Abstract
We used a two-stage radial diffusion assay to perform a structure-activity study of the antifungal effects of protegrin-1 (PG-1) on yeast-phase Candida albicans. While doing so, we computed MICs from the radial diffusion assay data by three methods and compared the respective values with results from colony count and broth microdilution assays. This allowed us to identify several technical modifications that improved the sensitivity and accuracy of radial diffusion assays. We found that both PG-1 and enantiomeric PG-1 (composed exclusively of d-amino acids) were potently fungicidal for yeast-phase C. albicans. The protegrins PG-2, -3, and -5, but not PG-4, were as effective as PG-1. At least one intramolecular disulfide bond was required to retain optimal candidacidal activity at physiological NaCl concentrations. Truncated variants of PG-1 that lacked its first four residues showed decreased candidacidal activity, although their activity against bacteria was substantially intact. Altering the β-turn region (residues 9 to 12) of PG-1 or its variants further decreased candidacidal activity. These studies suggest that only 12 residues are needed to endow protegrin molecules with strong antibacterial activity and that at least 4 additional residues are needed to add potent antifungal properties. Thus, the 16-residue protegrin PG-2 likely represents the minimal structure needed for broad-spectrum antimicrobial activity encompassing bacteria and fungi.
Protegrins, small β-sheet antimicrobial peptides found in porcine leukocytes (1, 4, 12), are members of the cathelicidin family (36, 38), a large group of structurally diverse antimicrobial peptides whose precursors contain a highly conserved cathelin domain (34). Because protegrins exert broad-spectrum antibacterial activity (12), show efficacy in experimental murine infections (24), and are readily prepared by solid-phase synthesis, they make attractive templates for analyzing the properties of β-sheet peptide antibiotics (8, 16, 20, 33). In addition to their antibacterial properties, protegrins can inactivate certain enveloped viruses (31) and show in vitro activity against Candida albicans (12). Unlike defensins, the larger β-sheet peptides found in the leukocytes and epithelial cells of humans and other animals (17), protegrins retain antimicrobial activity in the presence of extracellular NaCl concentrations (8).
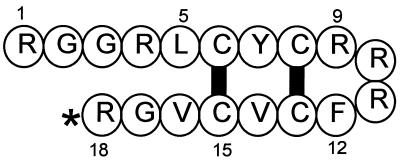
The solution structure of protegrin-1 (PG-1) was established by two-dimensional nuclear magnetic resonance spectroscopy (1, 4) and is shown schematically in Fig. 1. Its principal structural features include the antiparallel β-sheets formed by residues 5 to 8 and 13 to 16 and the four-residue turn made by residues 9 to 12. Except for a tyrosine residue, the β-sheet region contains apolar residues, while the positively charged arginines are present at both ends and clustered in the turn. The overall structure is strongly amphipathic.
FIG. 1.
Structure of PG-1. The two intramolecular disulfide bonds are represented by thick black bars. The asterisk denotes C-terminal amidation. Several residues are numbered.
We performed this study to examine how the various regions of a protegrin molecule contributed to in vitro activity against C. albicans. We made and tested more than 20 variants of PG-1. These included an enantiomeric PG-1 composed exclusively of d-amino acids, three alanine-substituted analogs that lacked one or both disulfide bonds, and various truncated forms with 10 to 16 residues and two disulfides.
MATERIALS AND METHODS
Peptides.
Rabbit α-defensin NP-2 was isolated from peritoneal exudate granulocytes (22). The protegrin PG-4 was a generous gift from IntraBiotics Pharmaceuticals, Inc. (Sunnyvale, Calif.). The other protegrin-related peptides used in this study were custom synthesized, either by SynPep Corporation (Dublin, Calif.) or by the University of Arizona Macromolecular Structure Facility (Tucson, Ariz.), and then purified in our laboratory. Table 1 shows the primary structures, masses, and numbers of residues of the protegrin peptides described in this report.
TABLE 1.
Description of the peptides used in this study
| Peptide | Sequence | No. of residues | Mass units |
|---|---|---|---|
| Full-length protegrins | |||
| PG-1 | RGGRLCYCRRRFCVCVGR* | 18 | 2,155 |
| PG-2 | RGGRLCYCRRRFCICV--* | 16 | 1,957 |
| PG-3 | RGGGLCYCRRRFCVCVGR* | 18 | 2,056 |
| PG-4 | RGGRLCYCRGWICVCVGR* | 18 | 2,052 |
| PG-5 | RGGRLCYCRPRFCVCVGR* | 18 | 2,098 |
| d-PG-1 | RGGRLCYCRRRFCVCVGR* | 18 | 2,155 |
| Disulfide variants of PG-1 | |||
| Snake | RGGRLAYARRRFAVAVGR* | 18 | 2,032 |
| Kite | RGGRLAYCRRRFCVAVGR* | 18 | 2,095 |
| Bullet | RGGRLCYARRRFAVCVGR* | 18 | 2,095 |
| Truncated variants of PG-1 | |||
| PC-13 | RGGRLCYCRRRFCVCV--* | 16 | 1,943 |
| PC-45 | RGGRLCYCRRRFCVC---* | 15 | 1,843 |
| PC-11 | ----LCYCRRRFCVCVGR* | 14 | 1,730 |
| PC-37 | -----CYCRRRFCVCVGR* | 13 | 1,616 |
| PC-17 | ----LCYCRRRFCVCV--* | 12 | 1,517 |
| PC-73 | -----CYCRRRFCVC---* | 10 | 1,304 |
| Turn variants of PC-17 | |||
| PC-17 | ----LCYCRRRFCVCV--* | 12 | 1,517 |
| PC-77 | ----LCYCRRRRCVCV--* | 12 | 1,526 |
| PC-78 | ----LCYCFRRRCVCV--* | 12 | 1,517 |
| PC-79 | ----LCYCRFRRCVCV--* | 12 | 1,517 |
| PC-80 | ----LCYCRRFRCVCV--* | 12 | 1,517 |
| PC-81 | ----LCYCRRFFCVCV--* | 12 | 1,508 |
Purification and folding.
Crude synthetic peptides were dissolved at 10 mg/ml at 52°C in an anaerobic reducing buffer (pH 8.07) that contained 6 M guanidine chloride, 0.02 M EDTA, 0.5 M Tris, and dithiothreitol (DTT) such that the molar ratio of DTT to peptide was 14:1. After 2.5 h, 50% more DTT was added to ensure complete reduction, and 45 min later, glacial acetic acid (5% final concentration) was added to stabilize the products. The reduced peptides were purified by reverse-phase high-pressure liquid chromatography and dissolved at 0.1 mg of peptide/ml in 0.1 M Tris buffer (pH 7.68). Intramolecular disulfide bonds were formed by stirring the peptides for 24 to 52 h at ambient temperature in room air. The “bullet” and “kite” forms of PG-1 (Table 1) were oxidized in a buffer that contained 10% (vol/vol) dimethyl sulfoxide to enhance disulfide bond formation (30).
