T from ATCC was grown in DMEM supplemented with 10% BCS, 1% L-Glutamine, and 1% Penicillin/Streptomycin. All cell lines were cultured in an incubator with 5% CO2 at 37°C and tested negative for Mycoplasma contamination.
CD11b+ myeloid cells were isolated from bone marrow by using CD11b microbeads (Miltenyi Biotec). T cells were isolated from spleen by using Dynabeads™ Untouched™ Mouse T Cells Kit (Invitrogen). Murine peritoneal macrophages were harvested 4 days after peritoneal injection of 4% thioglycollate (BD). Bone marrow-derived macrophages (BMDMs) were generated by culturing total bone marrow cells from femurs and tibiae in RPMI 1640 and 10% FBS supplemented with 30 ng/ml recombinant murine CSF1 (Peprotech) for 7 days. 40 ng/ml IL-4 (Peprotech), 20 ng/ml IL-13 (Peprotech), and 20 ng/ml CSF1 were added for 24 hours for polarization towards immunosuppressive macrophages. PLX3397 (Selleckchem) and GYY4137 (abcam) were used for inhibition of CSF1R and NF-κB, respectively. For in vitro deletion of Usp18, total bone marrow cells from Usp18f/f UBC-Cre-ERT2 mice were treated with β-estradiol (2 μM) for 48 hours. THP-1-derived macrophages were generated by culturing in the media containing PMA (50 ng/ml) for 48 hours. hUSP18 knockout THP-1 line was generated with CRISPR genome editing technology as previously described.19 sgRNA-resistant wild-type or C64S mutant hUSP18 was expressed in the hUSP18 knockout THP-1 cells.
For interferon treatment, mouse IFN-β (PBL Assay Science) and Universal Type I IFN (R&D Systems) were used for murine and human cell lines, respectively. For the analysis of downstream signaling of CSF1R, cells were treated with IFN-β (100 U/ml) for 24 hours followed by CSF1 (50 ng/ml) for 5 or 10 minutes after 4-hour starvation. For a protein stability assay, cells were treated with cycloheximide (CHX) at 50 μg/ml. Lactacystin (10 μM) was used as a proteasome inhibitor.
METHOD DETAILS
Genomic DNA extraction and PCR
Genomic DNA was extracted from tail with Allele-In-One Mouse Tail Direct Lysis Buffer (Allele Biotechnology) or from macrophage with TRIzol (Invitrogen). PCR was performed with Taq or Pfu enzymes generated in our lab.
Reverse transcription and quantitative PCR
Total RNA was extracted using TRIzol (Invitrogen) and reverse transcribed with First Strand cDNA Synthesis Kit (MCLAB) according to the manufacturer’s instructions. Quantitative PCR analysis was performed by using KAPA SYBR FAST (KAPA Biosystems) on the CFX96 thermal cycler (BIO-RAD). Primer sequences are listed in the key resources table.
Plasmid construction
mUsp18, hUSP18 and its mutants were cloned into pCX4-bsr retroviral vector. mCsf1r and hCSF1R cDNA were cloned into pCAG vector. Ubiquitin, mNedd4 and its mutants, and hNEDD4 cDNAs were cloned into pcDNA3 vector. hCSF1R cytoplasmic domain (543 – 972) and hNEDD4 were cloned into pGEX-6P-1 (cytiva). shRNA for hUSP18 was cloned into pSUPER.retro.puro vector (OligoEngine). GFP was cloned into pLKO.1 vector as a negative control for shRNA knockdown experiment. All the constructs were confirmed by DNA sequencing. shRNA pLKO.1 vectors for mNedd4 (TRCN0000092436), hNEDD4 (TRCN0000007550, TRCN0000007551), and hUBCH5C (TRCN0000038791, TRCN0000038793) were purchased from La Jolla Institute for Immunology.
Transfection and infection
Transfection was performed by using polyethylenimine (PEI). For retrovirus and lentivirus infection, HEK293T cells were co-transfected with plasmids encoding target genes and packaging vectors; pCL-10A1 or pCL-Eco for retrovirus, psPAX2 and pMD2.G for lentivirus. Viral particles from culture medium were collected and infected to target cells by spin infection (2,000 × g, 3 h, 30 °C) with hexadimethrine bromide (8 μg/ml). Infected cells were selected with appropriate selection drugs.
Immunoprecipitation and western blotting
Cells were lysed in lysis buffer composed of 25 mmol/L Tris-HCl, pH 8.0, 150 mmol/L NaCl, 1 mmol/L EDTA, 0.5% IgepalCA-630, and protease/phosphatase inhibitors (Roche). The cell lysates were centrifuged (20,000 × g) at 4 °C for 5 minutes. For co-immunoprecipitation assay, cell lysates were immunoprecipitated for 1 to 2 hours with FLAG M2 Affinity Gel for FLAG-tagged proteins or with primary antibodies as indicated followed by protein G/A-Agarose Suspension (EMD Millipore) for other proteins. Immunocomplexes were then adsorbed to the protein G/A-Agarose Suspension and washed three times. All samples were denatured in 1x sample buffer (50 mmol/L Tris-HCl, pH 6.8, 2% SDS, 5% 2-mercaptoethanol, 10% glycerol, and 0.01% bromophenol blue) for 5 minutes at 100 °C.
Proteins were electroblotted onto nitrocellulose membranes (cytiva) and incubated with primary antibodies. Li-Cor Fluorophoreconjugated secondary antibodies (Li-Cor) were used for detection by Odyssey system (Li-Cor). The following primary antibodies were used; anti-CSF1R, anti-NEDD4, anti-UBCH5C, anti-Akt, anti-Phospho-Akt, anti-p38, anti-Phospho-p38, anti-SAPK/JNK, and anti-Phospho-SAPK/JNK from Cell Signaling Technology, anti-β-actin, anti-α-tubulin, and anti-FLAG M2 from Sigma-Aldrich, anti-Myc and anti-Ubiquitin from Santa Cruz Biotechnology, anti-HA from Roche, anti-GFP from Invitrogen. Anti-ISG15 and anti-USP18 antibodies were previously described.14,15 Quantification was performed with LI-COR Image Studio software.
