Abstract
Cytokines are secreted or otherwise released polypeptide factors that exert autocrine and/or paracrine actions, with most cytokines acting in the immune and/or hematopoietic system. They are typically pleiotropic, controlling development, cell growth, survival, and/or differentiation. Correspondingly, cytokines are clinically important, and augmenting or attenuating cytokine signals can have deleterious or therapeutic effects. Besides physiological fine-tuning of cytokine signals, altering the nature or potency of the signal can be important in pathophysiological responses and can also provide novel therapeutic approaches. Here, we give an overview of cytokines, their signaling and actions, and the physiological mechanisms and pharmacologic strategies to fine-tune their actions. In particular, the differential utilization of STAT proteins by a single cytokine or by different cytokines and STAT dimerization versus tetramerization are physiological mechanisms of fine-tuning, whereas anticytokine and anticytokine receptor antibodies and cytokines with altered activities, including cytokine superagonists, partial agonists, and antagonists, represent new ways of fine-tuning cytokine signals.
Keywords: cytokine, fine-tuning, STAT protein, STAT tetramerization, partial agonist, superenhancers
INTRODUCTION
The term cytokine was coined by Stanley Cohen and colleagues in 1974 to refer to a molecule, typically in the immune system, that is produced by one immune cell and acts on another (1). Cytokines can act in an autocrine fashion on the same cell or in a paracrine fashion on another cell, with some cytokines exerting both autocrine and paracrine actions. Cytokines typically act locally but potentially can act at a distance as well.
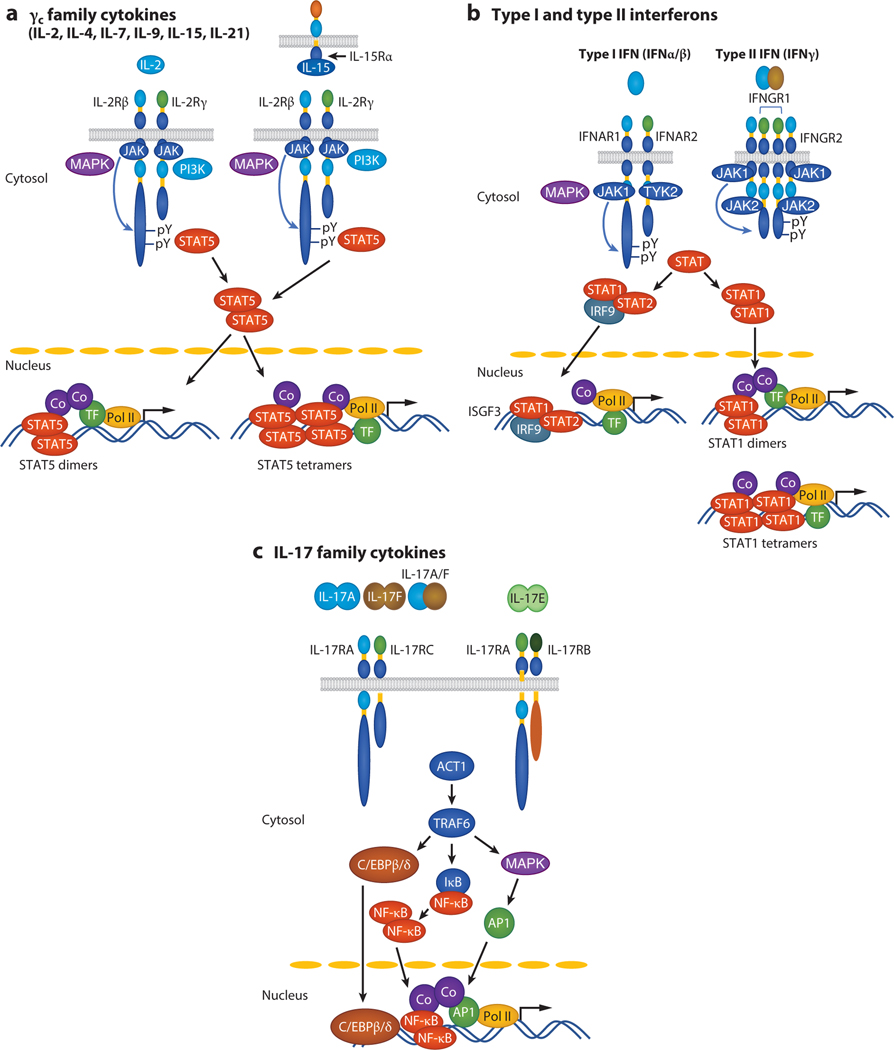
Cytokines can be grouped into multiple families, often on the basis of the structural considerations for the cytokines and their receptors as well as mechanisms of signaling. For example, type I cytokines have a four-α-helix bundle structure and include a large number of interleukins (see Table 1); typically their receptors are of the type I cytokine receptor family (historically also called hematopoietin receptors) and do not have intrinsic tyrosine kinase domains, although two exceptions exist: the stem cell factor (SCF) receptor, c-KIT; and the colony-stimulating factor 1 (CSF-1) receptor (2). Instead of containing intrinsic kinase domains, type I cytokine receptors utilize Janus family tyrosine kinases (JAKs) and signal in part by the activation of signal transducer and activator of transcription (STAT) proteins (Figure 1a,b). In addition, they typically activate phospho-inositol 3-kinase and ERK-dependent signaling mechanisms. Normally, type I cytokines are monomers, although IL-5 and granulocyte CSF (G-CSF) are homodimers (3,4) and the IL-12 family cytokines, IL-12 (p35 and p40), IL-23 (p19 and p40), IL-27 (p28 and EBI3), and IL-35 (p35 and EBI3), are heterodimers (5). Most type I cytokines bind to receptors that are homodimers or heterodimers (2, 5, 6), but more complex structures also exist (2, 6). Type II cytokines comprise the interferons—type I interferons (IFN-α’s and IFN-β), type II interferon (IFN-γ) (Figure 1b), and type III interferons (IFN-λ’s, also sometimes referred to as IL-28 and IL-29)—and IL-10 family cytokines (7–9) (see Table 1). Like type I cytokines, type II cytokines also use JAK-STAT signaling as a major signaling pathway (10). IL-1 family, IL-17 family, and TNF family cytokines are quite distinctive structurally from type I and type II cytokines, and their receptors are also very different from the receptors for type I and II cytokines. Correspondingly, none of these families of cytokines use JAK-STAT signaling, and they instead all share NF-κB as a major signaling mechanism (11–15) (see Figure 1c, showing IL-17 family cytokines as an example).
Table 1.
Cytokine families
| Type 1 cytokines (2, 5) | Type 2 cytokines (7–10) | TNF (12, 15) | IL-1 (11) | IL-17 (13, 14) | |||||
|---|---|---|---|---|---|---|---|---|---|
| γc | βc | gp130 | IL-12 | Others | IFNs | IL-10 | |||
| IL-2 | IL-3 | IL-6 | IL-12 | Growth hormone | IFN-α | IL-10 | TNF | IL-1α | IL-17A |
| IL-4 | IL-5 | IL-11 | IL-23 | Prolactin | IFN-β | IL-19 | LTα3 | IL-1β | IL-17B |
| IL-7 | GM-CSF | IL-31 | IL-27 | Erythropoietin | IFN-γ | IL-20 | LTα1β2 | IL-1RA | IL-17C |
| IL-9 | Oncostatin M | IL-35 | Thrombopoietin | IL-22 | OX40L | IL-18 | IL-17D | ||
| IL-15 | Leukemia inhibitory factor | M-CSF | IL-24 | CD40L | IL-33 | IL-17E (IL-25) | |||
| IL-21 | Ciliary neurotrophic factor | SCF | IL-26 | RANKL | IL-36α, β, γ | IL-17F | |||
| Cardiotropin-1 | TSLP | IL-28Aa | TWEAK | IL-36RA | |||||
| NNT-1/BSF-3 | IL-13 | IL-28Ba | APRIL | IL-37 | |||||
| Leptin | IL-29a | BAFF | IL-38 | ||||||
| G-CSF | LIGHTb | ||||||||
IL-28A, IL-28B, and IL-29 are also known as type III interferons or IFN-λs.
Homologous to lymphotoxin; exhibits inducible expression and competes with HSV glycoprotein D for binding to herpesvirus entry mediator; a receptor expressed on T lymphocytes.
Abbreviations: APRIL, a proliferation-inducing ligand; BAFF, B cell–activating factor; BSF-3, B cell–stimulating factor 3; CD40L, CD40 ligand; GM-CSF, granulocyte-macrophage colony-stimulating factor; G-CSF, granulocyte colony-stimulating factor; IL-1RA, IL-1 receptor antagonist; IL-36RA, IL-36 receptor antagonist; LTα1β2, lymphotoxin α1β2; LTα3, lymphotoxin α3; M-CSF, macrophage colony-stimulating factor; NNT-1, novel neurotrophin 1; SCF, stem cell factor; OX40L, OX40 ligand; RANKL, receptor activator of NF-κB; TNF, tumor necrosis factor; TSLP, thymic stromal lymphopoietin; TWEAK, TNF-related weak inducer of apoptosis.
Figure 1.
Illustrative signaling pathways for cytokines that are members of different cytokine families. (a) γc family cytokines, (b) type I and type II interferons, and (c) IL-17 family cytokines bind to their receptors, activate (a,b) JAK/STAT or (c) ACT1/TRAF6/NF-κB signaling pathways, and initiate cytokine-specific gene expression programs. (a) Of γc family cytokines (IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21), only IL-2 and IL-15 are shown, and these primarily signal in cis (IL-2) or in trans (IL-15). IL-4, IL-7, IL-9, and IL-21 are only known to signal in cis. In addition to JAK-STAT, MAPK and PI3K pathways are activated. Both STAT5 dimers and STAT5 tetramers are activated by IL-2 and IL-15. (b) Type I interferons (IFN-α/β) activate ISGF3 (STAT1/STAT2/IRF9), whereas type II interferon (IFN-γ) activates STAT1 dimers and tetramers. (c) Signaling via either IL-17RA/IL-17RC or IL-17RA/IL-17RB. The latter is used by IL-17E, which is also known as IL-25. Abbreviations: Co, co-activator (such as CBP or p300); TF, transcription factor; PI3K, phospho-inositol 3-kinase; Pol II, RNA polymerase II.
As noted above, cytokines tend to exert a wide range of actions, typically on more than one cell type; thus, they are usually pleiotropic. Moreover, it is common to find cytokines with overlapping or similar actions, suggesting that the cytokines might have partially redundant functions. However, the phenotypes resulting from mutations or deletions of cytokines or their receptors, as are found in certain human diseases or in genetically altered mice, often indicate that the actions of different cytokines are distinctive rather than truly redundant. Even if actions are similar in vitro, this may not necessarily translate into identical in vivo actions because of differences in the spatial location and/or kinetics of expression of cytokines and their receptors. In some cases, cytokines within a family have functional similarities, but cytokines with completely distinctive topological folds and signaling mechanisms can functionally overlap as well. For example, IL-1, IL-6, and TNF are all proinflammatory cytokines (11–15).
Because of the potency of cytokine signals,it is essential for cytokines to be carefully regulated. Whether in response to pathogens, tumors, or various antigenic insults, and whether involving the innate or adaptive immune system, careful control of cytokine signals is critical for a robust and effective immune system. Here we explore some of the ways in which such signals can be rationally modulated and fine-tuned by either natural cytokines or engineered agonists or antagonists. In this review, we focus on type I cytokines.
JAK-STAT SIGNALING
As noted above, JAK-STAT signaling pathways are critical mediators of the actions of type I and type II cytokines. There are four JAKs (JAK1, JAK2, JAK3, and TYK2) and seven STAT proteins (STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6); STAT5A and STAT5B are the most conserved STAT proteins (>91% identical at the amino acid level) and are encoded by adjacent genes, consistent with their having arisen by gene duplication (16). All seven STAT proteins share conserved functional domains [N-terminal, coiled-coil, DNA-binding, linker, Src homology 2 (SH2),and C-terminal transactivation domains] as well as a key tyrosine residue located between the SH2 and C-terminal transactivation domains that is a target for phosphorylation (17, 18). After a cytokine binds to its receptor, receptor-associated JAKs are activated and phosphorylate the receptor on tyrosine residues; some of these phosphorylated tyrosines and their flanking amino acids can then serve as docking sites for the SH2 domain of unphosphorylated STAT proteins and determine which STAT proteins can bind to the receptor (17, 18). The receptor-associated STAT(s) is then also phosphorylated by the JAKs, followed by interaction of the SH2 domain of one STAT molecule with the phosphorylated tyrosine of another and vice versa to form STAT homodimers and potentially heterodimers that then translocate to the nucleus, bind target sequences, and modulate expression of target genes (17, 18). All of the STAT proteins except STAT2 bind to GAS (IFN-γ-activated sequence) motifs (TTCN3GAA for STAT1, STAT3, STAT4, STAT5A, and STAT5B and TTCN4GAA for STAT6) (Figure 1a,b). STAT2 heterodimerizes with STAT1, and this complex additionally interacts with IRF9 (interferon regulatory factor 9) to form ISGF3 (interferon-stimulated gene factor 3), which binds to ISRE (interferon-stimulated response element) motifs (17, 18) (Figure 1b). Given the large number of type I and type II cytokines but small number (only four) of JAKs, it is evident that individual JAKs can be activated by many cytokines (Table 2); JAK3 is distinctive in that it is only activated by one family of type I cytokines— IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21—which share the common cytokine receptor γ chain, γc, which is the cytokine receptor subunit that uniquely associates with JAK3. Moreover, just as mutations in the gene encoding γc, IL2RG, cause X-linked severe combined immunodeficiency (XSCID; also known as SCID-X1) (19), JAK3 deficiency phenocopies the T−B+NK− phenotype [greatly diminished T and natural killer (NK) cells but normal B cell numbers] that is observed in XSCID (20,21). Mutations in IL7RA result in a T−B+NK+ immunodeficiency in humans; NK cell numbers are normal, given that IL-15 signaling is still intact (22–24). Although all cytokines that act via receptor homodimers only activate JAK2, other cytokines typically activate two or even three different JAKs; for example, the six γc family cytokines all activate JAK1 as well as JAK3 (Table 2). Given the relatively limited number of STAT proteins, each is also utilized by more than one cytokine. Some STATs are activated by relatively few cytokines: For example, STAT6 is only activated by IL-4 and IL-13; STAT2 is activated primarily by type I interferons; and STAT4 is activated primarily by IL-12, IL-23, and IL-35 (Table 2). In contrast, STAT1, STAT3, and STAT5 (i.e., STAT5A and STAT5B) are much more broadly activated (25–28) (Table 2).