Purification procedures were monitored by acid-urea polyacrylamide gel electrophoresis. Folding was confirmed by fast atom bombardment mass spectrometry, which showed a loss of 4 mass units after successful folding of protegrins that contained two disulfide bonds. For example, the calculated masses of reduced and oxidized PC-79 are 1,519.9 and 1,515.9 mass units, respectively. We found their measured masses to be 1,520.2 and 1,516.7 mass units, respectively. Protegrin concentrations were determined by spectrophotometry at 280 nm, using the formulae of Pace et al. (18). According to these, the theoretical molar extinction coefficients (ɛ) of PG-1 and PG-4 are 1,740 and 7,240, respectively. The actual molar extinction coefficient of PG-1 was within 8% of the theoretical value when we determined it by quantitative amino acid analysis (data not shown).
Fungi.
C. albicans UC820 is a clinical isolate that has been laboratory passaged for almost 30 years (13). Two recent clinical isolates of C. albicans, taken from vaginal and blood cultures, were provided by Elizabeth A. Wagar of the UCLA Clinical Microbiology Laboratory. Four American Type Culture Collection strains of C. albicans, ATCC 32354 (serotype A), ATCC 36802 (serotype B), ATCC 24433 (used to assay amphotericin B), and ATCC 14053, were purchased from the American Type Culture Collection (Rockville, Md.). Archival laboratory stocks of these strains were prepared by growing the cultures overnight at 37°C in Sabouraud dextrose broth (Difco, Detroit, Mich.) and transferring 0.5-ml volumes into 0.5 ml of 20% glycerol for frozen storage at −80°C.
Growth conditions.
Master plates were prepared every 3 weeks by streaking our archival laboratory stocks onto Sabouraud dextrose agar plates (Clinical Standard Laboratories, Rancho Dominguez, Calif.) and incubating them overnight at 37°C. The yeast-phase C. albicans used in experiments was obtained by picking a single colony, placing it into 50 ml of Sabouraud dextrose broth, and incubating the subculture in a shaking water bath at 37°C for 16 to 20 h.
Two-stage radial diffusion assay.
The rationale and methodology for the two-stage radial diffusion assay have been described elsewhere in detail (15, 25). Briefly, washed yeast-phase C. albicans cells were counted in a hemocytometer. Ten milliliters of molten (43°C) underlay gel solution was mixed with 4 × 106 CFU and poured into a 9- by 9-cm petri dish (Nunc, Naperville, Ill.), where it formed a 1.23-mm-high gel. Low-ionic-strength underlay gels contained 1% agarose (Sigma A-6013), 0.3 mg of Sabouraud dextrose broth powder (Difco 0382-17-9) per ml, and 10 mM sodium phosphate buffer, pH 7.4. High-ionic-strength underlay gels also contained 100 mM NaCl but were otherwise identical in composition. A regular array of 3.2-mm-diameter wells was punched into the underlay gel. Their potential capacity (9.9 μl) easily accommodated the introduction of 5-μl peptide samples that had been serially diluted in sterile acidified water (0.01% acetic acid), using 1.5-ml conical polypropylene microcentrifuge tubes (United Scientific Products, San Leandro, Calif.).
After the peptides had been added, the plates were inverted and incubated for 3 h at 37°C, during which time the peptides diffused into the underlay gel from the wells. A 43°C molten overlay gel (10 ml of 1% agarose, 60 mg of Sabouraud dextrose broth powder per ml, and 10 mM sodium phosphate buffer, pH 7.4) was poured, and the plates were incubated overnight at 37°C. The diameters of the clear zones surrounding the wells were measured to the nearest 0.1 mm with a ×7 magnifier. The zone boundaries were sharp. Core samples removed from the clear area and triturated in buffer yielded no colonies when spread over Sabouraud agar plates and cultured for 48 h.
The MIC was calculated by three methods. One involved performing a linear least-mean-squares fit to the data relating log10 peptide concentration (in micromolar or micrograms per milliliter) to zone size in units, as previously described (25). This value is called the MICa in this report. The second involved performing a least-mean-squares fit for data relating log10 peptide concentration to the zone diameter or area in square millimeters. A rationale for this calculation has been described previously (3), and the resulting value, called the MICb in this report, has also been called the critical concentration (3). Finally, we used the equations proposed by Hultmark and associates (9, 10) to calculate the lethal concentration (their terminology).
Studies with bacteria.
PG-1 and PC-17, a truncated variant of PG-1 that lacked residues 1, 2, 3, 4, 17, and 18, were also tested against several gram-positive and gram-negative bacteria in radial diffusion assays. These underlay gels contained 10 mM sodium phosphate buffer (pH 7.4) and 0.3 mg of Trypticase soy broth powder per ml, with or without 100 mM NaCl, and the overlay gel was double-strength (60-g/liter) Trypticase soy broth powder with 1% agarose. The peptides were serially diluted in 0.01% acetic acid with 0.1% human serum albumin (Sigma A-8763). Mid-logarithmic-phase bacteria were prepared as previously described and added to the underlay at a final concentration of 2 × 105 to 4 × 105 CFU/ml. MICa values were calculated as summarized above and elsewhere (25).
Colony count assays.
C. albicans, grown as for the radial diffusion assays, was washed twice in 10 mM sodium phosphate buffer (pH 7.4) and counted in a hemocytometer. Various final concentrations, from 2 × 103 to 2 × 106 CFU/ml, were exposed to a range of PG-1 concentrations in 200 μl of 10 mM sodium phosphate buffer (pH 7.4) containing 0.3 mg of Sabouraud dextrose broth powder per ml with or without 100 mM sodium chloride. At selected times, aliquots were removed and either plated directly on Sabouraud agar plates or diluted appropriately with 10 mM phosphate buffer and then transferred to the plates with a SpiralSpreader (SpiralTech, Rockville, Md.). Colonies were counted after incubation for 24 to 48 h at 37°C.
Broth microdilution assays.
The procedures described below correspond to the proposed standard reference method for broth dilution antifungal susceptibility testing (17a), performed in a “micro” format. Briefly, C. albicans UC820 was grown overnight in Sabouraud dextrose broth, washed in RPMI 1640 medium (Gibco BRL) containing 0.165 M 3-(N-morpholino) propanesulfonic acid (MOPS) buffer (pH 7.0), counted in a hemocytometer, and adjusted to 2.0 × 103/ml in RPMI-MOPS. The peptides were serially diluted in 0.01% acetic acid with 0.1% human serum albumin (Sigma A-8763). Two overlapping sets of serial twofold peptide dilutions were tested in each experiment. One series ranged from 37.0 to 1.16 μg/ml (final concentrations, after dilution), and the other ranged from 27.75 to 0.87 μg/ml. One hundred microliters of yeast suspension and 11 μl of 10×-concentrated peptide were mixed together and incubated for 46 to 50 h at 37°C in polypropylene 96-well plates (Costar 3790). After the plates were examined to determine the MICs, the entire content (111 μl) of all wells without evident growth was plated on Sabouraud agar plates and incubated for 24 to 48 h to determine the minimal fungicidal concentrations (MFCs).