In vitro ubiquitination assay
GST-CSF1R cytoplasmic domain (543 – 972) substrate protein and GST-NEDD4 E3 ligase were purified from E. coli BL21 by Glutathione Sepharose 4B (cytiva). GST was cleaved from NEDD4 by Prescission protease (cytiva). In vitro ubiquitination reactions were performed using Ubiquitinylation kit (Enzo Life Sciences) according to the manufacturer’s instructions. Briefly, Glutathione Sepharose 4B only, retained GST, or GST-CSF1R was incubated with UBE1, E2, and E3 enzymes in a buffer containing ATP and ubiquitin at 30°C for 1 hour. The GST-CSF1R were eluted from the resins and were subjected to western blot analysis.
Flow cytometry analysis
Total bone marrow cells from Usp18f/f UBC-Cre-ERT2 mice were harvested and cultured with or without β-estradiol (2 μM) for 48 hours. Cells were then stained with anti-CD11b antibody and propidium iodide for cell viability assay.
For immunophenotyping of tumor-bearing mice, single-cell suspension was prepared from tumor tissue and tumor-draining lymph nodes by passing through a 40 μm cell strainer. Single-cell suspension was then treated with ACK buffer for RBC lysis, and density gradient centrifugation on 40%/80% Percoll (cytiva) gradient was performed. After Fc blocking with anti-CD16/CD32 (eBioscience), cells were stained and analyzed on a BD FACS Canto II, BD LSR II, or Agilent NovoCyte Advanteon with standard lasers and optical filters. For single-cell RNA sequencing analysis, cells were sorted by BD FACS Aria II with standard lasers and optical filters. Fluorochrome-conjugated primary antibodies used in the study are listed in the key resources table. Propidium Iodide or Fixable Zombie (BioLegend) was used for viability staining. Foxp3/Transcription Factor Staining Buffer set (eBioscience) was used for fixation and permeabilization. Data were analyzed on FlowJo (FlowJo, LLC).
Single-cell RNA sequencing and analysis
Single-cell suspension of B16F10 melanoma from Usp18f/f or Usp18f/f LysM-Cre was prepared as described above. Cells were pooled from 3 mice (Usp18f/f) or 4 mice (Usp18f/f LysM-Cre). CD45+ cells were then sorted on BD FACS Aria. Cells were counted with Countess II FL (Invitrogen) and loaded onto the 10x Genomics Chromium controller. Libraries were prepared using Chromium Single Cell 3’Reagent Kit v3 (10x Genomics) as per the manufacturer’s protocol. The generated libraries were sequenced using an Illumina HiSeq 4000 at the Institute for Genomic Medicine at University of California, San Diego. Sequencing data were aligned using the 10x Genomics Cell Ranger pipeline83 (v3.0.1, mm10 reference genome) and subsequently analyzed using Seurat v4.3.0.84
For cell type annotation, an automated method with manual modification was employed. The clusters were first annotated with SingleR28 using prebuilt ImmGen database29 reference and then modified based on the gene expression and TAM classification from a published article30 as needed.
Pathway enrichment analysis was performed with Gene Set Enrichment Analysis (GSEA).31,32 First, differentially expressed genes (DEGs) were obtained by pesudobulk gene expression analysis87 using the DEseq286 model. Differentially expressed genes of each cluster were pre-ranked by differential test-statistic and analyzed by GSEA using the H: Hallmarks and C2: canonical pathways geneset collections. Metascape33 was also used for pathway enrichment analysis. For trajectory analysis, differentially expressed genes of TAM clusters (Cluster 1, 2, 6, 7, and 12) were analyzed by Monocle 3 using default and developer-recommended settings.41–43
Bulk RNA sequencing and analysis
RNA was extracted from untreated or IFN-I-treated (1000 U/ml for 6 hours) THP-1 WT and USP18−/− THP-1 cells by using TRIzol re-agent. All RNA-seq libraries were prepared by Novogene and sequenced using an Illumina Novaseq 6000 (PE150). STAR85 was used for alignment. Differential gene expression analysis was performed using the DESeq2.86 Genes with an adjusted P-value <0.05 were considered as differentially expressed.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Experiments were replicated two to four times. Data are presented as the means with S.D. or S.E.M. as indicated. Statistical significance was determined by Prism 8 (GraphPad). Two-tailed unpaired t-test and ordinary one-way ANOVA multiple comparison test with post-hoc Tukey test were conducted for comparisons of two groups and more than two groups, respectively. P value p <0.05 was considered to be statistically significant. *; p < 0.05, **; p < 0.01, ***; p < 0.001, ****; p < 0.0001.
Supplementary Material
Highlights.
