Summary
Sirt6, a member of the class III histone deacetylases (HDACs), functions in the regulation of genomic stability, DNA repair, cancer, metabolism and aging. Sirt6 deficiency is lethal, and newborn SIRT6-null cynomolgus monkeys show unfinished brain development. After the generation of a cortex-specific Sirt6 conditional knockout mouse model, we investigated the specific deletion of Sirt6 in NPCs at E10.5. This study found that Sirt6 deficiency causes excessive proliferation of neural precursor cells (NPCs) and retards differentiation. The results suggest that endogenous Sirt6 in NPCs regulates histone acetylation and limits stemness-related genes, including Notch1, in order to participate in NPC fate determination. These findings help elucidate Sirt6’s role in brain development and in NPC fate determination while providing data on species generality and differentiation.
Subject areas: Biological sciences, developmental genetics, developmental neuroscience
Graphical abstract

Highlights
-
•
Emx1-Cre was used to induce conditional Sirt6 knockout in the cerebral cortex
-
•
Sirt6 deficiency promotes the abnormal proliferation of neural progenitor cells
-
•
Sirt6 deficiency delays the development of the cerebral cortex
-
•
Sirt6 deficiency upregulated the Notch1 pathway through histone acetylation
Biological sciences; Developmental genetics; Developmental neuroscience
Introduction
A better understanding of neurodevelopment, the process by which the nervous system forms through the account of integration and regulation of a series of molecules, will lead to numerous opportunities for future diagnosis and treatment of neurological disorders, disabilities, and the cognitive effects of aging. Cerebral cortex development in the mouse telencephalon begins at E9.5 when neuroepithelial cells continuously expand in a symmetrical division mode and turn into neural progenitor cells (NPCs).1 NPCs located in the dorsal ventricular zone (VZ) differentiate into intermediate progenitor cells (IPCs) located in the subventricular zone (SVZ), after which these IPCs keep differentiating into neurons and glial cells, migrating and forming the hierarchical order of the cerebral cortex in an inside-out pattern. As NPCs in mice proliferate to complement the progenitor cell pool through symmetrical division, proper development maintains the balance between proliferation and differentiation. The cooperative regulation of transcription factors and post-transcriptional molecules is called NPCs’ fate determination.2
Histone deacetylases (HDACs) have an important role in embryo development and in NPC fate determination. Double knockout of HDAC1 and HDAC2 causes severe abnormalities in neuronal precursors in the differentiation into mature neurons and oligodendrocytes.3,4 HDAC2 and HDAC3 were also reported to bind to promoter regions of genes associated with neuronal and oligodendrocyte differentiation.5
As Class III HDACs, Sirtuins have been reported to act as regulators of longevity and to prevent age-related diseases.6,7,8,9 Among them, Sirt1 and Sirt6 prevent congenital anomalies in mammalian species,10,11 while the activation of Sirt1 suppresses the proliferation of NPCs and directs their differentiation toward the astroglial lineage.12 Research in 2006 determined that two enzyme activities, single ADP ribosyltransferase and deacetylase, were mainly responsible for Sirt6 function. Sirt6-deficient mice showed a smaller body size at 2–3 weeks, and subsequently exhibited several abnormalities, including severe lymphoid and subcutaneous fat reduction, lordosis and metabolic defects; mortality was significantly early, at approximately 4 weeks.13 Follow-up studies have shown that Sirt6 participates in DNA double-strand break repair, tumor suppression, and cellular metabolism by mediating single ADP ribosylation and histone deacetylation.14,15,16,17,18
Similar to other HDACs, Sirt6 has been correlated with nervous system development. Specifically, Sirt6 knockout cynomolgus monkeys with CRISPR/Cas9 resulted in acetylation in the ICR region of long noncoding RNA H19; differentiation of neurons was inhibited, apparently triggering cortical development delays.19 Nestin-Cre Sirt6-deficient mice, by the adult stage, had not only less brain volume than normal mice but also behavioral defects with major learning impairments.20 Similarly, in human patients diagnosed with Alzheimer’s disease, Sirt6 mRNA and protein levels were reduced,21 adding to evidence for a definite Sirt6-nervous system linkage, but that study did not focus on the cerebral cortex development stage. To exclude the secondary phenotype caused by heart defects in a conventional knockout animal or by hypothalamic hormone deficiency in Nestin-Cre-mediated conditional knockout mice (thus muddying Sirt6’s effect on NPC fate determination and neocortex formation), we constructed Emx1-Cre-induced conditional Sirt6 knockout mice. This cortex-specific configuration allowed us to examine the specific deletion of Sirt6 in NPCs at E10.5 just before cortical development.