RESULTS
Radial diffusion assays.
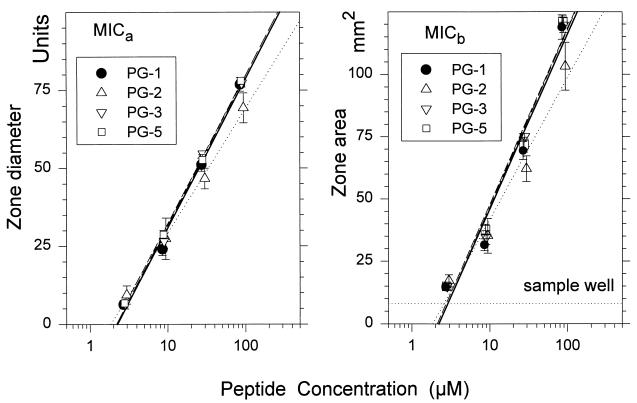
Table 2 shows the MICa, MICb, and lethal concentration values obtained for porcine protegrins PG-1 to -5. The results of radial diffusion assays for four of these protegrins (PG-1, -2, -3, and -5) are also shown graphically in Fig. 2, to illustrate the linearity of the assays used to obtain the data summarized in Table 2. In the left panel of Fig. 2, the MICa corresponds to the x intercept. In the right panel, the MICb corresponds to the x-axis coordinate of the point where the line representing zone area intersects the horizontal line whose y coordinate denotes the area (8.04 mm2) of the 3.2-mm-diameter sample wells.
TABLE 2.
Activities of protegrins against C. albicans UC820a
| Protegrin | MICa (μM) | MICb (μM) | Lethal concn (μM) |
|---|---|---|---|
| PG-1 | 2.29 ± 0.13 | 2.83 ± 0.11 | 0.38 ± 0.02 |
| PG-2 | 1.83 ± 0.35 | 2.50 ± 0.34 | 0.38 ± 0.02 |
| PG-3 | 2.21 ± 0.17 | 2.85 ± 0.12 | 0.36 ± 0.01 |
| PG-4 | 4.78 ± 0.74** | 4.90 ± 0.23** | 1.00 ± 0.07** |
| PG-5 | 2.10 ± 0.72 | 2.56 ± 0.18 | 0.35 ± 0.01 |
| d-PG-1 | 1.67 ± 0.21* | 2.35 ± 0.46 | 0.32 ± 0.01 |
All radial diffusion assays were performed with underlay gels containing 100 mM NaCl. Results are means ± SEMs from three experiments, except for PG-1 (n = 7). Values were compared to those obtained for PG-1 by unpaired t tests: ∗, P < 0.05, ∗∗, P < 0.01. The other differences were not statistically significant (P > 0.05). The r values (means ± SEMs) of the least-mean-square fits used to calculate the MICa values ranged from 0.995 ± 0.001 to 0.998 ± 0.001.
FIG. 2.
Two-stage radial diffusion assays. PG-1, -2, -3, and -5 were tested against C. albicans. The sample size was 5 μl, and no albumin was used in the diluent. Each symbol represents a mean value from three replicate experiments, except that for PG-1 (n = 7). Error bars show SEMs. The regression lines (least-mean-squares fits) are as follows: for PG-1 data, thick and solid, for PG-2 data, dotted; for PG-3 data, dashed; for PG-5 data, thin and solid. The left panel illustrates data that would be analyzed in a MICa calculation. The right panel shows the same data as they would be expressed to determine the MICb.
Whereas PG-1, -2, -3, and -5 had similar MICa and MICb values, PG-4 was only about half as potent. Since the primary structures of PG-1 and PG-4 differ only in their turn residues, which are RRRF in PG-1 and RGWI in PG-4, the reduced potency of PG-4 was our first indication that the turn structure influenced activity against C. albicans. The turn residues will be revisited below.
Table 2 also shows that enantiomeric PG-1, composed exclusively of d-amino acids, was at least as active as normal PG-1. Overall, the MICa values for PG-1 to -5 were 80.3 ± 3.9% of the corresponding MICb values (range, 71 to 98%). The lethal concentrations were considerably lower, only 18.4 ± 0.9 and 14.8 ± 1.1% of the corresponding MICa and MICb.
Broth microdilution assays.
We examined the activity of PG-1 against C. albicans UC820 in three broth microdilution assays, each performed on a different day. The assay results were identical. The MICs and MFCs for PG-1 were identical, with each being between 0.77 and 1.08 μM. The MICs and MFCs for PG-4 were about threefold higher, between 2.26 and 3.39 μM, confirming our findings in the radial diffusion assays (Table 2).
Colony count assays.
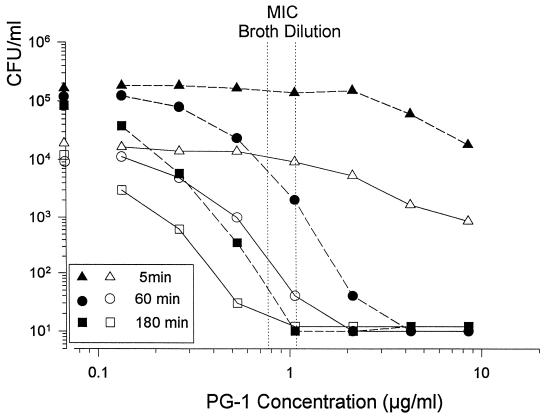
We were initially disconcerted to find that the MICa values obtained from our radial diffusion assays were more than double the MIC and MFC values obtained from broth dilution assays, and we turned to colony count experiments to see which values were correct. We found that when the broth microdilution assays and colony count experiments were performed in the same medium, their results corresponded perfectly (Fig. 3). Consequently, we reexamined our radial diffusion assay procedure to identify factors that caused it to underestimate potency.
FIG. 3.
Comparison of colony count and broth microdilution assays. The broth microdilution assays were performed with 2 × 103 CFU/ml in an RPMI 1640-based medium. The colony count assays used two different C. albicans concentrations (104 and 105 CFU/ml) and were performed in the high-salt medium (without agarose) that was used in radial diffusion assays. Three different incubation times (5, 60, and 180 min) were used in the colony count assays.