Deletion of USP18 in myeloid cells suppresses tumor progression
Deletion of USP18 increases anti-tumor macrophages in the tumor microenvironment
Deletion of USP18 in myeloid cells induces downregulation of CSF1R in macrophages
USP18 impairs degradation of CSF1R by interrupting the binding of E3 ligase NEDD4
ACKNOWLEDGMENTS
The UCSD Center for Computational Biology & Bioinformatics (CCBB) for Cell Ranger analysis was supported by Altman Clinical and Translational Research Institute (ACTRI) grant UL1TR001442. This study was supported by NIH R01CA232147 and R01CA177305 to D.-E.Z.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113560.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Gabrilovich DI, Ostrand-Rosenberg S, and Bronte V (2012). Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol 12, 253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mantovani A, Marchesi F, Malesci A, Laghi L, and Allavena P (2017). Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol 14, 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang QW, Liu L, Gong CY, Shi HS, Zeng YH, Wang XZ, Zhao YW, and Wei YQ (2012). Prognostic significance of tumor-associated macrophages in solid tumor: a meta-analysis of the literature. PLoS One 7, e50946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komohara Y, Jinushi M, and Takeya M (2014). Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci 105, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortese N, Donadon M, Rigamonti A, and Marchesi F (2019). Macrophages at the crossroads of anticancer strategies. Front. Biosci 24, 1271–1283. [DOI] [PubMed] [Google Scholar]
- 6.Biswas SK, and Mantovani A (2010). Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol 11, 889–896. [DOI] [PubMed] [Google Scholar]
- 7.Shen X, Zhou S, Yang Y, Hong T, Xiang Z, Zhao J, Zhu C, Zeng L, and Zhang L (2022). TAM-targeted reeducation for enhanced cancer immunotherapy: Mechanism and recent progress. Front. Oncol 12, 1034842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chawla-Sarkar M, Lindner DJ, Liu YF, Williams BR, Sen GC, Silverman RH, and Borden EC (2003). Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis 8, 237–249. [DOI] [PubMed] [Google Scholar]
- 9.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, and Kroemer G (2015). Type I interferons in anticancer immunity. Nat. Rev. Immunol 15, 405–414. [DOI] [PubMed] [Google Scholar]
- 10.Broz ML, Binnewies M, Boldajipour B, Nelson AE, Pollack JL, Erle DJ, Barczak A, Rosenblum MD, Daud A, Barber DL, et al. (2014). Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell 26. 938–652. [DOI] [PubMed] [Google Scholar]
- 11.Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, Archambault JM, Lee H, Arthur CD, White JM, Kalinke U, et al. (2011). Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J. Exp. Med 208, 1989–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novikov A, Cardone M, Thompson R, Shenderov K, Kirschman KD, Mayer-Barber KD, Myers TG, Rabin RL, Trinchieri G, Sher A, and Feng CG (2011). Mycobacterium tuberculosis triggers host type I IFN signaling to regulate IL-1beta production in human macrophages. J. Immunol 187, 2540–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bacher N, Raker V, Hofmann C, Graulich E, Schwenk M, Baumgrass R, Bopp T, Zechner U, Merten L, Becker C, and Steinbrink K (2013). Interferon-alpha suppresses cAMP to disarm human regulatory T cells. Cancer Res 73, 5647–5656. [DOI] [PubMed] [Google Scholar]
- 14.Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, and Zhang DE (2002). UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem 277, 9976–9981. [DOI] [PubMed] [Google Scholar]
- 15.Malakhova OA, Yan M, Malakhov MP, Yuan Y, Ritchie KJ, Kim KI, Peterson LF, Shuai K, and Zhang DE (2003). Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev 17, 455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arimoto KI, Löchte S, Stoner SA, Burkart C, Zhang Y, Miyauchi S, Wilmes S, Fan JB, Heinisch JJ, Li Z, et al. (2017). STAT2 is an essential adaptor in USP18-mediated suppression of type I interferon signaling. Nat. Struct. Mol. Biol 24, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KGS, Fuchs SY, Shuai K, and Zhang DE (2006). UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J 25, 2358–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou W, Kim JH, Handidu A, Li X, Kim KI, Yan M, Li J, and Zhang DE (2007). Microarray analysis reveals that Type I interferon strongly increases the expression of immune-response related genes in Ubp43 (Usp18) deficient macrophages. Biochem. Biophys. Res. Commun 356, 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arimoto KI, Miyauchi S, Troutman TD, Zhang Y, Liu M, Stoner SA, Davis AG, Fan JB, Huang YJ, Yan M, et al. (2023). Expansion of interferon inducible gene pool via USP18 inhibition promotes cancer cell pyroptosis. Nat. Commun 14, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burkart C, Arimoto K.i., Tang T, Cong X, Xiao N, Liu YC, Kotenko SV, Ellies LG, and Zhang DE (2013). Usp18 deficient mammary epithelial cells create an antitumour environment driven by hypersensitivity to IFN-lambda and elevated secretion of Cxcl10. EMBO Mol. Med 5, 1035–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duex JE, Comeau L, Sorkin A, Purow B, and Kefas B (2011). Usp18 regulates epidermal growth factor (EGF) receptor expression and cancer cell survival via microRNA-7. J. Biol. Chem 286, 25377–25386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan Y, Zhou G, Wang X, Chen W, and Gao H (2018). USP18 promotes breast cancer growth by upregulating EGFR and activating the AKT/Skp2 pathway. Int. J. Oncol 53, 371–383. [DOI] [PubMed] [Google Scholar]
