Abstract
The 90-kb virulence plasmid of Salmonella typhimurium encodes five spv genes which increase the growth rate of the bacteria within host cells within the first week of systemic infection of mice (P. A. Gulig and T. J. Doyle, Infect. Immun. 61:504–511, 1993). The presently described study was aimed at identifying the host cells associated with Spv-mediated virulence by manipulating the mouse host and the salmonellae. To test the effects of T cells and B cells on the Spv phenotype, salmonellae were orally inoculated into nude and SCID BALB/c mice. Relative to normal BALB/c mice, nude and SCID BALB/c mice were unaffected for splenic infection with either the Spv+ or Spv− S. typhimurium strains at 5 days postinoculation. When mice were pretreated with cyclophosphamide to induce granulocytopenia, there was a variable increase in total salmonella infection, but the relative splenic CFU of Spv+ versus Spv− S. typhimurium was not changed after oral inoculation. In contrast, depletion of macrophages from mice by treatment with cyclophosphamide plus liposomes containing dichloromethylene diphosphate resulted in equivalent virulence of Spv+ and Spv− salmonellae. To examine if the spv genes affected the growth of salmonellae in nonphagocytic cells, an invA::aphT mutation was transduced into Spv+ and Spv− S. typhimurium strains. InvA− Spv+ salmonellae were not significantly affected for splenic infection after subcutaneous inoculation compared with the wild-type strain, and InvA− Spv− salmonellae were only slightly attenuated relative to InvA+ Spv− salmonellae. Invasion-defective salmonellae still exhibited the Spv phenotype. Therefore, infection of nonphagocytes is not involved with the Spv virulence function. Taken together, these data demonstrate that macrophages are essential for suppressing the infection by Spv− S. typhimurium, by serving as the primary host cell for Spv-mediated intracellular replication and possibly by inhibiting the replication of salmonellae within other macrophages.
Nontyphoidal serovars of Salmonella spp. which possess related virulence plasmids have the potential to cause systemic disease, particularly in immunocompromised humans (65). In a mouse model, these virulence plasmids are essential for systemic infection within a week after oral inoculation (29). By genetic analysis of virulence genes on the plasmids, five spv genes, spvRABCD, were identified (29) and were sufficient to express the virulence phenotype of systemic infection for Salmonella typhimurium (26). We determined that the spv genes of S. typhimurium primarily enabled more rapid growth rate in mice but did not significantly affect killing or movement through tissues, by using a temperature-sensitive genetic marker to measure the relative number of bacterial cell divisions in vivo (30). In the natural infection, the bacteria enter the host by the oral route and invade the intestinal epithelial cells and/or M cells (3) in a plasmid-independent manner (28). Salmonellae then invade and proliferate in Peyer’s patches and mesenteric lymph nodes (3). The bacteria reach the liver and spleen through the lymphatics and blood. The virulence plasmid is not necessary for infection of the intestines, resistance to complement-mediated bacteriolysis of serum, resistance to phagocytosis and killing by macrophages, or adherence to, invasion into, and growth within certain cell lines in vitro (28, 29).
Since the spv genes affect the virulence of salmonellae primarily in lymphoid tissues, many investigators have proposed that the Spv phenotype is manifested in phagocytes, primarily macrophages. However, until recently (53), direct proof of this hypothesis has been lacking. In fact, irrespective of the role of the spv genes in salmonella virulence, the cellular location of salmonellae in the host has been controversial. Most reports support infection of macrophages as essential for salmonella virulence (15, 23, 68). However, others propose that salmonellae either are extracellular (35, 41) or infect nonphagocytic cells (6, 8) or polymorphonuclear leukocytes (PMNs) (11). Ultimately, a comprehensive histological analysis of infected tissues from mice that were inoculated in a relevant manner with a relevant inoculum will be required to settle these controversies.
We pursued a biological approach to examine the interaction of Spv+ and Spv− S. typhimurium with different populations of host cells. We used mice genetically deficient for lymphocytes, mice depleted of phagocytes by different drugs, and mutant S. typhimurium strains that were rendered defective for infecting nonphagocytic cells. Our results presented here indicate that invasion of nonphagocytes is irrelevant for virulence of either Spv+ or Spv− salmonellae during infection beyond the intestines, and that T cells and B cells have no detectable role in suppressing or enabling systemic infection by S. typhimurium within 5 days after oral inoculation. PMNs had a variable role in suppressing overall salmonella infection but did not differentially suppress Spv− salmonellae. However, quantitative depletion of macrophages from mice by using drugs rendered Spv+ and Spv− S. typhimurium equal for systemic infection. Together, these data indicate that within a week after oral inoculation of BALB/c mice the spv genes increase the growth rate of salmonellae within macrophages and suggest that macrophages are the only relevant host cells for replication of salmonellae beyond the intestines.
MATERIALS AND METHODS
Bacterial strains and culture.
The S. typhimurium strains used in this study are described in Table 1. Spv+ virulence plasmid-containing S. typhimurium SR-11, χ3456 (tetracycline resistant), χ3306 (nalidixic acid resistant), and isogenic Spv− virulence plasmid-cured χ3337 have been described elsewhere (28). A more isogenic Spv− strain was constructed in which the 6.3-kb ClaI fragment encoding spvRABCD′ was deleted from the virulence plasmid and replaced with the tet gene of pBR322. Construction of this strain, described elsewhere (43), involved construction of the mutation in a cloned spv sequence and recombining the mutation into χ3181 by using the suicide allelic exchange vector pCVD442 (10). The invA::aphT mutation originally constructed in S. typhimurium SB147 (21) was transduced into χ3456 and χ3337 by using generalized transduction with P22HTint (56) grown on SB136, yielding UF102 and UF103, respectively.
TABLE 1.
S. typhimurium strains used
| Strain | Plasmid | Genotype | Reference and/or comments |
|---|---|---|---|
| χ3181 | + | Wild type | 28 |
| χ3306 | + | gyrA1816 | Nalidixic acidr (28) |
| χ3337 | − | gyrA1816 | Nalidixic acidr (28) |
| χ3456 | + | pStSR100::Tnmini-tet | Tetracycliner (28) |
| UF102 | + | invA::aphT, pStSR100::Tnmini-tet | Inv− kanamycinr tetracycliner (this work) |
| UF103 | − | invA::aphT gyrA1816 | Inv− kanamycinr nalidixic acidr (this work) |
| UF110 | + | Δspv::tet | Tetracycliner (43) |
Bacteria were grown in L broth or on L agar (39) supplemented with antibiotics at the following concentrations as appropriate: chloramphenicol, 30 μg/ml; tetracycline, 12.5 μg/ml (7.5 μg/ml for UF110); kanamycin, 30 μg/ml; and nalidixic acid, 25 μg/ml.
Infection of mice.
All of these studies used specific-pathogen-free BALB/c mice, which are sensitive to infection by S. typhimurium because of the Itys mutation (57). Mice were orally inoculated with S. typhimurium as described previously (27). Approximately 108 CFU of S. typhimurium was fed to 7- to 11-week-old female BALB/c mice (Charles River, Wilmington, Mass., and University of Florida Department of Pathology, Immunology, and Laboratory Medicine Mouse Facility) after food and water deprivation and feeding of bicarbonate. Unless noted otherwise, 4 or 5 days later, spleens and livers were removed, homogenized in glass tissue homogenizers with phosphate-buffered saline (PBS) containing gelatin (BSG) (9), and plated to enumerate CFU. Nude BALB/c mice (Charles River) and SCID BALB/c mice (Jackson Laboratory, Bar Harbor, Maine; maintained at the University of Florida Department of Pathology, Immunology, and Laboratory Medicine Mouse Facility) were also used with oral inoculation. Alternatively, mice were inoculated subcutaneously (s.c.) into both hind footpads as described previously (30) with 105 CFU of salmonellae suspended in 0.02 ml of BSG per footpad. Three or four days later, spleens were removed, homogenized in BSG, and plated to enumerate CFU. Mice were either inoculated with a single salmonella strain or equal mixtures of two strains as described (30).
Treatment of mice with cyclophosphamide.
To examine the effects of PMNs, mice were treated with cyclophosphamide as described previously (45). Cyclophosphamide (Sigma Chemical Co., St. Louis, Mo.) was dissolved in sterile, pyrogen-free distilled water or saline and injected intraperitoneally (i.p.) at 3 days preinfection and on the day of infection at a dose of 150 mg/kg of body weight. Peripheral blood leukocytes (WBC) were enumerated by hemocytometer counts of EDTA-anticoagulated whole blood diluted in 3% (vol/vol) acetic acid. Differential WBC counts were performed on Wright-Giemsa (Camco Quik Stain II; Baxter, McGaw Park, Ill.)-stained smears. Absolute neutrophil counts were calculated at the time of necropsy by multiplying the total WBC by the percentage of neutrophils obtained from differential counts (45). We routinely observed at least 90% reduction of peripheral blood PMNs after cyclophosphamide treatment.
Treatment of mice with liposomes containing dCMdP.
To examine the effects of functional depletion of macrophages, mice were treated with liposomes containing dichloromethylene diphosphate (dCMdP) as described by van Rooijen (61). Briefly, liposomes consisted of 87% (wt/wt) phosphatidylcholine and 13% (wt/wt) cholesterol (Sigma). dCMdP (a gift of Boehringer Mannheim) was dissolved in PBS at a concentration of 18.9% (wt/vol) and was incorporated into liposomes by sonication. Liposomes were sized to 150 to 200 nm and washed in PBS before use in mice. Mice were injected intravenously (i.v.) in the lateral tail vein with approximately 4 mg of dCMdP contained in 0.2 ml of liposome suspension. For initial experiments using liposome-dCMdP alone, injection was on the day before inoculation with salmonellae. For subsequent experiments with liposome-dCMdP and experiments involving combined cyclophosphamide–liposome-dCMdP treatment, injection of liposomes was on the day of oral inoculation at the time of preinoculation food and water deprivation. Liposomes containing only PBS and occasionally PBS without liposomes were used as negative controls.
Immunohistochemical analysis of mouse tissues.
