Abstract
The field of dysmorphology has been changed by the use Artificial Intelligence (AI) and the development of Next Generation Phenotyping (NGP). The aim of this study was to propose a new NGP model for predicting KS (Kabuki Syndrome) on 2D facial photographs and distinguish KS1 (KS type 1, KMT2D-related) from KS2 (KS type 2, KDM6A-related). We included retrospectively and prospectively, from 1998 to 2023, all frontal and lateral pictures of patients with a molecular confirmation of KS. After automatic preprocessing, we extracted geometric and textural features. After incorporation of age, gender, and ethnicity, we used XGboost (eXtreme Gradient Boosting), a supervised machine learning classifier. The model was tested on an independent validation set. Finally, we compared the performances of our model with DeepGestalt (Face2Gene). The study included 1448 frontal and lateral facial photographs from 6 centers, corresponding to 634 patients (527 controls, 107 KS); 82 (78%) of KS patients had a variation in the KMT2D gene (KS1) and 23 (22%) in the KDM6A gene (KS2). We were able to distinguish KS from controls in the independent validation group with an accuracy of 95.8% (78.9–99.9%, p < 0.001) and distinguish KS1 from KS2 with an empirical Area Under the Curve (AUC) of 0.805 (0.729–0.880, p < 0.001). We report an automatic detection model for KS with high performances (AUC 0.993 and accuracy 95.8%). We were able to distinguish patients with KS1 from KS2, with an AUC of 0.805. These results outperform the current commercial AI-based solutions and expert clinicians.
Subject terms: Medical genetics, Genetic counselling, Computer science
Introduction
Kabuki syndrome (KS) is a rare genetic disorder, with an estimated prevalence of 1:86,000 to 1:32,0001–3. The typical KS face includes long palpebral fissures associated with eversion of the lateral third of the lower eyelid; long and heavy lashes giving the impression of made-up eyes; broad, arched and interrupted eyebrows; broad, depressed nasal tip; and prominent, cupped ears1,2,4. Extra-facial features include mild to moderate intellectual disability, visceral malformations, skeletal dysplasia and immunological manifestations5. KS has been described in all ethnic groups6,7.
More than 80% of KS patients have a pathogenic variant in the coding regions of KMT2D (KS type 1, KS1, OMIM147920), and around 10% of patients have a pathogenic variant in the KDM6A gene (KS type 2, KS2, OMIM300128)8–12.
Improving syndrome screening in clinical genetics is a crucial challenge in reducing diagnostic wandering. In France, the 7000 rare diseases identified to date represent 4.5% of the population, half of which affect children under the age of 5 with 10% of deaths between 0 and 5. Around 50% of patients are not diagnosed, and for the remaining 50%, diagnostic wandering reaches an average of 5 years13. Diagnostic wandering is defined by the failure to define the precise cause of a disease after having performed all available investigations. Applications of Artificial Intelligence (AI) are increasing in healthcare14–17. The field of dysmorphology has been changed by these new methods, under the name of Next Generation Phenotyping (NGP)18. Publications comparing human performances to NGP are flourishing19–22, and some suggest that digital tools do it better than human experts in terms of diagnosis: Dudding-Byth et al.23 showed a better performance of NGP compared to clinicians in a group of ten genetic syndromes, not including KS; Rouxel et al.5 compared the performance of the DeepGestalt technology18 using the Face2Gene online tool (FDNA Inc. Boston, MA, USA) to the performances of clinicians trained in the recognition of KS1 and KS2.
The aim of this study was to develop a NGP model for the diagnosis of KS and for distinguishing KS1 from KS2. We trained and validated the model on a large national and international multi-center cohort of patients of all ages and ethnicities. The specificity of this approach was the integration of lateral pictures, including the outline of the cranial vault and the position of the ears, as well as frontal pictures and the morphology of the external ear.
Materials and methods
The study was approved by the Comité Éthique et Scientifique pour les Recherches, les Études et les Évaluations dans le domaine de la Santé (CESREES), №4570023bis, the Commission Nationale Informatique et Libertés (CNIL), №MLD/MFI/AR221900, the Institutional Review Board, Faculty of Medicine, Chulalongkorn University (IRB 264/62), and in accordance with the 1964 Helsinki declaration and its later amendments. Informed and written consents were obtained from the legal representatives of each child or from the patients themselves if they were of age.
Photographic dataset
We included most pictures from the photographic database of the Maxillofacial surgery and Plastic surgery department of Hôpital Necker—Enfants Malades (Assistance Publique—Hôpitaux de Paris), Paris, France. This database contains 594,000 photographs from 22,000 patients, and all pictures since 1995 were taken by a professional medical photographer using a Nikon D7000 device in standardized positions.
We included retrospectively and prospectively, from 1995 to 2023, all frontal and lateral pictures of patients diagnosed with KS. The photographs were not calibrated. All patients had genetic confirmation of KS (KMT2D or KDM6A). We excluded all photographs taken after any surgerical procedure that could have modified the craniofacial morphology. Multiple photographs per patient corresponded to different ages of follow-up. Duplicates were excluded.
Controls were selected among patients admitted for lacerations, trauma, infection and various skin lesions, without any record of chronic conditions. More precisely, follow-up for any type of chronic disease was considered as an exclusion criterion. The reports were retrieved using the local data warehouse Dr Warehouse24. For each patient, the best lateral view was included.
Data from five other medical genetics departments were also included according to the same criteria: (1) Montpellier University Hospital (n = 32), (2) Grenoble University Hospital (n = 1), (3) Tours University Hospital (n = 1), (4) King Chulalongkorn Memorial Hospital Bangkok, Thailand (n = 8), and (5) Lausanne University Hospital, Lausanne, Switzerland (n = 1).
Validation set
For designs №1 and №2, we randomly selected a group of individuals corresponding to 10% of the number of patients with KS, and the equivalent number of control patients. These patients were removed from the training set. The two sets were therefore independent.
Landmarking
We used three different templates based on 105 landmarks for the frontal views, 73 for the lateral views and 41 for the external ear pictures. We developed an automatic annotation model for each template following a pipeline including: (1) detection of the Region Of Interest (ROI) and (2) automatic placement of the landmarks.
For ROI detection, a Faster Region-based Convolutional Neural Network (RCNN) model was trained after data augmentation (images and their + 10° and 10° rotations), with a learning rate of 0.001, a batch size of 4, a gamma of 0.05 and 2000 iterations, optimized and split into two stages: ROI detection and determination of profile laterality.
(1) ROI detection—Faster RNN trained on 15,633 images, after data augmentation (images and their + 10° and − 10° rotations): 6186 frontal images (2062 × 3) and 9447 right and left profile images (3159 × 3). The batch size was 2, learning rate was 0.0025, and the maximum number of iterations was 2800.
(2) Determination of profile laterality—Pre-trained ResNet50 network25 using the Pytorch library26. The training images included 1570 left profiles and 1579 right profiles. The batch size was 16, an Adam optimizer27 was used with a learning rate of 0.001, a step of 7, and a gamma of 0.1, trained over 25 epochs.