Table 2.
Cytokines and the JAKs and STATs they activate
| Cytokine family | Cytokines (2, 5, 7–9) | JAKs | STAT proteinsa |
|---|---|---|---|
| IFNs | IFN-α, IFN-β | JAK1, TYK2 | STAT1, STAT2 |
| IFN-γ | JAK1, JAK2 | STAT1 | |
| gp130 | IL-6, IL-11, OSM, LIF, CNTF, CT-1, NNT-1 | JAK1, JAK2, TYK2 | STAT3 |
| γc | IL-2, IL-7, IL-9, IL-15 | JAK1, JAK3 | STAT5A, STAT5B, STAT3, STAT1 |
| IL-4 | JAK1, JAK3 | STAT6, STAT5A, STAT5B | |
| IL-21 | JAK1, JAK3 | STAT3, STAT1, STAT5A, STAT5B | |
| TSLP | JAK1, JAK2 | STAT5A, STAT5B | |
| IL-13 | JAK1, JAK2 | STAT6 | |
| βc | IL-3, IL-5, GM-CSF | JAK2 | STAT5A, STAT5B |
| Cytokines whose receptors are homodimers | GH, PRL, EPO, TPO | JAK2 | STAT5A, STAT5B |
| IL-10 | IL-10 | JAK1, TYK2 | STAT3, STAT1, STAT5 |
| IL-19, IL-20, IL-24 | JAK1, JAK2 | STAT3, STAT1 | |
| IL-22 | JAK1, TYK2 | STAT3, STAT1, STAT5 | |
| IL-26 | JAK1, TYK2 | STAT1, STAT3 | |
| IL-28A, IL-28B, IL-29 | JAK1, TYK2 | STAT1, STAT2, STAT3, STAT5 | |
| IL-12 | IL-12 | JAK2, TYK2 | STAT4 |
| IL-23 | JAK2, TYK2 | STAT3, STAT4 | |
| IL-27 | JAK2, JAK1 | STAT3, STAT1 | |
| IL-35 | JAK2, JAK1 | STAT4, STAT1 |
STAT proteins preferentially activated by a given cytokine are indicated in bold.
Abbreviations: CNTF, ciliary neurotrophic factor; CT-1, cardiotrophin 1; EPO, erythropoietin; GH, growth hormone; GM-CSF, granulocyte-macrophage colony-stimulating factor; JAK, Janus family tyrosine kinases; LIF, leukemia inhibitory factor; NNT-1, novel neurotrophin 1; OSM, oncostatin M; PRL, prolactin; STAT, signal transducer and activator of transcription; TPO, thrombopoietin; TSLP, thymic stromal lymphopoietin.
Potential Involvement of JAK-STAT Polymorphisms in Human Diseases
In addition to gene mutations such as those found in JAK3-deficient patients that abrogate JAK-STAT signaling by γc-dependent cytokines, genome-wide association studies have revealed a number of polymorphisms present in genes for both JAKs and STATs, and some of the polymorphisms show significant association with susceptibility to autoimmune and inflammatory diseases and/or malignancies in humans (29–40) as well as with disease progression and response to the treatment (38, 41–44). For example, genetic variants in the TYK2 gene have been reported across a range of autoimmune disorders. Given that TYK2 is activated by type I interferons and select other cytokines (e.g., IL-12), this JAK is a potentially important target for modulating cytokine signaling. The absence of TYK2 is generally associated with immunodeficiency and severe infections, but a polymorphism was identified in the TYK2 gene that is associated with protection from a number of autoimmune diseases, whereas another correlates with protection from some but not for other autoimmune disorders (45). Interestingly, in psoriasis patients, noncoding polymorphisms potentially affecting STAT3 and STAT4 binding were identified at susceptibility loci (46), and polymorphisms associated with coronary artery disease and type 2 diabetes disrupt STAT1 binding to enhancers in the 9p21 gene desert, thereby affecting long-range chromatin interactions in this region and resulting in an impaired IFN-γ response (47). Thus, naturally arising polymorphisms in JAK/STAT signaling pathways that modulate signaling are indicative of additional ways to fine-tune cytokine signals, potentially with therapeutic benefit.
Physiological Fine-Tuning of Cytokine Signals
Fine-tuning the immune response by differential activation of multiple STATs by a single cytokine or by differential STAT protein activation by two different cytokines.
As noted above, STAT1, STAT3, and STAT5 proteins are activated by multiple cytokines, but normally only one or two of these STAT proteins are strongly activated by each cytokine. For example, IL-2, IL-7, IL-9, and IL-15 dominantly activate STAT5A and STAT5B, with less potent activation of STAT1 and STAT3 (48). In contrast, IL-21 most potently activates STAT3, with less potent activation of STAT1 and STAT5 proteins. For IL-21, STAT3 activation is also more sustained, whereas activation of STAT1 and STAT5 is more transient (28, 49).
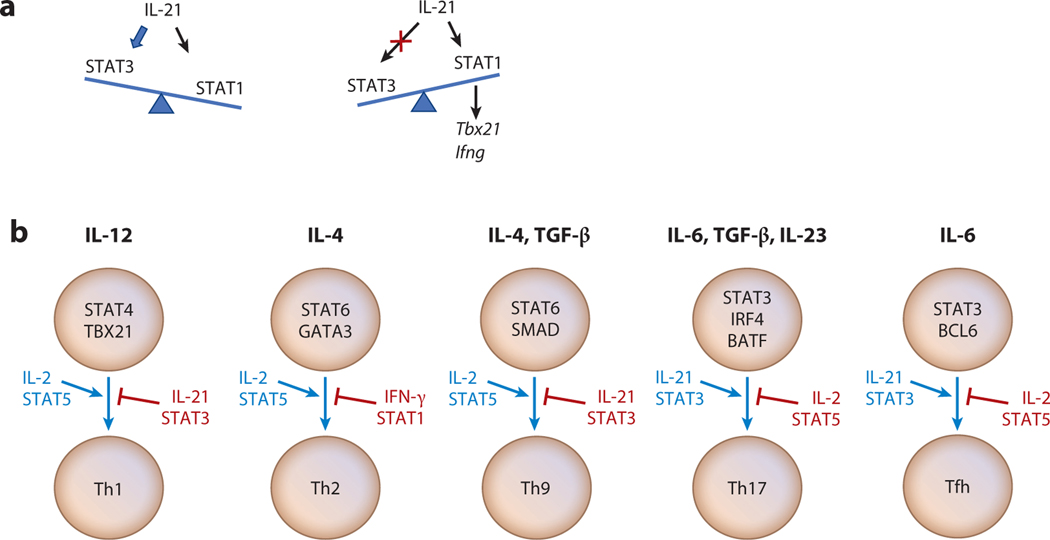
Although many STATs can bind to similar or even the same GAS motifs, the gene expression pattern mediated by each STAT varies. In the case of IL-21,more genes are dependent on STAT3, but IL-21-mediated STAT1 activation also serves a critical role (49).For example,IL-21 augments T-BET and IFN-γ expression via STAT1 in a T helper type 1 (Th1)-dependent fashion, and in fact IL-21-mediated activation of STAT3 can inhibit T-BET and IFN-γ induction, with higher IL-21-induced Tbx21 and Ifng gene expression in Stat3-deficient mice (49); this indicates that STAT3 indeed negatively regulates the expression of these genes (Figure 2a). Corresponding findings were observed for the human TBX21 and IFNG genes based on studies with cells from patients with autosomal dominant hyper-IgE syndrome (AD-HIES, also known as Job syndrome) (49), a disease with STAT3 deficiency caused by autosomal dominant mutations in STAT3 that result in a condition where cells are hypomorphic for STAT3 activation (50,51). Tuning STAT3 and STAT1 activity by Fas has also been demonstrated (52). In addition to its function in mediating apoptosis, Fas can increase autoimmunity not only by promoting pathogenic Th17 differentiation and stabilizing Th17 cells in a STAT3-dependent fashion but also by preventing Th1 differentiation via its binding to and sequestering STAT1. In Fas-deficient cells, IL-6-induced STAT1 activation and a Th1-associated transcription program are favored, whereas this is not observed in Fas-deficient cells that additionally lack STAT1. Thus, modulating Fas levels can fine-tune the balance of Th17 and Th1 differentiation by controlling available STAT3- versus STAT1-mediated gene activation. Accordingly, the IL-21 and Fas/IL-6 studies discussed above together indicate that the relative potency of the STAT3 versus STAT1 signal determines gene expression and represents a mechanism for fine-tuning the signal(s) induced by IL-21 and/or IL-6. There is now an increasing appreciation of the complexity of IL-21–STAT3 signaling, which involves the binding of multiple factors to AP1-IRF composite elements (AICEs) (53, 54), where STAT3 is associated with IRF4, AP-1, MAF, and p300 in a genome-wide fashion, and potentially other factors as well (55, 56). Because these AICEs do not always recruit the same FOS or JUN family proteins—for example, BATF versus FOSL and cJUN versus JUNB or JUND for the AP-1 elements (53, 54, 56, 57), it is conceivable that the specific transcription factors found in association with STAT3 and/or IRF4 help to determine the potency and/or specificity of the cytokine effect, potentially providing an additional mechanism for fine-tuning cytokine signaling.
Figure 2.
Fine-tuning cytokine signals by STAT proteins or other cytokines. (a) Multiple STAT proteins activated by a cytokine can determine its overall effect based on the potency of activation of each STAT; this is illustrated by the relative strength of STAT3 versus STAT1 activation by IL-21. (b) Different cytokines can either enhance (blue) or suppress (red) the actions of other cytokines. Abbreviations: Tfh, T follicular helper cell; Th1, T helper type 1 cell.
Fine-tuning of cytokine signaling can also be mediated by the relative potency of activation of two or more STATs by a single cytokine or by the balance of signaling by two different cytokines, each of which preferentially activates a different STAT protein (Figure 2b).For example,Th9 and T follicular helper (Tfh) differentiation processes are balanced by opposing actions of IL-2 and IL-21. In Th9 differentiation, IL-2 promotes and IL-21 inhibits the differentiation (58), whereas in Tfh differentiation, the situation is reversed (59,60). Thus, the relative potency of each cytokine signal affects the types of cells that are produced, and for both Th9 and Tfh differentiation, the balance of IL-2-activated STAT5 versus IL-21-activated STAT3 is critical.
Th17 differentiation is an IL-6-dependent process that is mediated in part via STAT3, and it involves an IL-21–IL-23 cytokine cascade (61–66) that promotes efficient Th17 differentiation in a STAT3-dependent manner. Interestingly, similar to Tfh differentiation, Th17 differentiation is also regulated by a balance between cytokines that activate STAT3 versus STAT5, with IL-2/STAT5 inhibiting Th17 differentiation (67). Mechanistically, it was suggested that IL-2-activated STAT5 inhibits IL-6-activated STAT3-mediated induction of the Il17a gene by competitive binding (67). In addition, IL-2 via STAT5 also inhibits gp130 expression, thus diminishing IL-6 signaling (68); moreover, by its STAT5-dependent induction of T-BET (68), IL-2 can potentially squelch Th17 differentiation by T-BET-mediated interaction with RUNX1, thereby potentially diminishing the interaction of RORγt with RUNX1 (69), which is required for Th17 differentiation (63). Thus, the type and extent of Th differentiation can be fine-tuned by the balance of STAT5 versus STAT3 activation by cytokines in the overall cellular milieu. Corresponding to these in vitro observations for Th17 differentiation, IL-23 production and IL-23R cell surface expression on T cells are increased in patients with systemic lupus erythematosus, and T cells from these patients have increased IL-17 production but decreased IL-2 production in response to IL-23 stimulation (70).
IL-27 is an IL-12 family heterodimeric cytokine, but unlike IL-12 and like IL-6, it uses gp130 as a receptor component and activates both STAT3 and STAT1 (71). As discussed above, IL-6 promotes Th17 differentiation but also suppresses Th1 differentiation, whereas IL-27 suppresses Th17 but promotes Th1 differentiation (5). Thus, the balance of IL-6 and IL-27 represents another mechanism for fine-tuning the T cell response. Interestingly, despite their opposing biological effects, most genes are commonly regulated at the transcriptional level by IL-6 and IL-27, which is explained by the finding that STAT3 controls the expression of genes commonly regulated by both cytokines (72).
Another example of fine-tuning relates to the balance of IL-12 and IL-23.IL-12 (a heterodimer of p35 and p40) is essential for Th1 differentiation (73), whereas IL-23 (a heterodimer of p19 and p40) promotes Th17 differentiation (74). The distinctive actions of IL-12 and IL-23 are likely at least in part due to their binding to distinctive receptor components (IL-12Rβ2 versus IL-23R) and activating different STAT proteins (STAT4 by IL-12 and STAT3 by IL-23), despite both cytokines sharing IL-12Rβ1 and activating JAK2 and TYK2 (5, 75). The action of IL-23 has been associated with a number of autoimmune and chronic inflammatory diseases (75) as well as certain cancers (76, 77; reviewed in 78), and identifying therapeutic agents to specifically block IL-23 action has become an area of interest related to autoimmune and inflammatory disorders (75, 79, 80). The structure of the IL-23–IL-23R complex has revealed that the N-terminal immunoglobulin domain on IL-23R and Trp157 of the IL-23 p19 subunit are critical for IL-23 binding with high affinity and provided a mechanistic basis for rationally designing agents to specifically block IL-23 action and suppress pathological Th17 responses (81), with implications for IL-12 as well.
Two cytokines that activate different STAT proteins can not only compete, but they can alternatively potentially cooperate to induce the net biological outcome (Figure 2b). For example, optimal Th2 differentiation requires both IL-2 and IL-4, which activate mainly STAT5 and STAT6, respectively, and these signals together cooperate in this context. Whereas IL-4-mediated STAT6 is the critical signal to drive Th2 differentiation (82), IL-2 and STAT5 are required for efficient priming of cells for Th2 differentiation, both by promoting open chromatin at the Il4 locus (83, 84) and by binding and activating transcription at the Il4ra locus, thereby enhancing IL-4Rα expression and cellular responsiveness to IL-4 (85). Analogously, for Th1 differentiation, IL-2 via STAT5 enhances T-BET and IL-12Rβ2 expression, thereby priming cells for IL-12 responsiveness (68), with associated STAT4 activation, which then can drive Th1 differentiation. Thus, cytokine fine-tuning can result from either opposing actions of two cytokines or cooperation of cytokines and sometimes involves cytokine cascades; an example can be seen in the context of Th17 differentiation for IL-6, IL-21, and IL-27 (62).