- 23.Mustachio LM, Lu Y, Kawakami M, Roszik J, Freemantle SJ, Liu X, and Dmitrovsky E (2018). Evidence for the ISG15-Specific Deubiquitinase USP18 as an Antineoplastic Target. Cancer Res 78, 587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clausen BE, Burkhardt C, Reith W, Renkawitz R, and Förster I (1999). Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 8, 265–277. [DOI] [PubMed] [Google Scholar]
- 25.Gubin MM, Esaulova E, Ward JP, Malkova ON, Runci D, Wong P, Noguchi T, Arthur CD, Meng W, Alspach E, et al. (2018). High-Dimensional Analysis Delineates Myeloid and Lymphoid Compartment Remodeling during Successful Immune-Checkpoint Cancer Therapy. Cell 175. 1443–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, Iracheta-Vellve A, Miller BC, Du PP, Yates KB, Dubrot J, et al. (2019). Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 565, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mould KJ, Jackson ND, Henson PM, Seibold M, and Janssen WJ (2019). Single cell RNA sequencing identifies unique inflammatory airspace macrophage subsets. JCI Insight 4, e126556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, et al. (2019). Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol 20, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heng TSP, and Painter MW; Immunological Genome Project Consortium (2008). The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol 9, 1091–1094. [DOI] [PubMed] [Google Scholar]
- 30.Ma RY, Black A, and Qian BZ (2022). Macrophage diversity in cancer revisited in the era of single-cell omics. Trends Immunol 43, 546–563. [DOI] [PubMed] [Google Scholar]
- 31.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet 34, 267–273. [DOI] [PubMed] [Google Scholar]
- 32.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun 10, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng S, Li Z, Gao R, Xing B, Gao Y, Yang Y, Qin S, Zhang L, Ouyang H, Du P, et al. (2021). A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 184, 792–809.e23. [DOI] [PubMed] [Google Scholar]
- 35.Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, Krishnan I, Maroni G, Meyerovitz CV, Kerwin CM, et al. (2019). Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity 50, 1317–1334.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molgora M, Esaulova E, Vermi W, Hou J, Chen Y, Luo J, Brioschi S, Bugatti M, Omodei AS, Ricci B, et al. (2020). TREM2 Modulation Remodels the Tumor Myeloid Landscape Enhancing Anti-PD-1 Immunotherapy. Cell 182, 886–900.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katzenelenbogen Y, Sheban F, Yalin A, Yofe I, Svetlichnyy D, Jaitin DA, Bornstein C, Moshe A, Keren-Shaul H, Cohen M, et al. (2020). Coupled scRNA-Seq and Intracellular Protein Activity Reveal an Immunosuppressive Role of TREM2 in Cancer. Cell 182, 872–885.e19. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y, Yang S, Ma J, Chen Z, Song G, Rao D, Cheng Y, Huang S, Liu Y, Jiang S, et al. (2022). Spatiotemporal Immune Landscape of Colorectal Cancer Liver Metastasis at Single-Cell Level. Cancer Discov 12, 134–153. [DOI] [PubMed] [Google Scholar]
- 39.Kemp SB, Steele NG, Carpenter ES, Donahue KL, Bushnell GG, Morris AH, The S, Orbach SM, Sirihorachai VR, Nwosu ZC, et al. (2021). Pancreatic cancer is marked by complement-high blood monocytes and tumor-associated macrophages. Life Sci. Alliance 4, e202000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pombo Antunes AR, Scheyltjens I, Lodi F, Messiaen J, Antoranz A, Duerinck J, Kancheva D, Martens L, De Vlaminck K, Van Hove H, et al. (2021). Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat. Neurosci 24, 595–610. [DOI] [PubMed] [Google Scholar]
- 41.Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mantovani A, Allavena P, Sica A, and Balkwill F (2008). Cancer-related inflammation. Nature 454, 436–444. [DOI] [PubMed] [Google Scholar]
- 45.Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, Olson OC, Quick ML, Huse JT, Teijeiro V, et al. (2013). CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med 19, 1264–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamilton JA (2008). Colony-stimulating factors in inflammation and autoimmunity. Nat. Rev. Immunol 8, 533–544. [DOI] [PubMed] [Google Scholar]
- 47.Svensson J, Jenmalm MC, Matussek A, Geffers R, Berg G, and Ernerudh J (2011). Macrophages at the fetal-maternal interface express markers of alternative activation and are induced by M-CSF and IL-10. J. Immunol 187, 3671–3682. [DOI] [PubMed] [Google Scholar]