For the automatic placement of landmarks, we used a patch-based Active Appearance Model (AAM) using the menpo library on Python 3.728. We have previously reported the relevance of this approach29. We used two-scale landmarking: the model for frontal pictures was trained on 904 manually annotated photographs, with a first stage of dimensioning (diagonal = 150), a patch shape of [(15, 15), (23, 23)] and 50 iterations and a second stage without resizing, with a patch shape of [(20, 20), (30, 30)] and 10 new iterations. The model for profile pictures was trained on 1,439 manually annotated photographs, with a first stage of dimensioning (diagonal = 150), a patch shape of [(15, 15), (23, 23)] and 25 iterations and a second stage without resizing, with a patch shape of [(15, 15), (23, 23)] and 5 new iterations. The model for ears was trained on 1221 manually annotated photographs, with a first stage of dimensioning (diagonal = 100), a patch shape of [(15, 15), (23, 23)] and 50 iterations and a second stage without resizing, with a patch shape of [(20, 20), (30, 30)] and 20 new iterations. All three models used the Lucas Kanade optimizer30.
Each automatically annotated photograph was checked by two authors blinded for the diagnosis, QH and MD, and landmarks were manually re-positioned when necessary, using landmarker.io31. The Intraclass Correlation Coefficient (ICC) was computed between the raters. ICC values greater than 0.9 corresponded to excellent reliability of the manual annotation32.
Geometric morphometrics
We performed Generalized Procrustes Analysis (GPA)33 on all landmark clouds using the geomorph package on R34. Since the data were uncalibrated photographs, ROI sizes were not available: shape parameters only were assessed and not centroid sizes. Procrustes coordinates were processed using Principal Component Analysis (PCA) for dimension reduction. We retained the principal components explaining 99% of the total variance in cumulative sum. The last 1% was considered as negligible information.
Texture extraction
We partitioned the frontal and profile pictures into key areas and applied textural feature extraction methods to each zone, allowing to check the results and determine which zone had contributed most to the diagnosis.
We defined 14 key areas that could potentially contribute to diagnosis: 11 on frontal views (right/left eyes, right/left eyebrows, glabella, forehead, nasal tip, philtrum, right/left cheeks, and chin) and 3 on lateral views (pre-auricular region, eye, and zygoma relief). Each zone was extracted automatically using the previously placed landmarks.
We used the Contrast Limited Adaptative Histogram Equalization (CLAHE) algorithm for histogram equalization, as previously reported before the use of feature extractors35,36. CLAHE enhanced contrast by evenly dispersing gray values37 and by reducing the influences of illumination during picture capture and of skin color. Kiflie et al. recommended CLAHE as a first choice equalization method38.
Gray-Level Co-occurrence Matrix (GLCM) methods, as proposed by Haralick39, are based on the estimation of the second-order joint conditional probability density functions, which characterize the spatial relationships between pixels. GLCM is commonly used in texture analysis40,41, for instance in radiomics on CT-scan or MRI images42–44 or for skin texture assessment45. In GLCM, the co-occurrence matrix contains information on entropy, homogeneity, contrast, energy and correlation between pixels. GLCM includes 28 features, taking into account the average and range for each item of information and for each zone, representing 28 × 14 = 394 textural features for each patient.
Stratification using metadata
The textural features and the geometric principal components were combined for further analysis. To consider associated metadata (age and gender) and the fact that we included more than one photograph per patient (that is the non-independence of the data), a mixed model was designed for each feature. The variables to be explained were the features (geometric and textural), with age, gender and ethnicity considered as explanatory variables. A random effect on age and individuals was introduced. The equation of the mixed model was:
where corresponded to a random slope for age per individual, and was a random error term. We did not use an interaction term between age and gender and age and ethnicity as it did not increase the likelihood of the model. Age, gender and ethnicity are significant factors in dysmorphology46,47.
The residuals of each feature were computed to consider potential biases linked to the metadata:
Classification model
The inputs to the model were the residuals from the linear models described above, for each geometric or textural feature. We used eXtreme Gradient Boosting (XGBoost), a supervised machine learning classifier, for all the analyses48. We chose a tree-based booster, and the loss function to be minimized was a logistic regression for binary classification. We set several hyperparameters to improve the performance and effect of the machine learning model: learning rate = 0.3, gamma = 0, maximum tree depth = 6. The model with the lowest error rate was chosen for analysis. We separated the dataset into a training set and a testing set, and a five-fold cross-validation was used to define the ideal number of iterations to avoid overfitting.
The chosen model with the ideal number of iterations was then used on the independent validation set to test performances, by plotting accuracy and AUC. The Receiver Operating Characteristics (ROC) curves were plotted in R using the plotROC package49. We used the DeepGestalt tool proposed by Face2Gene CLINIC on our validation set, to be able to compare its performance (accuracies).
Uniform Manifold Approximation and Projection (UMAP) representations
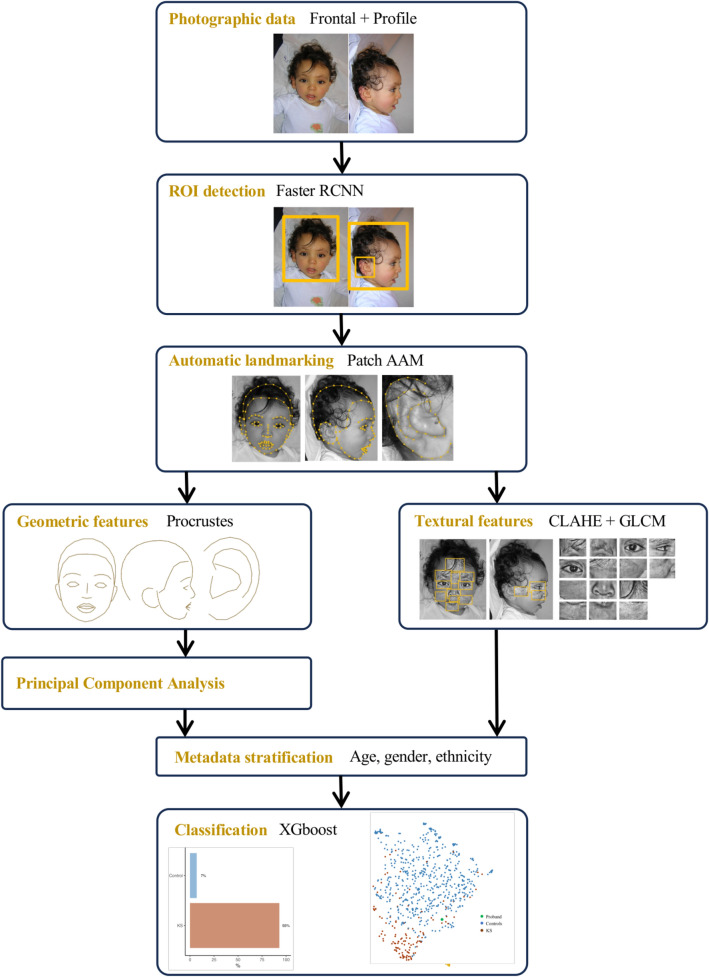
The residuals were represented using UMAP for visual clustering, a nonlinear dimension reduction technique50. We retained the residuals associated with features with a classification gain (in their cumulative sum) > 0.75 in the importance matrix associated with the XGboost model. A k (local neighborhood size) value of 15 was used. A cosine metric was introduced to compute distances in high dimensional spaces: the effective minimal distance between embedded points was . The three conditions of UMAP, namely uniform distribution, local constancy of the Riemannian metric and local connectivity were verified. UMAP analyses were performed using the package umap on R51 (Fig. 1).