Fine-tuning of cytokine signals via STAT tetramerization versus dimerization.
It is well established that STAT dimers are the basic core functional units that bind to consensus GAS motifs (17, 18), with high-resolution X-ray crystallographic structures for STAT1 and STAT3β dimers bound to DNA (86, 87). However, STAT proteins can also form tetramers that can bind to tandem GAS motifs, including lower-affinity nonconsensus motifs (88–90). Formation of tetramers depends on N-terminal domain–mediated interaction between two dimers, and tetramer formation then allows the cooperative binding of STAT proteins to DNA containing both consensus and nonconsensus GAS motifs (88–90).The key amino acids that mediate the N-terminal domain interaction in STAT4 were identified by X-ray crystallographic structure analysis (91), and these residues are highly conserved in the N-terminal domains of STAT1, STAT3, STAT4, STAT5A, STAT5B, and STAT6. Because the N-terminal domain interaction is not dependent on tyrosine phosphorylation, unphosphorylated STATs can form alternative dimers via N-terminal domain interactions (92–96). STAT protein tyrosine phosphorylation and nuclear translocation are additionally needed, as are two tandemly linked GAS or GAS-like motifs on DNA segments with favorable spacing to allow STAT tetramer formation (89, 90, 97, 98).
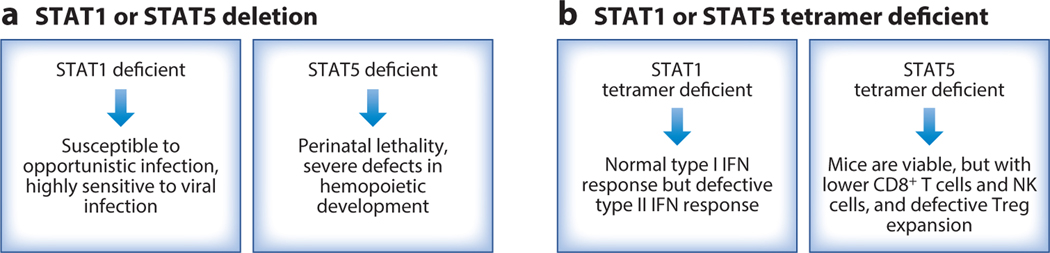
The in vivo biological importance of STAT5 and STAT1 tetramers has been studied using knock-in mice that contain mutations in critical residues in either the N-terminal domains of both STAT5A and STAT5B or the N-terminal domain of STAT1.Such mutations abolish STAT5 and STAT1 tetramer binding, respectively, without affecting tyrosine phosphorylation or binding as dimers (97, 99). STAT5 tetramer–deficient mice are viable and develop normally (97, 99), in contrast to Stat5a/Stat5b double knockout mice, which exhibit fetal lethality (100) (Figure 3a). Nevertheless, STAT5 tetramers are required for normal numbers of CD8+ T and NK cells, the expansion of antigen-specific CD8+ T cells in response to viral infection, NK cell maturation and survival, particularly after IL-15 withdrawal, and the normal function of regulatory T cells (Tregs), but they are not required for the development of normal numbers of B and CD4+ T cells (97, 98) (Figure 3b). In contrast to the situation for the STAT5 tetramer–deficient mice, there is severely impaired development and differentiation of lymphoid cells in Stat5a/Stat5b conditional double knockout mice (101), which lack both STAT5 dimers and tetramers. These results indicate essential, nonredundant roles of STAT5 tetramers and dimers. STAT5 dimers are essential for viability, and in a setting where STAT5 dimers exist, the addition of STAT5 tetramers allows further enhancement of cellular development and function.
Figure 3.
Fine-tuning cytokine signals by STAT dimers versus tetramers. (a) The essential functions of STAT1 and STAT5 are illustrated by the severe defects observed in mice in which the genes encoding STATs are deleted. (b) STAT tetramers serve critical functions, but the effect of preventing STAT tetramerization is less severe than the phenotype observed with complete deletion of the STAT proteins, indicating that STAT dimers alone can mediate certain core functions. Abbreviations: NK, natural killer; Treg, regulatory T cell.
In contrast to the markedly compromised interferon responses in Stat1 knockout mice (102) (Figure 3a), STAT1 tetramers are vital for type II interferon (IFN-γ) and antibacteria activities but are dispensable for at least some type I interferon (IFN-α/β) activity (99) (Figure 3b). Thus, whereas mice completely lacking STAT proteins typically have profound phenotypes, the phenotypes observed with tetramerization-deficient STAT proteins, for both STAT5 proteins and STAT1, are more modest. These results indicate that STAT tetramers have distinctive roles from STAT dimers and that the formation of STAT tetramers is a way of fine-tuning cytokine signaling. Selectively targeting tetramers without affecting dimers may be a rational approach for modulating cytokine responses.
It is interesting that at least for STAT5, STAT tetramers preferentially bind to DNA motifs that are separated by either a full helical turn or approximately 0.5 or 1.5 helical turns (97, 103). Whether there are differences in the signal mediated by STAT5 tetramers—either qualitative or quantitative—depending on the inter–GAS motif spacing is unknown, but this conceivably might represent another mode by which the potency or the nature of the tetramer signal can be fine-tuned.
Possible control of cytokine signals by unphosphorylated STAT proteins.
STAT proteins offer additional complexities, with possible roles for unphosphorylated STAT proteins related to influencing cytokine signals. For example, following interferon stimulation, STAT1 is tyrosine phosphorylated and translocates to the nucleus, but then the amount of unphosphorylated STAT1 increases as STAT1 is then dephosphorylated; and nuclear unphosphorylated STAT1 was reported to be complexed with unphosphorylated STAT2 and IRF9 and to serve to sustain the expression levels of interferon-induced genes (104, 105). Furthermore, unphosphorylated STAT5 proteins were detected in primary hematopoietic stem/progenitor cells colocalized with CTCF, potentially serving to suppress hematopoietic differentiation (106). Intriguingly, thrombopoietin-mediated phosphorylation of STAT5 not only induces its own gene expression program but also results in the loss of unphosphorylated STAT5, and it was proposed that the effect of thrombopoietin is influenced by the level of phosphorylated versus unphosphorylated STAT5 (106). Overall, these findings suggest that unphosphorylated STAT1 and STAT5 may provide additional mechanisms for fine-tuning the actions of cytokines.
Potential Roles of Mitochondrial STATs in Fine-Tuning Cytokine Actions
In addition to their critical roles in the regulation of expression of cytokine and growth factor target genes in the nucleus, some STAT proteins, including STAT1, STAT2, STAT3, STAT5, and STAT6, have also been detected in mitochondria (reviewed in 107, 108). The function of mitochondrial STAT proteins may vary, with STAT1 contributing to limiting mitochondrial biogenesis (107) and both STAT1 and STAT2 contributing to inhibition of mitochondrial RNA expression (107). Mitochondrial STAT3 has been reported to be involved in a number of metabolic and mitochondrial functions, including the regulation of cellular respiration, mitochondrial membrane potential, mitochondrial ROS production, ATP production, inhibition of mitochondrial permeability transition pore opening, protection against ischemia/reperfusion and cardiac injury, and TNF-induced necroptosis (reviewed in 107). Interestingly, serine phosphorylation of STAT3 on S727, but not tyrosine phosphorylation of STAT3, is required for STAT3 binding to mitochondrial protein GRIM-19 (genes associated with retinoid-interferon-induced mortality 19), a component of the electron transport chain (ETC), and their cotranslocation from cytosol to mitochondria and thus for their function (109–111). ETC activity is decreased in the absence of STAT3 and can be restored when STAT3 is expressed in mitochondria (108). Mitochondrial STAT3 localizes to the inner mitochondrial membrane and matrix and is required for optimal activity of the ETC (112) and for Ras-mediated oncogenic transformation by maintaining the glycolytic and oxidative phosphorylation levels in tumor cells (113). Consistent with these findings, targeting mitochondrial STAT3 can inhibit cancer cell growth (114, 115). In a colorectal cancer model, targeting STAT3 in intestinal epithelial cells can cause enhanced mitophagy, which leads to the uptake of complete antigen-MHC complexes by dendritic cells and their presentation to and activation of CD8+ T cells, a process that can promote antitumor immunity and host defense to viruses (116). In contrast to mitochondrial STAT3, much less is known about the importance of mitochondrial STAT5 and STAT6. The involvement of mitochondrial STAT proteins in cytokine actions, especially during the immune response, warrants further investigation, and this might provide additional means for fine-tuning cytokine actions.
Fine-Tuning of Cytokine Signals by Superenhancers
Superenhancers, also known as stretch enhancers, are regulatory elements that are composed of the extended regions with highly enriched binding of cohesin, Mediator, p300, CBP, and other factors associated with enhancer activity (117–119). Superenhancers are often associated with master regulator genes, oncogenes, and genes associated with cell identity, as well as locus control regions (119, 120). In addition, superenhancers have been associated with cytokine-induced and disease-associated genes (119,121–123). Importantly, when superenhancers were ranked based on STAT-binding intensity, there was a strong correlation between genes containing the most highly ranked STAT5-bound and STAT3-bound superenhancers and their inducibility by IL-2 and IL-21, respectively (122). Interestingly, the most highly ranked IL-2/STAT5-based superenhancer in mouse CD4+ and CD8+ T cells was located in the Il2ra gene, which encodes IL-2Rα (122), with similar high inducibility of the corresponding human IL2RA gene (122). Moreover, the Il2ra gene was also highly ranked based on p300-binding intensity under Th1, Th2, and Th17 conditions (121). Deletion of an autoimmune-associated enhancer element in the Il2ra gene by CRISPR-Cas9 only partially affected IL-2Rα expression in effector T cells, with a delayed T cell receptor– induced response (123). There was not a decrease in Tregs, but Tregs lacking the element had decreased viability at low concentrations of IL-2, and naive CD4+ T cells were more skewed toward proinflammatory Th17 differentiation than Treg differentiation (123). Importantly, although the mouse Il2ra superenhancer region had 13 principal STAT5-binding elements, deletions of three separate elements each substantially lowered Il2ra gene expression, indicating that the elements were not functionally redundant and allowing dissection of the elements of this superenhancer (122). Modulating IL-2Rα levels thus provides a means of tuning IL-2 responsiveness, and superenhancers thus provide a mechanism for sustaining and/or fine-tuning cellular responsiveness to this cytokine. In addition to the Il2ra superenhancer, IL-2/STAT5 superenhancers were identified in a range of other genes, including genes encoding other cytokines (e.g., Lta, Lif, and Osm); cell cycle–related genes (e.g., Cdk6); and a negative regulator of cytokine signaling, Cish (122). Thus, cytokine-induced STAT5-based superenhancers represent a mechanism for broadly affecting cytokine responsiveness, including both enhancing (e.g., by modulating Il2ra expression) and limiting (e.g., by modulating Cish expression) cytokine signaling. Together with competition among cytokines and STATs and STAT tetramerization, superenhancer-mediated regulation of key genes is another physiological mechanism for influencing and fine-tuning cytokine responses.
Pharmacological and/or Therapeutic Tuning of Cytokine Signals
Above, we have considered physiological mechanisms of tuning cytokine signals, including by the differential use of STAT proteins, JAK/STAT polymorphisms, STAT tetramerization, and superenhancers. However, there are a range of approaches for pharmacologically and potentially therapeutically fine-tuning cytokine signals.
Targeting the JAK-STAT pathway with JAK inhibitors.
As discussed above, the JAK-STAT pathway plays essential and critical roles in mediating cytokine signals and a range of cellular actions. Because there are only four JAKs and a large number of cytokines that use JAK-STAT signaling, JAK inhibitors cannot specifically inhibit a single cytokine, but they still exhibit partial specificity, affecting some cytokines but not others. In particular, JAK3 is used only by the six γc family cytokines (48), and the discovery of JAK3-deficient SCID led to the speculation that JAK3-specific inhibitors would be immunosuppressive (20), enhancing interest in the development of such agents. Indeed, tofacitinib (124) was approved by the US Food and Drug Administration (FDA) in 2012 for the treatment of patients with rheumatoid arthritis who do not respond to methotrexate and in 2018 for the treatment of adult patients with moderate to severe ulcerative colitis, and it is also in clinical trials for the treatment of patients with other inflammatory disorders, including psoriasis and juvenile idiopathic arthritis (125, 126). IL-21 has a clear association with autoimmune disease mouse models of type 1 diabetes, systemic lupus erythematosus, and uveitis (127–129) as well as with a range of human infectious, inflammatory, and autoimmune diseases based on genome-wide association studies (130, 131); thus, the effectiveness of JAK3 inhibition is likely due, at least in part, to its ability to inhibit the actions of IL-21.
Tofacitinib was developed as a JAK3 inhibitor, but it also inhibits JAK1 and JAK2 to some degree (124). Accordingly, there is considerable interest in more-selective JAK inhibitors. Recently, highly selective JAK3 inhibitors were identified that bind to JAK3 Cys909 near the ATP-binding site, a residue that is absent in other JAKs (132–134). Studies with the JAK3-specific inhibitor, JAK3i, indicated that there are two waves of STAT5 activation by CD4+ T cells in response to IL-2 and that JAK3i preferentially inhibits the second wave, which correlates with its inhibition of cyclin expression and cell cycle progression (135).As such,JAK3i is not only selective for γc family cytokines, which are the only cytokines that use JAK3, but it may also be somewhat selective with regard to part of the IL-2 signal. Another JAK3-specific inhibitor, PF-06651600, is a covalent irreversible inhibitor that binds to Cys909, and it also selectively inhibits γc family cytokine–induced STAT activation, without affecting cytokine-induced STAT activation via JAK1, JAK2, and TYK2 (136, 137). Although it is not yet clear whether selective JAK3 inhibitors are superior to tofacitinib from a clinical perspective, these agents may allow more precise fine-tuning and selective targeting of γc-cytokine-specific pathways.