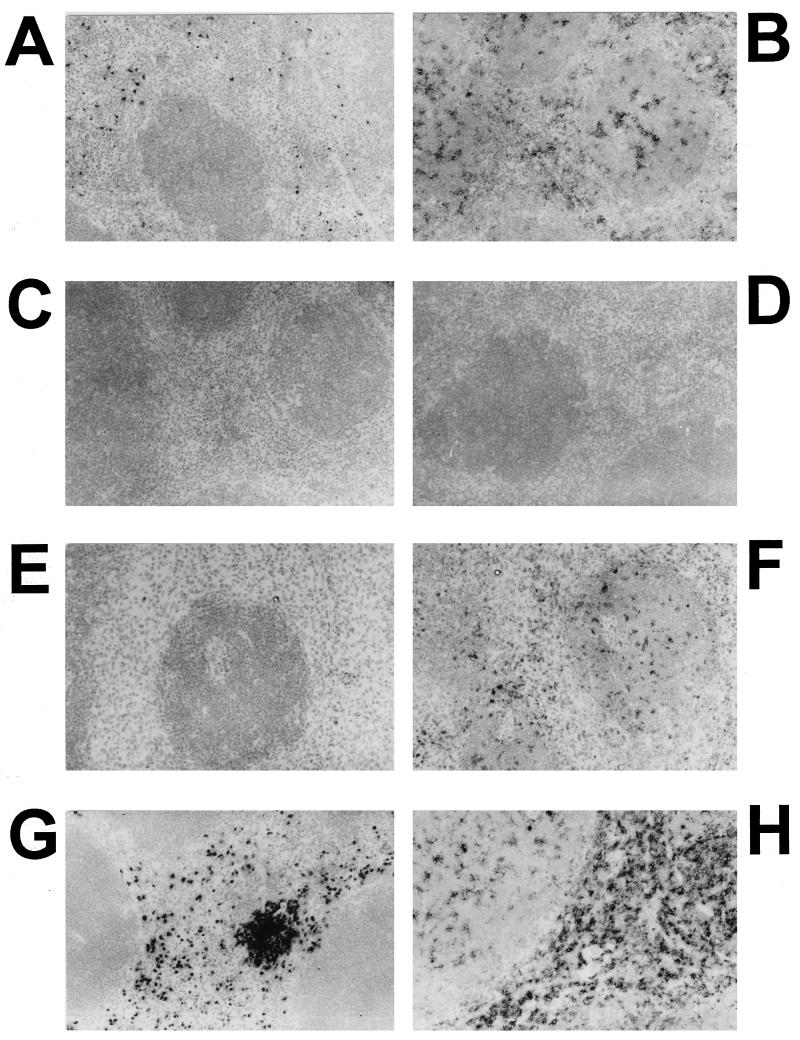
To confirm the effects of depleting mice of PMNs by using cyclophosphamide or depleting macrophages with liposome encapsulated dCMdP, a portion of infected mouse spleens and livers was quick frozen in OCT embedding medium (TisTek; Sakura Finetek, Torrance, Calif.), while the remainder was homogenized and plated as described above. Tissues were sectioned in 5-μm sections and mounted at the University of Florida Department of Pathology, Immunology, and Laboratory Medicine Diagnostic Referral Laboratory and stored at −70°C. On the day of analysis, sections were thawed and fixed in absolute methanol containing 1% (vol/vol) hydrogen peroxide to block endogenous peroxidase activity. Endogenous biotin was blocked with Biotin Blocking System (Dako Corp., Carpenteria, Calif.). Fc activity was blocked with 10% (vol/vol) normal porcine serum. Primary antibodies consisted of rabbit anti-mouse macrophage serum (Inter-Cell Technologies, Hopewell, N.J.) followed by porcine anti-rabbit immunoglobulin G (IgG) conjugated with peroxidase (Dako) and then rabbit peroxidase-antiperoxidase (Harlan Bioproducts, Indianapolis, Ind.); rat monoclonal antibody FA/11 directed against the CD68 pan-macrophage, macrophage-specific antigen (22, 51) followed by donkey anti-rat IgG conjugated with peroxidase (Jackson ImmunoResearch); or rat monoclonal antibody RB6-8C5 against granulocytes (Gr-1 antigen) (17) conjugated with biotin (CalTag) followed by avidin conjugated to peroxidase (Biostain Super ABC Basic Detector; Biomeda, Foster City, Calif.). Peroxidase-conjugated complexes were developed with diaminobenzidine (Metal Enhanced DAB Substrate kit; Pierce, Rockford, Ill.), and slides were counterstained with hematoxylin. Negative control antibodies were rat antidinitrophenol monoclonal antibody conjugated with biotin (CalTag) for RB6-8C5 and rat IgG1 monoclonal antibody R187 directed against the murine leukemia virus p30gag protein (5).
Statistical analysis.
Analysis of bacterial numbers as mean log10 ± standard deviation CFU was performed as described previously (2). In experiments in which individual mice were inoculated with single strains, the difference between the mean log10 CFU of different groups was determined by using the Student t test. For analysis of mixed infections, the Student t test was used to examine the mean paired difference in log10 CFU of the strains being different from 0. In all cases, two or three experiments were performed, and results were pooled for analysis when indicated.
RESULTS
Our previous results established that the spv genes of the S. typhimurium virulence plasmid primarily increase the growth rate of salmonellae during infection of host cells in infected mice (30). The central question was then which cells acted as the primary hosts for Spv-mediated increased growth and which cells, if any, acted as secondary effectors which suppressed the replication of the Spv− salmonellae. To gain insight into the function of the spv genes, we examined how the mouse host could be altered so that Spv− salmonellae would no longer be suppressed relative to Spv+ salmonellae, that is, so that the suppressive environment to which the Spv− bacteria were susceptible would be eliminated. The objective of most of the experiments described below, therefore, consisted of infecting mice depleted of certain cells, either pharmacologically or by genetic mutation, with Spv+ and/or Spv− S. typhimurium. If the primary or secondary host cells responsible for suppression of Spv− salmonellae were eliminated, the Spv− strain would be recovered from deep tissues such as the spleen and liver in the same or nearly the same numbers as the Spv+ parent.
Lymphocytes do not have a role in restricting the growth of Spv+ or Spv− S. typhimurium within 5 days after oral inoculation.
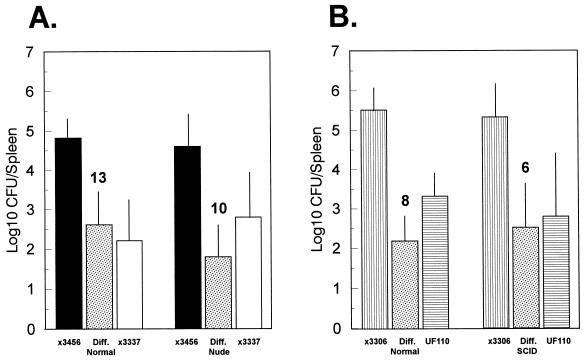
We considered the possibility that T cells and/or B cells were involved in suppressing the replication of Spv− S. typhimurium; i.e., the spv genes are involved either in increased replication within these lymphocytes or in overcoming lymphocyte-mediated suppression of salmonella replication within another host cell. To examine the role of T cells in differentially suppressing plasmid-cured S. typhimurium, nude BALB/c and normal BALB/c mice were inoculated orally with 108 CFU of both wild-type χ3456 and plasmid-cured χ3337 in mixed infections. Five days later, spleens were examined for total CFU of each strain (Fig. 1A). There was no effect of the nude BALB/c background on recovery of either Spv+ or Spv− S. typhimurium compared with normal BALB/c mice. Furthermore, the mean paired differences between plasmid-containing and plasmid-cured salmonellae were unchanged between normal and nude BALB/c mice. Therefore, T cells do not significantly affect splenic infection within 5 days of oral inoculation and have no role in the Spv phenotype.
FIG. 1.
Lack of effect of the nude and SCID mutations on the splenic infection of BALB/c mice inoculated orally with Spv+ and Spv− S. typhimurium. (A) Normal and nude BALB/c mice were orally inoculated with mixtures of wild-type, Spv+ χ3456 and virulence plasmid-cured, Spv− χ3337. (B) Normal and SCID BALB/c mice were orally inoculated with mixtures of wild-type χ3306 and Δspv::tet, Spv− UF110. Five days later, CFU of each strain in spleens were enumerated, and the mean paired difference (Diff.) between the Spv+ and Spv− strains was calculated. Standard deviations are shown as vertical lines, and n for each infection is shown above the data for the mean paired difference. These are the combined results of at least two experiments each for nude and SCID BALB/c mice. The log10 CFU recoveries for either salmonella strain between either nude and SCID BALB/c mice and the matched normal BALB/c mice were not significant (P > 0.2). The mean paired differences between Spv+ and Spv− strains were always significantly greater than 0 (P < 0.005 to P < 0.001).
To examine a more completely lymphocyte-deficient background for T cells, as well as B cells, SCID BALB/c mice were orally inoculated with a mixture of Spv+ (χ3306) and Spv− (UF110) S. typhimurium. Five days later, results essentially identical to those obtained with nude BALB/c mice were obtained: the SCID mutation had no effect on splenic infection by either Spv+ or Spv− S. typhimurium compared with normal BALB/c controls (Fig. 1B). Lymphocytes therefore have no role in the Spv phenotype, either as the primary cells in which the spv genes exert their effect or as secondary cells regulating the replication of salmonellae within a different primary cell.
Neutrophils have a variable role in suppressing Spv+ and Spv− S. typhimurium but do not affect the Spv phenotype.
It has been proposed that PMNs are the relevant site of intracellular residence for wild-type S. typhimurium in the spleens of mice shortly after i.v. infection (11) and are involved in suppressing infection of hepatocytes after i.v. inoculation (6, 8). To examine the role of PMNs in suppressing intracellular growth of plasmid-cured salmonellae relative to wild-type salmonellae, mice were treated i.p. with the chemotherapeutic agent cyclophosphamide to induce granulocytopenia (45). At necropsy, we routinely observed at least a 90% decrease in peripheral blood granulocytes after cyclophosphamide treatment compared to mice treated i.p. with PBS. Furthermore, no PMNs were detectable in spleens and livers of cyclophosphamide-treated mice, using monoclonal antibody RB6-8C5 directed against the granulocyte-specific antigen Gr-1 (Fig. 2E). Mice were orally inoculated with single or mixed strains of wild-type (χ3456 or χ3306) and/or Spv− (χ3337 or UF110) S. typhimurium and were examined for splenic infection 4 to 5 days after inoculation.