We found that with Sirt6 deletion NPCs may proliferate and thus delay neural differentiation and neural development in the early brain of mice. Here, “proliferation” means NPCs’ intention to generate more daughter progenitor cells instead of differentiating into neurons and glia. Combined with the RNA-seq results, the SIRT6 protein may regulate the Notch pathway to influence stem cell proliferation. Our study sheds new light on Sirt6’s crucial role in brain development.
Results
Sirt6 conditional knockout during the development of the mouse cortex increased histone acetylation levels
To investigate the role of Sirt6 in cortical development, we analyzed Sirt6 spatial and temporal expression patterns by in situ hybridization with a Sirt6 antisense probe at embryonic day 12.5 (E12.5), E14.5, E16.5, E18.5, and at postnatal day (P7). At these time points, Sirt6 was widely expressed in the cerebral cortex, and its expression level gradually decreased (Figure 1A). To ascertain whether this downward trend continued throughout life, we detected Sirt6 expression in age groups from E14.5 to 15 M by western blots and real-time PCR. The expression of Sirt6 gradually decreased with age (Figures 1B and 1C). Sirt6 expression was significantly higher in early brain development, as compared to the relatively low expression during postnatal development, suggesting its possible role in early neural development.
Figure 1.
The expression pattern of Sirt6 and construction of brain-specific knockout mice
(A) The expression pattern of Sirt6 mRNA during neocortical development (E12.5 to P7). Mouse brain sections were labeled with Sirt6 antisense probe and Hes5 antisense probe. Scale bar: 200 μm.
(B) The expression level of the Sirt6 protein in the brain with development and aging (E14.5 to 15M) was detected by Western blot (WB).
(C) Expression level of Sirt6 mRNA in the brain with development and aging (E14.5 to 15M) was detected by RT-qPCR.
(D) Construction strategy of Sirt6-Emx1-Cre conditional knockout mice. The yellow triangle represents the loxP sequence, while the downward-pointing arrow represents mating with the Emx1-Cre tool mouse. The upper line graph represents the full length of Sirt6 gene in wild-type mice, and lower line represents the genetic composition of knockout mice.
(E) The expression changes of Sirt6 and its targeted histone acetylation site in the cerebral cortex of P7 mice after the deletion of Sirt6 was detected by WB. The results are expressed as the mean ± SD. See also Figure S1.
To generate Sirt6 cKO mice, we used the Emx1-Cre mouse line, in which the Emx1 promoter and enhancer start to be activated in the dorsal cortex at E9.5, to mediate Sirt6 deletion at the early neocortical developmental stage (Figure 1D).22 Verification of knockout efficiency and compensatory effect was performed in the brain or NSC proteins at E14.5 and P7 (Figures S1B and S1C). Consistent with previous studies, the results showed that the Sirt6 expression level decreased significantly with increased levels of H3K9ac and H3K56ac in the P7 Sirt6 cKO cerebral cortex (Figure 1E).