- 48.Noy R, and Pollard JW (2014). Tumor-associated macrophages: from mechanisms to therapy. Immunity 41, 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rőszer T (2015). Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediators Inflamm 2015, 816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Genard G, Lucas S, and Michiels C (2017). Reprogramming of Tumor-Associated Macrophages with Anticancer Therapies: Radiotherapy versus Chemo- and Immunotherapies. Front. Immunol 8, 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mills CD, and Ley K (2014). M1 and M2 macrophages: the chicken and the egg of immunity. J. Innate Immun 6, 716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cannarile MA, Weisser M, Jacob W, Jegg AM, Ries CH, and Rüttinger D (2017). Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J. Immunother. Cancer 5, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boase NA, and Kumar S (2015). NEDD4: The founding member of a family of ubiquitin-protein ligases. Gene 557, 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nyman TA, Matikainen S, Sareneva T, Julkunen I, and Kalkkinen N (2000). Proteome analysis reveals ubiquitin-conjugating enzymes to be a new family of interferon-alpha-regulated genes. Eur. J. Biochem 267, 4011–4019. [DOI] [PubMed] [Google Scholar]
- 55.U’Ren L, Guth A, Kamstock D, and Dow S (2010). Type I interferons inhibit the generation of tumor-associated macrophages. Cancer Immunol. Immunother 59, 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawrence T, and Natoli G (2011). Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol 11, 750–761. [DOI] [PubMed] [Google Scholar]
- 57.Wang N, Liang H, and Zen K (2014). Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front. Immunol 5, 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coffelt SB, and de Visser KE (2015). Immune-mediated mechanisms influencing the efficacy of anticancer therapies. Trends Immunol 36, 198–216. [DOI] [PubMed] [Google Scholar]
- 59.Engblom C, Pfirschke C, and Pittet MJ (2016). The role of myeloid cells in cancer therapies. Nat. Rev. Cancer 16, 447–462. [DOI] [PubMed] [Google Scholar]
- 60.Ruffell B, and Coussens LM (2015). Macrophages and therapeutic resistance in cancer. Cancer Cell 27, 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaneda MM, Messer KS, Ralainirina N, Li H, Leem CJ, Gorjestani S, Woo G, Nguyen AV, Figueiredo CC, Foubert P, et al. (2016). PI3Kgamma is a molecular switch that controls immune suppression. Nature 539, 437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tong Y, Zhou L, Yang L, Guo P, Cao Y, Qin FXF, and Liu J (2019). Concomitant type I IFN and M-CSF signaling reprograms monocyte differentiation and drives pro-tumoral arginase production. EBioMedicine 39, 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rovida E, Paccagnini A, Del Rosso M, Peschon J, and Dello Sbarba P (2001). TNF-alpha-converting enzyme cleaves the macrophage colony-stimulating factor receptor in macrophages undergoing activation. J. Immunol 166, 1583–1589. [DOI] [PubMed] [Google Scholar]
- 64.Wilhelmsen K, and van der Geer P (2004). Phorbol 12-myristate 13-acetate-induced release of the colony-stimulating factor 1 receptor cytoplasmic domain into the cytosol involves two separate cleavage events. Mol. Cell Biol 24, 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Glenn G, and van der Geer P (2007). CSF-1 and TPA stimulate independent pathways leading to lysosomal degradation or regulated intramembrane proteolysis of the CSF-1 receptor. FEBS Lett 581, 5377–5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Glenn G, and van der Geer P (2008). Toll-like receptors stimulate regulated intramembrane proteolysis of the CSF-1 receptor through Erk activation. FEBS Lett 582, 911–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, Yeung YG, and Stanley ER (1999). CSF-1 stimulated multiubiquitination of the CSF-1 receptor and of Cbl follows their tyrosine phosphorylation and association with other signaling proteins. J. Cell. Biochem 72, 119–134. [PubMed] [Google Scholar]
- 68.Wang Y, Yeung YG, Langdon WY, and Stanley ER (1996). c-Cbl is transiently tyrosine-phosphorylated, ubiquitinated, and membrane-targeted following CSF-1 stimulation of macrophages. J. Biol. Chem 271, 17–20. [DOI] [PubMed] [Google Scholar]
- 69.Wilhelmsen K, Burkhalter S, and van der Geer P (2002). C-Cbl binds the CSF-1 receptor at tyrosine 973, a novel phosphorylation site in the receptor’s carboxy-terminus. Oncogene 21, 1079–1089. [DOI] [PubMed] [Google Scholar]
- 70.Lee PS, Wang Y, Dominguez MG, Yeung YG, Murphy MA, Bowtell DD, and Stanley ER (1999). The Cbl protooncoprotein stimulates CSF-1 receptor multiubiquitination and endocytosis, and attenuates macrophage proliferation. EMBO J 18, 3616–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen C, and Matesic LE (2007). The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev 26, 587–604. [DOI] [PubMed] [Google Scholar]