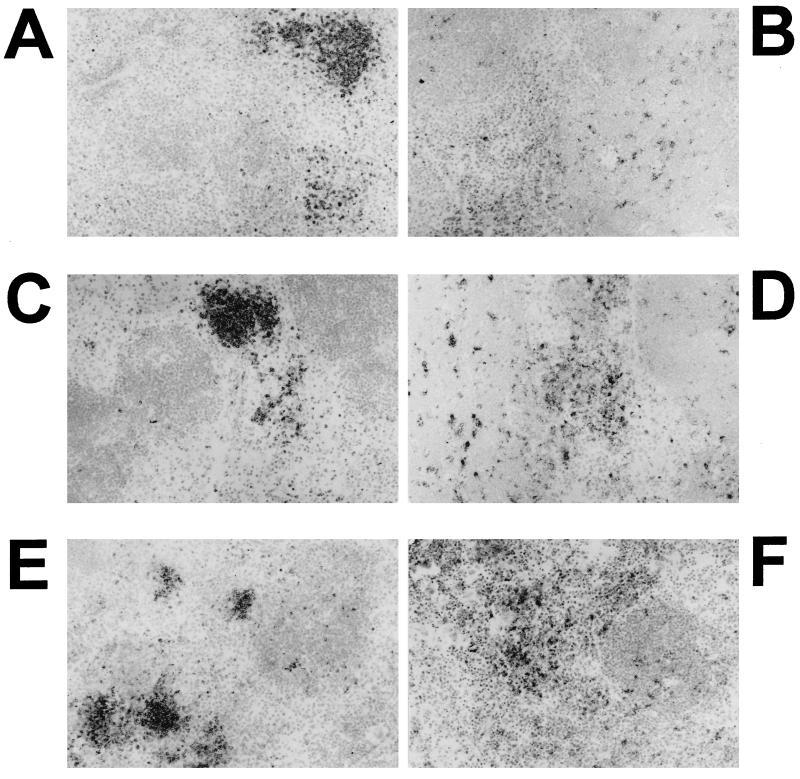
FIG. 2.
Immunochemical analysis of PMNs and macrophages in spleens of normal and cyclophosphamide-treated mice infected with a mixture of Spv+ and Spv− S. typhimurium. Mice were treated with cyclophosphamide or PBS and orally inoculated with S. typhimurium as described in the text. Four days later, spleens and livers (not shown) were frozen in OCT and sectioned for immunochemical analysis of PMNs and macrophages as described in the text. Hamster anti-mouse granulocyte monoclonal antibody was RB6-8C5 (A, E, and G), and rat anti-mouse macrophage monoclonal antibody was FA/11 (B, F, and H). RB6-8C5 was conjugated with biotin and detected with avidin-horseradish peroxidase. FA/11 was detected using donkey anti-rat IgG conjugated with peroxidase. Reactions were developed with diaminobenzidine. Negative control antibodies were rat anti-murine leukemia virus p30gag protein (5) (C) and rat antidinitrophenol (D) and consistently produced clean reactions. Sections were counterstained with hematoxylin. All images are at a magnification of ×200. (A to D) Normal mouse spleen demonstrating specificity of the primary antibodies and lack of reactivity for the negative control antibodies; (E and F) spleen from infected mouse treated with cyclophosphamide for depletion of PMNs; (G and H) spleen from infected mouse injected with PBS in place of cyclophosphamide. Note the absence of PMNs in panel E and the continued presence of macrophages in panel F.
The data in Fig. 3A are combined results of two experiments each for mixed inoculation with wild-type χ3306 and Δspv::tet UF110. Cyclophosphamide-treated mice possessed few, if any, detectable PMNs in spleens (Fig. 2E) and livers (not shown), even during the course of systemic salmonella infection. Furthermore, as expected, the presence of macrophages, detected with monoclonal antibody FA/11, was not decreased by treatment of mice with cyclophosphamide (Fig. 2F). As above, peripheral blood granulocyte levels were decreased by at least 90% by cyclophosphamide treatment. Even though three of eight cyclophosphamide-treated mice died from causes other than salmonellae, the splenic and hepatic CFU of wild-type S. typhimurium were not changed by depletion of PMNs from mice, and no significant differences in splenic and hepatic CFU of the isogenic Spv− strain were observed (Fig. 3A). Similar results were obtained in at least three other experiments using wild-type χ3456 and virulence plasmid-cured χ3337 in mixed and single strain infections of cyclophosphamide-treated mice (data not shown).
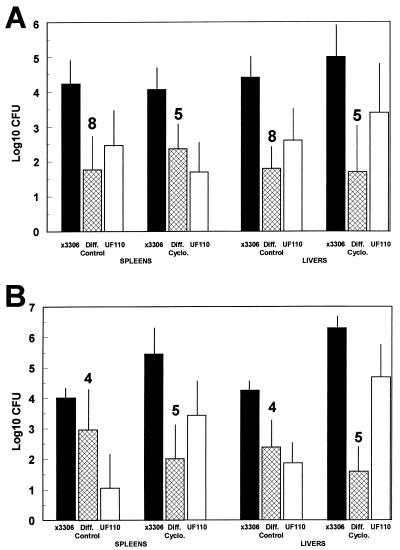
FIG. 3.
Depletion of PMNs by cyclophosphamide treatment does not affect the Spv virulence phenotype of S. typhimurium after oral inoculation of mice. Cyclophosphamide (Cyclo.) at a dose of 150 mg/kg of body weight or PBS (Control) was injected i.p. at 3 days preinfection and on the day of infection. Mice were orally inoculated with Spv+ χ3306 and Δspv::tet Spv− UF110 at a dose of 108 CFU. Four days later, spleens and livers were examined for salmonella CFU, with the exception of two cyclophosphamide-treated mice in panel B, which were harvested 18 h earlier. Data shown are for total CFU/spleen or CFU/gram of liver. The mean paired difference (Diff.) was calculated between the salmonella strains for each mouse. Each bar represents the mean log10 CFU with standard deviation shown. The number of mice is indicated above the mean paired difference. The mean paired differences between Spv+ and Spv− salmonellae were significantly greater than 0 in every case (P < 0.05 to P < 0.001). (A) Representative data from a series of three experiments in which treatment with cyclophosphamide did not result in a significant change in CFU of either Spv+ or Spv− salmonellae or the mean paired difference between the strains (P > 0.1 to P > 0.5). (B) Representative data from a series of three experiments in which treatment with cyclophosphamide resulted in significant increases in CFU for both Spv+ and Spv− salmonellae (P < 0.01 to P < 0.001), but the mean paired difference between the strains was not significantly changed (P ≥ 0.2).
We performed a semiquantitative analysis using immunohistology of the relative numbers of macrophages in spleen and liver sections from cyclophosphamide-treated and control-treated mice infected with S. typhimurium to examine if increases in macrophages could have compensated for lack of PMNs (data not shown). There was no observable effect on the numbers of hepatic macrophages between cyclophosphamide-treated and control mice; however, there was a tendency for increased infiltration by splenic macrophages into the white pulp of spleens of cyclophosphamide-treated mice versus control-treated mice as a result of salmonella infection. Because of the density of macrophages in the red pulp, we could make no conclusions about total numbers of macrophages in spleens. These results indicated that granulocytes, in particular PMNs, were not essential for restricting systemic infection by S. typhimurium within 5 days after oral inoculation and that PMNs do not have a detectable role in suppressing Spv− salmonellae relative to Spv+ salmonellae. The resident macrophages in tissues appeared to be capable of both suppressing salmonella infection in general and exerting differential suppression against the Spv− strain.
Several months later, we repeated this series of experiments exactly, except that two of five cyclophosphamide-treated mice infected with salmonellae were killed 18 h earlier than the regular harvest at 4 days postinoculation. We obtained results somewhat different from those of the first set of experiments (Fig. 3B). The splenic CFU for both Spv+ and Spv− salmonellae increased with treatment of mice with cyclophosphamide (32-fold [P = 0.01] and 250-fold [P < 0.02], respectively); however, the mean paired difference between the strains was not significantly affected by cyclophosphamide treatment (P > 0.2). Similarly, hepatic CFU for both strains were increased by cyclophosphamide treatment of mice (125-fold [P < 0.001] and 630-fold [P < 0.002], respectively, for Spv+ and Spv−), but the mean paired difference was not significantly affected (P = 0.2). Similar results were obtained on two other contemporary repetitions of this experiment. These latter experiments demonstrated a significant effect on systemic salmonella infection by depletion of PMNs; however, as in the previous set of experiments, the Spv phenotype was not affected by severe granulocytopenia. Therefore, PMNs are not likely to be involved in specific suppression of Spv− S. typhimurium, but macrophages were candidates for such a role.
Macrophages are essential for suppression of Spv− S. typhimurium.
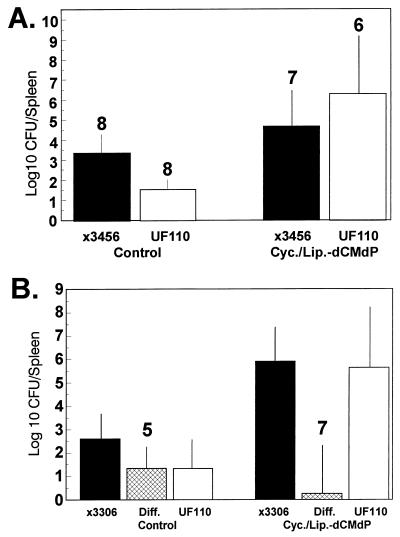
To examine the role of macrophages in suppressing the growth of Spv− S. typhimurium, mice were treated in a variety of ways which have been used by several investigators to physically or functionally deplete mice of macrophages. These treatments included i.v. injection of silica (1, 50) and i.v. injection of liposomes containing dCMdP (59, 61). After treatment of mice to deplete macrophages, mice were orally inoculated with Spv+ and/or Spv− S. typhimurium, and splenic CFU were measured 4 to 5 days later. With these treatments, we observed that both Spv+ and Spv− strains were increased for splenic infection 10- to 100-fold. However, the recoveries of the two strains relative to each other either remained the same or the difference increased—approximately 100-fold-higher numbers of Spv+ than Spv− salmonellae (data are provided for liposome-dCMdP treatment in Fig. 4). Injection of liposome-PBS resulted in splenic infection no different than injection of PBS alone (Fig. 4). The mean paired difference between Spv+ χ3456 and Spv− χ3337 was unchanged as a result of liposome-dCMdP treatment (Fig. 4). Our initial interpretation was that macrophages were essential for suppression of systemic disease by S. typhimurium but that macrophages were not involved in differential suppression of replication of Spv− S. typhimurium. However, when we stained macrophages in frozen sections of infected mouse tissues with rabbit anti-murine macrophage antiserum and later with monoclonal antibody FA/11 (22, 51), we observed that treatment with silica or liposome-dCMdP failed to completely deplete infected spleens of macrophages (data are shown for liposome-dCMdP in Fig. 5A and B). We further reasoned that although the treatments may have initially depleted resident macrophages, the increased systemic infection with salmonellae may have elicited monocytes and macrophages from the bone marrow. In fact, Samsom et al. showed that liposome-dCMdP treatment does not inhibit the elicitation of macrophages as part of an inflammatory response (55). We therefore treated mice with liposome-dCMdP on the day of inoculation and 2 days later to deplete any elicited macrophages. However, even with this double treatment, spleens still contained readily detectable macrophages, and the differential growth yields persisted between Spv+ and Spv− salmonellae (data not shown).