Figure 1.
Analysis pipeline, from the initial photograph to diagnostic probability. ROI, Region Of Interest; AAM, active appearance model; Faster RCNN, Faster Region-based Convolutional Neural Network; CLAHE, Contrast Limited Adaptative Histogram Equalization; GLCM, Gray-Level Co-occurrence Matrix; XGboost, eXtreme Gradient Boosting.
Classification designs
Design №1, syndrome diagnosis support: KS was tested against controls in a binary classification.
Design №2, genotype–phenotype correlations: KS1 and KS2 were tested in binary classifications.
Design №3, genotype–phenotype correlations: KS1 Protein-Altering Variants (PAVs) and Protein-Truncating Variants (PTVs) were tested in binary classifications.
Ethics approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the CESREES (17/06/2021, 4570023).
Consent to participate
Written informed consent was obtained from the parents.
Consent to publish
The authors affirm that human research participants provided informed consent for publication of the images in Figs. 1, 4 and 7.
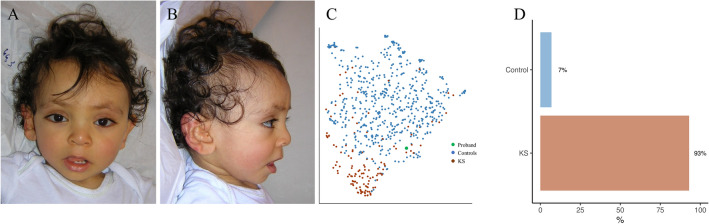
Figure 4.
Classification using design №1 for proband 3 of the validation set. (A) and (B) Frontal and profile views of proband 3. (C) UMAP representation of the training data according to the two groups, with positioning of proband 3. (D) Histogram of predictions by the model. This child was also detected as KS by Face2Gene CLINIC. KS, Kabuki Syndrome.
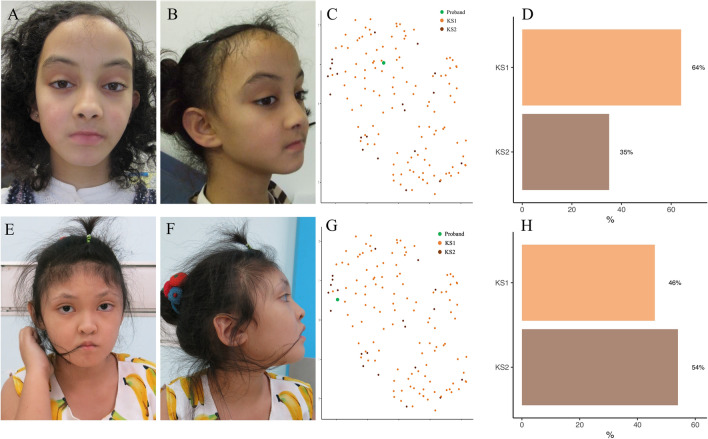
Figure 7.
Classification using design №2 for two probands of the training set. (A, B, E and F) Frontal and profile views of the two probands. (C and G) UMAP representations of the training data according to the two groups, with positioning of probands 3. (D and H) Histograms of predictions by the model. The phenotype included a reduced height of the midface, a thicker upper lip, and a vertical elongation of the external ear in the KS2 group (E and F). KS, Kabuki Syndrome.
Results
Population description
Ranging between 1998 and 2023, we included 1448 frontal and lateral facial photographs, corresponding to 634 patients. The mean age was 7.2 ± 4.2 years and ranged from 0 to 40.2 years; 52% were girls. Ethnicity was 92% Caucasian, 6% African or Caribbean, and 3% Asian.
The control group comprised 1084 photographs, corresponding to 527 patients with a mean age of 7.0 ± 4.6 years. Fifty-four percent were girls and ethnicities were 93% Caucasian, 5% African/Caribbean, and 2% Asian.
The KS group comprised 364 photographs, corresponding to 107 patients with a mean age of 7.8 ± 6.7 years. Forty-two percent were girls and ethnicities were 85% Caucasian, 7% African/Caribbean, and 8% Asian. Seventy-eight percent of patients were KS1 (Table 1).
Table 1.
Clinical description of the cohort.
| Total | Controls | KS | ||
|---|---|---|---|---|
| N | ||||
| Consultations | 724 | 542 (75%) | 182 (25%) | |
| Photographs | 1448 | 1084 (75%) | 364 (35%) | |
| Patients | 634 | 527 (83%) | 107 (17%) | |
| Gender | ||||
| Female | 331 (52%) | 286 (54%) | 45 (42%) | |
| Male | 303 (48%) | 241 (46%) | 62 (58%) | |
| Age (years) | ||||
| Mean ± SD | 7.2 ± 4.2 | 7.0 ± 4.6 | 7.8 ± 6.7 | |
| Median | 6.8 | 7.1 | 6.0 | |
| Minimum | 0 | 0.1 | 0 | |
| Maximum | 40.2 | 22.1 | 40.2 | |
| Ethnicity | ||||
| African/Caribbean | 35 (6%) | 27 (5%) | 8 (7%) | |
| Asian | 18 (3%) | 10 (2%) | 8 (7%) | |
| Caucasian | 581 (92%) | 490 (93%) | 91 (85%) | |
| Genetic variation | ||||
| KMT2D (KS1) | 82 (78%) | |||
| KDM6A (KS2) | 23 (22%) | |||
SD, standard deviation; KS, Kabuki Syndrome; KMT2D, Lysine (K)-specific methyltransferase 2D; KDM6A, Lysine (K)-specific demethylase 6A.
Two patients had a genetically confirmed diagnosis of KS, but we had no information on the causal gene. We thus collected information on genetic variation for 105 KS individuals with 82 (78%) and 23 (22%) with variations in KMT2D (KS1) and KDM6A (KS2) respectively.
In the KS1 group, 74% of variants were PTVs, with 49% nonsense variants leading to a premature stop codon (24% non-sense, 24% frameshift) and 26% splice donor site variants. Eighteen percent were PAVs, with 17% missense variants and 1% in-frame indel.
In the KS2 group, 78% of variants were PTVs, with 43% nonsense variants leading to a premature stop codon (30% non-sense, 13% frameshift), 30% splice donor site variants and 4% a large deletion. Nine percent were missense PAVs (Table 2).
Table 2.