Beyond autoimmune diseases, JAK inhibitors can be potentially therapeutic in cancer, given that constitutively activated JAK-STAT pathways are associated with certain malignancies. This was first observed in the context of human T cell lymphotropic virus I (HTLV-I) (138), v-Abl (139), and the Src oncogene (140). In HTLV-I-induced adult T cell leukemia, the transition from the cytokine-dependent proliferation to cytokine-independent proliferation has been correlated with the acquisition of an activated JAK-STAT pathway (138,141).Gain-of-function mutations in the JAK-STAT pathway can result in excessive inflammation and/or malignancies (142), and targeting JAK-STAT signaling can be therapeutic (125,142,143).In the chronic/smoldering form of adult T cell leukemia, the activated JAK-STAT pathway is driven by cytokines, and JAK inhibitors diminish this proliferation in vitro (142), suggesting that this may represent a rational therapeutic approach.
Besides the JAK3 inhibitors noted above, there are a range of inhibitors of other JAKs (144). Ruxolitinib preferentially inhibits JAK1 and JAK2 and was approved by the FDA for the treatment of patients with intermediate- or high-risk myelofibrosis (145) and polycythemia vera (146), myeloproliferative diseases associated with gain-of-function mutations in JAK2. Relatively specific JAK2 inhibitors are also being developed and evaluated to better restrict the pathways affected, such as CHZ868, which is a type II JAK2 inhibitor (147). It will be interesting to determine whether more specific inhibition of JAK2, analogous to JAK3 inhibitors, results in better fine-tuning of cytokine responses and whether this offers therapeutic advantages over ruxolitinib. In addition to ruxolitinib, another JAK inhibitor with specificity for JAK1 and JAK2, baricitinib, was approved by the FDA in 2018 for the treatment of adults with moderately to severely active rheumatoid arthritis who do not sufficiently respond to or cannot tolerate methotrexate. More selective JAK1 inhibitors, filgotinib and upadacitinib (ABT-494), are also in various phases of clinical trials for rheumatoid arthritis, Crohn disease, ulcerative colitis, atopic dermatitis, and moderate to severe plaque psoriasis (125, 148).
Tuning the JAK-STAT pathway with STAT inhibitors.
Inhibiting STAT proteins is another mechanism for controlling cytokine signaling. With only seven STATs and many cytokines, inhibition of a STAT will not be cytokine specific, but inhibiting a STAT potentially has the ability to fine-tune the signal from a particular cytokine (e.g., inhibiting the STAT1 or the STAT3 component of IL-21 signaling while leaving the others intact). Moreover, gain-of-function mutations in STAT proteins, particularly in STAT3 and STAT5B, have been reported in patients with both solid and hematopoietic malignancies (149). Most of the somatic mutations in STAT3 and STAT5B found in patients with hematopoietic malignancies occur in the SH2 domains (150–154),making the mutant STAT proteins more resistant to dephosphorylation and thus resulting in sustained STAT activation. Identifying molecules that block STAT dimer formation by targeting the SH2 domain could potentially be effective in controlling the STAT and could be of therapeutic interest as well (155, 156). For STAT3, a range of different types of inhibitors have been developed, including an antisense oligonucleotide targeting the STAT3 3’ untranslated region (AZD9150), a cyclic STAT3 decoy oligonucleotide (157), and small-molecule inhibitors (C188–9, OPB-31121, and OPB-51602) that block STAT3 dimer formation by either inhibiting tyrosine phosphorylation or binding to the SH2 domains. In addition to STAT3 inhibitors, there now are also a range of STAT5 inhibitors under evaluation (158–161). Such agents offer potential for degrees of specificity in regulating signaling by a range of different cytokines, with associated tuning of the biological response.
Therapeutic approaches for CRISPR-Cas9 as an approach to correct immunodeficiencies that are diseases of defective cytokine signaling.
A range of primary immunodeficiency diseases result from mutations or deletions in genes encoding cytokines, cytokine receptors, or signaling molecules, including IL2RA, IL2RG, IL7R, IL21, IL21R, JAK3, STAT1, STAT3, and STAT5B (19–22, 162–166). Accordingly, these are diseases of defective cytokine signaling, and the nature of disease corresponds to the particular pathways that are disrupted. For patients with well-matched donors, allogeneic hematopoietic stem cell transplantation resultsin survival rates of approximately 90% (167–169), but for those lacking such donors, gene therapy can be effective (170–173). Although retroviral-insertion-mediated T cell leukemia occurred in several patients with XSCID (174, 175), newer gene therapy approaches such as lentiviral vectors or self-inactivating retroviral vectors driven by the human elongation factor (EFα1) promoter without viral enhancers can reconstitute T cell numbers in XSCID patients (176).
CRISPR-Cas9 genome-editing technology is an approach to potentially precisely correct the mutation without the risk of gene insertion and mutation (177), or it can be used to introduce a normal cDNA into the gene locus. Considerable advances have been made with this approach, e.g., for correcting the gene defect in sickle cell disease (178), with work for XSCID and JAK3 deficiency underway (179). Select diseases including XSCID, JAK3-deficient SCID, and IL7Rdeficient SCID are ideal models for gene therapy in that corrected cells have a selective growth and survival advantage. Whereas correcting or replacing the defective gene to augment signaling is not a mechanism of fine-tuning per se, it provides the basic machinery for cytokine signaling that previously was absent in the affected patients. Interestingly, hypomorphic immunodeficiencies can exist, as for example was identified related to XSCID. One family had a moderate form of X-linked combined immunodeficiency (XCID) wherein T cells develop but their function is not normal (180, 181). Like XSCID, XCID results from a mutation in γc (182, 183), but signaling and immune function are diminished rather than abrogated. XCID can be viewed as a naturally occurring type of partial agonism (discussed below) and is consistent with the idea that function might be diminished for potential therapeutic benefit without being completely eliminated.
Strategies to improve chimeric antigen receptors by borrowing JAK-STAT protein docking sites from cytokine receptors.
CD19 chimeric antigen receptor (CAR)-T cells are effective in treating hematologic/immunologic malignancies, like B cell lymphoma, but they are less effective for solid organ tumors (184). To improve its anti–solid organ tumor activity, researchers have developed a novel CAR construct [28-IL2RB-z(YXXQ)] that contains the CD28 costimulatory domain, a truncated IL-2Rβ cytoplasmic domain, the TCR ζ chain cytoplasmic domain, and a STAT3 docking site (YXXQ) derived from IL-21R (185). STAT3 and STAT5 are activated in 28-IL2RB-z(YXXQ) CAR-T cells in response to antigen stimulation, and the cells show potent proliferation without significant terminal differentiation. Interestingly, the gene expression signature in these CAR-T cells resembles those seen in IL-21-treated cells. As a result, the 28-ΔIL2RB-z(YXXQ) CAR-T cells show persistent and markedly better in vivo antihematologic and anti–solid organ tumor activity and represent a way of enhancing and expanding what is in part a cytokine-related signal without cytokine stimulation.
Strategies for targeted therapies by modulating cytokine responses: therapy with IL-2 and IL-2–anti-IL-2 conjugates.
The early findings that IL-2 can potently expand effector T cells and enhance NK cell cytotoxicity in vitro led to studies that showed that administration of high-dose IL-2 could inhibit the growth in mice of established pulmonary metastases from B16 melanoma and subcutaneous MCA-105 sarcoma (186). Indeed, infusion of high-dose recombinant human IL-2 was the first cytokine therapy to achieve durable cancer regression, with some patients achieving long-term complete remissions (187, 188), and IL-2 was approved for treatment of patients with metastatic renal cancer and melanoma by the FDA in 1992 and 1998, respectively. Although effective in some patients, IL-2 therapy can have severe side effects, including capillary leak syndrome (188). IL-2’s ability to potently expand T cells that retain responsiveness also led to the development of adoptive cell therapy (ACT) (189,190). ACT has been performed with autologous tumor-infiltrating lymphocytes, with patient T cells that are genetically engineered with chimeric antigen receptors (184, 188, 191), and with tumor-reacting autologous T cells from patients that were expanded in vitro using IL-2 (188, 192).
Tregs maintain self-tolerance and suppress excessive immune responses to protect the host from harm (193, 194). In addition to its ability to potently expand effector T cells, IL-2 is essential for the normal development and maintenance of Treg numbers (195), with IL-2 acting via the high-affinity IL-2 receptors that are constitutively expressed on these cells (196, 197). Accordingly, Tregs can respond to an approximately tenfold-lower dose of IL-2 than is needed to expand effector or memory T cells (198), which allows the preferential in vivo expansion of Tregs by low-dose IL-2. Administration of low-dose IL-2 can expand Tregs in patients with chronic graft-versus-host disease who have dysfunctional Tregs and are refractory to glucocorticoid therapy (199, 200) as well as in patients with autoimmune disorders including type 1 diabetes (198, 201, 202) and systemic lupus erythematosus (203, 204). Although IL-2 has a very short half-life in vivo (188), complexing anti-IL-2 antibodies to IL-2 can prolong IL-2’s half-life and enhance its potency, both in vitro and in vivo (205, 206). Interestingly, anti-IL-2 monoclonal antibody JES6–1 when complexed with IL-2 mimics low-dose IL-2 and preferentially expands Tregs, whereas anti-IL-2 S4B6 monoclonal antibody complexed with IL-2 mimics high-dose IL-2 and potently expands effector T cells (206). Although both monoclonal antibodies extend the half-life of IL-2 in vivo, they differentially affect the affinity of ligand-receptor subunit interactions by binding to different epitopes of IL-2 (207). When complexed to IL-2, JES6–1 monoclonal antibody sterically blocks the interaction of IL-2 with IL-2Rβ and γc, thereby lowering the binding of IL-2 to IL-2Rα on IL-2Rαlow effector cells but favoring IL-2Rαhigh Tregs. In contrast, S4B6 monoclonal antibody sterically blocks the interaction of IL-2 with IL-2Rα, augmenting the affinity and stability of the IL-2:IL-2Rβ interaction and potently expanding IL-2Rβhigh effector T cells (207). Thus, using low-dose versus high-dose IL-2 and using IL-2 combined with JES6–1 versus S4B6 monoclonal antibodies are approaches for differentially controlling the effect of IL-2 on Tregs versus T effector cells. Similar to anti-mouse IL-2 monoclonal antibody JES6–1, a fully human anti-IL-2 antibody (F5111.2) has been identified, and when complexed with IL-2, it preferentially activates STAT5 in Tregs and expands Tregs in vivo (208). Importantly, F5111.2–IL-2 complexes can induce remission of type 1 diabetes in a mouse model, reduce the severity of experimental autoimmune encephalomyelitis, and protect mice against xenogeneic graft-versus-host disease (208).
Soluble IL-15–IL-15Rα complexes have also been developed and can potently and selectively expand memory CD8+ T cells, whereas soluble anti-IL-2Rα suppresses IL-2-induced expansion of memory CD8+ T cells (209).Moreover, a single-chain IL-15–IL-15Rα fusion protein, denoted RLI (receptor linker IL-15) (210, 211), acts as a superagonist, with more sustained STAT5 activation compared with IL-15.RLI preferentially expands memory phenotype CD8+CD44high T cells but not CD4+ T cells or Tregs in vivo (212). Interestingly, in a mouse tumor model, RLI exhibits antitumor activity when administered early but is not effective unless combined with anti-PD-L1 when administered late, due to its inability by itself to expand exhausted CD8+ T cells (212).
Th2 cytokines, including IL-4, IL-5, IL-13, and thymic stromal lymphopoietin (TSLP), play major roles in allergic diseases, such as atopic dermatitis and asthma (213, 214). IL-4Rα is a shared signaling component of IL-4 and IL-13 receptors, and either a mutant IL-4 or a blocking IL-4Rα monoclonal antibody can markedly suppress IL-4 and IL-13 responses (215, 216). A number of monoclonal antibodies to either Th2 cytokines or their receptors have been developed, some of which are effective for treating patients with allergic disorders. For example, the human anti-IL-4Rα monoclonal antibody, dupilumab, is effective in treating patients with persistent asthma (217, 218) and is approved by the FDA for the treatment of adults with moderate to severe atopic dermatitis (219). Anti-IL-5 (mepolizumab and reslizumab) and antiIL-5Rα (benralizumab) monoclonal antibodies are also approved by the FDA for the treatment of patients with severe asthma that is associated with an eosinophilic phenotype (220).
Fine-tuning by super-IL-2, IL-2–anti-IL-2 conjugates, and IL-2 partial agonists.
As noted above, cytokines signal by inducing dimerization (either homodimerization or heterodimerization) of the receptor, which results in activation of JAKs and other signaling pathways. In the case of IL-2, there are three receptor components, IL-2Rα, IL-2Rβ, and IL-2Rγ (IL-2Rγ is also known as the common cytokine receptor γ chain, γc). These three proteins in different combinations form three classes of IL-2 receptors: The low-affinity IL-2 receptors (Kd ∼ 10−8 M) consist solely of IL-2Rα, intermediate-affinity receptors (Kd ∼ 10−9 M) contain IL-2Rβ+γc, and high-affinity receptors (Kd ∼ 10−11 M) contain IL-2Rα+ IL-2Rβ+γc. Some resting lymphocytes—particularly NK cells and CD8+ T cells—constitutively express intermediate-affinity receptors, but after antigen-mediated activation, IL-2Rα is potently induced, resulting in high-affinity receptors; because IL-2Rα is expressed in excess of IL-2Rβ and γc, activated T cells express low-affinity receptors as well. Interestingly, Tregs constitutively express all three IL-2 receptor chains and thus also express high-affinity receptors. The functional receptors are the intermediate- and high-affinity receptors; these forms of receptors both contain IL-2Rβ and γc, and heterodimerization of their cytoplasmic domains results in signaling.