Sirt6 deletion promoted proliferation and delayed differentiation in neural progenitor cells in vivo
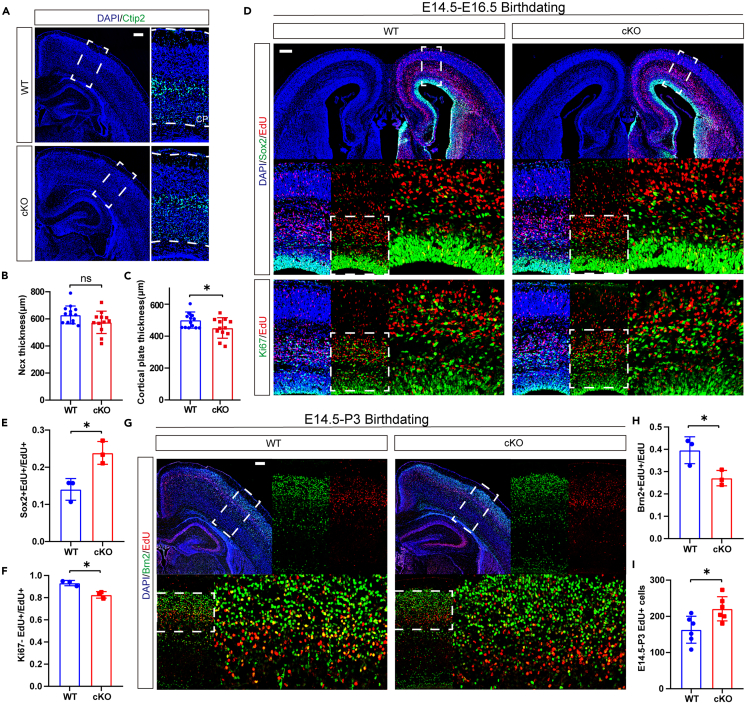
During mouse neocortex development, NPCs gradually change from symmetrical division to differentiation and produce cortical neurons.23 Most layer Ⅵ neurons are produced at E12.5; layer V (E13.5), layer IV (E14.5), and layers III and II (E15.5-E16.5) are produced from inside out.24 We observed that Sirt6-deficient mice had thinner cortical plates than their counterparts at P3. However, their neocortical thickness and layer proportion did not show significant changes (Figures 2A–2C and S2A–S2C). This situation in layer proportion is consistent with the finding in the cynomolgus monkey research, which showed that Sirt6 deficiency causes exorbitant NPC proliferation and retards cortical development.19 To test whether Sirt6 knockout in mice affected NPC proliferation and differentiation, we performed a series of EdU birth dating experiments. By labeling and tracing proliferating cells at E14.5–16.5 in the EdU birth dating cortex, we found that the Sirt6 cKO group, compared with the WT group, showed a significantly higher proportion of Sox2+ EdU-labeled cells without a significant increase in EdU+ cells. This finding suggests that during this time, more NPCs are reserved but do not generate many daughter cells (Figures 2D, 2E, and S2D). To identify the possibility of a decreased differentiation rate, we stained Ki67, a marker of the cell cycle. The significantly lower proportion of EdU+ Ki67-cells that exited the cell cycle suggests that Sirt6 deletion may promote NPC proliferative activity (Figure 2F). However, the results between WT and cKO groups after 30 min EdU labeling (Figures S2E–S2G) were not statistically significantly different, which could be because the time window was too short. We also performed EdU+BrdU co-labeling and Sox2+Tbr2 co-labeling experiments and found no significant difference between WT and cKO groups, indicating that there was no change in DNA synthesis and intermediate precursor cells generation (Figures S3A–S3D). To further investigate whether Sirt6 deficiency caused a quiescent or senescent phenotype, we performed a p27+Tbr2 co-labeling experiment and observed p27 expression in the upper layer of Tbr2, but the proportion of co-labeling cells did not change significantly (Figures S3E and S3F).
Figure 2.
Sirt6 deficiency causes an excessive proliferation of NPCs but disturbs timeline of cortical layer generation in vivo
(A) Immunostaining of the deep layer marker Ctip2 in P3 brain tissue sections shows cortical plate’s outline. Scale bars: 200 μm.