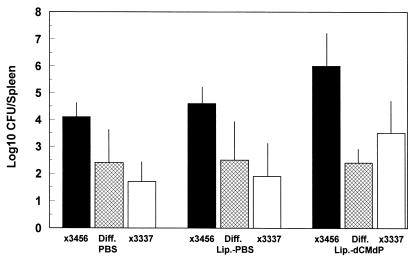
FIG. 4.
Treatment of mice with liposome-dCMdP alone increases splenic infection by Spv+ and Spv− S. typhimurium but does not relieve differential suppression of Spv− S. typhimurium. Mice were injected i.v. with PBS, liposome (Lip.)-PBS, or liposome-dCMdP 1 day before oral inoculation with mixtures of Spv+ χ3456 and Spv− χ3337 as described in the text. Four days later, spleens were removed, and CFU were enumerated. Data are mean log10 CFU with standard deviations shown, and the mean paired difference between χ3456 and χ3337 (Diff.) is shown (n = 4 for all groups). In all three sets, the mean paired difference is significantly greater than 0 (P < 0.05 to P < 0.005). Liposome-dCMdP treatment resulted in significantly increased splenic infection for both χ3456 and χ3337 compared with PBS treatment or liposome-PBS treatment (P < 0.05), except for χ3337, which was not significantly increased compared with liposome-PBS. Note that as shown in Fig. 5, liposome-dCMdP treatment did not completely eliminate splenic macrophages.
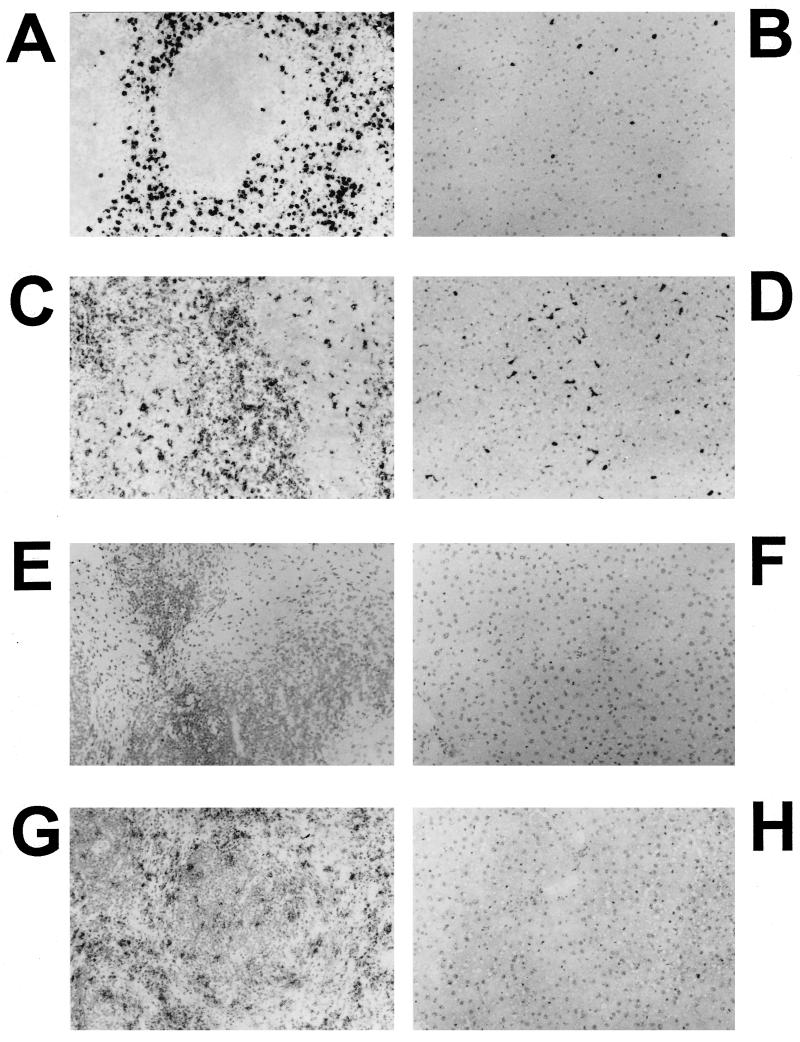
FIG. 5.
Immunochemical analysis of PMNs and macrophages in spleens of liposome-dCMdP and control treated mice infected with wild-type S. typhimurium. Spleens and livers (not shown) were prepared as described for Fig. 2 with rat anti-mouse granulocyte monoclonal antibody RB6-8C5 (A, C, and E) and rat anti-mouse macrophage monoclonal antibody FA/11 (B, D, and F). All images are at a magnification of ×200. (A and B) Spleen from infected mouse treated with PBS; (C and D) spleen from infected mouse treated with liposome-PBS; (E and F) spleen from infected mouse treated with liposome-dCMdP for depletion of macrophages. Note the continued presence of PMNs and macrophages with all treatments. Reactions from the negative control antibodies were clean (data not shown).
Since it was possible that monocytes and macrophages were still being recruited from the bone marrow as a result of the infection process, we attempted depletion of these phagocytes by combining cyclophosphamide and liposome-dCMdP treatment. The cyclophosphamide would destroy the macrophage-regenerative function of the bone marrow, and the liposome-dCMdP would kill peripheral macrophages. These mice would also be depleted of PMNs; however, results described above indicated that PMNs did not affect the Spv phenotype. Immunochemical staining of spleens and livers indicated that the efficiency of depletion of macrophages by cyclophosphamide–liposome-dCMdP treatment was sometimes variable, whereas tissues were consistently depleted of PMNs (Fig. 6). Reading depletion of macrophages was complicated by the fact that once macrophages had been destroyed, FA/11-staining debris remained in tissues, presumably because there were no phagocytes to clear the material. However, examination of tissues at a magnification of ×1,000 (Fig. 6I to K) and with immunofluorescence (data not shown) enabled the distinction between residual intact macrophages and debris due the lack of nuclei associated with debris and any cellular morphology. We made no conclusions as to the viability of intact-appearing macrophages.
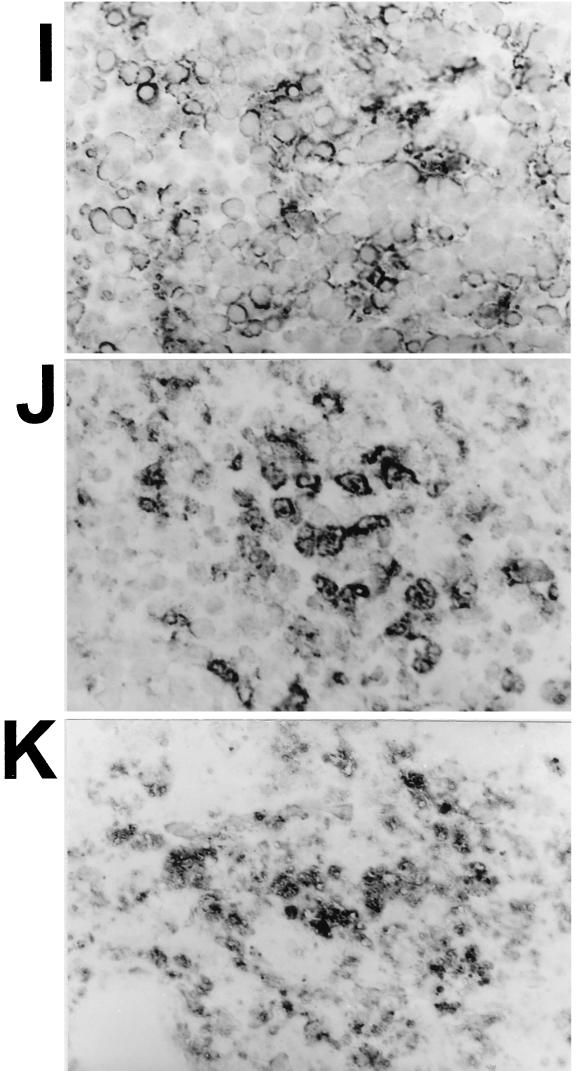
FIG. 6.
Immunochemical analysis of PMNs and macrophages in spleens and livers of cyclophosphamide–liposome-dCMdP and control treated mice. Mice were treated with cyclophosphamide–liposome-dCMdP or PBS–liposome-PBS and orally infected with Spv+ χ3456 or Spv− UF110 (not shown) as described in the text. Three days later, spleens and livers were prepared as described for Fig. 2 with rat anti-mouse granulocyte monoclonal antibody RB6-8C5 (A, B, E, and F) and rat anti-mouse macrophage monoclonal antibody FA/11 (C, D, and G to K). Magnifications: A to H, are ×200; I to K, ×1,000. Images show spleens (A, C, and J) and livers (B and D) from infected mice treated with PBS/liposome-PBS, spleens (E, G, and K) and livers (F and H) from infected mice treated with cyclophosphamide–liposome-dCMdP for depletion of macrophages, and spleen from a normal, uninfected mouse (I). Note the absence of PMNs in panels E and F and the lack of macrophages in panel H. The FA/11-staining material in panel G is debris, not intact macrophages, as determined by examination at higher power in panel K. Note the lack of nuclei associated with the FA/11-staining debris in panel K compared with the associated nuclei and cellular staining in panels I and J.
Initial experiments using the cyclophosphamide–liposome-dCMdP depletion procedure involved inoculation of mice with either Spv+ or Spv− S. typhimurium. The cyclophosphamide–liposome-dCMdP-treated mice were extremely sick and had to be sacrificed as soon as 3 days postinoculation, even without infection with salmonellae. The recoveries of Spv+ and Spv− salmonellae from spleens and livers were greatly increased, but extremely large ranges of recoveries were observed (Fig. 7A). The relative level of macrophage depletion correlated with the yield of salmonellae. In fact, mice which did not contain intact macrophages or PMNs in spleens and livers possessed similar levels of Spv+ and Spv− S. typhimurium, and the highest three splenic recoveries were from mice infected with Spv− UF110. When cellular depletion of mice was less efficient, the recoveries of both strains were reduced, and the recovery of Spv+ was higher than for Spv− salmonellae.
FIG. 7.