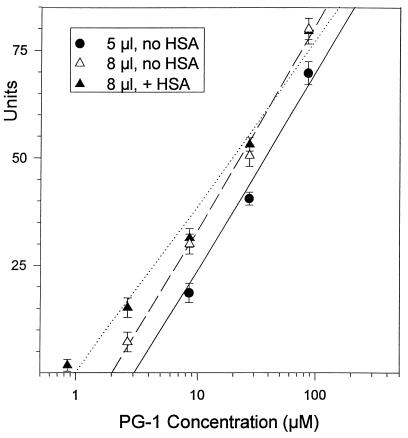
We identified two technical factors whose effects were largely responsible for this. They proved to be a matter of simple geometry. Although the height of the underlay gel (1.23 mm) and the diameter of the well (3.2 mm) gave the wells a capacity of 9.89 μl, we had added only 5 μl of sample. Consequently, when the peptide solutions entered the underlay gel, the peptides not only diffused outward (radially) but also diffused upward to occupy the full height of the underlay gel. In effect, the upward diffusion effectively diluted the peptide concentration placed into the well by about half (5/9.89 = 50.5%). To confirm the consequences of this effect, we compared MICa values obtained after applying 5- and 8-μl samples from the same dilution series to seven replicate plates. Samples larger than 8 μl were difficult to instill without causing occasional overflows. Tests performed with the 5-μl samples yielded a MICa of 3.23 ± 0.199 μM (mean ± standard deviation), whereas those performed with 8-μl samples yielded a MICa of 2.02 ± 0.084 μM. The ratio of these mean values (2.02 ÷ 3.23 = 0.67) was remarkably close to the theoretical value (0.625) expected from the ratio (5 μl/8 μl) of the sample volumes.
The other technical factor was, in retrospect, even more obvious. By preparing serial peptide dilutions in the absence of a carrier protein, amphipathic peptide was lost from solution via nonspecific adsorption to the plastic tubes. This effect disproportionately affected the lowest concentrations of peptides in a dilution series and could largely be prevented by adding 0.1% albumin to the diluting solution to block the tubes. When we performed the radial diffusion assays with 8-μl samples and used peptides diluted in 0.01% acetic acid with 0.1% albumin, the resulting MICa of 1.07 ± 0.075 μM (mean ± standard error of the mean [SEM]) was completely consistent with the broth microdilution and colony count assays. These experimental results are shown in Fig. 4. When these same data were calculated by the other formulae described in Materials and Methods, the MICb was 1.11 ± 0.136 μM and the lethal concentration was 0.505 ± 0.014 μM (means ± SEMs). Because these technical epiphanies occurred after most of the radial diffusion data had already been collected, the marginal benefits of redoing the experiments did not justify the cost and effort that this would entail.
FIG. 4.
Effects of sample volume and an albumin carrier. Seven replicate radial diffusion experiments were done, using either 5- or 8-μl samples that had been serially diluted in the presence or absence of 0.1% human serum albumin (HSA). Note the effects of these changes on the MIC, which corresponds to the x intercept. The regression lines for the data points (least-mean-squares fit) are as follows: solid line, 5 μl without HSA; dashed line, 8 μl without HSA; dotted line, 8 μl with HSA.
Kinetics.
In other colony count studies, we examined the effect of the medium on the kinetics of candidacidal activity. At a given concentration of PG-1, killing occurred more rapidly in a low-salt medium (10 mM buffer) than when the medium contained buffer plus 100 mM NaCl. For example, whereas addition of 4.35 μM PG-1 to 1.48 × 106 C. albicans CFU/ml reduced the colony count in 5 min by >4 log10 units in low-salt medium, it achieved only a 1.9-log10-unit reduction in 5 min in the high-salt medium. By 30 min, the same >4-log10-unit decrease was also achieved in the medium comprised of buffer plus 100 mM NaCl. Candidacidal activity occurred even more slowly in the RPMI-MOPS medium. For example, when the inoculum (2 × 103 CFU/ml) was exposed to 2.1 μM PG-1 in RPMI-MOPS, 25% of the yeast cells remained viable after 60 min, as determined by quantitative subculture, and 1% were still viable after 120 min. No survivors were found at 3, 4, 5, or 6 h. When 2.1 μM PG-1 was added to the same concentration of C. albicans in our customary underlay medium (buffer plus 100 mM NaCl), no fungi survived after a 1-h exposure. When we tested higher concentrations (>106 CFU/ml) of C. albicans in the 100 mM NaCl medium, 2.45 to 4.5 μM PG-1 sterilized the inoculum by 30 min, and 8.9 μM PG-1 caused a 4-log10-unit kill within 5 min.
Other Candida strains.
Although we used C. albicans UC820 in most of our studies, we also tested PG-1 against six other C. albicans strains, including two that were fresh clinical isolates (Table 3). Since all seven strains showed equal susceptibility to PG-1, we concluded that UC820 was representative of the species.
TABLE 3.
Effect of salinity on activity of PG-1 against C. albicans
| Candida strain | MICb (μM)a
|
|||
|---|---|---|---|---|
| PG-1
|
NP-2
|
|||
| Buffer only | Buffer + 100 mM NaCl | Buffer only | Buffer + 100 mM NaCl | |
| UC820 | 1.87 ± 0.10 | 1.95 ± 0.13 | 1.91 ± 0.60 | >65.0 |
| ATCC 36802 | 1.51 ± 0.17 | 1.69 ± 0.16 | 1.70 ± 0.10 | >65.0 |
| ATCC 14053 | 1.60 ± 0.11 | 1.78 ± 0.10 | 1.55 ± 0.03 | >20.6 |
| ATCC 24433 | 1.77 ± 0.07 | 1.85 ± 0.04 | 1.63 ± 0.21 | >65.0 |
| ATCC 32354 | 2.05 ± 0.07 | 1.81 ± 0.01 | 2.05 ± 0.14 | >65.0 |
| UC751 (vaginal) | 1.96 ± 0.07 | 2.07 ± 0.10 | 1.78 ± 0.20 | >65.0 |
| UC430 (blood) | 2.02 ± 0.07 | 2.16 ± 0.04 | 1.66 ± 0.08 | >65.0 |
| Composite | 1.83 ± 0.12 | 2.59 ± 0.08 | 1.75 ± 0.07 | >65.0 |
Underlays contained 10 mM phosphate buffer (pH 7.4) and 0.3 mg of Sabouraud dextrose broth powder per ml, with or without 100 mM NaCl. The data represent means ± SEMs (n = 3). The MICb values obtained with the various isolates did not differ significantly by unpaired t tests from values obtained for strain UC820. The r values of the regressions used to calculate the MICb values shown above ranged from 0.964 ± 0.004 to 0.997 ± 0.001.
Effect of added cations.
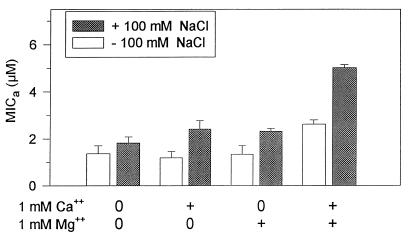
Although most of the data presented in this paper were obtained from assays performed at physiologic NaCl concentrations (100 mM), we also tested the peptides in low-salt underlays. Table 3 also shows that whereas 100 mM NaCl did not affect the candidacidal activity of PG-1, rabbit α-defensin NP-2 lost its effectiveness (MICb > 65 μM) even though it was slightly more potent than PG-1 on a molar basis (MICb = 1.75 ± 0.07 μM) when tested in low-salt (buffer-only) underlays. As we had previously noted that divalent cations adversely affect the candidacidal activity of defensins in low-salt media (14), we tested the effects of Ca2+ and Mg2+ on protegrins. Addition of either 1 mM Ca2+ or 1 mM Mg2+ to low- or high-salt underlays had very little effect on anticandidal activity (Fig. 5), but in combination, these divalent cations did reduce the efficacy of PG-1, especially in 100 mM NaCl.