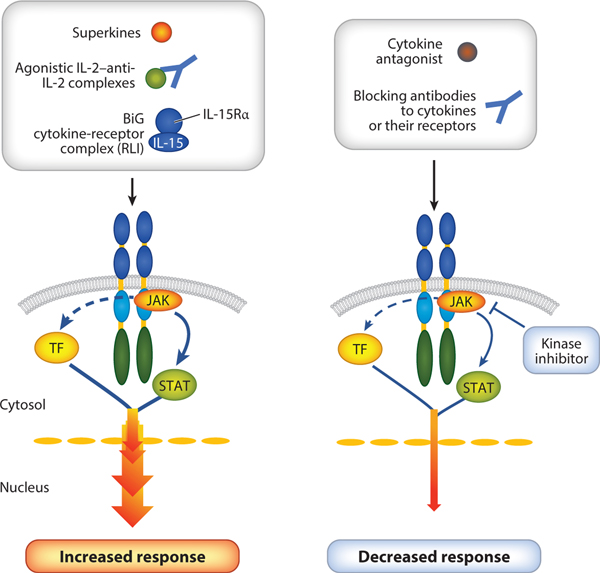
The formation of the high-affinity IL-2–IL-2 receptor quaternary complex is kinetically driven by the binding of IL-2. IL-2 first interacts with IL-2Rα, which has rapid on and off rates (221). This interaction results in a conformational change in IL-2, which then allows IL-2 to efficiently interact with IL-2Rβ, and γc is then recruited to the complex (222). An IL-2 superkine, also denoted as super-IL-2 or H9, was identified based on a high-throughput protein-engineering approach using yeast display in which a library of IL-2 mutants was conjugated to the A-agglutinin-binding subunit to allow stable and soluble expression of proteins on yeast (223).H9 has mutations in IL-2 that obviate the need for its interaction with IL-2Rα in order to achieve the conformation that allows efficient interaction with IL-2Rβ (223). As a result, H9 exhibits a 200-fold-higher binding affinity to IL-2Rβ in the absence of IL-2Rα than is observed with wild-type IL-2, with correspondingly greater effects at lower concentrations of protein. In part for this reason, H9 has lower toxicity, mediating diminished vascular leak syndrome and thus reduced pulmonary edema (223).
Besides its augmented binding and activity, the importance of H9 stems in part from its representing a backbone on which additional mutations can be made. Indeed, a range of such novel partial agonist molecules has been created. By exhibiting markedly enhanced binding (i.e., superbinding) to IL-2Rβ, such molecules can outcompete and effectively block the actions of endogenous IL-2 and confer to the system their own activities, serving in part as receptor signaling clamps (224) (Figure 4).Actions of H9-based partial agonists can range from H9 super-IL-2 itself, which functions as a potent agonist, to molecules such as H9-RETR, which contains mutations in four amino acids, essentially abrogating its ability to interact with and recruit γc (224). As a result, no IL-2Rβ-γc heterodimerization occurs, and signaling is therefore defective. Indeed,H9-RETR is a potent inhibitor of IL-2, and because IL-2Rβ is also a key component of the IL-15 receptor, H9-RETR can block the actions of both IL-2 and IL-15, including for example their effects on increasing T cell proliferation and boosting NK cytolytic activity. Moreover, H9-RETR inhibits the development of graft-versus-host disease in an animal model as well as the spontaneous proliferation of malignant T cells from patients with the chronic-smoldering forms of HTLV-I-induced adult T cell leukemia (224). Interestingly, however, there are molecules between super-IL-2 and H9-RETR that might function as immunomodulatory partial agonists. For example, it was observed that H9-RET, a molecule with three of the four mutations present in H9-RETR, had defective recruitment of γc; interestingly, like H9-RETR, H9-RET was incapable of driving the proliferation of freshly isolated CD8+ T cells, but in contrast to H9-RETR it could drive the proliferation of freshly isolated preactivated CD8+ T cells (224). In principle, it is possible to generate a range of molecules with varied abilities to recruit γc. Such molecules might include ones that affect some but not all IL-2-activated signaling pathways or that differentially affect different lineages of lymphocytes, potentially providing novel research molecules that might lead to new therapeutics. The basic concept of partial agonism is relatively new for type I cytokines but is a well-established principle in the context of drugs for targeting G protein–coupled receptors or channels (225). Whereas the H9 superagonist exhibits maximum activity at a lower concentration than is observed with wild-type IL-2, the partial agonists induce submaximal effects.
Figure 4.
Rationally fine-tuning cytokine signals. Cytokine signals can be fine-tuned by changing a few amino acids of the cytokine to either promote better binding (superagonist) or diminish the binding (antagonist) of a receptor chain. Fine-tuning can also be achieved by the use of a ligand-receptor complex (e.g., BiG, which is an IL-15–IL-15Rαfusion protein) or antibodies to either cytokines or cytokine receptors. Orthogonal cytokine-cytokine receptor systems can be used to selectively expand a given set of cells. Abbreviations: RLI, receptor linker IL-15; TF, transcription factor.
In a study related to IL-15,a molecule denoted BiG was generated by covalently linking mutant IL-15 Q108D with IL-15Rα (226) (Figure 4); BiG exhibits increased affinity for IL-2Rβ due to its slower off rate (211), but there is impaired recruitment of γc due to the Q108D mutation in IL-15 (227). Like H9-RETR, BiG efficiently inhibited the actions of IL-2 and IL-15 on primary CD8+ T and NK cells, in vitro and in vivo. Interestingly, however, BiG had no impact on IL2-induced Treg proliferation, possibly because it does not interfere with the binding of IL-2 to IL-2Rα, whereas H9-RETR potently inhibited the differentiation of Th1 cells, Th2 cells, and induced Tregs and additionally inhibited IL-2-induced STAT5 phosphorylation of induced Tregs (224).
The approaches outlined above for generating biologically interesting cytokine partial agonists can be applied to other cytokine systems as well. SCF binds to dimers of c-Kit (the receptor for SCF) to promote hematopoietic progenitor cell expansion; it also can expand and activate mast cells, resulting in anaphylaxis (228, 229). A two-step strategy was used to generate a SCF partial agonist that exhibits markedly increased binding affinity for c-Kit monomers but greatly decreased binding affinity for c-Kit dimers (230). This SCF partial agonist favors the expansion of hematopoietic progenitor cells over the activation of mast cells in in vitro and in vivo mouse models (230). Thus, rationally engineered cytokine partial agonists can be valuable tools for finetuning the actions of a pleiotropic cytokine and can favor beneficial over unwanted activities.
Moreover, in a patient with severe anemia, a naturally occurring mutation in erythropoietin (EPO), EPO R150Q, exhibited diminished activation of JAK2 (231). Surprisingly, phosphorylation of STAT5 was only minimally affected, but phosphorylation of JAK2, STAT1, and STAT3 induced by EPO R150Q was impaired, an effect that was phenocopied when ruxolitinib was added or when a more specific JAK2 inhibitor was used (231). EPO R150Q provides an example of a naturally occurring cytokine partial agonist, where a single amino acid change in the cytokine sufficiently perturbs the efficiency of EPO-mediated homodimerization of its receptor, resulting in altered cytosolic JAK2 activity, with altered activation of STAT5 versus STAT1 and STAT3. Such observations underscore the concept that artificial molecules with a range of interesting activities can be generated.
Fine-tuning by using novel engineered cytokine-cytokine receptor systems.
As noted above, dimerization of receptors is critical for signaling by type I cytokines. For IL-2 signaling, the importance of receptor heterodimerization was demonstrated by studies using chimeric receptors (232, 233). When the extracellular domains of IL-2Rβ and γc were replaced by the extracellular domain of IL-2Rα, signaling could be induced by an anti-IL-2Rα monoclonal antibody (particularly when cross-linked with a secondary antibody), and when the extracellular domain of γc was replaced by that of IL-2Rα but IL-2Rβ was left intact, signaling could be triggered by an engineered bispecific antibody with one Fab specific for IL-2Rα and the other for IL-2Rβ (232). Similarly, when the extracellular domains of both IL-2Rβ and γc were replaced by the extracellular domain of c-Kit, signaling could be induced by SCF, and when the extracellular domains of IL-2Rβ and γc were replaced by the extracellular domains of granulocyte-macrophage CSF receptor α chain (GM-CSFRα) and GM-CSFRβ, respectively, signaling could be induced by stimulation with GM-CSF (233). These results indicated that the cytoplasmic signaling machinery of IL-2Rβ and γc can be artificially activated to provide an “IL-2” signal even in the absence of IL-2 itself. Indeed, this underlying principle corresponds to the creation of a novel orthogonal system wherein the extracellular domain of IL-2Rβ was mutated based on the known structure (222) to create orthogonal IL-2Rβ (orthoIL-2Rβ) so that it could no longer bind native IL-2, and IL-2 was mutated and selected by phage display to create orthogonal IL-2 (orthoIL-2) that no longer bound native IL-2Rβ but could bind orthoIL-2Rβ (234). orthoIL-2 could then be used to selectively trigger cells into which the orthoIL-2Rβ had been inserted. In such fashion, cells of interest could be specifically targeted, with the idea of minimizing off-target effects of IL-2. Such an approach could lower toxicity, as only engineered cells can respond to orthoIL-2. Interestingly, more than one orthoIL-2 variant were created, with differential sensitivity of each variant to the presence of IL-2Rα (234). Importantly, when orthoIL-2 was used with orthoIL-2Rβ-transduced pmel-1 transgenic T cells, there indeed was effective antitumor activity against B16 melanoma in a mouse model (234).
CONCLUSIONS
A tremendous amount of knowledge about cytokine biology and cytokine signaling mechanisms has been learned from basic research employing cell culture–based research, from animal models, and from the phenotypes of patients with mutations in cytokines, their receptors, or in signaling proteins. This information has in turn been used to advance clinical applications, with the development of new therapeutic advances for treating patients with immunodeficiency, allergy, autoimmunity, and cancer. Some of the mutations in genes affecting cytokines, their receptors, or signaling proteins can cause immunodeficiency or malignancies, underscoring the clinical importance of these molecules, and successful treatment with high-dose IL-2 of patients with metastatic melanoma has indicated that cytokine-based therapies can be clinically effective. This has led to the development of improved strategies to treat immunodeficiencies, allergy, autoimmunity, and cancer. Monoclonal antibodies and antagonists to cytokines and their receptors, as well as inhibitors of JAKs and STAT proteins, can be effective in inhibiting cytokine responses. Because of high homology among JAKs and among STAT proteins, finding specific inhibitors for these signaling molecules has been challenging. The recent exciting developments in identifying selective JAK3 inhibitors and generating various superagonists, partial agonists, and antagonists for IL-2 allow one to rationally fine-tune cytokine signals. Moreover, other protein-engineering approaches such as orthogonal cytokine-cytokine receptors might offer distinctive strategies for selectively expanding tumor-specific T cells. Finally, the rapid advances in CRISPR-Cas9 and other gene-editing approaches will likely allow advances in the goal to cure Mendelian diseases such as XSCID. These new strategies have considerable promise for fine-tuning cytokine signals, with better manipulation of the immune system in health and disease.
ACKNOWLEDGMENTS
This work was supported by the Division of Intramural Research; National Heart, Lung, and Blood Institute; National Institutes of Health, Bethesda, MD, USA. We thank Dr. Rosanne Spolski for critical comments.
DISCLOSURE STATEMENT
W.J.L. is an inventor on patents/patent applications related to IL-2, IL-21, and IL-2 partial agonists.