Combined treatment with cyclophosphamide–liposome-dCMdP results in equivalent splenic infection by Spv+ and Spv− S. typhimurium. Mice were treated with cyclophosphamide–liposome-dCMdP (Cyc./Lip.-dCMdP) or PBS–liposome-PBS (Control) as described in the text and infected with Spv+ and/or Spv− S. typhimurium. (A) Single-strain inoculation. Mice were infected with either Spv+ χ3456 or Spv− UF110. Three days later, splenic CFU were examined. Mean with standard deviation of log10 splenic CFU for χ3456 or UF110 are shown; n for each group is shown above the bars. In control-treated mice, χ3456 was significantly higher than UF110 for splenic CFU (P < 0.05); however, in cyclophosphamide–liposome-dCMdP-treated mice, recovery of the Spv− strain was insignificantly higher than the Spv+ strain (P > 0.05). Similar results were obtained for livers (data not shown). (B) Mixed-strain inoculation. Mice were infected with a mixture of Spv+ χ3306 and Spv− UF110. Two to three days later, CFU of each strain in spleens were examined. Mean log10 CFU with standard deviation for χ3306, UF110, and the mean paired difference (Diff.) are shown. In control-treated mice, the mean paired difference between χ3306 and UF110 was significantly greater than 0 (P < 0.05); however, in cyclophosphamide–liposome-dCMdP-treated mice the mean paired difference between χ3306 and UF110 was not significantly greater than 0 (P > 0.5). Similar results were obtained for livers (data not shown). Numbers of mice in each group are shown above the bar for the mean paired difference.
To examine the relative infection with Spv+ and Spv− salmonellae together in the same mouse experiencing the same macrophage background in light of variable phagocyte depletion noted above, we inoculated cyclophosphamide–liposome-dCMdP-treated mice with mixtures of Spv+ χ3306 and Spv− UF110 (Fig. 7B). The cyclophosphamide–liposome-dCMdP treatment for the mixed infection was more efficient than the previous attempts and made the mice so sick that the experiment had to be terminated between 2 and 3 days postinoculation. In control-treated mice, the mean paired difference between Spv+ and Spv− S. typhimurium was 101.3 (P < 0.05 for mean paired difference greater than 0), while in the macrophage-depleted mice, the mean paired difference was 100.3 (P > 0.5 for mean paired difference greater than 0). Similar results were obtained with livers from the same mice (data not shown). These results demonstrate that macrophages are essential for mice to differentially suppress the replication of Spv− S. typhimurium, either by serving as the primary host cells in which Spv-mediated intracellular replication occurs or by acting as secondary cells which suppress infection of a different, primary host cell by Spv− salmonellae.
Invasion of nonphagocytic host cells is not necessary for the functional expression of the Spv phenotype or systemic infection by S. typhimurium after s.c. inoculation.
The experiments described above left open the possibility that Spv-mediated growth occurred within a nonphagocytic host cell, e.g., hepatocyte. In fact, Conlan and North (8) reported that S. typhimurium proliferates within hepatocytes of i.v.-inoculated mice. To examine if infection of nonphagocytic host cells was important in the Spv phenotype of increased intracellular growth rate, salmonellae were mutated so as to be defective for intracellular infection of nonphagocytes. This was accomplished by transduction with the invA::aphT mutation, which inhibits invasion into, but not adherence to, nonphagocytic cells (21). We confirmed that InvA− Spv+ UF102 and InvA− Spv− UF103 were approximately 1,000-fold inhibited for invasion into Henle-407 cells and that the invasion defective genotype could be complemented in trans by plasmid pYA2217 encoding the S. typhimurium inv region (20) (data not shown). To examine if InvA− salmonellae could still be phagocytosed by macrophages after opsonization with complement, which would most closely mimic the in vivo condition, J774.1 cells or peritoneal macrophages were infected with InvA+ and InvA− S. typhimurium. We detected a slight difference in the abilities of the bacteria to adhere to and be taken up by macrophages in the absence of complement. However, opsonization with 10% normal rat serum more than compensated for any deficiency of InvA− salmonellae to enter macrophages (data not shown).
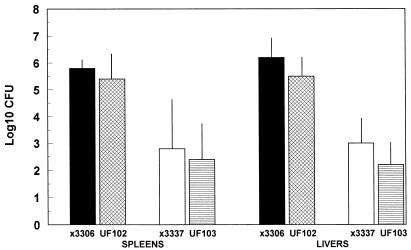
Since InvA− S. typhimurium strains are attenuated by the oral route (20), mice were injected s.c. with 105 CFU of χ3456, χ3337, UF102, or UF103 by single or mixed inoculation. We previously showed that the Spv virulence function is required for systemic infection after s.c. inoculation in our model system (30). Five days later, spleens and livers were examined for log10 CFU of each strain (Fig. 8). The invA::aphT mutation resulted in an average 0.4-log decrease in splenic CFU for both virulence plasmid-containing and cured salmonellae, P > 0.1 to 0.5 for the differences being >0. However, there was still a 1,000-fold difference in the splenic recoveries between Spv+ and Spv− S. typhimurium, regardless of the invA genotype (P < 0.01 to 0.005). Similar results were obtained for hepatic infection; however, the invA::aphT mutation resulted in slightly higher, yet still insignificant, decreases in hepatic recovery (Fig. 8). In other experiments, we noted either no decrease in splenic infection at all for either Spv background or as high as 2.2-log decreases for plasmid-cured recovery from spleens (although very rare) in InvA− salmonellae. However, in every case, inhibition of invasion into nonphagocytes did not affect the Spv phenotype of increased recovery of Spv+ versus Spv− salmonellae from spleens. Therefore, invasion of nonphagocytic cells is irrelevant to the Spv virulence function of increased intracellular growth rate. Furthermore, these results suggest that invasion of nonphagocytic cells by S. typhimurium is not essential for systemic infection if inoculation bypasses the intestines. Therefore, the only cells identified in this study as being related to the Spv virulence function were macrophages.
FIG. 8.
Preventing invasion of nonphagocytes by the invA::aphT mutation does not significantly affect systemic infection by either Spv+ or Spv− S. typhimurium after s.c. inoculation of mice. Mice were inoculated s.c. with Spv+/InvA+ χ3306, Spv+/InvA− UF102, Spv−/InvA+ χ3337, or Spv−/InvA− UF103. Five days later, splenic and hepatic CFU were enumerated. Data are shown as mean with standard deviation for log10 CFU/spleen or CFU/gram of liver. All groups contain four mice. Differences between χ3306 and χ3337 or UF102 and UF103 were significant (P < 0.02 to P < 0.005); however, differences between χ3306 and UF102 or χ3337 and UF103 were not significant (P > 0.1 to P > 0.5).
DISCUSSION
Molecular genetic analysis of the spv genes of the salmonella virulence plasmid has proceeded at a rapid pace (reviewed in references 25 and 29). Although a region of the virulence plasmid encoding five genes, spvRABCD, has been identified through cloning, mutagenesis, and DNA sequencing, the exact roles of most of these genes in virulence are not known (29). The SpvR protein is a positive regulator of spvABCD (29). Regulation of the spv genes is dependent on the alternative sigma factor RpoS (14, 48, 66), and the genes are expressed within host cells (16, 52, 66). We have determined that the spv genes do not require a cessation of bacterial growth for induction but instead can be greatly induced in minimal medium mimicking the intracellular environment of animal cells (66, 67).
The elucidation of the virulence plasmid phenotype has undergone significant evolution since the first description of the plasmid as being associated with virulence (36). Initial studies suggested that the plasmid of S. typhimurium was involved with adherence and invasion into tissue culture cells, serum resistance, and lipopolysaccharide biosynthesis; however, each of these hypotheses has not proven reproducible (29). Our own data demonstrated that plasmid-cured S. typhimurium was deficient in recovery from lymphoid tissues after oral inoculation of BALB/c mice but was capable of surviving in the lymph nodes and spleen for extended periods (28). These results were not consistent with the plasmid affecting resistance to complement or survival within phagocytes. Indeed, short-term survival rates of virulence plasmid-containing and cured salmonellae were equivalent in murine macrophages (28). Others have confirmed this result and extended in vitro infection of macrophages to as long as 24 h without demonstrable differences between wild-type and cured strains (54). In only a single study by Libby et al. (40) was a moderate divergence achieved in recoveries of plasmid-containing and cured S. dublin in cultured bovine macrophages. However, the difficulty in reproducing the Spv phenotype in vitro does not definitively disprove a role for the spv genes in these phenotypes in vivo. In vitro models may not sufficiently mimic the complexities of the host-pathogen interaction which occurs in vivo. We therefore concentrated our studies of the Spv phenotype by using the mouse model. By using genetic markers (temperature-sensitive antibiotic resistance plasmid pHSG422) and genetic manipulation (ΔaroA), we previously demonstrated that the spv genes primarily increase the growth rate of S. typhimurium in BALB/c mice without significantly affecting either the killing or movement of the bacteria in the mice (30). The resistance to in vivo-administered gentamicin by both virulence plasmid-containing and cured S. typhimurium strongly suggested that both strains were located intracellularly within host cells. The equal yields of both strains from the extracellular environment of peritoneal chamber implants supported this conclusion. However, the relevant host cell(s) for either permitting the intracellular proliferation of Spv+ S. typhimurium or conversely suppressing the intracellular growth of the Spv− derivative was not identified. Our study described here was executed to probe the roles of different host cells in the salmonella Spv-mediated phenotype of increased intracellular growth rate.
Lymphocytes are not involved with Spv-mediated virulence.
Since T cells can affect the abilities of macrophages and other cells to control intracellular pathogens such as S. typhimurium (37) and since S. typhimurium has been shown to infect lymphocyte cell lines in vitro (62), we examined the effect of lymphocytes on the Spv phenotype in BALB/c mice by using homozygous nude BALB/c mice and SCID BALB/c mice. By infection of normal and lymphocyte-deficient mice with mixtures of Spv+ and Spv− S. typhimurium, we observed no significant effect on recoveries of either bacterial strain from spleens of orally inoculated mice within 5 days of inoculation (Fig. 1). We also demonstrated that T cells and B cells do not have a significant role in the Spv phenotype of increased intracellular growth rate. This is not surprising since the Spv phenotype is exerted as soon as 3 days after oral inoculation (28), and T cells probably do not have time to be stimulated and have any effect at this early time of the disease process. Our results are most consistent with those of Weintraub et al. (64) and Guilloteau et al. (24). Weintraub et al. performed a comprehensive analysis of the roles of αβ and γδ T cells in both Ityr and Itys mice which were genetically deficient in T-cell subsets (64). In Itys C57BL/6 mice, infection with wild-type S. dublin was unaffected within 4 days of oral inoculation even when αβ- and γδ-T cells were lacking. Guilloteau et al. found that splenic and hepatic infection of SCID CB-17/IcrCru mice after i.p. inoculation was unaffected compared with normal BALB/c mice until 11 days postinoculation. Others have used a variety of congenic mouse strains, reconstitution, and depletion procedures to show in Ityr mice that T cells are not important in suppressing the initial net replication of S. typhimurium after i.v. inoculation (34, 42, 49). In contrast, Emoto et al. (13) reported that mice genetically deficient for γδ T cells were more resistant to i.p. infection with S. choleraesuis. Mixter et al. (46) depleted BALB/c mice of T-cell subsets by using monoclonal antibodies to the αβ and γδ T-cell receptors and reported that depletion of αβ T cells decreased the oral 50% lethal dose (LD50) of S. enteritidis 104-fold, whereas depletion of γδ T cells decreased the LD50 300-fold. The oral LD50 determinations using S. enteritidis involved infection for as long as 14 days. A protective role for αβ T cells within a week of i.p. inoculation of BALB/c mice with S. choleraesuis was reported by Matsumoto et al. (44), who depleted mice by using a monoclonal antibody. We feel that most emphasis should be placed on the use of genetically deficient mice and that care should be taken in interpreting results obtained by depleting mice of cell populations with antibodies. Phagocytes such as macrophages could become occupied clearing lysed host cells in tissues and thereby be less available for inhibiting the salmonella infection.