FIG. 5.
Effects of divalent cations on activity against C. albicans. The bars show MICa values (means ± SEMs) from three experiments that were performed with underlay gels containing 10 mM phosphate buffer with or without 100 mM NaCl. The gels were supplemented with 1 mM Ca2+ and/or 1 mM Mg2+ as shown.
Disulfide structure.
Native protegrins, like PG-1, contain two intramolecular disulfide bonds that pair Cys6:Cys15 and Cys8:Cys13 (Fig. 1). To learn if these disulfides contributed to activity against C. albicans, we made alanine-substituted protegrins that lacked one or both cysteine pairs. The disulfide-free (“snake”) protegrin variant lacked candidacidal activity in 100 mM NaCl. In contrast, the “bullet” protegrin variant, which contained only the Cys6:Cys15 disulfide bond, retained about 70% of the potency of PG-1, and the “kite” variant, with only a Cys8:Cys13 disulfide bond, retained about 50% (Table 4). All three disulfide variants showed excellent activity against C. albicans when their activities were tested in low-salt underlays. The MICa values (mean ± SEM, n = 3) for the respective peptides were as follows: snake, 1.03 ± 0.27 μM; kite, 0.93 ± 0.16 μM; and bullet, 0.89 ± 0.02 μM.
TABLE 4.
Activities of disulfide variants against C. albicans UC820a
| Prote- grin | No. of SS bonds | MICa (μM) | MICb (μM) | Lethal concn (μM) |
|---|---|---|---|---|
| PG-1 | 2 | 1.68 ± 0.10 | 2.08 ± 0.08 | 0.28 ± 0.02 |
| Snake | 0 | >28.6** | >28.6** | |
| Kite | 1 | 3.49 ± 0.21** | 4.86 ± 0.25** | 0.63 ± 0.02** |
| Bullet | 1 | 2.53 ± 0.24** | 3.09 ± 0.25** | 0.38 ± 0.10 (NS) |
Radial diffusion assays were performed as described in Table 2, footnote a. Values represent means ± SEMs (n = 7 for PG-1; n = 3 for all others). Values were compared with those obtained for PG-1 by unpaired t tests: ∗∗, P < 0.01. The mean values for the snake variant were well outside the 99% confidence limits for PG-1. The r values for the regression lines used to calculate the MICa values ranged from 0.992 ± 0.004 to 0.995 ± 0.001.
Optimal length.
Native protegrins contain 18 amino acid residues, except for PG-2, which has 16 (Table 1). Except for its Val14→Ile14 substitution and its deletion of residues 17 and 18 (Gly-Arg), PG-2 is identical to PG-1. To determine how truncations of PG-1 affected its activity against C. albicans, we prepared a series of deletion mutants (Tables 1 and 5). Removal of two C-terminal residues (PC-13) was well tolerated, but elimination of three C-terminal residues (PC-45) reduced potency by 70%. The N-terminal residues were clearly even more important, since deletion of residues 1 to 4 (PC-11) or residues 1 to 5 (PC-37) reduced the potency against C. albicans by 73 and 90%, respectively. When we deleted four N-terminal and two C-terminal residues (PC-17), potency was decreased by 76% relative to that of PG-1. The indifferent performance of PC-17 against C. albicans stands in marked contrast to its strong antibacterial effects (Table 6).
TABLE 5.
Activities of truncated protegrins against C. albicans UC820a
| Prote- grin | Deletions (positions) | MICa (μM) | MICb (μM) | Lethal concn (μM) | Ratio |
|---|---|---|---|---|---|
| PG-1 | None | 1.68 ± 0.10 | 1.53 ± 0.08 | 0.28 ± 0.02 | 1.00 |
| PC-13 | 17, 18 | 2.23 ± 0.13* | 2.77 ± 0.18** | 0.70 ± 0.03** | 0.75 |
| PC-45 | 16–18 | 5.60 ± 0.43** | 5.54 ± 0.40** | 1.15 ± 0.07** | 0.30 |
| PC-11 | 1–4 | 6.27 ± 0.30** | 6.92 ± 0.30** | 1.24 ± 0.05** | 0.27 |
| PC-37 | 1–5 | 17.1 ± 0.44** | 16.0 ± 0.55** | 5.46 ± 0.19** | 0.10 |
| PC-17 | 1–4, 17, 18 | 7.04 ± 0.46** | 7.58 ± 0.39** | 1.36 ± 0.04** | 0.24 |
| PC-73 | 1–5, 16–18 | >103.7** | >103.7** | >103.7** | <0.02 |
Radial diffusion assays were performed as described in Table 2, footnote a. Values represent means ± SEMs (n = 3 experiments, except for PG-1 [n = 7]). Deleted residue positions are keyed to PG-1. See Table 1 for more details. The ratio was calculated by dividing the MICa of PG-1 by the MICa of the peptide of interest, shown in column 1. Values were compared with those for PG-1 by unpaired t tests: ∗, P < 0.05; ∗∗, P < 0.01. The mean values for PC-73 were well outside the 99% confidence limits for PG-1. The r values for the regression lines used to calculate the MICa values ranged from 0.979 ± 0.012 to 0.999 ± 0.000.
TABLE 6.
Antibacterial activities of PG-1 and PC-17
| Bacterium | MICa (μM)a
|
|||
|---|---|---|---|---|
| PG-1
|
PC-17
|
|||
| Phosphate | Phosphate + 100 mM NaCl | Phosphate | Phosphate + 100 mM NaCl | |
| Escherichia coli ML-35p | 0.32 ± 0.20 | 0.21 ± 0.06 | 0.28 ± 0.10 | 0.26 ± 0.07 |
| Listeria monocytogenes EGD | 0.28 ± 0.02 | 0.24 ± 0.02 | 0.26 ± 0.04 | 0.55 ± 0.12 |
| Pseudomonas aeruginosa MR 3007 | 0.15 ± 0.03 | 0.30 ± 0.03* | 0.20 ± 0.06 | 0.29 ± 0.01 |
| Bacillus subtilis | 0.27 ± 0.01 | 0.25 ± 0.03 | 0.22 ± 0.03 | 0.21 ± 0.04 |
| Staphylococcus aureus 930918-3 | 0.41 ± 0.13 | 0.32 ± 0.05 | 0.33 ± 0.09 | 0.40 ± 0.07 |
Values are means ± SEMs (n = 3). The MICs obtained for each organism in buffer and high-salt buffer were compared by unpaired t tests: ∗, P < 0.05 (all other differences were not statistically significant). The r values for the regression lines used to calculate the MICa values ranged from 0.942 ± 0.019 to 0.988 ± 0.005.