Molecular description of the cohort.
| Amino-acids | Nucleotides | Exon | N (%) | ||||
|---|---|---|---|---|---|---|---|
| KMT2D | 82 (78%) | ||||||
| PTV | 61 (74%) | ||||||
| Nonsense | 40 (49%) | ||||||
| Nonsense | 20 (24%) | ||||||
| p.Cys247* | c.741T>A | 7 | 1 | ||||
| p.Ser286* | c.855_856del | 7 | 1 | ||||
| p.Gln1773* | c.5317C>T | 22 | 1 | ||||
| p.Gln1949* | c.5845C>T | 27 | 1 | ||||
| p.Gln2109* | c.6325C>T | 31 | 1 | ||||
| p.Arg2645* | c.7933C>T | 31 | 1 | ||||
| p.Arg2801* | c.8401C>T | 34 | 1 | ||||
| p.Gln3910* | c.11728C>T | 39 | 1 | ||||
| p.Gln3942* | c.11824C>T | 39 | 1 | ||||
| p.Gln4223* | c.12667C>T | 39 | 1 | ||||
| p.Gln4230* | c.12688C>T | 39 | 2 | ||||
| p.Arg4484* | c.13450C>T | 39 | 1 | ||||
| p.Arg4904* | c.14710C>T | 48 | 1 | ||||
| p.Arg5282* | c.15844C>T | 49 | 1 | ||||
| p.Tyr5321* | c.15963T>G | 50 | 1 | ||||
| Frameshift | 20 (24%) | ||||||
| p.Leu656Profs*12 | c.1966dup | 10 | 1 | ||||
| p.Glu1224Argfs*26 | c.3669dup | 11 | 1 | ||||
| p.Gly1235Valfs*95 | c.3699del | 11 | 1 | ||||
| p.Met1379Valfs*52 | c.4135_4136del | 14 | 1 | ||||
| p.Asp1876Glyfs*38 | c.5627_5630del | 25 | 1 | ||||
| p.Ser2039Glnfs*8 | c.6115del | 29 | 2 | ||||
| p.Ala2119Argfs*36 | c.6354dup | 31 | 1 | ||||
| p.Pro2330Serfs*47 | c.6987_6988insT | 31 | 1 | ||||
| p.Phe3672Leufs*76 | c.11016_11019del | 39 | 1 | ||||
| p.Met3894Trpfs*85 | c.11679del | 39 | 1 | ||||
| p.Glu4039Glyfs*17 | c.12116_12117del | 39 | 1 | ||||
| p.Ser4138Cysfs*29 | c.12413_12414del | 39 | 1 | ||||
| p.Leu4483Cysfs*36 | c.13446del | 39 | 1 | ||||
| p.Tyr5113Leufs*25 | c.15337dup | 48 | 1 | ||||
| p.Lys5139Gly | c.15415_15418del | 48 | 1 | ||||
| p.Leu5318Serfs*14 | c.15953_15956del | 50 | 1 | ||||
| Splice donor site | 21 (26%) | ||||||
| c.674-1G>A | 1 | ||||||
| c.1258+1G>A | 1 | ||||||
| c.2797+1G>C | 1 | ||||||
| c.13531-2A>C | 1 | ||||||
| c.14516-1G>C | 1 | ||||||
| PAV | 15 (18%) | ||||||
| Missense | 14 (17%) | ||||||
| p.Ala2182Thr | 10 | 1 | |||||
| p.Glu1391Lys | c.4171G>A | 14 | 1 | ||||
| p.Arg5048Cys | c.15142C>T | 48 | 1 | ||||
| p.Arg5048His | c.15143G>A | 48 | 1 | ||||
| p.Arg5154Gln | c.15461G>A | 48 | 2 | ||||
| p.Arg5179His | c.15536G>A | 48 | 2 | ||||
| p.Arg5214Cys | c.15640C>T | 48 | 1 | ||||
| p.Gly5295Ala | c.15884G>C | 49 | 1 | ||||
| p.Arg5340Gln | c.16019G>A | 50 | 3 | ||||
| p.Arg5432Trp | c.16294C>T | 51 | 1 | ||||
| Indel | 1 (1%) | ||||||
| p.Val275Ser | c.822_825delinsGTAGGCT | 7 | 1 | ||||
| KDM6A | 23 (22%) | ||||||
| PTV | 18 (78%) | ||||||
| Nonsense | 10 (43%) | ||||||
| Nonsense | 7 (30%) | ||||||
| p.Tyr109* | c.327_333del | 3 | 1 | ||||
| p.Arg172* | c.514C>T | 6 | 1 | ||||
| p.Gln692* | c.2074C>T | 17 | 1 | ||||
| p.Gln1037* | c.3109C>T | 20 | 1 | ||||
| p.Trp1221* | c.3663G>A | 25 | 1 | ||||
| p.Arg1279* | c.3835C>T | 26 | 2 | ||||
| Frameshift | 3 (13%) | ||||||
| p.Gln607Alafs*25 | c.1818_1819del | 16 | 1 | ||||
| p.Thr613Tyrfs*8 | c.1846_1849del | 16 | 1 | ||||
| p.Ser1091Metfs*12 | c.3270_3273del | 17 | 1 | ||||
| Splice donor site | 7 (30%) | ||||||
| c.564+1G>T | 1 | ||||||
| c.619+6T>C | 1 | ||||||
| c.620-2A>G | 1 | ||||||
| c.875+1G>A | 1 | ||||||
| c.2939-1G>T | 1 | ||||||
| c.2988+1G>C | 1 | ||||||
| c.3366-8_3366-4del | 1 | ||||||
| Large deletion | 1 (4%) | ||||||
| exons 1 and 2 | 1 | ||||||
| PAV | 2 (9%) | ||||||
| Missense | 2 (9%) | ||||||
| p.Arg481His | c.1442G>A | 14 | 1 | ||||
| p.Arg1255Trp | c.3763C>T | 26 | 1 | ||||
KMT2D , Lysine (K)-specific methyltransferase 2D; KDM6A, Lysine (K)-specific demethylase 6A; PTV, protein-truncating variant; PAV, protein-altering variant.
Design №1 : KS vs controls
Phenotype
We confirmed the usual characteristics described in KS: high and arched eyebrows, long palpebral fissures, and large and prominent ears (Fig. 2).
-
2.
Classification
Figure 2.
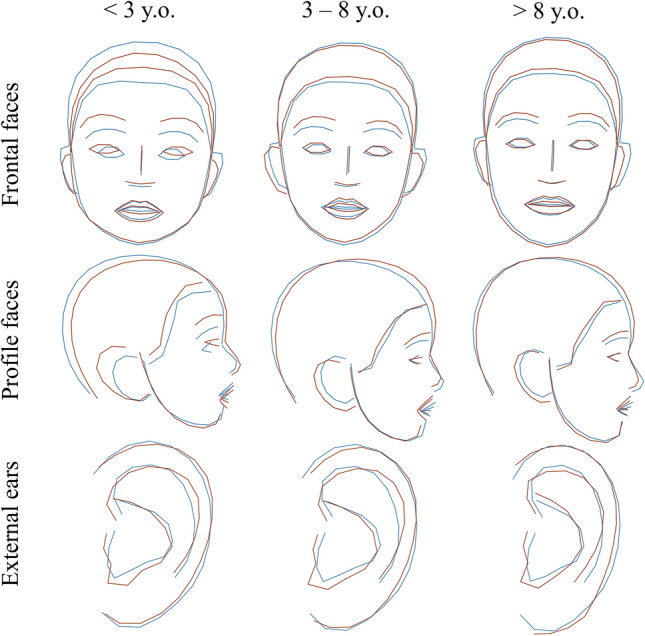
Average shapes in KS and controls and comparisons after Procrustes superimposition of frontal views, profile views, and external ears for three age groups. Blue = controls, Dark red = KS.