LITERATURE CITED
- 1.Cohen S, Bigazzi PE, Yoshida T. 1974. Similarities of T cell function in cell-mediated immunity and antibody production. Cell Immunol. 12:150–59 [DOI] [PubMed] [Google Scholar]
- 2.Leonard WJ. 2013. Type I cytokines and interferons, and their receptors. In Fundamental Immunology, ed. Paul WE, pp. 601–38. Philadelphia, PA: Wolters Kluwer Lippincott Williams Wilkins [Google Scholar]
- 3.Milburn MV, Hassell AM, Lambert MH, Jordan SR, Proudfoot AE, et al. 1993. A novel dimer configuration revealed by the crystal structure at 2.4 A resolution of human interleukin-5. Nature 363:172–76 [DOI] [PubMed] [Google Scholar]
- 4.Tamada T, Honjo E, Maeda Y, Okamoto T, Ishibashi M, et al. 2006. Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex. PNAS 103:3135–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vignali DA, Kuchroo VK. 2012. IL-12 family cytokines: immunological playmakers. Nat. Immunol 13:722–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Lupardus P, Laporte SL, Garcia KC. 2009. Structural biology of shared cytokine receptors. Annu. Rev. Immunol 27:29–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Negishi H, Taniguchi T, Yanai H. 2017.The Interferon (IFN) class of cytokines and the IFN regulatory factor (IRF) transcription factor family. Cold Spring Harb. Perspect. Biol In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Wong K, Ouyang W, Rutz S. 2017. Targeting IL-10 family cytokines for the treatment of human diseases. Cold Spring Harb. Perspect. Biol In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudakov JA, Hanash AM, van den Brink MR. 2015. Interleukin-22: immunobiology and pathology. Annu. Rev. Immunol 33:747–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renauld JC. 2003. Class II cytokine receptors and their ligands: key antiviral and inflammatory modulators. Nat. Rev. Immunol 3:667–76 [DOI] [PubMed] [Google Scholar]
- 11.Dinarello CA. 2018. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol. Rev 281:8–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croft M, Siegel RM. 2017. Beyond TNF: TNF superfamily cytokines as targets for the treatment of rheumatic diseases. Nat. Rev. Rheumatol 13:217–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwakura Y, Ishigame H, Saijo S, Nakae S. 2011. Functional specialization of interleukin-17 family members. Immunity 34:149–62 [DOI] [PubMed] [Google Scholar]
- 14.Monin L, Gaffen SL. 2018. Interleukin 17 family cytokines: signaling mechanisms, biological activities, and therapeutic implications. Cold Spring Harb. Perspect. Biol 10:a028522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallach D. 2017. The tumor necrosis factor family: family conventions and private idiosyncrasies. Cold Spring Harb. Perspect. Biol In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin JX, Mietz J, Modi WS, John S, Leonard WJ. 1996. Cloning of human Stat5B: reconstitution of interleukin-2-induced Stat5A and Stat5B DNA binding activity in COS-7cells. J. Biol. Chem 271:10738–44 [PubMed] [Google Scholar]
- 17.Leonard WJ, O’Shea JJ. 1998. Jaks and STATs: biological implications. Annu. Rev. Immunol 16:293–322 [DOI] [PubMed] [Google Scholar]
- 18.Levy DE, Darnell JE Jr. 2002. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol 3:651–62 [DOI] [PubMed] [Google Scholar]
- 19.Noguchi M, Yi H, Rosenblatt HM, Filipovich AH, Adelstein S, et al. 1993. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell 73:147–57 [DOI] [PubMed] [Google Scholar]
- 20.Russell SM, Tayebi N, Nakajima H, Riedy MC, Roberts JL, et al. 1995. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science 270:797–800 [DOI] [PubMed] [Google Scholar]
- 21.Macchi P, Villa A, Giliani S, Sacco MG, Frattini A, et al. 1995. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID). Nature 377:65–68 [DOI] [PubMed] [Google Scholar]
- 22.Puel A, Ziegler SF, Buckley RH, Leonard WJ. 1998. Defective IL7R expression in T−B+NK+ severe combined immunodeficiency. Nat. Genet 20:394–97 [DOI] [PubMed] [Google Scholar]
- 23.Roifman CM, Zhang J, Chitayat D, Sharfe N. 2000. A partial deficiency of interleukin-7Rαis sufficient to abrogate T-cell development and cause severe combined immunodeficiency. Blood 96:2803–7 [PubMed] [Google Scholar]
- 24.Giliani S, Mori L, de Saint Basile G, Le Deist F, Rodriguez-Perez C, et al. 2005. Interleukin-7 receptor alpha (IL-7Rα) deficiency: cellular and molecular bases; analysis of clinical, immunological, and molecular features in 16 novel patients. Immunol. Rev 203:110–26 [DOI] [PubMed] [Google Scholar]
- 25.Lin JX, Migone TS, Tsang M, Friedmann M, Weatherbee JA, et al. 1995. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity 2:331–39 [DOI] [PubMed] [Google Scholar]
- 26.Greenlund AC, Morales MO, Viviano BL, Yan H, Krolewski J, Schreiber RD. 1995. Stat recruitment by tyrosine-phosphorylated cytokine receptors: an ordered reversible affinity-driven process. Immunity 2:677–87 [DOI] [PubMed] [Google Scholar]
- 27.Weber-Nordt RM, Riley JK, Greenlund AC, Moore KW, Darnell JE, Schreiber RD. 1996. Stat3 recruitment by two distinct ligand-induced, tyrosine-phosphorylated docking sites in the interleukin-10 receptor intracellular domain. J. Biol. Chem 271:27954–61 [DOI] [PubMed] [Google Scholar]
- 28.Zeng R, Spolski R, Casas E, Zhu W, Levy DE, Leonard WJ. 2007. The molecular basis of IL-21mediated proliferation. Blood 109:4135–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franke A, Balschun T, Karlsen TH, Hedderich J, May S, et al. 2008. Replication of signals from recent studies of Crohn’s disease identifies previously unknown disease loci for ulcerative colitis. Nat. Genet 40:713–15 [DOI] [PubMed] [Google Scholar]
- 30.Int. Consort. Syst. Lupus Erythematosus Genet., Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, et al. 2008. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet 40:204–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson CA, Massey DC, Barrett JC, Prescott NJ, Tremelling M, et al. 2009. Investigation of Crohn’s disease risk loci in ulcerative colitis further defines their molecular relationship. Gastroenterology 136:523–29.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kilpivaara O, Mukherjee S, Schram AM, Wadleigh M, Mullally A, et al. 2009. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat. Genet 41:455–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee YH, Woo JH, Choi SJ, Ji JD, Song GG. 2010. Association between the rs7574865 polymorphismof STAT4 and rheumatoid arthritis: a meta-analysis. Rheumatol. Int 30:661–66 [DOI] [PubMed] [Google Scholar]
- 34.Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, et al. 2012. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet 44:1341–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura M, Nishida N, Kawashima M, Aiba Y, Tanaka A, et al. 2012. Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am. J. Hum. Genet 91:721–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellinghaus D, Ellinghaus E, Nair RP, Stuart PE, Esko T, et al. 2012. Combined analysis of genome-wide association studies for Crohn disease and psoriasis identifies seven shared susceptibility loci. Am. J. Hum. Genet 90:636–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirino Y, Bertsias G, Ishigatsubo Y, Mizuki N, Tugal-Tutkun I, et al. 2013. Genome-wide association analysis identifies new susceptibility loci for Behcet’s disease and epistasis between HLA-B*51 and ERAP1. Nat. Genet 45:202–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slattery ML, Lundgreen A, Hines LM, Torres-Mejia G, Wolff RK, et al. 2014. Genetic variation in the JAK/STAT/SOCS signaling pathway influences breast cancer-specific mortality through interaction with cigarette smoking and use of aspirin/NSAIDs: the Breast Cancer Health Disparities Study. Breast Cancer Res. Treat 147:145–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tapper W, Jones AV, Kralovics R, Harutyunyan AS, Zoi K, et al. 2015. Genetic variation at MECOM, TERT, JAK2 and HBS1L-MYB predisposes to myeloproliferative neoplasms. Nat. Commun 6:6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McIntosh LA, Marion MC, Sudman M, Comeau ME, Becker ML, et al. 2017.Genome-wide association meta-analysis reveals novel juvenile idiopathic arthritis susceptibility loci. Arthritis Rheumatol. 69:2222–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito N, Eto M, Nakamura E, Takahashi A, Tsukamoto T, et al. 2007. STAT3 polymorphism predicts interferon-alfa response in patients with metastatic renal cell carcinoma. J. Clin. Oncol 25:2785–91 [DOI] [PubMed] [Google Scholar]
- 42.Kreil S, Waghorn K, Ernst T, Chase A, White H, et al. 2010. A polymorphism associated with STAT3 expression and response of chronic myeloid leukemia to interferon alpha. Haematologica 95:148–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamana A, Balsa A, Rueda B, Ortiz AM, Nuno L, et al. 2012.The TT genotype of the STAT4 rs7574865 polymorphism is associated with high disease activity and disability in patients with early arthritis. PLOS ONE 7:e43661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conigliaro P, Ciccacci C, Politi C, Triggianese P, Rufini S, et al. 2017.Polymorphisms in STAT4, PTPN2, PSORS1C1 and TRAF3IP2 genes are associated with the response to TNF inhibitors in patients with rheumatoid arthritis. PLOS ONE 12:e0169956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dendrou CA, Cortes A, Shipman L, Evans HG, Attfield KE, et al. 2016. Resolving TYK2 locus genotype-to-phenotype differences in autoimmunity. Sci. Transl. Med 8:363ra149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swindell WR, Stuart PE, Sarkar MK, Voorhees JJ, Elder JT, et al. 2014. Cellular dissection of psoriasis for transcriptome analyses and the post-GWAS era. BMC Med. Genom 7:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, et al. 2011. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature 470:264–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin JX, Leonard WJ. 2017. The common cytokine receptor γ chain family of cytokines. Cold Spring Harb. Perspect. Biol 10:a028449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wan CK, Andraski AB, Spolski R, Li P, Kazemian M, et al. 2015. Opposing roles of STAT1 and STAT3 in IL-21 function in CD4+ T cells. PNAS 112:9394–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, et al. 2007. STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med 357:1608–19 [DOI] [PubMed] [Google Scholar]
- 51.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, et al. 2007. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 448:1058–62 [DOI] [PubMed] [Google Scholar]
- 52.Meyer zu Horste G, Przybylski D, Schramm MA, Wang C, Schnell A, et al. 2018.Fas promotes T helper17cell differentiation and inhibits Thelper 1 cell development by binding and sequestering transcription factor STAT1. Immunity 48:556–69.e7 [DOI] [PubMed] [Google Scholar]
- 53.Li P, Spolski R, Liao W, Wang L, Murphy TL, et al. 2012. BATF-JUN is critical for IRF4-mediatedtranscription in T cells. Nature 490:543–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, et al. 2012. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science 338:975–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim HP, Oh J, et al. 2009. Analysis of interleukin-21induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity 31:941–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ciofani M, Madar A, Galan C, Sellars M, Mace K, et al. 2012. A validated regulatory network for Th17 cell specification. Cell 151:289–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tussiw and R, Lee WL, Murphy TL, Mashayekhi M, Kc W, et al. 2012. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 490:502–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao W, Spolski R, Li P, Du N, West EE, et al. 2014. Opposing actions of IL-2 and IL-21 on Th9 differentiation correlate with their differential regulation of BCL6 expression. PNAS 111:3508–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. 2012. STAT5 is a potent negative regulator of TFH cell differentiation. J. Exp. Med 209:243–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oestreich KJ, Mohn SE, Weinmann AS. 2012. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat. Immunol 13:405–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manel N, Unutmaz D, Littman DR. 2008. The differentiation of human TH-17 cells requires transforming growth factor-βand induction of the nuclear receptor RORγt. Nat. Immunol 9:641–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, et al. 2007.IL-6 programs TH-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol 8:967–74 [DOI] [PubMed] [Google Scholar]
- 63.Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, et al. 2008. TGF-β-induced Foxp3 inhibits TH17 cell differentiation by antagonizing RORγt function. Nature 453:236–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Korn T, Bettelli E, Gao W, Awasthi A, Jager A, et al. 2007. IL-21 initiates an alternative pathway to induce proinflammatory TH17 cells. Nature 448:484–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, et al. 2007. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature 448:480–83 [DOI] [PubMed] [Google Scholar]
- 66.McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, et al. 2009. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat. Immunol 10:314–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, et al. 2007. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26:371–81 [DOI] [PubMed] [Google Scholar]
- 68.Liao W, Lin JX, Wang L, Li P, Leonard WJ. 2011. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat. Immunol 12:551–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, et al. 2011. T-bet represses TH17 differentiation by preventing Runx1-mediated activation of the gene encoding RORγt. Nat. Immunol 12:96–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dai H, He F, Tsokos GC, Kyttaris VC. 2017.IL-23 limits the production of IL-2 and promotes autoimmunity in lupus. J. Immunol 199:903–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aparicio-Siegmund S, Garbers C. 2015. The biology of interleukin-27 reveals unique pro- and anti-inflammatory functions in immunity. Cytokine Growth Factor Rev. 26:579–86 [DOI] [PubMed] [Google Scholar]
- 72.Hirahara K, Onodera A, Villarino AV, Bonelli M, Sciume G, et al. 2015. Asymmetric action of STAT transcription factors drives transcriptional outputs and cytokine specificity. Immunity 42:877–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robinson DS, O’Garra A. 2002. Further checkpoints in Th1 development. Immunity 16:755–58 [DOI] [PubMed] [Google Scholar]
- 74.Bettelli E, Korn T, Oukka M, Kuchroo VK. 2008. Induction and effector functions of TH17 cells. Nature 453:1051–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patel DD, Kuchroo VK. 2015. Th17 cell pathway in human immunity: lessons from genetics and therapeutic interventions. Immunity 43:1040–51 [DOI] [PubMed] [Google Scholar]
- 76.Langowski JL, Zhang X, Wu L, Mattson JD, Chen T, et al. 2006.IL-23 promotes tumour incidence and growth. Nature 442:461–65 [DOI] [PubMed] [Google Scholar]
- 77.Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, et al. 2012. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491:254–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yan J, Smyth MJ, Teng MWL. 2018. Interleukin (IL)-12 and IL-23 and their conflicting roles in cancer. Cold Spring Harb. Perspect. Biol 10:a028530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Teng MW, Bowman EP, McElwee JJ, Smyth MJ, Casanova JL, et al. 2015. IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat. Med 21:719–29 [DOI] [PubMed] [Google Scholar]
- 80.Fragoulis GE, Siebert S, McInnes IB. 2016. Therapeutic targeting of IL-17 and IL-23 cytokines in immune-mediated diseases. Annu. Rev. Med 67:337–53 [DOI] [PubMed] [Google Scholar]