When we deleted five N-terminal and three C-terminal residues from PG-1, the potency of the resulting decapeptide peptide (PC-73) was reduced by >98% relative to that of PG-1. From these data, we concluded that each region of the PG molecule, with the exception of residues 17 and 18, contributed substantially to activity against C. albicans. In contrast, only 12 residues (residues 5 to 16) and their two associated disulfide bonds (Fig. 1) sufficed to endow PG-1 with strong antibacterial properties (Table 6).
Nature and placement of turn residues.
As shown in Table 1, the turn regions (residues 9 to 12) of PG-1, -2, and -3 are identical (RRRF). They differ slightly from that in PG-5 (RPRF) but markedly from that in PG-4 (RGWI). To explore the consensus RRRF turn motif, we constructed and successfully refolded six variants that were based on the structure of PC-17, a truncated form of PG-1 (Table 1). In three variants (PC-78, -79, and -80), we maintained the turn’s composition (three arginines and one phenylalanine) but varied the placement of the residues. All three variants were substantially less effective than PC-17 (Table 7). PC-77, with four arginine residues in its turn, showed about half the potency of PC-17, and PC-81, which had an RRFF turn, was completely inactive. Additional members of this series containing only arginine and phenylalanine residues in the turn region were synthesized but could not be folded. Taken together with the reduced candidacidal activity shown by PG-4 (Table 2), these results indicate that the turn region residues play an important part in activity against C. albicans.
TABLE 7.
Activities of turn variants against C. albicans UC820a
| Protegrin | Turn | MICa (μM) | MICb (μM) | Lethal concn (μM) |
|---|---|---|---|---|
| PC-17 | RRRF | 8.24 ± 0.40 | 8.0 ± 0.49 | 1.32 ± 0.09 |
| PC-77 | RRRR | 12.4 ± 1.76 (NS) | 14.2 ± 2.17* | 2.31 ± 0.32* |
| PC-78 | FRRR | >28.2** | >28.2** | |
| PC-79 | RFRR | >89.1** | >89.1** | |
| PC-80 | RRFR | >28.2** | >28.2** | |
| PC-81 | RRFF | >89.1** | >89.1** |
Radial diffusion assays were performed as described in Table 2, footnote a. The values represent means ± SEMs (n = 3 experiments). Values were compared with those for PC-17 by unpaired t tests or by determining the 99% confidence limits for the mean MIC or lethal concentration of PC-17: ∗, P < 0.05; ∗∗, P < 0.01. The r values for the regression lines used to calculate the MICa values ranged from 0.986 ± 0.009 to 0.987 ± 0.014.
Retention of susceptibility to PG-1.
We performed the following experiment to determine if repeated exposures to PG-1 induced or selected resistant C. albicans. The organisms (suspended in 10 mM phosphate buffer plus 100 mM NaCl plus 1% [vol/vol] Sabouraud broth) were exposed for 3 h at 37°C to PG-1 (1.45, 2.9, 5.8, or 11.6 μM). The initial experiment was performed with 0.7 × 106 CFU/ml. Only 102 CFU/ml survived exposure to 1.45 μM PG-1, and none survived higher concentrations. Four surviving colonies (named clones A, B, C, and D) were picked individually and grown overnight. We exposed approximately 1.5 × 106 CFU/ml derived from clones A to D to 1.45 to 11.6 μM PG-1 for 3 h as described above. Over 99% were killed by 2.9 μM PG-1, and all were killed by the higher PG-1 concentrations. In the third experiment, 50 colonies that had survived exposure to 2.9 μM PG-1 were picked from each of the four clones (A to D), pooled, and used to inoculate four fresh overnight cultures. After exposure to PG-1, over 99% of the progeny of each clone were killed by 2.9 μM, and all organisms were killed by 5.8 μM. In the fourth and final experiment, 50 colonies were derived from each clone that had survived their third exposure to 2.9 μM PG-1. These were pooled and used to inoculate four fresh overnight cultures. Once again, over 99% of each clone’s progeny were killed by 2.9 μM PG-1, and all organisms were killed by 5.8 μM. Thus, at least under these experimental conditions, repeated exposure to PG-1 did not appear to induce or select for resistant mutants.
DISCUSSION
The prominence of methodology in this report requires a few brief comments to open the discussion. We have relied upon two-stage radial diffusion assays in our natural-product research because they are quantitative and sensitive and consume minimal quantities of the molecules being studied. Our usual practice has been to use 5-μl samples and to calculate x intercepts from plots that relate log10 peptide concentrations to the resulting zone diameters. This practice yielded the MICa calculations shown in this report. Although the use of 5-μl samples was peptide sparing and produced internally consistent data, it yielded values that were about twice as high as those obtained from conventional broth microdilution assays, for reasons discussed in Results. Simply by filling up the sample wells closer to their capacity, i.e., by using 8-μl samples instead of 5-μl samples, the MICa values agreed much more closely to those from broth microdilution assays. The other improvements involved steps to decrease peptide loss via nonspecific adsorption: using polypropylene tubes and plates and adding 0.1% albumin to the acidified water diluent.
Several alternative methods, in addition to the one we described (15, 25), have been used to calculate MICs from radial diffusion data. One of these calculates the MIC from the relationship between the zone area and the log10 peptide concentration. This value has been named the critical concentration by others (3) and was called the MICb in this report. When we calculated it from our data, the resulting values were very close to the MICa. The lethal concentration formulae (9, 10) were developed by Hultmark and associates from theoretical considerations, and their application to our data yielded MICs that were about twofold lower than MICa or MICb values obtained under optimal experimental conditions (i.e., with 8-μl samples, 0.01% albumin diluent, and polypropylene containers). These lethal concentrations were also below the MICs that we obtained from colony counting and broth microdilution assays (Fig. 3). Overall, the three calculation methods (MICa, MICb, and lethal concentration) agreed reasonably well with each other and with the widely used broth microdilution assay. When the radial diffusion assays were performed with 5-μl samples and without an albumin-containing diluent, the lethal-concentration calculation tended to overestimate protegrin potency, whereas the MICa calculation underestimated it.
Mounting evidence indicates that antimicrobial peptides play key roles in the innate immune defenses of vertebrates, invertebrates, and plants. Protegrins and the structurally analogous tachyplesins (11), found in horseshoe crabs, are distinctive among β-sheet peptides because of their structural simplicity, their unusually potent and broad-spectrum antimicrobial properties, and their retention of activity in normal or elevated concentrations of NaCl. Although porcine leukocytes lack defensins, they contain a multitude of cathelicidins, including up to five protegrins, plus at least two α-helical (27, 32), and three proline-rich (6, 7, 28, 37) peptides. Porcine neutrophil proprotegrins lack microbicidal activity (19) until the cathelin and protegrin domains are separated by limited proteolysis. Whereas the larger and structurally more complex β-sheet defensins of human leukocytes are sequestered in cytoplasmic granules whose contents are delivered preferentially to intracellular phagosomes, cathelicidins are typically found in secretory granules of neutrophils and are readily released to the plasma (6).