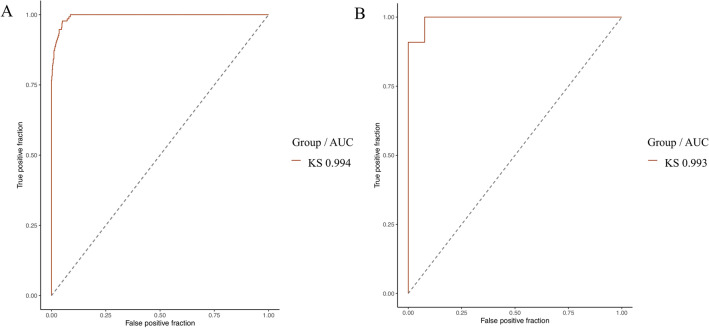
We were able to distinguish KS vs controls in the independent validation group with an accuracy of 95.8% (78.9–99.9%, p < 0.001). AUCs were comparable in the training set (0.994) and in the validation set (0.993) (Fig. 3, Table 3).
Figure 3.
(A) Empirical ROC curves (training set) for KS with AUC in design №1. (B) ROC curves (validation set) for KS with AUC in design №1. AUC, area under the curve; KS, Kabuki Syndrome.
Table 3.
Classification performances for design №1 (KS vs controls) in the validation group.
| Design №1 | ||
|---|---|---|
| Accuracy | 0.958 [0.789–0.999] | p < 0.001* |
| AUC | 0.993 [0.974–1.000] | p < 0.001* |
| F1 score | 0.963 |
AUC, area under the curve.
*Statistically significant (p < 0.05).
Ten out of eleven patients were correctly predicted as KS with our model, and this performance was the same using Face2Gene CLINIC (Supp. Table 1). In addition, we were able to predict all control patients (Fig. 4, Table 4).
Table 4.
Confusion matrix for design №1 (KS versus controls) in the validation group.
| Reference | |||
|---|---|---|---|
| Control | KS | ||
| Prediction | Control | 13 | 1 |
| KS | 0 | 10 | |
Bold values: True Positives (TP).
KS, Kabuki Syndrome.
Design №2 : KS1 vs KS2
Phenotype
KS2 individuals had a rounder face (HP:0000311), a shorter nose (HP:0003196), a thicker upper lip (HP:0000215), anteverted nostrils (HP:0000463), and a shorter midface (HP:0011800). There was no obvious difference in the eyebrows and eyes. The external ears were more elongated vertically in KS2 (HP:0400004), with a hypoplastic lobe (HP:0000385), and with a counter-clockwise rotation. The conch seemed more vertical in KS1 (Fig. 5).
-
2.
Classification
Figure 5.
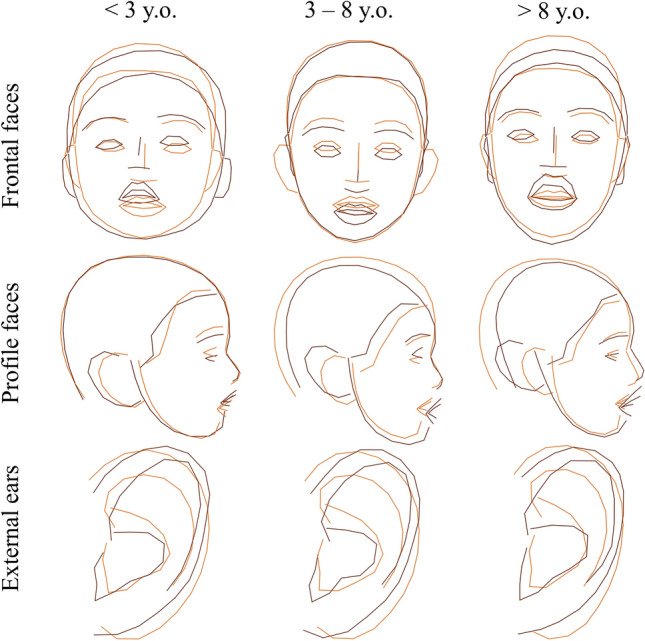
Average shapes in KS1 and KS2 and comparisons after Procrustes superimposition of frontal views, lateral views, and external ears for three age groups. Orange = KS1, Dark red = KS2.
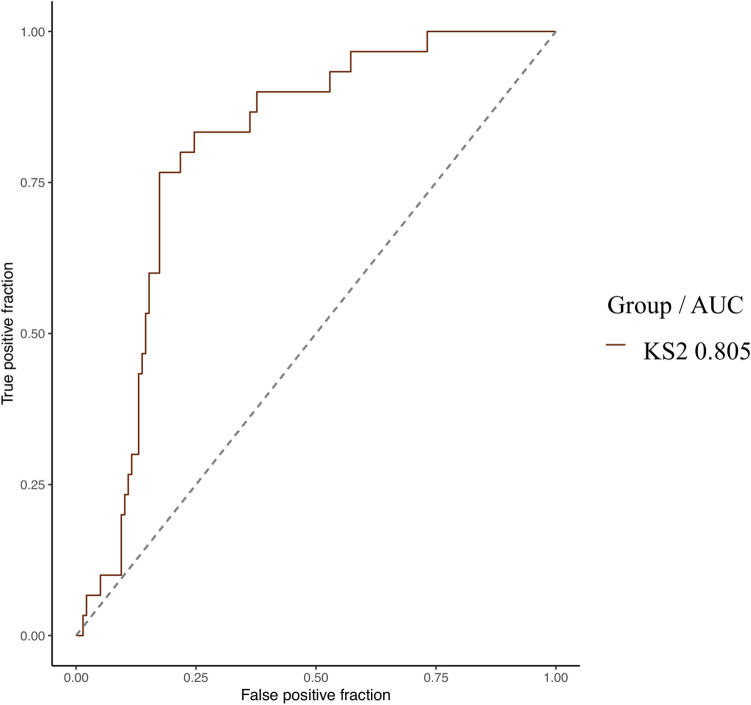
The model was able to distinguish KS1 from KS2 with an empirical AUC of 0.805 (0.729–0.880, p < 0.001) (Figs. 6, 7). This trend was found in the validation group, with an accuracy of 70% without reaching the significance threshold (Tables 5 and 6).
Figure 6.
Empirical ROC curve (training set) for KS2 with AUC in design №2. AUC, Area Under the Curve; KS, Kabuki Syndrome.
Table 5.
Classification performances for design №2 (KS1 versus KS2) in the validation group.
| Design №2 | ||
|---|---|---|
| Accuracy | 0.700 [0.348–0.933] | p = 0.172 |
| AUC | 0.660 [0.314–1.000] | p = 0.221 |
| F1 score | 0.727 |
Significant values are in [italics].
AUC, area under the curve.
Table 6.
Confusion matrix for design №2 (KS1 versus KS2) in the validation group.
| Reference | |||
|---|---|---|---|
| KS1 | KS2 | ||
| Prediction | KS1 | 4 | 2 |
| KS2 | 1 | 3 | |
Bold values: True Positives (TP).
KS, Kabuki Syndrome.