(B) Comparison of neocortical thickness between P3 wild type and Sirt6 cKO mice. n = 14 for control brains and n = 12 for Sirt6 cKO brains.
(C) Comparison of cortical plate thickness between P3 wild type and Sirt6 cKO mice. n = 14 for control brains and n = 12 for Sirt6 cKO brains.
(D) Immunostaining of Sox2 and Ki67 in brain tissue sections of E16.5 WT and Sirt6 cKO mice that EdU were injected at E14.5. Scale bars: 200 μm.
(E and F) Quantification analysis for the proportion of Sox2+ or Ki67-cells in EdU-labeled cells (F, n = 3; G, n = 3).
(G) Immunostaining of Brn2 in brain tissue sections of P3 WT and Sirt6 cKO mice that EdU were injected at E14.5. Scale bars: 200 μm.
(H and I) Quantification analysis for the proportion of Brn2+ cells in EdU-labeled cells (n = 3) and the number of E14.5-P3 EdU+ cells (n = 6). The results are expressed as the mean ± SD. Comparisons were performed by 2-tailed unpaired Student’s t test, and the statistically significant P-values are shown as ns (p > 0.05) and ∗ (p < 0.05). See also Figures S2 and S3.
To explore the Sirt6 knockout effect on NPC differentiation, we performed long-term (E14.5-P3) EdU birth dating and immunofluorescence staining were performed to detect Brn2, a marker of layer 2/3/5. E14.5 is the time point when the superficial layer starts growth, including layer 2/3/4 (Figure S3G). Compared with the WT cortex, the Sirt6 cKO cortex showed decreased EdU distribution in the fourth bin, that is, in the corresponding position of the third layer of the neocortex (the cortex was divided into 10 bins averaged from outside to the inside.). (Figure S3H). Combined with the lower position of labeling cells, the costained proportion of L2/3 Brn2+ and EdU+ decreased significantly, suggesting that the labeled NPCs at E14.5 retained their fate in their deeper layer and that they had generated fewer L2/3 neurons than their WT counterpart (Figures 2G and 2H). We also observed an increased number of EdU+ cells in the cKO group, suggesting that from E14.5 to P3 these NPCs eventually generated more daughter cells (Figure 2I).
Sirt6 deficiency promoted neural stem cells proliferation and delayed differentiation in vitro
Further verification of the gene function of Sirt6 at the cellular level was performed after isolating neural stem cells (NSCs) of E14.5 and P7 based on Sirt6’s effect on the proliferation and differentiation of NPCs in vivo. Cells were subcultured to the second generation for functional experiments. Under a light microscope, larger neurospheres exhibited an obvious defect in differentiation pattern in the Sirt6 cKO group (Figures 3A and 3B). The proliferation of P7 NSCs was evaluated with cultured cells in proliferation medium using cell slide staining and the MTS assay. At 7 days, the proliferation activity of the cKO NSCs was significantly higher than that of the control NSCs (Figure 3C), and the proportion of cells costained with the NSC marker Sox2+ and EdU+ was also increased (Figures 3D and 3E). A similar phenomenon in E14.5 NPCs was observed (Figures S4A–S4C).
Figure 3.
Sirt6 deficiency delays the differentiation of neural stem cells in vitro
(A and B) Representative images of P7 NSCs proliferated for 7 days and differentiated for 3 days under a light microscope and quantification of neurosphere diameters. Scale bars: 100 μm.
(C) The MTS assay was used to measure the proliferation efficiency of P7 NSCs (n = 3).
(D) Immunostaining of Sox2 in the P7 NSCs that have proliferated for 7 days. Scale bars: 100 μm.
(E) Quantification of EdU+ cells positive for Sox2 (n = 5).
(F) The expression of Sox2 in proliferating P7 NSCs at 7 days was detected by WB.
(G) Immunostaining of GFAP and Map2 in NSCs that have differentiated for 3 and 7 days (left), and Sox2 and Oligo2 in NSCs that have differentiated for 5 days (right). Scale bars: 100 μm.