Although many endogenous antimicrobial peptides are known, relatively few of them manifest antifungal properties, especially in the presence of physiological extracellular concentrations of sodium chloride (data not shown). The findings of this study underlined the importance of the intramolecular disulfide bonds that stabilize the β-sheet structure of protegrins for activity against yeast-phase C. albicans. When both disulfide bonds were eliminated by substituting alanines for all four regularly conserved cysteines, the resulting linearized (snake) peptide lost its activity to kill C. albicans. While the retention of both intramolecular disulfide bonds was optimal, the kite and bullet variants of PG-1 retained 50 to 70% of the activity of the parent molecule, even though each contained only one disulfide bond. As we previously obtained similar results in studies with gram-positive and gram-negative bacteria and with model membranes (8), it is clear that retention of the β-sheet structure is important for antifungal activity in the extracellular environment.
Several natural antimicrobial peptides with single intramolecular disulfide bonds have been described, including two dodecapeptides found in bovine (21) and ovine (2, 26) leukocytes. Both of these peptides are produced on cathelin-containing precursors, and their sequences are quite similar: RLCRIVVIRVCR (bovine) and RICRIIFLRVCR (ovine). The uni-disulfide kite and bullet protegrins also show some resemblance to other naturally occurring peptides, including thanatin (GSKKPVPIIY CNRRTGKCQR M) and brevinin-1 (FLPVLAGIAA KVVPALFCKI TKKC). Thanatin is a 21-residue peptide that appears in the hemolymph of the insect Podisus maculiventris after immune challenge (5), and brevinin-1 is one of many such peptides found in the skin of frogs (23, 29). In both of these peptides, a C-terminally located disulfide bridge delineates a cationic loop that, along with its flanking residues, is important for antimicrobial activity. Both thanatin and brevinin-1 have bactericidal and fungicidal properties.
While it should prove possible to modify the structure of protegrins to alter their pharmacokinetic properties or to improve their biocompatibility, the present results suggest that the native protegrin design is already a remarkably economical one. Indeed, the 16-residue protegrin PG-2 may already approach the minimal structure that is capable of exerting strong activity against gram-positive bacteria, gram-negative bacteria, and fungi in an extracellular environment.
ACKNOWLEDGMENTS
The late Sylvia Harwig contributed substantially to the design and purification of the protegrin variants. We thank Debbie Steinberg of IntraBiotics for suggesting the use of albumin in radial diffusion assays.
This work was supported by Public Health Service grants AI 37945 and AI 22839.
REFERENCES
- 1.Aumelas A, Mangoni M, Roumestand C, Chiche L, Despaux E, Grassy G, Calas B, Chavanieu A. Synthesis and solution structure of the antimicrobial peptide protegrin-1. Eur J Biochem. 1996;237:575–583. doi: 10.1111/j.1432-1033.1996.0575p.x. [DOI] [PubMed] [Google Scholar]
- 2.Bagella L, Scocchi M, Zanetti M. cDNA sequences of three sheep myeloid cathelicidins. FEBS Lett. 1995;376:225–228. doi: 10.1016/0014-5793(95)01285-3. [DOI] [PubMed] [Google Scholar]
- 3.Barry A L. Procedures for testing antibiotics in agar media: theoretical considerations. In: Lorian V, editor. Antibiotics in laboratory medicine. Baltimore, Md: Williams and Wilkins; 1980. pp. 1–23. [Google Scholar]
- 4.Fahrner R L, Dieckmann T, Harwig S S L, Lehrer R I, Feigon J. Solution structure of protegrin 1, a broad spectrum antimicrobial peptide from procine leukocytes. Chem Biol. 1996;3:543–550. doi: 10.1016/s1074-5521(96)90145-3. [DOI] [PubMed] [Google Scholar]
- 5.Fehlbaum P, Bulet P, Chernysh S, Briand J P, Roussel J P, Letellier L, Hetru C, Hoffmann J A. Structure-activity analysis of thanatin, a 21-residue inducible insect defense peptide with sequence homology to frog skin antimicrobial peptides. Proc Natl Acad Sci USA. 1996;93:1221–1225. doi: 10.1073/pnas.93.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gudmundsson G H, Magnusson K P, Chowdhary B P, Johansson M, Andersson L, Boman H G. Structure of the gene for porcine peptide antibiotic PR-39, a cathelin gene family member: comparative mapping of the locus for the human peptide antibiotic FALL-39. Proc Natl Acad Sci USA. 1995;92:7085–7089. doi: 10.1073/pnas.92.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harwig S S, Kokryakov V N, Swiderek K M, Aleshina G M, Zhao C, Lehrer R I. Prophenin-1, an exceptionally proline-rich antimicrobial peptide from porcine leukocytes. FEBS Lett. 1995;27:362:65–69. doi: 10.1016/0014-5793(95)00210-z. [DOI] [PubMed] [Google Scholar]
- 8.Harwig S S, Waring A, Yang H J, Cho Y, Tan L, Lehrer R I. Intramolecular disulfide bonds enhance the antimicrobial and lytic activities of protegrins at physiological sodium chloride concentrations. Eur J Biochem. 1996;240:352–357. doi: 10.1111/j.1432-1033.1996.0352h.x. [DOI] [PubMed] [Google Scholar]
- 9.Hultmark D, Engstrom A, Andersson K, Steiner H, Bennich H, Boman H G. Insect immunity: attacins, a family of antibacterial proteins from Hyalophora cecropia. EMBO J. 1983;2:571–576. doi: 10.1002/j.1460-2075.1983.tb01465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hultmark D, Engstrom A, Bennich H, Kapur R, Boman H G. Insect immunity: isolation and structure of cecropin D and four minor antibacterial components from Cecropia pupae. Eur J Biochem. 1982;127:207–217. doi: 10.1111/j.1432-1033.1982.tb06857.x. [DOI] [PubMed] [Google Scholar]
- 11.Iwanaga S, Muta T, Shigenaga T, Seki N, Kawano K, Katsu T, Kawabata S. Structure-function relationships of tachyplesins and their analogues. Ciba Found Symp. 1994;186:160–174. doi: 10.1002/9780470514658.ch10. [DOI] [PubMed] [Google Scholar]
- 12.Kokryakov V N, Harwig S S, Panyutich E A, Shevchenko A A, Aleshina G M, Shamova O V, Korneva H A, Lehrer R I. Protegrins: leukocyte antimicrobial peptides that combine features of corticostatic defensins and tachyplesins. FEBS Lett. 1993;327:231–236. doi: 10.1016/0014-5793(93)80175-t. [DOI] [PubMed] [Google Scholar]