- 81.Bloch Y, Bouchareychas L, Merceron R, Skladanowska K, Van den Bossche L, et al. 2018. Structural activation of pro-inflammatory human cytokine IL-23 by cognate IL-23 receptor enables recruitment of the shared receptor IL-12Rβ1. Immunity 48:45–58.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu J, Guo L, Watson CJ, Hu-Li J, Paul WE. 2001. Stat6 is necessary and sufficient for IL-4’s role inTh2 differentiation and cell expansion. J. Immunol 166:7276–81 [DOI] [PubMed] [Google Scholar]
- 83.Zhu J, Cote-Sierra J, Guo L, Paul WE. 2003. Stat5 activation plays a critical role in Th2 differentiation. Immunity 19:739–48 [DOI] [PubMed] [Google Scholar]
- 84.Cote-Sierra J, Foucras G, Guo L, Chiodetti L, Young HA, et al. 2004. Interleukin 2 plays a central role in Th2 differentiation. PNAS 101:3880–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liao W, Schones DE, Oh J, Cui Y, Cui K, et al. 2008. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression. Nat. Immunol 9:1288–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE Jr., Kuriyan J. 1998. Crystal structure of atyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93:827–39 [DOI] [PubMed] [Google Scholar]
- 87.Becker S, Groner B, Müller CW. 1998. Three-dimensional structure of the Stat3βhomodimer bound to DNA. Nature 394:145–51 [DOI] [PubMed] [Google Scholar]
- 88.Xu X, Sun YL, Hoey T. 1996. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science 273:794–97 [DOI] [PubMed] [Google Scholar]
- 89.Vinkemeier U, Cohen SL, Moarefi I, Chait BT, Kuriyan J, Darnell JE Jr. 1996. DNA binding of in vitro activated Stat1α, Stat1β and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. 15:5616–26 [PMC free article] [PubMed] [Google Scholar]
- 90.Soldaini E, John S, Moro S, Bollenbacher J, Schindler U, Leonard WJ.2000.DNA binding site selection of dimeric and tetrameric Stat5 proteins reveals a large repertoire of divergent tetrameric Stat5a binding sites. Mol. Cell. Biol 20:389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen X, Bhandari R, Vinkemeier U, Van Den Akker F, Darnell JE Jr., Kuriyan J.2003.A reinterpretation of the dimerization interface of the N-terminal domains of STATs. Protein Sci. 12:361–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ota N, Brett TJ, Murphy TL, Fremont DH, Murphy KM. 2004. N-domain-dependent nonphosphorylated STAT4 dimers required for cytokine-driven activation. Nat. Immunol 5:208–15 [DOI] [PubMed] [Google Scholar]
- 93.Mao X, Ren Z, Parker GN, Sondermann H, Pastorello MA, et al. 2005. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol. Cell 17:761–71 [DOI] [PubMed] [Google Scholar]
- 94.Mertens C, Zhong M, Krishnaraj R, Zou W, Chen X, Darnell JE Jr. 2006. Dephosphorylation of phos-photyrosine on STAT1 dimers requires extensive spatial reorientation of the monomers facilitated by the N-terminal domain. Genes Dev. 20:3372–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wenta N, Strauss H, Meyer S, Vinkemeier U.2008.Tyrosine phosphorylation regulates the partitioningof STAT1 between different dimer conformations. PNAS 105:9238–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhong M, Henriksen MA, Takeuchi K, Schaefer O, Liu B, et al. 2005. Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-inactivation cycle. PNAS 102:3966–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lin JX, Li P, Liu D, Jin HT, He J, et al. 2012. Critical role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function. Immunity 36:586–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lin JX, Du N, Li P, Kazemian M, Gebregiorgis T, et al. 2017. Critical functions for STAT5 tetramers in the maturation and survival of natural killer cells. Nat. Commun 8:1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Begitt A, Droescher M, Meyer T, Schmid CD, Baker M, et al. 2014. STAT1-cooperative DNA binding distinguishes type 1 from type 2 interferon signaling. Nat. Immunol 15:168–76 [DOI] [PubMed] [Google Scholar]
- 100.Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, et al. 2004. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol 24:8037–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, et al. 2006. Stat5a/b are essential for normal lymphoid development and differentiation. PNAS 103:1000–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Durbin JE, Hackenmiller R, Simon MC, Levy DE. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443–50 [DOI] [PubMed] [Google Scholar]
- 103.Sathyanarayana BK, Li P, Lin JX, Leonard WJ, Lee B. 2016. Molecular models of STAT5A tetramers complexed to DNA predict relative genome-wide frequencies of the spacing between the two dimer binding motifs of the tetramer binding sites. PLOS ONE 11:e0160339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cheon H, Stark GR. 2009. Unphosphorylated STAT1 prolongs the expression of interferon-inducedimmune regulatory genes. PNAS 106:9373–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cheon H, Holvey-Bates EG, Schoggins JW, Forster S, Hertzog P, et al. 2013. IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J.32:2751–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Park HJ, Li J, Hannah R, Biddie S, Leal-Cervantes AI, et al. 2016. Cytokine-induced megakaryocytic differentiation is regulated by genome-wide loss of a uSTAT transcriptional program. EMBO J.35:580–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Meier JA, Larner AC. 2014. Toward a new STATe: the role of STATs in mitochondrial function. Semin. Immunol 26:20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Garama DJ, White CL, Balic JJ, Gough DJ. 2016. Mitochondrial STAT3: powering up a potent factor.Cytokine 87:20–25 [DOI] [PubMed] [Google Scholar]
- 109.Lufei C, Ma J, Huang G, Zhang T, Novotny-Diermayr V, et al. 2003. GRIM-19, a death-regulatory gene product, suppresses Stat3 activity via functional interaction. EMBO J. 22:1325–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang J, Yang J, Roy SK, Tininini S, Hu J, et al. 2003. The cell death regulator GRIM-19 is an inhibitorof signal transducer and activator of transcription 3. PNAS 100:9342–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tammineni P, Anugula C, Mohammed F, Anjaneyulu M, Larner AC, Sepuri NB. 2013. The import of the transcription factor STAT3 into mitochondria depends on GRIM-19, a component of the electron transport chain. J. Biol. Chem 288:4723–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, et al. 2009. Function of mitochondrial Stat3 in cellular respiration. Science 323:793–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. 2009. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 324:1713–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mackenzie GG, Huang L, Alston N, Ouyang N, Vrankova K, et al. 2013. Targeting mitochondrialSTAT3 with the novel phospho-valproic acid (MDC-1112) inhibits pancreatic cancer growth in mice. PLOS ONE 8:e61532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Genini D, Brambilla L, Laurini E, Merulla J, Civenni G, et al. 2017. Mitochondrial dysfunction inducedby a SH2 domain-targeting STAT3 inhibitor leads to metabolic synthetic lethality in cancer cells. PNAS 114:E4924–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ziegler PK, Bollrath J, Pallangyo CK, Matsutani T, Canli O, et al. 2018. Mitophagy in intestinal epithelial cells triggers adaptive immunity during tumorigenesis. Cell 174: 88–101.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, et al. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153:307–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, et al. 2013. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. PNAS 110:17921–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, et al. 2013. Super-enhancers in the control of cell identity and disease. Cell 155:934–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470:279–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vahedi G, Kanno Y, Furumoto Y, Jiang K, Parker SC, et al. 2015. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520:558–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li P, Mitra S, Spolski R, Oh J, Liao W, et al. 2017. STAT5-mediated chromatin interactions in superenhancers activate IL-2 highly inducible genes: functional dissection of the Il2ra gene locus. PNAS 114:12111–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Simeonov DR,Gowen BG,Boontanrart M,Roth TL,Gagnon JD,et al.2017.Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature 549:111–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Changelian PS,Flanagan ME, Ball DJ, Kent CR, Magnuson KS, et al.2003.Preventionoforganallograftrejection by a specific Janus kinase 3 inhibitor. Science 302:875–78 [DOI] [PubMed] [Google Scholar]
- 125.Winthrop KL. 2017. The emerging safety profile of JAK inhibitors in rheumatic disease. Nat. Rev. Rheumatol 13:234–43 [DOI] [PubMed] [Google Scholar]
- 126.Sandborn WJ, Su C, Sands BE, D’Haens GR, Vermeire S, et al. 2017. Tofacitinib as induction and maintenance therapy for ulcerative colitis. N. Engl. J. Med 376:1723–36 [DOI] [PubMed] [Google Scholar]
- 127.Wang L, Yu CR, Kim HP, Liao W, Telford WG, et al. 2011.Key role for IL-21 in experimental autoimmune uveitis. PNAS 108:9542–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Spolski R, Kashyap M, Robinson C, Yu Z, Leonard WJ. 2008. IL-21 signaling is critical for the development of type I diabetes in the NOD mouse. PNAS 105:14028–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bubier JA, Sproule TJ, Foreman O, Spolski R, Shaffer DJ, et al. 2009. A critical role for IL-21 receptor signaling in the pathogenesis of systemic lupus erythematosus in BXSB-Yaa mice. PNAS 106:1518–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Langlais D, Fodil N, Gros P. 2017. Genetics of infectious and inflammatory diseases: overlapping discoveries from association and exome-sequencing studies. Annu. Rev. Immunol 35:1–30 [DOI] [PubMed] [Google Scholar]
- 131.Roederer M, Quaye L, Mangino M, Beddall MH, Mahnke Y, et al. 2015. The genetic architecture of the human immune system: a bioresource for autoimmunity and disease pathogenesis. Cell 161:387–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.London N, Miller RM, Krishnan S, Uchida K, Irwin JJ, et al. 2014. Covalent docking of large libraries for the discovery of chemical probes. Nat. Chem. Biol 10:1066–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Goedken ER, Argiriadi MA, Banach DL, Fiamengo BA, Foley SE, et al. 2015. Tricyclic covalent inhibitors selectively target Jak3 through an active site thiol. J. Biol. Chem 290:4573–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tan L,Akahane K,McNally R,Reyskens KM,Ficarro SB,et al.2015.Development of selective covalent Janus kinase 3 inhibitors. J. Med. Chem 58:6589–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Smith GA, Uchida K, Weiss A, Taunton J. 2016. Essential biphasic role for JAK3 catalytic activity inIL-2 receptor signaling. Nat. Chem. Biol 12:373–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Telliez JB, Dowty ME, Wang L, Jussif J, Lin T, et al. 2016. Discovery of a JAK3-selective inhibitor: functional differentiation of JAK3-selective inhibition over pan-JAK or JAK1-selective inhibition. ACS Chem. Biol 11:3442–51 [DOI] [PubMed] [Google Scholar]
- 137.Thorarensen A, Dowty ME, Banker ME, Juba B, Jussif J, et al. 2017. Design of a Janus kinase 3 (JAK3) specific inhibitor 1-((2S,5R)-5-((7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino)-2-methylpiperidin-1yl)prop-2-en-1-one (PF-06651600) allowing for the interrogation of JAK3 signaling in humans. J. Med. Chem 60:1971–93 [DOI] [PubMed] [Google Scholar]
- 138.Migone TS, Lin JX, Cereseto A, Mulloy JC, O’Shea JJ, et al. 1995. Constitutively activated Jak-STAT pathway in T cells transformed with HTLV-I. Science 269:79–81 [DOI] [PubMed] [Google Scholar]
- 139.Danial NN, Pernis A, Rothman PB. 1995. Jak-STAT signaling induced by the v-abl oncogene. Science 269:1875–77 [DOI] [PubMed] [Google Scholar]
- 140.Yu CL, Meyer DJ, Campbell GS, Larner AC, Carter-Su C, et al. 1995. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science 269:81–83 [DOI] [PubMed] [Google Scholar]
- 141.Nicot C,Mulloy JC, Ferrari MG,Johnson JM,Fu K, et al. 2001. HTLV-1 p12I protein enhances STAT5 activation and decreases the interleukin-2 requirement for proliferation of primary human peripheral blood mononuclear cells. Blood 98:823–29 [DOI] [PubMed] [Google Scholar]
- 142.Waldmann TA, Chen J. 2017. Disorders of the JAK/STAT pathway in T cell lymphoma pathogenesis: implications for immunotherapy. Annu. Rev. Immunol 35:533–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Banerjee S, Biehl A, Gadina M, Hasni S, Schwartz DM. 2017. JAK-STAT signaling as a target for inflammatory and autoimmune diseases: current and future prospects. Drugs 77:521–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Musumeci F, Greco C, Giacchello I, Fallacara AL, Ibrahim MM, et al. 2018. An update on JAK inhibitors. Curr. Med. Chem In press [DOI] [PubMed] [Google Scholar]
- 145.Quintas-Cardama A, Kantarjian H, Cortes J, Verstovsek S. 2011. Janus kinase inhibitors for thetreatment of myeloproliferative neoplasias and beyond. Nat. Rev. Drug Discov 10:127–40 [DOI] [PubMed] [Google Scholar]
- 146.Vannucchi AM, Kiladjian JJ, Griesshammer M, Masszi T, Durrant S, et al. 2015. Ruxolitinib versus standard therapy for the treatment of polycythemia vera. N. Engl. J. Med 372:426–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Meyer SC, Keller MD, Chiu S, Koppikar P, Guryanova OA, et al. 2015. CHZ868, a type II JAK2 inhibitor, reverses type I JAK inhibitor persistence and demonstrates efficacy in myeloproliferative neoplasms. Cancer Cell 28:15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Weisshof R, El Jurdi K, Zmeter N, Rubin DT.2018.Emerging therapies for inflammatory bowel disease. Adv. Ther 35:1746–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shahmarvand N,Nagy A, Shahryari J, Ohgami RS. 2018. Mutations in the signal transducer and activator of transcription family of genes in cancer. Cancer Sci. 109:926–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rajala HL, Porkka K, Maciejewski JP, Loughran TP Jr., Mustjoki S. 2014. Uncovering the pathogenesis of large granular lymphocytic leukemia-novel STAT3 and STAT5b mutations. Ann. Med 46:114–22 [DOI] [PubMed] [Google Scholar]
- 151.Rajala HL, Eldfors S, Kuusanmaki H, van Adrichem AJ, Olson T, et al. 2013. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood 121:4541–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Andersson EI, Tanahashi T, Sekiguchi N, Gasparini VR, Bortoluzzi S, et al. 2016. High incidence of activating STAT5B mutations in CD4-positive T-cell large granular lymphocyte leukemia. Blood 128:2465–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Forbes SA, Beare D, Bindal N, Bamford S, Ward S, et al. 2016. COSMIC: high-resolution cancer genetics using the catalogue of somatic mutations in cancer. Curr. Protoc. Hum. Genet 91:10.11.1–37 [DOI] [PubMed] [Google Scholar]