(H) Quantification of Map2+ cells and Gfap+ cells was performed (n = 3).
(I and J) Quantification of the percentage of Sox2+ cells and Olig2+ cells was performed (n>=3).
(K) The expressions of GFAP, Sox2, and Olig2 in P7 NSCs were detected by WB. The results are expressed as the mean ± SD. Comparisons were performed by 2-tailed unpaired Student’s t test, and the statistically significant P-values are shown as ns (p > 0.05), ∗ (p < 0.05), and ∗∗ (p < 0.01). See also Figure S4.
As shown by Western blot or qPCR analyses, both E14.5 and P7 cKO NSCs had higher levels of the stemness marker Sox2/Pax6 (Figures 3F, S4D, and S4H), leading us to question the change in their differentiation ability. By cell culture in differentiation medium and cell slide staining, we found that in 3 days the proportion of neurons (MAP2+) and astrocytes (GFAP+) in the Sirt6 deficient NSCs decreased significantly as did the relative total differentiation index, but these neurons and astrocytes did catch up with those in the WT group within 7 days (Figures 3G, 3H, and S4J). For analysis of the differentiation timeline, staining for 5 days revealed that the Sirt6 deficiency group retained a higher proportion of Sox2+ and Olig2+ pluripotent cells than the WT group (Figures 3G, 3I, and 3J). The Western blot results were consistent with the above assumption (Figure 3K). Interestingly, an aging-related marker, p27, showed up-regulation in the cKO group of P7 neural stem cells, indicating that Sirt6 deficiency may cause cell senescence early in life (Figure S4K).
In conclusion, these observations indicated that Sirt6 deletion maintains the stemness of NSCs over the short term by delaying differentiation, which was consistent with the experimental results in vivo.
Overexpression of neural sirtuin 6 made little difference in neural stem cell fate determination
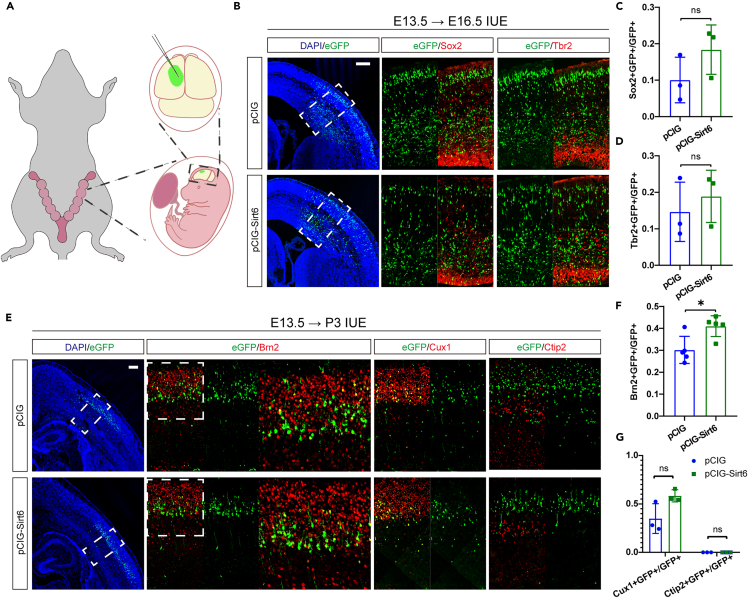
To test whether Sirt6 overexpression (OE) affected NPC proliferation and differentiation, we amplified the Sirt6 gene coding sequence and cloned it into the pCIG vector to allow Sirt6 to be highly expressed in vivo. In utero electroporation experiments introduced the Sirt6 OE plasmid or empty vector into periventricular NPCs at E13.5 (Figure 4A). The expression efficiency was proven by the SIRT6 antibody test (Figure S5A). The collected embryos, at E16.5 and P3, indicated that GFP-expressing (GFP+) cells were electroporated cells in the neocortices. To explore Sirt6 OE’s effect on NPC fate determination, we assessed Sirt6 OE tissues from E13.5 to E16.5 by immunofluorescence assays (Figure 4B), but no significant difference was found in the costained proportion of NPCs (Sox2+) (Figure 4C) and IPCs (Tbr2+) (Figure 4D) with GFP+ between the OE group and the control group. To explore the long-term effects of Sirt6 OE on NPC differentiation, we performed an immunofluorescence assay (Figure 4E) in P3 Sirt6 OE tissues, with the finding that the costained proportion of Brn2+ with GFP+ increased (Figure 4F). We observed no change by Cux1+ or Ctip2+ staining (Figure 4G). Perhaps because of the high expression of Sirt6 at this early stage of neural development, overexpression did not stimulate extra activity and effect.