Design №3: PTV vs PAV in KS1
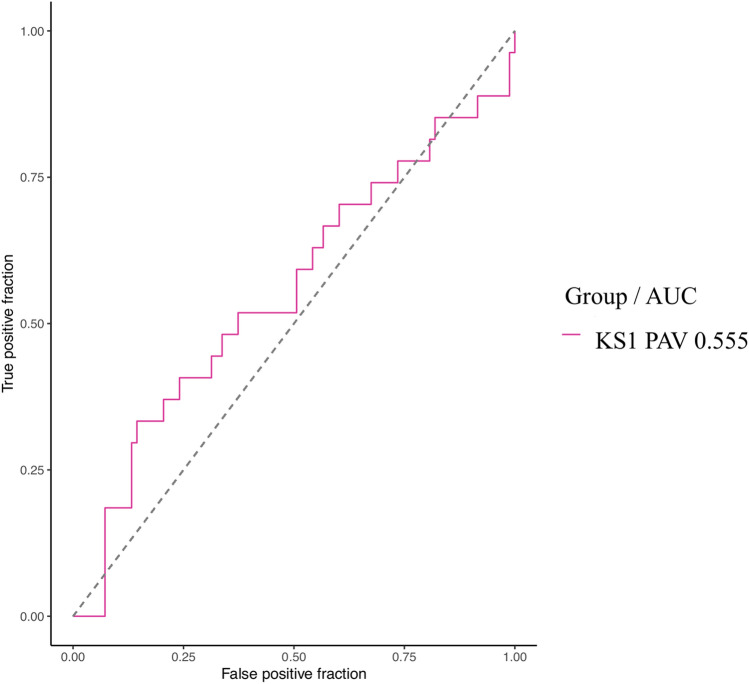
The model was unable to detect a difference in facial phenotype between KS1 patients with a PTV compared to KS1 patients with a PAV (0.555 [0.419–0.690], p = 0.786) (Fig. 8).
Figure 8.
Empirical ROC curve (training set) for KS1 PAV with AUC in design №3. AUC, Area Under the Curve; KS, Kabuki Syndrome; PAV, protein-altering variant.
Discussion
The model we report distinguished KS from controls in the independent validation group with an accuracy of 95.8% (78.9–99.9%, p < 0.001). Only 1 patient out of 24 was classified as ‘control’ while she had KS (accuracy 96%). In the KS group, 10 out of 11 patients were correctly classified (accuracy 91%). Using the Face2Gene CLINIC tool on KS patients (because DeepGestalt technology is not capable of recognizing non-syndromic patients) 1 patient out of 11 could not be analyzed and could not be classified as KS (accuracy 91%). Performances were therefore comparable. Interestingly, the patient not recognized by our model and by Face2Gene CLINIC was of African ethnicity, highlighting the lack of training data for non-Caucasian patients. The distribution of ethnic groups varies greatly from one center to another, which is why we believe it is important to encourage international collaborations in the field of Next Generation Phenotyping.
The model we report was also capable to distinguish KS1 from KS2 with an empirical AUC of 0.805 (0.729–0.880, p < 0.001). Rouxel et al.5 showed that the Face2Gene RESEARCH tool distinguished KS1 from KS2 in a cohort of 66 patients with an AUC of 0.722 (p = 0.022). The same team showed a classification accuracy of 61% (20/33) by clinical genetics experts between KS1 and KS2. The performance of our model was at least comparable to Face2Gene RESEARCH and seemed to outperform that of clinical experts.
Rouxel et al.5 explained that KS1 patients had a longer face and nose, a thin upper lip vermilion and a longer midface in comparison to KS2 patients, who have a rounder face, a thicker vermilion and anteverted nostrils. Our study reports new phenotypic features not seen on frontal images alone for KS2, such as a particular morphology of the external ear, longer along the vertical axis and with counter-clockwise rotation.
Phenotype-genotype correlations have been reported in KS for extra-facial anomalies. Cardiovascular abnormalities, namely ventricular septal defects, coarctation of the aorta, atrial septal defects, bicuspid aortic valve, patent ductus arteriosus, and hypoplastic left heart syndrome52,53,53–55 are more prevalent in KS2 compared to KS11,56. Persistent hypoglycemia due to pituitary hormone deficiency, adrenal insufficiency, growth hormone deficiency and dysregulated insulin secretion by the pancreatic β-cells57,58 are also more frequent in KS210, possibly because the inhibition of KDM6A increases the release of insulin from pancreatic islet cells, as suggested by mouse models1,59. Urinary tract anomalies, such as horseshoe kidneys and renal hypoplasia, seem to be more frequent in KS1, and genital defects such as cryptorchidism and hypospadias could be more frequent in KS256,60,61.
Rouxel et al.5 underline the lack of Asian patients in their evaluation, and proposed that larger series were needed to better define phenotypical differences between KS1 and KS2, and the general dependance of the phenotype with ethnicity6,12. The collaboration with an Asian clinical genetics center (Bangkok) is thus a strong point of this study.
The use of textural feature extraction allowed our model account for typical KS characteristics not recognized by geometric analysis (Procrustes) alone. The lateral sparsening of the eyebrows and heavy lashes giving the impression of make-up eyes were thus included into in the classification.
Barry et al.1 reported a large meta-analysis including 152 articles and 1369 individuals with KS and assessed the prevalence of the different types of pathogenic variation per gene. The majority of KMT2D variants were truncating (non-sense 34%, frameshift 34%), then missense (23%) and finally splice site variants (9%). The majority of KDM6A variants were truncating (frameshift 36% > non-sense 27%), followed by splice site (20%), and missense (18%). We found similar results, with a higher prevalence of truncating non-sense variants for both genes. There was a higher prevalence of splice donor site variants, with 26% for KMT2D and 30% for KDM6A. Some authors report a more severe clinical outcomes in patients with non-sense variants than in patients with a frameshift variant1. Faundes et al.56 found more severe neurodevelopmental anomalies in patients with protein-truncating mutations in the KS2 group. Shah et al.62 reported ophthalmological anomalies such as strabismus, blue sclerae, microphthalmia and refractive anomalies that were more severe in patients with a non-sense variant, and less frequent in patients with a frameshift variant. Our model did not find any significant difference in facial phenotype between PTV and PAV.
Conclusion
Here we report an automatic detection model for KS including the face, profiles and ears, with performances (AUC 0.993 and accuracy 95.8%) comparable to those of Face2Gene, on an independent validation set. These performances were achieved using an international cohort of 107 patients with a confirmed molecular diagnosis of KS. Using the same model, we were able to separate patients with KS1 (KMT2D) from KS2 (KDM6A), with an AUC of 0.805. These results seem to at least outperform Face2Gene and support the possibility of using a phenotype-first strategy to diagnose KS and detect its two causal genes.
Supplementary Information
Author contributions
All authors read and approved the final manuscript. Conceptualization: Q.H., RH.K., N.G., D.G. Data curation: Q.H., M.D., T.B., T.B., Formal analysis: Q.H., J.F., T.B., T.B. Funding acquisition: N.G., RH.K., S.L., V.CD., Investigation: Q.H., RH.K., N.G., D.G. Methodology: Q.H., N.G., D.G., RH.K., J.F., K.Y., F.R. Project administration: S.L., N.G., M.R., J.A. Resources: A.T., D.G., M.W., K.D., S.A., T.P., F.G., V.S., P.C. Software: Q.H., T.B., T.B. Supervision: Q.H., RH.K., N.G., D.G. Validation: Q.H., RH.K., N.G., D.G. Visualization : Q.H., RH.K., N.G., D.G. Writing-original draft: Q.H. Writing-review & editing: Q.H., RH.K., N.G., D.G., A.T., D.G., M.W., K.D., S.A., T.P., F.G., V.S., P.C.