The fact that mixing of Spv+ and Spv− strains in infection of mice does not result in either the functional complementation of the mutant strain or adverse consequences to the wild-type strain (28, 30) suggests that the spv genes do not exert their effect via induction or suppression of global immune responses in mice. Instead, the spv genes probably affect the host-pathogen interaction at the individual cell level. Consistent with this hypothesis, we found that depletion of gamma interferon and/or tumor necrosis factor alpha from BALB/c mice did not increase the virulence of Spv− S. typhimurium relative to the Spv+ strain (32). These cytokines therefore are not responsible for differential suppression of growth of Spv− salmonellae. This result is consistent with a lack of effect of T cells on the Spv phenotype, since T cells would be expected to produce gamma interferon as well as tumor necrosis factor alpha. Contrary to our hypothesized lack of involvement of the spv genes in global regulation of immune stimulation, Emoto et al. (12) reported data suggesting that the virulence plasmid of S. choleraesuis inhibited the elicitation of γδ T cells during infection of mice. However, Guilloteau et al. (24) and we (45a) could not reproduce this result in studies using S. dublin and S. typhimurium, respectively.
Macrophages are essential for Spv-mediated virulence.
We next examined the roles of phagocytes in selecting against the growth of Spv− S. typhimurium. We envisioned that either phagocytes were the permissive cell type for Spv-mediated intracellular growth or these cells suppressed Spv− salmonellae within nonphagocytic cells. The potential roles of phagocytes in Spv-mediated virulence were examined by depletion of phagocyte populations from mice. At necropsy, cyclophosphamide treatment depleted circulating PMNs by more than 90%, depleted spleens and livers of PMNs, but did not decrease tissue macrophages. However, PMN depletion had no significant effect on the differential recovery of Spv+ S. typhimurium compared with Spv− S. typhimurium from the spleens and livers of orally inoculated mice within 5 days of inoculation (Fig. 3). Between two sets of experiments performed months apart, we obtained different results on the effects of granulocytopenia on recovery of salmonellae from spleens and livers of orally inoculated mice. In the first set of experiments, there was no effect of granulocytopenia on either Spv+ or Spv− salmonellae; however, in the second set of experiments, there was a significant increase in CFU recovered from systemic sites for both strains. These results suggest that PMNs may have a role in suppressing systemic infection by S. typhimurium but are not involved with or necessary for the suppression of Spv− salmonellae. At the present, we cannot explain the differences between the two sets of experiments. In addition to PMNs, cyclophosphamide treatment would be expected to affect other rapidly dividing cell populations, including elicited monocytes that are generated from bone marrow precursors. Additionally, it is possible that the intestinal mucosa of the mice reacted differently between the two sets of experiments and that in the latter set, salmonellae were able to invade into deeper tissues more easily. We did not examine for effects on other cell populations, other than demonstrating that macrophages were still present in spleens and livers of cyclophosphamide-treated mice. Alternatively, the time course of functional PMN depletion may have been different between the experiments, and in those experiments in which recovery of salmonellae was increased by granulocytopenia, the PMN depletion may have occurred more rapidly than in those in which no significant effects on systemic infection were observed. The results from the first set of granulocytopenic mice (Fig. 3A) differ from, while those from the second set (Fig. 3B) agree with, results of Conlan (6) and Conlan and North (7), who used the same monoclonal antibody that we used for immunocytochemistry, RB6-8C5, to eliminate PMNs from mice. In these studies, which involved i.v. inoculation of mice with salmonellae, significant increases in systemic infection by S. typhimurium were noted in PMN-depleted mice. As discussed below, results from others examining effects on systemic salmonella infection with total body irradiation of mice, which induced granulocytopenia (24, 33), were as mixed as our own data shown in Fig. 3. A more definitive analysis of the role of PMNs in salmonella infection to reconcile all of these observations will require more detailed analysis utilizing granulocytopenia induced by both chemical (e.g., cyclophosphamide) and other (e.g., antigranulocyte monoclonal antibody and irradiation) treatments.
Many investigators have postulated that the virulence plasmid affects the interaction of salmonellae within murine macrophages because the plasmid affects the bacterial recoveries from lymphoid tissues. However, only a single reported study achieved differential infection of cultured macrophages by plasmid-containing and cured salmonellae in vitro (40). To examine the role of macrophages, we attempted to deplete mice of macrophages by using numerous procedures, including injection of silica and liposome-dCMdP; however, histological analysis of tissues from treated and infected mice consistently revealed large numbers of residual macrophages (Fig. 5). It is important to note that the choice of macrophage-specific antibody had a large bearing on the sensitivity of this analysis. Initial use of rabbit anti-murine macrophage antiserum failed to sensitively identify residual macrophages since white pulp macrophages were not stained, even in normal mouse tissues (data not shown). However, when we obtained monoclonal antibody FA/11 (22, 51), greater sensitivity and specificity were possible, thereby demonstrating the lack of effectiveness of the aforementioned macrophage depletion procedures. It is believed that treatment of mice with silica may stimulate and occupy macrophages (58, 63); however, some have stated that this treatment kills macrophages (1). The more recently developed treatment with liposome-dCMdP has been documented to kill and clear macrophages from various mouse organs and tissues, depending on the route of administration (59–61). Macrophages phagocytose the liposomes, degrade the vesicles, and thereby release the toxic dCMdP to kill themselves. Injection i.v. of liposome-dCMdP primarily clears the spleen and liver of different macrophage populations for at least 1 week (60). However, when we used this treatment with mice orally inoculated with S. typhimurium, numerous macrophages were still detectable in livers and spleens 4 to 5 days postinoculation (Fig. 5). In retrospect, this was not unexpected since liposome-dCMdP treatment does not clear circulating monocytes or prevent inflammatory responses by monocytic cells (55). With all of the aforementioned treatments to clear mice of macrophages, the splenic and hepatic recoveries of both Spv+ and Spv− S. typhimurium were increased as much as 100-fold; however, the 100-fold difference in recoveries between the Spv+ and Spv− strains remained. Clearly, macrophages were important for suppressing systemic infection by both Spv+ and Spv− S. typhimurium after oral inoculation; however, their role in specifically suppressing Spv− salmonellae relative to Spv+ salmonellae was unclear. The residual macrophages in infected tissues were a serious concern. Since we reasoned that the macrophages observed during the course of the salmonella infection were recruited from the bone marrow, we performed experiments in which the bone marrow was depleted of precursor cells by cyclophosphamide followed by depletion of peripheral macrophages with liposome-dCMdP. This double treatment often produced spleens and livers devoid of intact macrophages (Fig. 6). There was still considerable material which stained with macrophage-specific anti-CD68 monoclonal antibody FA/11; however, this would be expected since there would be no phagocytic cells to clear lysed macrophages. Most important, when depletion of macrophages was complete or nearly so, Spv+ and Spv− S. typhimurium strains were equally virulent in terms of splenic and hepatic infection after oral inoculation. In fact, in phagocyte-depleted mice infected with single salmonella strains, Spv− salmonellae produced the highest levels of infection (Fig. 7A). In mixed strain inoculations of cyclophosphamide–liposome-dCMdP-treated mice, there was not a significant difference between Spv+ and Spv− S. typhimurium for splenic and hepatic infection (Fig. 7B).
These results argue that macrophages, as opposed to PMNs, are required for the Spv phenotype of increased intracellular growth rate in systemic infection of mice. Macrophages could either act as the primary host cell in which Spv-mediated replication occurs or suppress the Spv− salmonellae in some other host cell. The fact that the cyclophosphamide-plus-liposome-dCMdP-treated mice were also depleted of PMNs is of note. However, the lack of effect on the Spv phenotype by quantitatively depleting mice of tissue as well as circulating PMNs with cyclophosphamide treatment strongly argues that the simultaneous depletion of PMNs did not significantly contribute to the lack of Spv phenotype seen on the double treatment of mice. We cannot completely disprove this possibility. As noted above, we speculate that the continued Spv phenotype in mice treated only with liposome-dCMdP could be due to the elicitation of monocytes and macrophages from the bone marrow after the resident macrophages were cleared by the liposome-dCMdP treatment. If this is true, it would suggest that inflammatory, elicited macrophages are Spv−-suppressive cells. Since mice treated only with cyclophosphamide should contain only resident macrophages with no PMNs and elicited macrophages, the continued difference in systemic infection between Spv+ and Spv− salmonellae argues that resident macrophages are capable of differentially suppressing Spv− salmonellae as well as elicited macrophages. Two other groups have performed studies on phagocytes which are relevant to our study. Heffernan et al. (33) examined i.p. infection of irradiated BALB/c mice with virulence plasmid-cured S. dublin. During the first week after irradiation and infection, when resident macrophages would be present in tissues but there would be no elicited monocytes, hepatic infection with virulence plasmid-cured S. dublin was unchanged compared with normal mice. After the resident macrophages were expected to have been depleted from tissues, the plasmid-cured strain began to proliferate and kill the mice. Since plasmid-containing S. dublin was not examined in irradiated mice, a more complete analysis with respect to the Spv phenotype is not possible from that study. The equivalent infection by plasmid-cured S. dublin during the first week of infection argues that radiosensitive host cells such as PMNs are dispensable for suppressing salmonellae, in agreement with our data in Fig. 3A. Guilloteau et al. (24) similarly infected irradiated BALB/c mice i.p. with virulence plasmid-containing and cured S. dublin but obtained results very different from those of Heffernan et al. (33). During the first week after irradiation and infection, both plasmid-containing and cured S. dublin strains were significantly increased for splenic and hepatic infection, but the differences between the recoveries of the two salmonella strains were maintained in the irradiated mice. These results argue that radiosensitive host cells such as PMNs suppress salmonella infection and that radioresistant cells such as macrophages are capable of differentially suppressing Spv− salmonellae relative to Spv+ salmonellae. This is consistent with our data in Fig. 3B. Between the studies by Heffernan et al. (33), Guilloteau et al. (24), and us, the complexities of oral versus i.p. inoculation of S. typhimurium versus S. dublin, and the different means of depleting phagocytes, make comparison of the data difficult. However, all of the data are consistent with macrophages being suppressors of salmonella infection in general and mediators of the differential suppression of Spv− salmonellae resulting in the Spv phenotype.