Figure 4.
Overexpression of Sirt6 makes little difference in NPCs’ fate determination
(A) Schematic diagram of embryo in utero electroporation.
(B) Immunostaining of Sox2 and Tbr2 in brain tissue sections from E16.5 mice that were electroporated with pCIG or pCIG-Sirt6 at E13.5. Scale bars: 200 μm.
(C and D) Quantitative analysis for the proportion of Sox2+ cells or Tbr2+ cells among all eGFP+ cells (C, n = 3; D, n = 3).
(E) Immunostaining of Brn2, Cux1, and Ctip2 in brain tissue sections of P3 mice that were electroporated with pCIG or pCIG-Sirt6 at E13.5. Scale bars: 200 μm.
(F and G) Quantitative analysis for the proportion of the layer markers among all eGFP+ cells (n>=3). The results are expressed as the mean ± SD. Comparisons were performed by 2-tailed unpaired Student’s t test, and the statistically significant P-values are shown as ns (p > 0.05) and ∗ (p < 0.05). See also Figure S5.
Transcriptomics analysis revealed genetic changes related to neurodevelopment
The molecular mechanism of Sirt6 in neural development was further investigated by a genome-wide RNA-seq analysis. IGV software revealed that in the knockout group the read distributions on Sirt6 exons 2 and 3 were significantly reduced, indicating an effective knockout strategy (Figure 5A). In all, 571 genes were differentially expressed between the knockout group and the control group, with a difference of more than 1.5 times, p < 0.05. Among them, 273 genes were significantly downregulated and 298 genes were significantly upregulated (Figure 5B). From cluster analysis, differentially expressed genes could be well distinguished between the control and knockout groups (Figure 5C). Gene Ontology (GO) analyses of these differentially expressed genes revealed significant enrichment of cell adhesion, cell cycle, cell division and proliferation, and neural development-related categories (Figures 5D–5F). KEGG pathway analysis showed significant enrichment in the focal adhesion kinase pathway, oxytocin signaling pathway, cell cycle and Notch signaling pathway (Figure 5G). Based on the GO and KEGG analysis, differentially expressed genes were screened, and RT-PCR was performed to verify genes related to cell division and differentiation (Figure 5H). Using the published database,25,26,27 we discovered that Sirt6 and the Notch pathway may have a regulatory relationship (Figure S6). The Notch pathway was significantly enriched in the KEGG results, and it has been reported to be regulated by Sirt6 deacetylation, so we considered it to be the downstream target of Sirt6.28
Figure 5.
Transcriptomics analysis revealed that Sirt6 regulates genes related to neurodevelopment
(A) RNA-sequencing revealed the efficiency of Sirt6 knockout. The red rectangle identifies the missing sequence of 2–3 exons.
(B) Up-regulated and down-regulated genes are shown in a volcano plot.
(C) Clustering analysis of differentially expressed genes.
(D–F) GO enrichment analysis of the differentially expressed genes.
(G) KEGG pathway enrichment analysis of the differentially expressed genes.