Funding
This work was supported by the ‘Agence Nationale de la Recherche’, ’Investissements d’Avenir’ program (ANR-10-IAHU-01), France 2030 grant “Face4Kids” (ANR-21-PMRB-0004), Health Systems Research Institute (66-101, 66-122), Thailand Science Research and Innovation Fund Chulalongkorn University, and National Research Council of Thailand (N42A650229).
Data availability
The code is available to readers on the website https://framagit.org/imagine-plateforme-bdd/mfdm/. The datasets (photographs) supporting the current study have not been deposited in a public repository because of their identifiable nature.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: David Geneviève and Roman H. Khonsari.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-52691-3.
References
- 1.Barry KK, et al. From genotype to phenotype—a review of Kabuki syndrome. Genes (Basel) 2022;13:1761. doi: 10.3390/genes13101761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niikawa N, et al. Kabuki make-up (Niikawa-Kuroki) syndrome: A study of 62 patients. Am. J. Med. Genet. 1988;31:565–589. doi: 10.1002/ajmg.1320310312. [DOI] [PubMed] [Google Scholar]
- 3.White SM, et al. Growth, behavior, and clinical findings in 27 patients with Kabuki (Niikawa-Kuroki) syndrome. Am. J. Med. Genet. A. 2004;127A:118–127. doi: 10.1002/ajmg.a.20674. [DOI] [PubMed] [Google Scholar]
- 4.Kuroki Y, Suzuki Y, Chyo H, Hata A, Matsui I. A new malformation syndrome of long palpebral fissures, large ears, depressed nasal tip, and skeletal anomalies associated with postnatal dwarfism and mental retardation. J. Pediatr. 1981;99:570–573. doi: 10.1016/S0022-3476(81)80256-9. [DOI] [PubMed] [Google Scholar]
- 5.Rouxel F, et al. Using deep-neural-network-driven facial recognition to identify distinct Kabuki syndrome 1 and 2 gestalt. Eur. J. Hum. Genet. 2022;30:682–686. doi: 10.1038/s41431-021-00994-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adam MP, Hudgins L. Kabuki syndrome: A review. Clin. Genet. 2005;67:209–219. doi: 10.1111/j.1399-0004.2004.00348.x. [DOI] [PubMed] [Google Scholar]
- 7.Bögershausen N, et al. Mutation update for Kabuki syndrome genes KMT2D and KDM6A and further delineation of X-Linked Kabuki syndrome subtype 2. Hum. Mutat. 2016;37:847–864. doi: 10.1002/humu.23026. [DOI] [PubMed] [Google Scholar]
- 8.Lederer D, et al. Deletion of KDM6A, a Histone Demethylase Interacting with MLL2, in three patients with Kabuki Syndrome. Am. J. Hum. Genet. 2012;90:119–124. doi: 10.1016/j.ajhg.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paděrová J, et al. Molecular genetic analysis in 14 Czech Kabuki syndrome patients is confirming the utility of phenotypic scoring. Clin. Genet. 2016;90:230–237. doi: 10.1111/cge.12754. [DOI] [PubMed] [Google Scholar]
- 10.Banka S, et al. Novel KDM6A (UTX) mutations and a clinical and molecular review of the X-linked Kabuki syndrome (KS2) Clin. Genet. 2015;87:252–258. doi: 10.1111/cge.12363. [DOI] [PubMed] [Google Scholar]
- 11.Bögershausen N, Wollnik B. Unmasking Kabuki syndrome. Clin. Genet. 2013;83:201–211. doi: 10.1111/cge.12051. [DOI] [PubMed] [Google Scholar]
- 12.Ng SB, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DGOS. Les maladies rares. Ministère de la Santé et de la Préventionhttps://sante.gouv.fr/soins-et-maladies/prises-en-charge-specialisees/maladies-rares/article/les-maladies-rares (2023).
- 14.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N. Engl. J. Med. 2019;380:1347–1358. doi: 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- 15.Choy G, et al. Current applications and future impact of machine learning in radiology. Radiology. 2018;288:318–328. doi: 10.1148/radiol.2018171820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novoa RA, Gevaert O, Ko JM. Marking the path toward artificial intelligence-based image classification in dermatology. JAMA Dermatol. 2019;155:1105–1106. doi: 10.1001/jamadermatol.2019.1633. [DOI] [PubMed] [Google Scholar]
- 17.Loftus TJ, et al. Artificial Intelligence and surgical decision-making. JAMA Surg. 2020;155:148–158. doi: 10.1001/jamasurg.2019.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurovich Y, et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat. Med. 2019;25:60–64. doi: 10.1038/s41591-018-0279-0. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, et al. Molecular and phenotypic expansion of alström syndrome in Chinese patients. Front. Genet. 2022;13:808919. doi: 10.3389/fgene.2022.808919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javitt MJ, Vanner EA, Grajewski AL, Chang TC. Evaluation of a computer-based facial dysmorphology analysis algorithm (Face2Gene) using standardized textbook photos. Eye. 2022;36:859–861. doi: 10.1038/s41433-021-01563-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latorre-Pellicer A, et al. Evaluating Face2Gene as a tool to identify cornelia de lange syndrome by facial phenotypes. Int. J. Mol. Sci. 2020;21:E1042. doi: 10.3390/ijms21031042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishima H, et al. Evaluation of Face2Gene using facial images of patients with congenital dysmorphic syndromes recruited in Japan. J. Hum. Genet. 2019;64:789–794. doi: 10.1038/s10038-019-0619-z. [DOI] [PubMed] [Google Scholar]
- 23.Dudding-Byth T, et al. Computer face-matching technology using two-dimensional photographs accurately matches the facial gestalt of unrelated individuals with the same syndromic form of intellectual disability. BMC Biotechnol. 2017;17:90. doi: 10.1186/s12896-017-0410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcelon N, et al. A clinician friendly data warehouse oriented toward narrative reports: Dr Warehouse. J. Biomed. Inform. 2018;80:52–63. doi: 10.1016/j.jbi.2018.02.019. [DOI] [PubMed] [Google Scholar]
- 25.Koonce, B. ResNet 50 63–72 (Springer, 2021). 10.1007/978-1-4842-6168-2_6.
- 26.Paszke, A. et al.PyTorch: An Imperative Style, High-Performance Deep Learning Library. 10.48550/arXiv.1912.01703 (2019).
- 27.Kingma, D. P. & Ba, J. Adam: A Method for Stochastic Optimization. 10.48550/arXiv.1412.6980 (2017).
- 28.Alabort-i-Medina, J., Antonakos, E., Booth, J., Snape, P. & Zafeiriou, S. Menpo: A comprehensive platform for parametric image alignment and visual deformable models. In Proceedings of the 22nd ACM international conference on Multimedia 679–682 (ACM, 2014). 10.1145/2647868.2654890.