Another interpretation to the equalized infectivities of Spv+ and Spv− S. typhimurium in cyclophosphamide plus liposome-dCMdP-treated mice in our studies is that all of the bacteria were extracellular at the systemic sites. We did not examine this possibility. There would have been no phagocytes for uptake of the bacteria, and our results reported here and for studies using immunofluorescence (31) demonstrate that invasion of nonphagocytes is not important during infection beyond the intestines. Most recently, Richter-Dahlfors et al. (53) used confocal microscopy to demonstrate that most salmonellae reside within macrophages in livers of i.v.-inoculated mice; hepatocytes were not significantly infected. The extracellular infection in phagocyte-depleted mice is reminiscent of our previous study in which Spv+ and Spv− S. typhimurium strains were maintained in an extracellular environment within a porous i.p. chamber implant and demonstrated equivalent growth yields (30). In any case, it is clear that the presence of macrophages is critical for the Spv phenotype of increased intracellular replication to occur.
Two technical points are worth emphasizing with regard to depletion of macrophages since these are becoming highly used procedures. First, the procedures previously used (i.e., silica and liposome-dCMdP) were not sufficient in themselves to quantitatively deplete mice of macrophages in the face of systemic salmonella infection. Second, the method used for analysis of residual macrophages in tissues is critical. When we used an antimacrophage serum, we failed to detect white pulp macrophages. However, the use of anti-CD68 monoclonal antibody FA/11 gave a greatly different result. Many investigators have used a variety of monoclonal antibodies, including F4/80, MAC-1, MOMA-1 and MOMA-2, and others, to detect macrophages (18, 22, 38). It is important to note that these antibodies recognize subpopulations of macrophages depending on their tissue localization or stage of activation/differentiation. It is our opinion that FA/11 is the best pan-macrophage marker available.
Invasion of nonphagocytes is not involved with Spv-mediated virulence and systemic infection beyond the intestines.
Since we could not deplete mice of nonphagocytic cells, as we did for phagocytes and lymphocytes, we denied the bacteria access to the intracellular location of nonphagocytes with a mutation known to inhibit invasion, invA::aphT (21). invA is the 5′ gene of the invABC operon located in pathogenicity island I at centisome 63 on the S. typhimurium chromosome (19). A polar mutation in invA, which abolished the expression of downstream inv genes, severely impeded the invasive phenotype of virulent strains of S. typhimurium and caused a significant increase in the LD50 when administered orally to BALB/c mice (20). Our intended use of an invA mutation required that InvA− salmonellae not invade any nonphagocytes in vivo and that InvA− salmonellae could still be phagocytosed by phagocytes. Inv− S. typhimurium is unable to invade hepatocytes in vitro (19a). We (data not shown) and others (4, 47) have confirmed that Inv− salmonellae are phagocytosed by macrophages in vitro in nearly normal levels. We therefore assume that InvA− S. typhimurium used in our studies have access to the intracellular environment of murine macrophages in vivo after s.c. inoculation. In fact, we have confirmed this hypothesis by using immunofluorescence analysis of tissues from infected mice (31).
When S. typhimurium was denied access to the intracellular environment of nonphagocytic cells after s.c. inoculation, there was very little effect on splenic or hepatic infection by Spv+ salmonellae, and there was only a slight effect on Spv− salmonellae. The former result argues that salmonellae probably do not infect nonphagocytes during infection after s.c. inoculation or that if invasion of nonphagocytes does occur, it is without significance for systemic infection. Our results differ with those of Conlan and North (8), who investigated the location of S. typhimurium and other intracellular pathogens in the livers of i.v.-inoculated mice. Based on microscopic analysis of infected mouse tissues, they reported that salmonellae replicated within hepatocytes, and that PMNs acted to prevent this replication. The major difference between our study and that of Conlan and North (8) is the route of inoculation. We believe that i.v. injection of broth-grown salmonellae into mice enables an artifactual infection of nonphagocytic cells. However, it is clear from our results that salmonellae do not have to invade nonphagocytes to exert the Spv phenotype of increased intracellular replication. This finding argues that the relevant host cell for Spv-mediated intracellular growth is a phagocyte, and the results discussed above indicate that this phagocyte is a macrophage.
ACKNOWLEDGMENTS
This work was supported by NIH grant AI28421, USDA grant 9502044, and American Heart Association-Florida Affiliate grants 89GIA81 and 92GIA868 to P.A.G., who is an American Heart Association Established Investigator with funds provided in part by the American Heart Association-Florida Affiliate.
We thank Jorge E. Galán for providing S. typhimurium SB136 with the invA::aphT mutation and for his advice on these studies. We are extremely grateful to Siamon Gordon for providing monoclonal antibody FA/11, which has been indispensable to this work. We thank Steven Roberts and Vince A. Chiodo for expert technical assistance. We thank Andreas Baumler and Michael Clare-Salzler for review of the manuscript. We thank the reviewers of the original study and manuscript, whose comments resulted in our development of the improved techniques described in this paper and greatly changed the conclusions of the study.
REFERENCES
- 1.Allison A C. Fluorescence microscopy of lymphocytes and mononuclear phagocytes and the use of silica to eliminate the latter. In: Bloom B, David J, editors. In vitro methods in cell mediated and tumor immunity. New York, N.Y: Academic Press Inc.; 1976. pp. 395–404. [Google Scholar]
- 2.Caldwell A L, Gulig P A. The Salmonella typhimurium virulence plasmid encodes a positive regulator of a plasmid-encoded virulence gene. J Bacteriol. 1991;173:7176–7185. doi: 10.1128/jb.173.22.7176-7185.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter P B, Collins F M. The route of enteric infection in normal mice. J Exp Med. 1974;139:1189–1203. doi: 10.1084/jem.139.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L M, Kaniga K, Galán J E. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 5.Chesebro B, Britt W, Evans L, Wehrly K, Nishio J, Cloyd M. Characterization of monoclonal antibodies reactive with murine leukemia viruses: Use in analysis of strains of Friend MCF and Friend ecotropic murine leukemia virus. Virology. 1983;127:134–148. doi: 10.1016/0042-6822(83)90378-1. [DOI] [PubMed] [Google Scholar]
- 6.Conlan J W. Neutrophils prevent extracellular colonization of the liver microvasculature by Salmonella typhimurium. Infect Immun. 1996;64:1043–1047. doi: 10.1128/iai.64.3.1043-1047.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conlan J W. Critical roles of neutrophils in host defense against experimental systemic infections of mice by Listeria monocytogenes, Salmonella typhimurium, and Yersinia enterocolitica. Infect Immun. 1997;65:630–635. doi: 10.1128/iai.65.2.630-635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conlan J W, North R J. Early pathogenesis of infection in the liver with the facultative intracellular bacteria Listeria monocytogenes, Francisella tularensis, and Salmonella typhimurium involves lysis of infected hepatocytes by leukocytes. Infect Immun. 1992;60:5164–5171. doi: 10.1128/iai.60.12.5164-5171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtiss R., III . Gene transfer. In: Gerhardt P, Murray R G E, Costilow R, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C: American Society for Microbiology; 1981. pp. 243–265. [Google Scholar]
- 10.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunlap N E, Benjamin W H, Jr, Berry A K, Eldridge J H, Briles D E. A ‘safe site’ for Salmonella typhimurium is within splenic polymorphonuclear cells. Microb Pathog. 1992;13:181–190. doi: 10.1016/0882-4010(92)90019-k. [DOI] [PubMed] [Google Scholar]
- 12.Emoto M, Danbara H, Yoshikai Y. Induction of gamma/delta T cells in murine salmonellosis by an avirulent but not by a virulent strain of Salmonella choleraesuis. J Exp Med. 1992;176:363–372. doi: 10.1084/jem.176.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emoto M, Nishimura H, Sakai T, Hiromatsu K, Gomi H, Itohara S, Yoshikai Y. Mice deficient in γδ T cells are resistant to lethal infection with Salmonella choleraesuis. Infect Immun. 1995;63:3736–3738. doi: 10.1128/iai.63.9.3736-3738.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang F C, Libby S J, Buchmeier N A, Loewen P C, Switala J, Harwood J, Guiney D G. The alternative sigma factor KatF (RpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fields P I, Swanson R V, Haidaris C G, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fierer J, Eckmann L, Fang F, Pfeifer C, Finlay B B, Guiney D. Expression of the Salmonella virulence plasmid gene spvB in cultured macrophages and nonphagocytic cells. Infect Immun. 1993;61:5231–5236. doi: 10.1128/iai.61.12.5231-5236.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleming T J, Fleming M L, Malek T R. Selective expression of Ly-6G on myeloid lineage cells in mouse bone marrow. RB6-8C5 mAb to granulocyte-differentiation antigen (Gr-1) detects members of the Ly-6 family. J Immunol. 1993;151:2399–2408. [PubMed] [Google Scholar]
- 18.Fraser I, Doyle A, Hughes D, Gordon S. Use of surface molecules and receptors for studying macrophages and mononuclear phagocytes. J Immunol Methods. 1994;174:95–102. doi: 10.1016/0022-1759(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 19.Galán J E. Molecular genetic bases of Salmonella entry into host cells. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 19a.Galán, J. E. Personal communication.
- 20.Galán J E, Curtiss R., III Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galán J E, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;174:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon S, Lawson L, Rabinowitz S, Crocker P R, Morris L, Perry V H. Antigen markers of macrophage differentiation in murine tissues. Curr Top Microbiol Immunol. 1992;181:1–37. doi: 10.1007/978-3-642-77377-8_1. [DOI] [PubMed] [Google Scholar]
- 23.Groisman E A, Saier M H J. Salmonella virulence: new clues to intramacrophage survival. Trends Biochem Sci. 1990;15:30–33. doi: 10.1016/0968-0004(90)90128-x. [DOI] [PubMed] [Google Scholar]
- 24.Guilloteau L A, Lax A J, MacIntyre S, Wallis T S. The Salmonella dublin virulence plasmid does not modulate early T-cell responses in mice. Infect Immun. 1996;64:222–229. doi: 10.1128/iai.64.1.222-229.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guiney D G, Libby S, Fang F C, Krause M, Fierer J. Growth-phase regulation of plasmid virulence genes in Salmonella. Trends Microbiol. 1995;3:275–279. doi: 10.1016/s0966-842x(00)88944-1. [DOI] [PubMed] [Google Scholar]
- 26.Gulig P A, Caldwell A L, Chiodo V A. Identification, genetic analysis, and DNA sequence of a 7.8-kilobase virulence region of the Salmonella typhimurium virulence plasmid. Mol Microbiol. 1992;6:1395–1411. doi: 10.1111/j.1365-2958.1992.tb00860.x. [DOI] [PubMed] [Google Scholar]
- 27.Gulig P A, Chiodo V A. Genetic and DNA sequence analysis of the Salmonella typhimurium virulence plasmid gene encoding the 28,000-molecular-weight protein. Infect Immun. 1990;58:2651–2658. doi: 10.1128/iai.58.8.2651-2658.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gulig P A, Curtiss R., III Plasmid-associated virulence of Salmonella typhimurium. Infect Immun. 1987;55:2891–2901. doi: 10.1128/iai.55.12.2891-2901.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gulig P A, Danbara H, Guiney D G, Lax A J, Norel F, Rhen M. Molecular analysis of virulence genes of the salmonella virulence plasmids. Mol Microbiol. 1993;7:825–830. doi: 10.1111/j.1365-2958.1993.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 30.Gulig P A, Doyle T J. The Salmonella typhimurium virulence plasmid increases the growth rate of salmonellae in mice. Infect Immun. 1993;61:504–511. doi: 10.1128/iai.61.2.504-511.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gulig, P. A., S. Roberts, and T. J. Doyle. Salmonella typhimurium resides primarily within macrophages in spleens and livers after oral and subcutaneous inoculation of BALB/c mice, but does not infect hepatocytes except after intravenous inoculation. In Abstracts of the 1998 General Meeting of the American Society for Microbiology 1998, in press. American Society for Microbiology, Washington, D.C.