- 13.Lehrer R I, Cline M J. Interaction of Candida albicans with human leukocytes and serum. J Bacteriol. 1969;98:996–1004. doi: 10.1128/jb.98.3.996-1004.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehrer R I, Ganz T, Szklarek D, Selsted M E. Modulation of the in vitro candidacidal activity of human neutrophil defensins by target-cell metabolism and divalent cations. J Clin Invest. 1988;81:1829–1835. doi: 10.1172/JCI113527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehrer R I, Rosenman M, Harwig S S L, Jackson R, Eisenhauer P. Ultrasensitive assays for endogenous antimicrobial polypeptides. J Immunol Methods. 1991;137:167–173. doi: 10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
- 16.Mangoni M E, Aumelas A, Charnet P, Roumestand C, Chiche L, Despaux E, Grassy G, Calas B, Chavanieu A. Change in membrane permeability induced by protegrin 1: implication of disulphide bridges for pore formation. FEBS Lett. 1996;383:93–98. doi: 10.1016/0014-5793(96)00236-0. [DOI] [PubMed] [Google Scholar]
- 17.Martin E, Ganz T, Lehrer R I. Defensins and other endogenous peptide antibiotics of vertebrates. J Leukocyte Biol. 1995;58:128–136. doi: 10.1002/jlb.58.2.128. [DOI] [PubMed] [Google Scholar]
- 17a.National Committee for Clinical Laboratory Standards. Document M-27-P. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1992. [Google Scholar]
- 18.Pace C N, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panyutich A, Shi J, Boutz P L, Zhao C, Ganz T. Porcine polymorphonuclear leukocytes generate extracellular microbicidal activity by elastase-mediated activation of secreted proprotegrins. Infect Immun. 1997;65:978–985. doi: 10.1128/iai.65.3.978-985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu X D, Harwig S S, Shafer W M, Lehrer R I. Protegrin structure and activity against Neisseria gonorrhoeae. Infect Immun. 1997;65:636–639. doi: 10.1128/iai.65.2.636-639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romeo D, Skerlavaj B, Bolognesi M, Gennaro R. Structure and bactericidal activity of an antibiotic dodecapeptide purified from bovine neutrophils. J Biol Chem. 1988;263:9573–9575. [PubMed] [Google Scholar]
- 22.Selsted M E, Szklarek D, Lehrer R I. Purification and antibacterial activity of antimicrobial peptides of rabbit granulocytes. Infect Immun. 1984;45:150–154. doi: 10.1128/iai.45.1.150-154.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simmaco M, Mignogna G, Barra D, Bossa F. Antimicrobial peptides from skin secretions of Rana esculenta. Molecular cloning of cDNAs encoding esculentin and brevinins and isolation of new active peptides. J Biol Chem. 1994;269:11956–11961. [PubMed] [Google Scholar]
- 24.Steinberg D A, Hurst M A, Fujii C A, Kung A H C, Ho J F, Cheng F C, Loury D J, Fiddes J F. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob Agents Chemother. 1997;41:1738–1742. doi: 10.1128/aac.41.8.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinberg D, Lehrer R I. Designer assays for antimicrobial peptides. In: Shafer W M, editor. Methods in molecular biology. Totowa, N.J: Humana Press; 1997. pp. 169–188. [DOI] [PubMed] [Google Scholar]
- 26.Storici P, Del Sal G, Schneider C, Zanetti M. cDNA sequence analysis of an antibiotic dodecapeptide from neutrophils. FEBS Lett. 1992;314:187–190. doi: 10.1016/0014-5793(92)80971-i. [DOI] [PubMed] [Google Scholar]
- 27.Storici P, Scocchi M, Toss I A, Gennaro R, Zanetti M. Chemical synthesis and biological activity of a novel antibacterial peptide deduced from a pig myeloid cDNA. FEBS Lett. 1994;337:303–307. doi: 10.1016/0014-5793(94)80214-9. [DOI] [PubMed] [Google Scholar]
- 28.Strukelj B, Pungercar J, Kopitar G, Renko M, Lenarcic B, Berbic S, Turk V. Molecular cloning and identification of a novel porcine cathelin-like antibacterial peptide precursor. Biol Chem Hoppe-Seyler. 1995;376:507–510. doi: 10.1515/bchm3.1995.376.8.507. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki S, Ohe Y, Okubo T, Kakegawa T, Tatemoto K. Isolation and characterization of novel antimicrobial peptides, rugosins A, B, and C, from the skin of the frog, Rana rugosa. Biochem Biophys Res Commun. 1995;212:249–254. doi: 10.1006/bbrc.1995.1963. [DOI] [PubMed] [Google Scholar]
- 30.Tam J P, Wu C R, Liu W, Zang J W. Disulfide bond formation in peptides by dimethyl sulfoxide. Scope and application. J Am Chem Soc. 1991;113:6657–6662. [Google Scholar]
- 31.Tamamura H, Murakami T, Horiuchi S, Sugihara K, Otaka A, Takada W, Ibuka T, Waki M, Yamamoto N, Fujii N. Synthesis of protegrin-related peptides and their antibacterial and anti-human immunodeficiency virus activity. Chem Pharm Bull. 1995;43:853–858. doi: 10.1248/cpb.43.853. [DOI] [PubMed] [Google Scholar]
- 32.Tossi A, Scocchi M, Zanetti M, Storici P, Gennaro R. PMAP-37, a novel antibacterial peptide from pig myeloid cells. cDNA cloning, chemical synthesis and activity. Eur J Biochem. 1995;228:941–946. doi: 10.1111/j.1432-1033.1995.tb20344.x. [DOI] [PubMed] [Google Scholar]
- 33.Yasin B, Lehrer R I, Harwig S S, Wagar E A. Protegrins: structural requirements for inactivating elementary bodies of Chlamydia trachomatis. Infect Immun. 1996;64:4863–4866. doi: 10.1128/iai.64.11.4863-4866.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zanetti M, Gennaro R, Romeo D. Cathelicidins: a novel protein family with a common proregion and a variable C-terminal antimicrobial domain. FEBS Lett. 1995;374:1–5. doi: 10.1016/0014-5793(95)01050-o. [DOI] [PubMed] [Google Scholar]
- 35.Zanetti M, Storici P, Tossi A, Scocchi M, Gennaro R. Molecular cloning and chemical synthesis of a novel antibacterial peptide derived from pig myeloid cells. J Biol Chem. 1994;269:7855–7858. [PubMed] [Google Scholar]
- 36.Zhao C, Liu L, Lehrer R I. Identification of a new member of the protegrin family by cDNA cloning. FEBS Lett. 1994;346:285–288. doi: 10.1016/0014-5793(94)00493-5. [DOI] [PubMed] [Google Scholar]
- 37.Zhao C, Ganz T, Lehrer R I. Structures of genes for two cathelin-associated antimicrobial peptides prophenin-2 and PR-39. FEBS Lett. 1995;376:130–134. doi: 10.1016/0014-5793(95)01237-3. [DOI] [PubMed] [Google Scholar]
- 38.Zhao C, Ganz T, Lehrer R I. The structure of porcine protegrin genes. FEBS Lett. 1995;368:197–202. doi: 10.1016/0014-5793(95)00633-k. [DOI] [PubMed] [Google Scholar]