(H) Differentially expressed genes in the candidate signaling pathway were measured by q-PCR. The results are expressed as the mean ± SD. Comparisons were performed by 2-tailed unpaired Student’s t test, and the statistically significant P-values are shown as ∗ (p < 0.05), and ∗∗ (p < 0.01). See also Figure S6.
Neural sirtuin 6 may regulate cell proliferation via the Notch signaling pathway
To further confirm whether Sirt6 regulates NSC fate determination through the Notch signaling pathway, we detected the expression of the Notch1 and NICD proteins in three pairs of P7 NSCs by WB, and we found that they were upregulated in the cKO group (Figure 6A). The mRNA levels of Notch1 and downstream genes were also detected by qPCR (Figure 6B). By performing ChIP-qPCR, we found an up-regulation of H3K56ac on the promoter region of Notch1, indicating a possible regulatory mechanism that Sirt6 acts as a histone deacetylase to limit the expression of Notch1 (Figure 6C). A dual luciferase reporter gene assay found that Sirt6 knockdown significantly increased luciferase activity compared with that of the control group, whereas overexpression of Sirt6 significantly reduced luciferase activity (Figure 6D), indicating that Sirt6 could influence Notch pathway activity. Notch receptors can be cleaved by γ secretase to generate NICD and can subsequently activate the classical Notch target genes Myc, P21, and HES family members to regulate NPC proliferation and differentiation.29,30,31 To verify the influence of Sirt6 and NICD influence on NSCs, we treated NSCs with the Notch pathway inhibitor DAPT to inhibit γ secretase. We detected the proliferation of NSCs during MTS and EdU labeling experiments and found that NSC proliferation decreased after DAPT treatment (Figures 6E–6G). The DAPT treatment for 2 days resulted in similar proliferative activity between the WT group and the KO + DAPT group, while the treatment for 5 days did not show the same effect. This may be due to a diminished inhibitory effect of DAPT or the activation of other compensatory pathways. When we tested other Notch signaling pathway-related proteins in P7 NSCs with WB, the results were consistent with the change in NICD levels (Figure 6H), strongly suggesting that the enhancement of NSC proliferation activity after Sirt6 knockout may be caused by the Notch pathway.
Figure 6.
Sirt6 regulates cell proliferation via Notch signal pathway
(A) Upregulation of Notch1 and NICD was detected by WB in three pairs of P7 NSCs.
(B) Upregulation of the mRNA levels of Notch1 and downstream genes was detected by qPCR.
(C) The level of H3K56ac on the promoter region of Notch1 was detected by ChIP-qPCR.
(D) Verification of the relationship between Sirt6 and the Notch signal pathway by luciferase assay. Si781 was used to knock down Sirt6.
(E) The proliferation efficiency of WT and Sirt6 KO NSC after DAPT treatment was measured by MTS assay (n = 3).
(F) Immunostaining of Sox2 in the P7 NSC which had been proliferated and treated with DAPT. Scale bars: 100 μm.
(G) Quantification of the ratio of EdU+ cells that are positive for Sox2 (n>=3).
(H) Upregulation of other stemness-related genes was detected by WB in P7 NSCs. The results are expressed as the mean ± SD. Comparisons were performed by 2-tailed unpaired Student’s t test, and the statistically significant P-values are shown as ns (p > 0.05), ∗ (p < 0.05), and ∗∗ (p < 0.01). See also Figure S7.
Discussion
Scientists, to adequately explain the high level of neocortical expansion in primates, have indicated the importance of progenitor cell increases during development.32 Sirt6 deficiency in primates, including humans, causes severe embryonic lethality and brain abnormalities, but allows rodents to survive. This finding may be due to their simple brain structure. NPCs from SIRT6-null cynomolgus monkeys exhibit arrested neuronal differentiation.10,19 We observed a similar phenotype in mouse NPCs, where loss of the histone deacetylase SIRT6 disrupted the balance in NPC fate determination, retarding growth by promoting proliferation and inhibiting differentiation. This finding is due to the upregulation of proliferation-related genes, including Notch1. Our findings fill the research gap regarding the mechanism of Sirt6’s influence on neural progenitor cells, highlighting cortex development differences across species.