- 72.Huang X, Chen J, Cao W, Yang L, Chen Q, He J, Yi Q, Huang H, Zhang E, and Cai Z (2019). The many substrates and functions of NEDD4–1. Cell Death Dis 10, 904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo H, Qiao G, Ying H, Li Z, Zhao Y, Liang Y, Yang L, Lipkowitz S, Penninger JM, Langdon WY, and Zhang J (2012). E3 ubiquitin ligase Cbl-b regulates Pten via Nedd4 in T cells independently of its ubiquitin ligase activity. Cell Rep 1, 472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qiao G, Li Z, Molinero L, Alegre ML, Ying H, Sun Z, Penninger JM, and Zhang J (2008). T-cell receptor-induced NF-kappaB activation is negatively regulated by E3 ubiquitin ligase Cbl-b. Mol. Cell Biol 28, 2470–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang B, Gay DL, MacLeod MKL, Cao X, Hala T, Sweezer EM, Kappler J, Marrack P, and Oliver PM (2008). Nedd4 augments the adaptive immune response by promoting ubiquitin-mediated degradation of Cbl-b in activated T cells. Nat. Immunol 9, 1356–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fang DF, He K, Wang N, Sang ZH, Qiu X, Xu G, Jian Z, Liang B, Li T, Li HY, et al. (2014). NEDD4 ubiquitinates TRAF3 to promote CD40-mediated AKT activation. Nat. Commun 5, 4513. [DOI] [PubMed] [Google Scholar]
- 77.Liu Q, Zhang S, Chen G, and Zhou H (2017). E3 ubiquitin ligase Nedd4 inhibits AP-1 activity and TNF-alpha production through targeting p38alpha for polyubiquitination and subsequent degradation. Sci. Rep 7, 4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pei G, Buijze H, Liu H, Moura-Alves P, Goosmann C, Brinkmann V, Kawabe H, Dorhoi A, and Kaufmann SHE (2017). The E3 ubiquitin ligase NEDD4 enhances killing of membrane-perturbing intracellular bacteria by promoting autophagy. Autophagy 13, 2041–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nuro-Gyina PK, Tang N, Guo H, Yan C, Zeng Q, Waldschmidt TJ, and Zhang J (2021). HECT E3 Ubiquitin Ligase Nedd4 Is Required for Antifungal Innate Immunity. J. Immunol 207, 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kawabe H, Neeb A, Dimova K, Young SM Jr., Takeda M, Katsurabayashi S, Mitkovski M, Malakhova OA, Zhang DE, Umikawa M, et al. (2010). Regulation of Rap2A by the ubiquitin ligase Nedd4–1 controls neurite development. Neuron 65, 358–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Duan Z, and Luo Y (2021). Targeting macrophages in cancer immunotherapy. Signal Transduct. Target. Ther 6, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Osiak A, Utermöhlen O, Niendorf S, Horak I, and Knobeloch KP (2005). ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol. Cell Biol 25, 6338–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hafemeister C, and Halbritter F (2023). Single-cell RNA-seq differential expression tests within a sample should use pseudo-bulk data of pseudo-replicates. bioRxiv. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell and bulk RNAseq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| InVivoMAb anti-mouse IFNAR-1 (Clone MAR1-5A3) | Bio X Cell | Cat #: BE0241; RRID: AB_2687723 |
| InVivoMAb mouse IgG1 isotype control (Clone MOPC-21) | Bio X Cell | Cat #: BE0083; RRID: AB_1107784 |
| Rat monoclonal anti-mouse CD16/CD32 (Clone: 93) | Thermo Fisher Scientific (eBioscience) | Cat #: 14-0161-82; RRID: AB_467133 |
| Mouse monoclonal anti-mouse CD45.2 (clone 104) Brilliant Violet 605 | BioLegend | Cat #: 109841; RRID: AB_2563485 |
| Mouse monoclonal anti-mouse CD45.2 (clone 104) PerCP-Cyanine5.5 | BioLegend | Cat #: 109828; RRID: AB_893350 |
| Mouse monoclonal anti-mouse CD45.2 (clone 104) Alexa Fluor 700 | BioLegend | Cat #: 109822; RRID: AB_493731 |
| Rat monoclonal anti-mouse/human CD11b (clone M1/70) Alexa Fluor 700 | BD Biosciences | Cat #: 557960; RRID: AB_396960 |
| Rat monoclonal anti-mouse/human CD11b (clone M1/70) FITC | BD Biosciences | Cat #: 553310; RRID: AB_394774 |
| Rat monoclonal anti-mouse/human CD11b (clone M1/70) PE-Cyanine7 | BioLegend | Cat #: 101216; RRID: AB_312799 |
| Armenian Hamster monoclonal anti-mouse CD11c (clone N418) Pacific Blue | BioLegend | Cat #: 117322; RRID: AB_755988 |
| Rat monoclonal anti-mouse F4/80 (clone BM8) Brilliant Violet 605 | BioLegend | Cat #: 123133; RRID: AB_2562305 |
| Rat monoclonal anti-mouse F4/80 (clone BM8) APC | BioLegend | Cat #: 123116; RRID: AB_893481 |
| Rat monoclonal anti-mouse Gr-1 (Ly6G/Ly6C) (clone RB6-8C5) PE-Cyanine7 | BD Biosciences | Cat #: 565033; RRID: AB_2739049 |
| Rat monoclonal anti-mouse CD115 (CSF-1R) (clone AFS98) APC | BioLegend | Cat #: 135510; RRID: AB_2085221 |
| Rat monoclonal anti-mouse CD115 (CSF-1R) (clone AFS98) PE | BioLegend | Cat #: 135506; RRID: AB_1937253 |
| Rat monoclonal anti-mouse CD3 (clone 17A2) Brilliant Violet 510 | BioLegend | Cat #: 100234; RRID: AB_2562555 |
| Armenian Hamster monoclonal anti-mouse CD3e (clone 145-2C11) Brilliant Violet 650 | BD Biosciences | Cat #: 564378; RRID: AB_2738779 |
| Rat monoclonal anti-mouse CD4 (clone RM4-5) Pacific Blue | BD Biosciences | Cat #: 558107; RRID: AB_397030 |
| Rat monoclonal anti-mouse CD4 (clone RM4-5) Brilliant Violet 570 | BioLegend | Cat #: 100542; RRID: AB_2563051 |
| Rat monoclonal anti-mouse CD8a (clone 5H10) Pacific Orange | Thermo Fisher Scientific (Invitrogen) | Cat #: MCD0830; RRID: AB_10376311 |
| Rat monoclonal anti-mouse CD8a (clone 53-6.7) PE-Cyanine5 | BioLegend | Cat #: 100710; RRID: AB_312749 |
| Rat monoclonal anti-mouse Ly6G (clone 1A8) APC-Cyanine7 | BioLegend | Cat #: 127623; RRID: AB_10645331 |
| Rat monoclonal anti-mouse Ly6C (clone HK1.4) PE-Dazzle594 | BioLegend | Cat #: 128043; RRID: AB_2566576 |