- 154.Kucuk C, Jiang B, Hu X, Zhang W, Chan JK, et al. 2015. Activating mutations of STAT5B and STAT3 in lymphomas derived from γδ-T or NK cells. Nat. Commun 6:6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Miklossy G, Hilliard TS, Turkson J. 2013. Therapeutic modulators of STAT signalling for human diseases. Nat. Rev. Drug Discov 12:611–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Johnson DE, O’Keefe RA, Grandis JR. 2018. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol 15:234–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sen M, Thomas SM, Kim S, Yeh JI, Ferris RL, et al. 2012. First-in-human trial of a STAT3 decoy oligonucleotide in head and neck tumors: implications for cancer therapy. Cancer Discov. 2:694–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Toniolo PA, Liu S, Yeh JE, Moraes-Vieira PM, Walker SR, et al. 2015. Inhibiting STAT5 by the BET bromodomain inhibitor JQ1 disrupts human dendritic cell maturation. J. Immunol 194:3180–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Elumalai N, Berg A, Rubner S, Blechschmidt L, Song C, et al. 2017. Rational development of Stafib-2: a selective, nanomolar inhibitor of the transcription factor STAT5b. Sci. Rep 7:819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rondanin R, Simoni D, Maccesi M, Romagnoli R, Grimaudo S, et al. 2017. Effects of pimozide derivatives on pSTAT5 in K562 cells. Chem. Med. Chem 12:1183–90 [DOI] [PubMed] [Google Scholar]
- 161.Liu LJ, Wang W, Kang TS, Liang JX, Liu C, et al. 2016. Antagonizing STAT5B dimerization with an osmium complex. Sci. Rep 6:36044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Casanova JL, Holland SM, Notarangelo LD. 2012. Inborn errors of human JAKs and STATs. Immunity 36:515–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kotlarz D, Zietara N, Milner JD, Klein C. 2014. Human IL-21 and IL-21R deficiencies: two novel entities of primary immunodeficiency. Curr. Opin. Pediatr 26:704–12 [DOI] [PubMed] [Google Scholar]
- 164.Milner JD, Holland SM. 2013. The cup runneth over: lessons from the ever-expanding pool of primary immunodeficiency diseases. Nat. Rev. Immunol 13:635–48 [DOI] [PubMed] [Google Scholar]
- 165.Sharfe N, Dadi HK, Shahar M, Roifman CM. 1997. Human immune disorder arising from mutation of the alpha chain of the interleukin-2 receptor. PNAS 94:3168–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nadeau K, Hwa V, Rosenfeld RG. 2011. STAT5b deficiency: an unsuspected cause of growth failure, immunodeficiency, and severe pulmonary disease. J. Pediatr 158:701–8 [DOI] [PubMed] [Google Scholar]
- 167.Fischer A, Hacein-Bey-Abina S, Cavazzana-Calvo M. 2010. 20 years of gene therapy for SCID. Nat. Immunol 11:457–60 [DOI] [PubMed] [Google Scholar]
- 168.Thrasher AJ, Williams DA. 2017. Evolving gene therapy in primary immunodeficiency. Mol. Ther 25:1132–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kuo CY, Kohn DB. 2016. Gene therapy for the treatment of primary immune deficiencies. Curr. Allergy Asthma Rep 16:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Aiuti A, Slavin S, Aker M, Ficara F, Deola S, et al. 2002. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science 296:2410–13 [DOI] [PubMed] [Google Scholar]
- 171.Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, et al. 2000. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science 288:669–72 [DOI] [PubMed] [Google Scholar]
- 172.Gaspar HB, Parsley KL, Howe S, King D, Gilmour KC, et al. 2004. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet 364:2181–87 [DOI] [PubMed] [Google Scholar]
- 173.Hacein-Bey-Abina S, Hauer J, Lim A, Picard C, Wang GP, et al. 2010. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med 363:355–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, et al. 2008. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Investig 118:3143–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, et al. 2008. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Investig 118:3132–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hacein-Bey-Abina S, Pai SY, Gaspar HB, Armant M, Berry CC, et al. 2014. A modified gamma-retrovirus vector for X-linked severe combined immunodeficiency. N. Engl. J. Med 371:1407–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Komor AC, Badran AH, Liu DR. 2017. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168:20–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, et al. 2016. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature 539:384–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chang CW, Lai YS, Westin E, Khodadadi-Jamayran A, Pawlik KM, et al. 2015. Modeling human severe combined immunodeficiency and correction by CRISPR/Cas9-enhanced gene targeting. Cell Rep. 12:1668–77 [DOI] [PubMed] [Google Scholar]
- 180.Brooks EG, Schmalstieg FC, Wirt DP, Rosenblatt HM, Adkins LT, et al. 1990. A novel X-linked combined immunodeficiency disease. J. Clin. Investig 86:1623–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Schmalstieg FC, Wirt DP, Adkins LT, Brooks EG, Stansberry SD, et al. 1992. Postnatal development of T lymphocytes in a novel X-linked immunodeficiency disease. Clin. Immunol. Immunopathol 64:71–77 [DOI] [PubMed] [Google Scholar]
- 182.Russell SM, Johnston JA, Noguchi M, Kawamura M, Bacon CM, et al. 1994. Interaction of IL-2R beta and gamma c chains with Jak1 and Jak3: implications for XSCID and XCID. Science 266:1042–45 [DOI] [PubMed] [Google Scholar]
- 183.Schmalstieg FC, Leonard WJ, Noguchi M, Berg M, Rudloff HE, et al. 1995. Missense mutation in exon 7 of the common gamma chain gene causes a moderate form of X-linked combined immunodeficiency. J. Clin. Investig 95:1169–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC. 2018. CAR T cell immunotherapy for human cancer. Science 359:1361–65 [DOI] [PubMed] [Google Scholar]
- 185.Kagoya Y, Tanaka S, Guo T, Anczurowski M, Wang CH, et al. 2018. A novel chimeric antigen receptor containing a JAK-STAT signaling domain mediates superior antitumor effects. Nat. Med 24:352–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rosenberg SA, Mule JJ, Spiess PJ, Reichert CM, Schwarz SL.1985. Regression of established pulmonary metastases and subcutaneous tumor mediated by the systemic administration of high-dose recombinant interleukin 2. J. Exp. Med 161:1169–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rosenberg SA, Lotze MT, Muul LM, Leitman S, Chang AE, et al. 1985. Observations on the systemic administration of autologous lymphokine-activated killer cells and recombinant interleukin-2 to patients with metastatic cancer. N. Engl. J. Med 313:1485–92 [DOI] [PubMed] [Google Scholar]
- 188.Rosenberg SA. 2014. IL-2: the first effective immunotherapy for human cancer. J. Immunol 192:5451–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Rosenberg SA. 2011. Cell transfer immunotherapy for metastatic solid cancer—what clinicians need to know. Nat. Rev. Clin. Oncol 8:577–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kalos M, June CH. 2013. Adoptive T cell transfer for cancer immunotherapy in the era of synthetic biology. Immunity 39:49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Barrett DM, Singh N, Porter DL, Grupp SA, June CH. 2014. Chimeric antigen receptor therapy for cancer. Annu. Rev. Med 65:333–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tran E, Robbins PF, Rosenberg SA. 2017. ‘Final common pathway’ of human cancer immunotherapy: targeting random somatic mutations. Nat. Immunol 18:255–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Li MO, Rudensky AY. 2016. T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat. Rev. Immunol 16:220–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ohkura N, Kitagawa Y, Sakaguchi S. 2013. Development and maintenance of regulatory T cells. Immunity 38:414–23 [DOI] [PubMed] [Google Scholar]
- 195.Malek TR, Yu A, Vincek V, Scibelli P, Kong L. 2002. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rβ-deficient mice: implications for the nonredundant function of IL-2. Immunity 17:167–78 [DOI] [PubMed] [Google Scholar]
- 196.Malek TR, Castro I. 2010. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity 33:153–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Liao W, Lin JX, Leonard WJ. 2013. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity 38:13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yu A, Snowhite I, Vendrame F, Rosenzwajg M, Klatzmann D, et al. 2015. Selective IL-2 responsiveness of regulatory T cells through multiple intrinsic mechanisms supports the use of low-dose IL-2 therapy in type 1 diabetes. Diabetes 64:2172–83 [DOI] [PubMed] [Google Scholar]
- 199.Koreth J, Matsuoka K, Kim HT, McDonough SM, Bindra B, et al. 2011. Interleukin-2 and regulatory T cells in graft-versus-host disease. N. Engl. J. Med 365:2055–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Asano T, Meguri Y, Yoshioka T, Kishi Y, Iwamoto M, et al. 2017. PD-1 modulates regulatory T-cell homeostasis during low-dose interleukin-2 therapy. Blood 129:2186–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hartemann A, Bensimon G, Payan CA, Jacqueminet S, Bourron O, et al. 2013. Low-dose interleukin 2 in patients with type 1 diabetes: a phase 1/2 randomised, double-blind, placebo-controlled trial. Lancet Diabetes Endocrinol. 1:295–305 [DOI] [PubMed] [Google Scholar]
- 202.Rosenzwajg M, Churlaud G, Mallone R, Six A, Derian N, et al. 2015. Low-dose interleukin-2 fosters a dose-dependent regulatory T cell tuned milieu in T1D patients. J. Autoimmun 58:48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.He J, Zhang X, Wei Y, Sun X, Chen Y, et al. 2016. Low-dose interleukin-2 treatment selectively modulates CD4+ T cell subsets in patients with systemic lupus erythematosus. Nat. Med 22:991–93 [DOI] [PubMed] [Google Scholar]
- 204.von Spee-Mayer C, Siegert E, Abdirama D, Rose A, Klaus A, et al. 2016. Low-dose interleukin-2 selectively corrects regulatory T cell defects in patients with systemic lupus erythematosus. Ann. Rheum. Dis 75:1407–15 [DOI] [PubMed] [Google Scholar]
- 205.Letourneau S, van Leeuwen EM, Krieg C, Martin C, Pantaleo G, et al. 2010. IL-2/anti-IL-2 antibody complexes show strong biological activity by avoiding interaction with IL-2 receptor alpha subunit CD25. PNAS 107:2171–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Boyman O, Kovar M, Rubinstein MP, Surh CD, Sprent J. 2006. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science 311:1924–27 [DOI] [PubMed] [Google Scholar]
- 207.Spangler JB,Tomala J, Luca VC, Jude KM, Dong S, et al. 2015. Antibodies to interleukin-2 elicit selective T cell subset potentiation through distinct conformational mechanisms. Immunity 42:815–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Trotta E, Bessette PH, Silveria SL, Ely LK, Jude KM, et al. 2018. A human anti-IL-2 antibody that potentiates regulatory T cells by a structure-based mechanism. Nat. Med 24:1005–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Rubinstein MP, Kovar M, Purton JF, Cho JH, Boyman O, et al. 2006. Converting IL-15 to a superagonist by binding to soluble IL-15Rα. PNAS 103:9166–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bouchaud G, Garrigue-Antar L, Sole V, Quemener A, Boublik Y, et al. 2008. The exon-3-encoded domain of IL-15Rαcontributes to IL-15 high-affinity binding and is crucial for the IL-15 antagonistic effect of soluble IL-15Rα. J. Mol. Biol 382:1–12 [DOI] [PubMed] [Google Scholar]
- 211.Mortier E, Quemener A, Vusio P, Lorenzen I, Boublik Y, et al. 2006. Soluble interleukin-15 receptorα(IL-15Rα)-sushi as a selective and potent agonist of IL-15 action through IL-15Rβ/γ; hyperagonist IL-15·IL-15Rαfusion proteins. J. Biol. Chem 281:1612–19 [DOI] [PubMed] [Google Scholar]
- 212.Desbois M, Le Vu P, Coutzac C, Marcheteau E, Beal C, et al. 2016. IL-15 trans-signaling with the superagonist RLI promotes effector/memory CD8+ T cell responses and enhances antitumor activity of PD-1 antagonists. J. Immunol 197:168–78 [DOI] [PubMed] [Google Scholar]
- 213.Kay AB. 2001. Allergy and allergic diseases: first of two parts. N. Engl. J. Med 344:30–37 [DOI] [PubMed] [Google Scholar]
- 214.Rochman Y, Spolski R, Leonard WJ. 2009. New insights into the regulation of T cells by γc family cytokines. Nat. Rev. Immunol 9:480–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tony HP, Shen BJ, Reusch P, Sebald W. 1994. Design of human interleukin-4 antagonists inhibiting interleukin-4-dependent and interleukin-13-dependent responses in T-cells and B-cells with high efficiency. Eur. J. Biochem 225:659–65 [DOI] [PubMed] [Google Scholar]
- 216.Andrews AL, Holloway JW, Holgate ST, Davies DE. 2006. IL-4 receptor alpha is an important modulator of IL-4 and IL-13 receptor binding: implications for the development of therapeutic targets. J. Immunol 176:7456–61 [DOI] [PubMed] [Google Scholar]
- 217.Wenzel S, Castro M, Corren J, Maspero J, Wang L, et al. 2016. Dupilumab efficacy and safety in adults with uncontrolled persistent asthma despite use of medium-to-high-dose inhaled corticosteroids plus a long-acting β2 agonist: a randomised double-blind placebo-controlled pivotal phase 2b dose-ranging trial. Lancet 388:31–44 [DOI] [PubMed] [Google Scholar]
- 218.Wenzel S, Ford L, Pearlman D, Spector S, Sher L, et al. 2013. Dupilumab in persistent asthma with elevated eosinophil levels. N. Engl. J. Med 368:2455–66 [DOI] [PubMed] [Google Scholar]
- 219.Shirley M. 2017. Dupilumab: first global approval. Drugs 77:1115–21 [DOI] [PubMed] [Google Scholar]
- 220.Sheridan C. 2018. Drugmakers cling to dual IL-13/IL-4 blockbuster hopes. Nat. Biotechnol 36:3–5 [DOI] [PubMed] [Google Scholar]
- 221.Lowenthal JW, Greene WC. 1987. Contrasting interleukin 2 binding properties of the alpha (p55) and beta (p70) protein subunits of the human high-affinity interleukin 2 receptor. J. Exp. Med 166:1156–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wang X, Rickert M, Garcia KC. 2005. Structure of the quaternary complex of interleukin-2 with its α, β, and γc receptors. Science 310:1159–63 [DOI] [PubMed] [Google Scholar]
- 223.Levin AM, Bates DL, Ring AM, Krieg C, Lin JT, et al. 2012.Exploiting a natural conformational switch to engineer an interleukin-2 ‘superkine’. Nature 484:529–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mitra S, Ring AM, Amarnath S, Spangler JB, Li P, et al. 2015. Interleukin-2 activity can be fine tuned with engineered receptor signaling clamps. Immunity 42:826–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Strange PG. 2008. Agonist binding, agonist affinity and agonist efficacy at G protein-coupled receptors.Br. J. Pharmacol 153:1353–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Meghnem D, Morisseau S, Frutoso M, Trillet K, Maillasson M, et al. 2017. Cutting edge: differential fine-tuning of IL-2– and IL-15–dependent functions by targeting their common IL-2/15Rβ/γc receptor. J. Immunol 198:4563–68 [DOI] [PubMed] [Google Scholar]
- 227.Pettit DK, Bonnert TP, Eisenman J, Srinivasan S, Paxton R, et al. 1997. Structure-function studies of interleukin 15 using site-specific mutagenesis, polyethylene glycol conjugation, and homology modeling. J. Biol. Chem 272:2312–18 [DOI] [PubMed] [Google Scholar]
- 228.Broudy VC. 1997. Stem cell factor and hematopoiesis. Blood 90:1345–64 [PubMed] [Google Scholar]
- 229.Lyman SD, Jacobsen SE. 1998. c-kit ligand and Flt3 ligand:stem/progenitor cell factors with overlapping yet distinct activities. Blood 91:1101–34 [PubMed] [Google Scholar]
- 230.Ho CCM, Chhabra A, Starkl P, Schnorr PJ, Wilmes S, et al. 2017. Decoupling the functional pleiotropy of stem cell factor by tuning c-kit signaling. Cell 168:1041–52.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kim AR,Ulirsch JC,Wilmes S, Unal E,Moraga I, et al.2017. Functional selectivity in cytokine signaling revealed through a pathogenic EPO mutation. Cell 168:1053–64.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Nakamura Y,Russell SM,Mess SA,Friedmann M, Erdos M,et al. 1994. Heterodimerization of the IL-2 receptor β- and γ-chain cytoplasmic domains is required for signalling. Nature 369:330–33 [DOI] [PubMed] [Google Scholar]
- 233.Nelson BH, Lord JD, Greenberg PD. 1994. Cytoplasmic domains of the interleukin-2 receptor β and γ chains mediate the signal for T-cell proliferation. Nature 369:333–36 [DOI] [PubMed] [Google Scholar]
- 234.Sockolosky JT, Trotta E, Parisi G, Picton L, Su LL, et al. 2018. Selective targeting of engineered T cells using orthogonal IL-2 cytokine-receptor complexes. Science 359:1037–42 [DOI] [PMC free article] [PubMed] [Google Scholar]