- 32.Gulig P A, Doyle T J, Clare-Salzler M J, Maiese R L, Matsui H. Systemic infection of mice by wild-type but not Spv−Salmonella typhimurium is enhanced by neutralization of gamma interferon and tumor necrosis factor alpha. Infect Immun. 1997;65:5191–5197. doi: 10.1128/iai.65.12.5191-5197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heffernan E J, Fierer J, Chikami G, Guiney D. Natural history of oral Salmonella dublin infection in BALB/c mice: effect of an 80-kilobase-pair plasmid on virulence. J Infect Dis. 1987;155:1254–1259. doi: 10.1093/infdis/155.6.1254. [DOI] [PubMed] [Google Scholar]
- 34.Hormaeche C E, Mastroeni P, Arena A, Uddin J, Joysey H S. T cells do not mediate the initial suppression of a Salmonella infection in the RES. Immunology. 1990;70:247–250. [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu H S. Pathogenesis and immunity in murine salmonellosis. Microbiol Rev. 1989;53:390–409. doi: 10.1128/mr.53.4.390-409.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones G W, Rabert D K, Svinarich D M, Whitfield H J. Association of adhesive, invasive, and virulent phenotypes of Salmonella typhimurium with autonomous 60-megadalton plasmids. Infect Immun. 1982;38:476–486. doi: 10.1128/iai.38.2.476-486.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufmann S H. Immunity to intracellular bacteria. Annu Rev Immunol. 1993;11:129–163. doi: 10.1146/annurev.iy.11.040193.001021. [DOI] [PubMed] [Google Scholar]
- 38.Leenen P J, de Bruijn M F, Voerman J S, Campbell P A, van Ewijk W. Markers of mouse macrophage development detected by monoclonal antibodies. J Immunol Methods. 1994;174:5–19. doi: 10.1016/0022-1759(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 39.Lennox E S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 40.Libby S J, Adams L G, Ficht T A, Allen C, Whitford H A, Buchmeier N A, Bossie S, Guiney D G. The spv genes on the Salmonella dublin virulence plasmid are required for severe enteritis and systemic infection in the natural host. Infect Immun. 1997;65:1786–1792. doi: 10.1128/iai.65.5.1786-1792.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin F R, Hsu H S, Mumaw V R, Moncure C W. Confirmation of destruction of salmonellae within murine peritoneal exudate cells by immunocytochemical technique. Immunology. 1989;67:394–400. [PMC free article] [PubMed] [Google Scholar]
- 42.Maskell D J, Hormaeche C E, Harrington K A, Joysey H S, Liew F Y. The initial suppression of bacterial growth in a salmonella infection is mediated by a localized rather than a systemic response. Microb Pathog. 1987;2:295–305. doi: 10.1016/0882-4010(87)90127-6. [DOI] [PubMed] [Google Scholar]
- 43.Matsui, H., T. J. Doyle, C. M. Bacot, W. A. Garlington, and P. A. Gulig. Unpublished data. [DOI] [PMC free article] [PubMed]
- 44.Matsumoto Y, Emoto M, Usami J, Maeda K, Yoshikai Y. A protective role of extrathymic alpha beta TcR cells in the liver in primary murine salmonellosis. Immunology. 1994;81:8–14. [PMC free article] [PubMed] [Google Scholar]
- 45.McIntyre K W, Unowsky J, DeLorenzo W, Benjamin W. Enhancement of antibacterial resistance of neutropenic, bone marrow-suppressed mice by interleukin-1α. Infect Immun. 1989;57:48–54. doi: 10.1128/iai.57.1.48-54.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a.Menon, A., and P. A. Gulig. Unpublished data.
- 46.Mixter P F, Camerini V, Stone B J, Miller V L, Kronenberg M. Mouse T lymphocytes that express a gamma/delta T-cell antigen receptor contribute to resistance to Salmonella infection in vivo. Infect Immun. 1994;62:4618–4621. doi: 10.1128/iai.62.10.4618-4621.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monack D M, Raupach B, Hromockyj A E, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Norel F, Robbe Saule V, Popoff M Y, Coynault C. The putative sigma factor KatF (RpoS) is required for the transcription of the Salmonella typhimurium virulence gene spvB in Escherichia coli. FEMS Microbiol Lett. 1992;78:271–276. doi: 10.1016/0378-1097(92)90039-q. [DOI] [PubMed] [Google Scholar]
- 49.O’Brien A D, Metcalf E S. Control of early Salmonella typhimurium growth in innately Salmonella-resistant mice does not require functional T lymphocytes. J Immunol. 1982;129:1349–1351. [PubMed] [Google Scholar]
- 50.O’Brien A D, Scher I, Formal S B. Effect of silica on the innate resistance of inbred mice to Salmonella typhimurium infection. Infect Immun. 1979;25:513–520. doi: 10.1128/iai.25.2.513-520.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rabinowitz S S, Gordon S. Macrosialin, a macrophage-restricted membrane sialoprotein differentially glycosylated in response to inflammatory stimuli. J Exp Med. 1991;174:827–836. doi: 10.1084/jem.174.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhen M, Riikonen P, Taira S. Transcriptional regulation of Salmonella enterica virulence plasmid genes in cultured macrophages. Mol Microbiol. 1993;10:45–56. doi: 10.1111/j.1365-2958.1993.tb00902.x. [DOI] [PubMed] [Google Scholar]
- 53.Richter-Dahlfors A, Buchan A M J, Finlay B B. Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J Exp Med. 1997;186:569–580. doi: 10.1084/jem.186.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riikonen P, Mäkelä P H, Saarilahti H, Sukupolvi S, Taira S, Rhen M. The virulence plasmid does not contribute to growth of Salmonella in cultured murine macrophages. Microb Pathog. 1992;13:281–291. doi: 10.1016/0882-4010(92)90038-p. [DOI] [PubMed] [Google Scholar]
- 55.Samsom J N, Annema A, Groeneveld P H, van Rooijen N, Langermans J A, van Furth R. Elimination of resident macrophages from the livers and spleens of immune mice impairs acquired resistance against a secondary Listeria monocytogenes infection. Infect Immun. 1997;65:986–993. doi: 10.1128/iai.65.3.986-993.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmeiger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 57.Schultz L D, Sidman C L. Genetically determined murine models of immunodeficiency. Annu Rev Immunol. 1987;5:367–403. doi: 10.1146/annurev.iy.05.040187.002055. [DOI] [PubMed] [Google Scholar]
- 58.Souvannavong V, Adam A. Macrophages from C3H/HeJ mice require an additional step to produce monokines: synergistic effects of silica and poly(I:C) in the release of interleukin 1. J Leukocyte Biol. 1990;48:183–192. doi: 10.1002/jlb.48.2.183. [DOI] [PubMed] [Google Scholar]
- 59.van Rooijen N. The liposome-mediated macrophage ‘suicide’ technique. J Immunol Methods. 1989;124:1–6. doi: 10.1016/0022-1759(89)90178-6. [DOI] [PubMed] [Google Scholar]
- 60.van Rooijen N, Kors N, Kraal G. Macrophage subset repopulation in the spleen: differential dinetics after liposome-mediated elimination. J Leukocyte Biol. 1989;45:97–104. doi: 10.1002/jlb.45.2.97. [DOI] [PubMed] [Google Scholar]
- 61.van Rooijen N, Sanders A. Liposome mediated depletion of macrophages: mechanism of action, preparation of liposomes and applications. J Immunol Methods. 1994;174:83–93. doi: 10.1016/0022-1759(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 62.Verjans G M, Ringrose J H, van Alphen L, Feltkamp T E, Kusters J G. Entrance and survival of Salmonella typhimurium and Yersinia enterocolitica within human B- and T-cell lines. Infect Immun. 1994;62:2229–2235. doi: 10.1128/iai.62.6.2229-2235.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vreden S G, Sauerwein R W, Verhave J P, van Rooijen N, Meuwissen J H, Van Den Broek M F. Kupffer cell elimination enhances development of liver schizonts of Plasmodium berghei in rats. Infect Immun. 1993;61:1936–1939. doi: 10.1128/iai.61.5.1936-1939.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weintraub B C, Eckmann L, Okamoto S, Hense M, Hedrick S M, Fierer J. Role of αβ and γδ T cells in the host response to Salmonella infection as demonstrated in T-cell-receptor-deficient mice of defined Ity genotypes. Infect Immun. 1997;65:2306–2312. doi: 10.1128/iai.65.6.2306-2312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilkins E G L, Roberts C. Extraintestinal salmonellosis. Epidemiol Infect. 1988;100:361–368. doi: 10.1017/s095026880006711x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilson J A, Doyle T J, Gulig P A. Exponential phase expression of spvA of the Salmonella typhimurium virulence plasmid: induction in intracellular salts medium and intracellularly in mice and cultured mammalian cells. Microbiology. 1997;143:3827–3839. doi: 10.1099/00221287-143-12-3827. [DOI] [PubMed] [Google Scholar]
- 67.Wilson, J. A., and P. A. Gulig. Regulation of the spvR gene of the Salmonella typhimurium virulence plasmid during exponential phase growth in intracellular salts medium and at stationary phase in L broth. Microbiology, in press. [DOI] [PubMed]
- 68.Zwilling B S, Eisenstein T K. Macrophage-pathogen interactions. New York, N.Y: Marcel Dekker, Inc.; 1994. [Google Scholar]