| Mouse monoclonal anti-mouse NK-1.1 (clone PK136) PE | BioLegend | Cat #: 108708; RRID: AB_313395 |
| Rat monoclonal anti-mouse/human CD45R/B220 (clone RA3-6B2) Alexa Fluor 700 | BioLegend | Cat #: 103232; RRID: AB_493717 |
| Rat monoclonal anti-mouse CD25 (clone PC61) PE-Cyanine7 | BioLegend | Cat #: 102016; RRID: AB_312865 |
| Rat monoclonal anti-mouse Foxp3 (clone MF23) Alexa Fluor 647 | BD Biosciences | Cat #: 560401; RRID: AB_1645201 |
| Rat monoclonal anti-mouse Foxp3 (clone FJK-16s) APC | Thermo Fisher Scientific (eBioscience) | Cat #: 17-5773-82; RRID: AB_469457 |
| Rat monoclonal anti-mouse/human CD44 (clone IM7) FITC | BioLegend | Cat #: 103006; RRID: AB_312957 |
| Rat monoclonal anti-mouse CD62L (clone MEL-14) PerCP-Cyanine5.5 | BioLegend | Cat #: 104432; RRID: AB_2285839 |
| Armenian Hamster monoclonal anti-mouse CD69 (clone H1.2F3) PE-Dazzle594 | BioLegend | Cat #: 104536; RRID: AB_2565583 |
| Rat monoclonal anti-mouse IFN-γ (clone XMG1.2) PE | BioLegend | Cat #: 505807; RRID: AB_315401 |
| Rat monoclonal anti-mouse Perforin (clone S16009A) PE-Dazzle594 | BioLegend | Cat #: 154315; RRID: AB_2922482 |
| Rat monoclonal anti-mouse CD206 (MMR) (clone C068C2) PE | BioLegend | Cat #: 141705; RRID: AB_10896421 |
| Rabbit polyclonal anti-CSF-1R/M-CSF-R | Cell Signaling Technology | Cat #: 3152; RRID: AB_2085233 |
| Rabbit monoclonal anti-CSF-1R/M-CSF-R (clone D3O9X) | Cell Signaling Technology | Cat #: 67455; RRID: AB_2799725 |
| Rabbit polyclonal anti-NEDD4 | Cell Signaling Technology | Cat #: 2740; RRID: AB_2149312 |
| Rabbit monoclonal anti-UbcH5C (clone D60E2) | Cell Signaling Technology | Cat #: 4330; RRID: AB_10544697 |
| Rabbit polyclonal anti-Akt | Cell Signaling Technology | Cat #: 9272; RRID: AB_329827 |
| Rabbit monoclonal anti-Phospho-Akt (Ser473) (clone D9E) | Cell Signaling Technology | Cat #: 4060; RRID: AB_2315049 |
| Rabbit polyclonal anti-p38 MAPK | Cell Signaling Technology | Cat #: 9212; RRID: AB_330713 |
| Rabbit monoclonal anti-Phospho-p38 (Thr180/Tyr182) (clone 12F8) | Cell Signaling Technology | Cat #: 4631; RRID: AB_331765 |
| Rabbit monoclonal anti-SAPK/JNK (clone 56G8) | Cell Signaling Technology | Cat #: 9258; RRID: AB_2141027 |
| Rabbit polyclonal anti-Phospho-SAPK/JNK (Thr183/Tyr185) | Cell Signaling Technology | Cat #: 9251; RRID: AB_331659 |
| Mouse monoclonal anti-β-actin (clone AC-15) | Sigma-Aldrich | Cat #: A1978; RRID: AB_476692 |
| Mouse monoclonal anti-α-tubulin (clone DM1A) | Sigma-Aldrich | Cat #: T6199; RRID: AB_477583 |
| Mouse monoclonal anti-FLAG M2 | Sigma-Aldrich | Cat #: F3165; RRID: AB_259529 |
| Mouse monoclonal anti-Myc (clone 9E10) | Santa Cruz Biotechnology | Cat #: sc-40; RRID: AB_627268 |
| Rat monoclonal anti-HA (clone 3F10) | Roche | Cat #: 11867423001; RRID: AB_390918 |
| Rabbit polyclonal anti-GFP | Thermo Fisher Scientific (Invitrogen) | Cat#: A-11122; RRID: AB_221569 |
| Anti-ISG15 | Malakhov et al.14 | N/A |
| Anti-USP18 | Malkahova et al.15 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| BD DIFCO™ Thioglycollate Medium, Brewer Modified | Becton Dickinson | Cat#: 211716 |
| Recombinant Murine M-CSF | Peprotech | Cat#: 315-02 |
| Recombinant Murine IL-4 | Peprotech | Cat#: 214-14 |
| Recombinant Murine IL-13 | Peprotech | Cat#: 210-13 |
| PMA (Phorbol 12-Myristate 13-Acetate) | Sigma-Aldrich | Cat#: P1585 |
| Mouse IFN-Beta, Mammalian | PBL Assay Science | Cat#: 12405 |
| Universal Type I IFN Protein | R&D Systems | Cat#: 11200-2 |
| Cycloheximide | Sigma-Aldrich | Cat#: 01810 |
| Lactacystin | Sigma-Aldrich | Cat #: L6785 |
| β-Estradiol | Sigma-Aldrich | Cat #: E2758 |
| Polyinosinic-polycytidylic acid sodium salt | Sigma-Aldrich | Cat#: P0913 |
| Pexidartinib (PLX3397) | Selleckchem | Cat#: S7818 |
| GYY4137, H2S donor | abcam | Cat#: ab142145 |
| Percoll™ | cytiva | Cat #: 17089101 |
| Propidium Iodide | Roche | Cat#: 11348639001 |
| eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set | Thermo Fisher Scientific (Invitrogen) | Cat #: 00–5523-00 |
| Allele-In-One Mouse Tail Direct Lysis Buffer | Allele Biotechnology | Cat #: ABP-PP-MT01500 |
| TRIzol™ Reagent | Thermo Fisher Scientific (Invitrogen) | Cat#: 15596018 |
| First Strand cDNA Synthesis Kit | MCLAB | Cat #: FSCS-200 |
| KAPA SYBR FAST qPCR Kit Master Mix (2X) Universal | KAPA Biosystems | Cat #: KK4602 |
| cOmplete™, EDTA-free Protease Inhibitor Cocktail | Sigma-Aldrich (Roche) | Cat#: 11873580001 |
| PhosSTOP™ | Sigma-Aldrich (Roche) | Cat #: 4906837001 |
| Protein G Plus/Protein A Agarose Suspension | EMD Millipore | Cat #: IP05 |
| Anti-FLAG M2 Affinity Gel | Sigma-Aldrich | Cat #: A2220 |
| PEI (Polyethylenimine, branched) | Sigma-Aldrich | Cat #: 408727 |
| Hexadimethrine bromide | Sigma-Aldrich | Cat #: H9268 |
| Glutathione Sepharose 4B | cytiva | Cat #: 17075601 |
| PreScission Protease | cytiva | Cat #: 27084301 |
| Critical commercial assays | ||
| CD11b MicroBeads, human and mouse | Miltenyi Biotec | Cat #: 130-049-601 |
| Dynabeads™ Untouched™ Mouse T Cells Kit | Invitrogen | Cat#: 11413D |
| Chromium Single Cell 3’Reagent Kit v3 | 10x Genomics | Cat#: PN-1000075 |
| Zombie Aqua™ Fixable Viability Kit | BioLegend | Cat#: 423101 |
| Zombie NIR™ Fixable Viability Kit | BioLegend | Cat#: 423106 |