- 29.Hennocq Q, et al. An automatic facial landmarking for children with rare diseases. Am. J. Med. Genet. Part A. 2022;2022:145. doi: 10.1002/ajmg.a.63126. [DOI] [PubMed] [Google Scholar]
- 30.Lucas B, Kanade T. An Iterative Image Registration Technique with an Application to Stereo Vision (IJCAI) Springer; 1981. [Google Scholar]
- 31.landmarker.io. The Menpo Project. https://www.menpo.org/landmarkerio/ (2022).
- 32.Bartko JJ. The intraclass correlation coefficient as a measure of reliability. Psychol. Rep. 1966;19:3–11. doi: 10.2466/pr0.1966.19.1.3. [DOI] [PubMed] [Google Scholar]
- 33.Rohlf FJ, Slice D. Extensions of the procrustes method for the optimal superimposition of landmarks. Syst. Zool. 1990;39:40–59. doi: 10.2307/2992207. [DOI] [Google Scholar]
- 34.Baken EK, Collyer ML, Kaliontzopoulou A, Adams DC. geomorph v4.0 and gmShiny: Enhanced analytics and a new graphical interface for a comprehensive morphometric experience. Methods Ecol. Evol. 2021;12:2355–2363. doi: 10.1111/2041-210X.13723. [DOI] [Google Scholar]
- 35.Avcı H, Karakaya J. A novel medical image enhancement algorithm for breast cancer detection on mammography images using machine learning. Diagn. (Basel) 2023;13:348. doi: 10.3390/diagnostics13030348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anifah L, Purnama IKE, Hariadi M, Purnomo MH. Osteoarthritis classification using self organizing map based on Gabor Kernel and contrast-limited adaptive histogram equalization. Open Biomed. Eng. J. 2013;7:18. doi: 10.2174/1874120701307010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang C, Li X, Wen Y. AN OTSU image segmentation based on fruitfly optimization algorithm. Alexandr. Eng. J. 2021;60:183–188. doi: 10.1016/j.aej.2020.06.054. [DOI] [Google Scholar]
- 38.Kiflie A, Tesema Tufa G, Salau AO. Sputum smears quality inspection using an ensemble feature extraction approach. Front. Public Health. 2023;10:1032467. doi: 10.3389/fpubh.2022.1032467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973;3:610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 40.Mohanaiah P, Sathyanarayana P, GuruKumar L. Image Texture Feature Extraction Using GLCM Approach. Springer; 2013. [Google Scholar]
- 41.Löfstedt T, Brynolfsson P, Asklund T, Nyholm T, Garpebring A. Gray-level invariant Haralick texture features. PLoS One. 2019;14:e0212110. doi: 10.1371/journal.pone.0212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mundt P, et al. Periaortic adipose radiomics texture features associated with increased coronary calcium score-first results on a photon-counting-CT. BMC Med. Imaging. 2023;23:97. doi: 10.1186/s12880-023-01058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adelsmayr G, et al. Three dimensional computed tomography texture analysis of pulmonary lesions: Does radiomics allow differentiation between carcinoma, neuroendocrine tumor and organizing pneumonia? Eur. J. Radiol. 2023;165:110931. doi: 10.1016/j.ejrad.2023.110931. [DOI] [PubMed] [Google Scholar]
- 44.Peng B, et al. Preoperative computed tomography-based tumoral radiomic features prediction for overall survival in resectable non-small cell lung cancer. Front. Oncol. 2023;13:1131816. doi: 10.3389/fonc.2023.1131816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ou X, Pan W, Xiao P. In vivo skin capacitive imaging analysis by using grey level co-occurrence matrix (GLCM) Int. J. Pharm. 2014;460:28–32. doi: 10.1016/j.ijpharm.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 46.Muenke M, Adeyemo A, Kruszka P. An electronic atlas of human malformation syndromes in diverse populations. Genet. Med. 2016;18:1085–1087. doi: 10.1038/gim.2016.3. [DOI] [PubMed] [Google Scholar]
- 47.Burchard EG, et al. The importance of race and ethnic background in biomedical research and clinical practice. N. Engl. J. Med. 2003;348:1170–1175. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- 48.Chen, T. & Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 785–794 (Association for Computing Machinery, 2016). 10.1145/2939672.2939785.
- 49.Sachs MC. plotROC: A tool for plotting ROC curves. J. Stat. Softw. 2017;79:2. doi: 10.18637/jss.v079.c02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McInnes, L., Healy, J. & Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. 10.48550/arXiv.1802.03426 (2020).
- 51.R Core Team. European Environment Agency. https://www.eea.europa.eu/data-and-maps/indicators/oxygen-consuming-substances-in-rivers/r-development-core-team-2006 (2020).
- 52.Hughes HE, Davies SJ. Coarctation of the aorta in Kabuki syndrome. Arch. Dis. Child. 1994;70:512–514. doi: 10.1136/adc.70.6.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Digilio MC, et al. Congenital heart defects in molecularly proven Kabuki syndrome patients. Am. J. Med. Genet. A. 2017;173:2912–2922. doi: 10.1002/ajmg.a.38417. [DOI] [PubMed] [Google Scholar]
- 54.Cheon C-K, Ko JM. Kabuki syndrome: Clinical and molecular characteristics. Korean J. Pediatr. 2015;58:317–324. doi: 10.3345/kjp.2015.58.9.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon JK, et al. The strong association of left-side heart anomalies with Kabuki syndrome. Korean J. Pediatr. 2015;58:256–262. doi: 10.3345/kjp.2015.58.7.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Faundes V, et al. Clinical delineation, sex differences, and genotype-phenotype correlation in pathogenic KDM6A variants causing X-linked Kabuki syndrome type 2. Genet. Med. 2021;23:1202–1210. doi: 10.1038/s41436-021-01119-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yap KL, et al. Congenital hyperinsulinism as the presenting feature of Kabuki syndrome: Clinical and molecular characterization of 9 affected individuals. Genet. Med. 2019;21:233–242. doi: 10.1038/s41436-018-0013-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gole H, Chuk R, Coman D. Persistent hyperinsulinism in Kabuki syndrome 2: Case report and literature review. Clin. Pract. 2016;6:848. doi: 10.4081/cp.2016.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gibson CE, et al. Congenital hyperinsulinism in infants with turner syndrome: Possible association with monosomy X and KDM6A haploinsufficiency. Horm. Res. Paediatr. 2018;89:413–422. doi: 10.1159/000488347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Courcet J-B, et al. Clinical and molecular spectrum of renal malformations in Kabuki syndrome. J. Pediatr. 2013;163:742–746. doi: 10.1016/j.jpeds.2013.02.032. [DOI] [PubMed] [Google Scholar]
- 61.Cetinkaya E, Misirlioğlu ED, Vidinlisan S, Baydar Z, Ozhan ZR. Hypospadias in a patient with Kabuki make-up (Niikawa-Kuroki) syndrome. J. Pediatr. Endocrinol. Metab. 2001;14:803–805. doi: 10.1515/JPEM.2001.14.6.803. [DOI] [PubMed] [Google Scholar]
- 62.Shah SS, et al. Insights into the genotype-phenotype relationship of ocular manifestations in Kabuki syndrome. Am. J. Med. Genet. A. 2023;191:1325–1338. doi: 10.1002/ajmg.a.63155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code is available to readers on the website https://framagit.org/imagine-plateforme-bdd/mfdm/. The datasets (photographs) supporting the current study have not been deposited in a public repository because of their identifiable nature.