| Ubiquitinylation kit | Enzo Life Sciences | Cat #: BML-UW9920-0001 |
| Deposited data | ||
| Single-cell RNAseq data | This paper | GEO: GSE173705 |
| Bulk RNAseq data | Arimoto et al.19 | GEO: GSE165428 |
| Experimental models: Cell lines | ||
| Mouse: B16F10 | Dr. Michiko Fukuda (Sanford Burnham Prebys Institute) | RRID: CVCL_0159 |
| Mouse: EL4 | American Type Culture Collection (ATCC) | Cat #: TIB-39; RRID: CVCL_0255 |
| Mouse: Lewis Lung Carcinoma (LLC) | National Cancer Institute | RRID: CVCL_4358 |
| Human: THP-1 | ATCC | Cat #: TIB-202; RRID: CVCL_0006 |
| Human: THP-1 USP18 knockout | Arimoto et al.19 | N/A |
| Human: MOLM13 | Dr. Lee Grimes (Cincinnati Children’s Hospital) | RRID:CVCL_2119 |
| Human: OCI-AML2 | Dr. Suming Huang (University of Florida) | RRID:CVCL_1619 |
| Human: HEK293T | ATCC | Cat #: CRL-3216; RRID: CVCL_0063 |
| Experimental models: Organisms/strains | ||
| Mouse: B6.129P2-Lyz2tm1(cre)Ifo/J | The Jackson Laboratory | Strain #: 004781; RRID: IMSR_JAX:004781 |
| Mouse: B6.Cg-Ndor1Tg(UBC-cre/ERT2)1Ejb/1J | The Jackson Laboratory | Strain #: 008085; RRID: IMSR_JAX:008085 |
| Mouse: Usp18f/f | Arimoto et al.19 | N/A |
| Mouse: Isg15 knockout | Osiak et al.82 | N/A |
| Oligonucleotides | ||
| Primer: mouse Gapdh Forward: TATGTCGTGGAGTCTACTGG |
This paper | N/A |
| Primer: mouse Gapdh Reverse: GAGTTGTCATATTTCTCGTG |
This paper | N/A |
| Primer: mouse Usp18 Forward: CAGCCCTCATGGTCTGGTTG |
This paper | N/A |
| Primer: mouse Usp18 Reverse: GCACTCCGAGGCACTGTTAT |
This paper | N/A |
| Primer: mouse Csf1r Forward: AATGCTAACGCCACCGAGA |
This paper | N/A |
| Primer: mouse Csf1r Reverse: CATGGAAAGTTCGGACACAGG |
This paper | N/A |
| Primer: mouse Nedd4 Forward: ACCAGCGTGCAGACAAAAAC |
This paper | N/A |
| Primer: mouse Nedd4 Reverse: AAAAGAATGCGGTGTCGCTG |
This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pCL-10A1 | Novus | Cat #: NBP2-29542 |
| Plasmid: pCL-Eco | Novus | Cat #: NBP2-29540 |
| Plasmid: psPAX2 | Dr. Didier Trono (Addgene) | RRID: Addgene_12260 |
| Plasmid: pMD2.G | Dr. Didier Trono (Addgene) | RRID: Addgene_12259 |
| Plasmid: pCX4-bsr | Dr. Tsuyoshi Akagi (KAN Research Institute) | N/A |
| Plasmid: pcDNA™3.1 (+) Mammalian Expression Vector | Thermo Fisher Scientific (Invitrogen) | Cat #: V79020 |
| Plasmid: pCX4-bsr-mouse Usp18 | This paper | N/A |
| Plasmid: pCX4-bsr -FLAG-mouse Usp18 | This paper | N/A |
| Plasmid: pCX4-bsr-human USP18 (sgRNA resistant) | This paper | N/A |
| Plasmid: pCX4-bsr-human USP18 C64S (sgRNA resistant) |
This paper | N/A |
| Plasmid: pCAG-mouse Csf1r-FLAG | This paper | N/A |
| Plasmid: pCAG-human CSF1R-FLAG | This paper | N/A |
| Plasmid: pcDNA-Myc-human NEDD4 | This paper | N/A |
| Plasmid: pcDNA -HA-mouse Nedd4 | This paper | N/A |
| Plasmid: pcDNA -HA-mouse Nedd4 (C744A mutant) | This paper | N/A |
| Plasmid: pcDNA -HA-mouse Nedd4 (HECT domain deletion mutant) | This paper | N/A |
| Plasmid: pcDNA-FLAG-mouse Nedd4 | This paper | N/A |
| Plasmid: pcDNA-Myc-Ubiquitin | This paper | N/A |
| Plasmid: pEGFP-N1 | Clontech | Cat #: 6085-1 |
| Plasmid: pSUPER.retro.puro | OligoEngine | Cat #: VEC-PRT-0002 |
| Plasmid: pSUPER.retro.puro-human USP18 shRNA | This paper | N/A |
| Plasmid: pLKO.1 Negative control (GFP) | This paper | N/A |
| Plasmid: pLKO.1-human UBCH5C shRNA | La Jolla Institute for Immunology | Cat #: TRCN0000038791 |
| Plasmid: pLKO.1-human UBCH5C shRNA | La Jolla Institute for Immunology | Cat #: TRCN0000038793 |
| Plasmid: pLKO.1-mouse Nedd4 shRNA | La Jolla Institute for Immunology | Cat #: TRCN0000092436 |
| Plasmid: pLKO.1-human NEDD4 shRNA | La Jolla Institute for Immunology | Cat #: TRCN0000007550 |
| Plasmid: pLKO.1-human NEDD4 shRNA | La Jolla Institute for Immunology | Cat #: TRCN0000007551 |
| Plasmid: pGEX-6P-1 | cytiva | Cat #: 28954648 |
| Plasmid: pGEX-6P-1-human CSF1R (543 – 972) | This paper | N/A |
| Plasmid: pGEX-6P-1-human NEDD4 | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad | N/A |
| FlowJo 10 | FlowJo, LLC | N/A |
| LI-COR Image Studio™ Software | LI-COR | N/A |
| 10x Genomics Cell Ranger (v3.0.1) | Zheng et al.83 | N/A |
| Seurat (v4.3.0) | Hao et al.84 | https://satijalab.org/seurat/ |
| STAR | Dobin et al.85 | N/A |
| DESeq2 | Love et al.86 | N/A |
| SingleR | Aran et al.28 | N/A |
| ImmGen (Immunological Genome Project) | Heng et al.29 | N/A |
| Gene Set Enrichment Analysis (GSEA) | Mootha et al.31
Subramanian et al.32 |
https://www.gsea-msigdb.org |
| Metascape | Zhou et al.33 | https://metascape.org |
| Monocle 3 | Cao et al.41
Qiu et al.42 Trapnell et al.43 |
N/A |
| Other | ||
| scil Vet abc Plus hematology analyzer | scil animal care company | N/A |
| MycoAlert PLUS Mycoplasma Detection Kit | Lonza | Cat#: LT07-710 |
| BD FACS Canto II Flow Cytometer | BD Biosciences | N/A |
| BD LSR II Flow Cytometer | BD Biosciences | N/A |
| BD FACS Aria II Cell Sorter | BD Biosciences | N/A |
| NovoCyte Advanteon Flow Cytometer | Agilent Technologies | N/A |
| CFX96 Thermal cycler | Bio-Rad Laboratories | N/A |
| Odyssey Imaging System | LI-COR Biotechnology | N/A |
| Countess II FL Automated Cell Counter | Thermo Fisher Scientific (Invitrogen) | N/A |
| Chromium controller | 10x Genomics | N/A |