Because we recognized that Sirt6 deletion resulted in a brain volume decrease but greater body weight in Nestin-Cre knockout mice,20 we focused on adult brain morphology in 17-month-old Emx1-Cre knockout mice. Consequently, we discovered a significant loss of brain volume in the Sirt6 knockout group (data not given). Since we did not observe obesity, we reason that the weight of the Emx1-Cre knockout mice was lower than that of the WT mice, possibly because of a difference in Cre enzymes. Since Nestin-Cre expression covered the region of the hypothalamus related to metabolism, while Emx1-Cre was only expressed in the ventricular zone of the dorsal cortex, our experiment, after excluding the possible effects of hypothalamus hormone secretion, proposes another viewpoint on the importance of Sirt6.
For a deeper understanding of the underlying cellular and molecular mechanisms, a cell-level experiment with NSCs collected at the E14.5 and P7 development points was conducted. Sirt6 knockout P7 NSCs showed a more significant change in acetylation than did E14.5 NSCs, possibly a consequence of the Emx1-Cre expression pattern. Construction of conditional knockout mice in this study involved the Cre-loxP system in which the Cre enzyme mediated by the Emx1 promoter starts to be expressed at E9.5 in the dorsal telencephalon.33 However, considering our observations, the Sirt6 expression pattern was too high in the early development of the cerebral cortex and may not be completely exhausted at E14.5. The Western blot results confirmed our hypothesis by showing residues of SIRT6 at E14.5 NSCs with the result that P7 NSCs were chosen to complete our research about related signaling pathways.
Using previous studies as a starting point, we focused on abnormal histone acetylation caused by Sirt6 knockout. This situation affects various stem cells including embryo stem cells (ESCs), hematopoietic stem cells (HSCs) and mesenchymal stem cells (MSCs). Sirt6 deacetylation generally reduces target gene expression and keeps stem cells relatively quiescent. Researchers found that the deacetylation of H3K9ac and H3K56ac inhibited Oct4, Sox2 and Nanog expression in ESCs,34 while the increase in 5hmc levels led to the upregulation of neuron-related gene expression.35 The absence of Sirt6 in HSCs leads to decreased cell quiescence, which is crucial to the stem cell bank long-term maintenance.36 Since Sirt6-deficient MSCs show signs of osteoblast damage, chondrocyte differentiation and cell aging,37,38 our findings suggest the broad upregulation of H3K56ac and H3K9ac. This outcome could be why stemness-related genes cannot be shut down during late cortex development.
Our results showed that the expression level of Sirt6 decreased significantly in the knockout mice, while the expression levels of other SIRT family proteins remained unchanged, except that SIRT2 and SIRT5 increased slightly. SIRT5 is a protein that regulates oxidative stress by acting on metabolic enzymes in mitochondria, and SIRT2 is distributed in both the cytoplasm and nucleus.39 SIRT2, such as SIRT6, is also a histone deacetylase related with aging,40,41 which makes it a highly promising compensatory factor. As suggested in our study, histone deacetylation caused by SIRT6 could restrict the chromatin accessibility of Notch1 and downregulate its expression not only in podocytes,28 but also in NSCs. The histone deacetylase SIRT1 has also been reported to modulate neuronal differentiation and to act as a negative modulator of Notch1 signaling by opposing acetylation-induced NICD stabilization in endothelial cells.42,43,44 This function reveals another way in which deacetylases may modulate the Notch signaling pathway. What is more, research has found that SIRT6 activates the Notch pathway in tumor cells.45 Taking this into account, the Sirtuin family regulate the Notch signaling pathway in a diverse and complex manner. Our work primarily focuses on the role of Sirt6 as a transcription factor in neural precursor cells.
In conclusion, we explored Sirt6 function in early neurogenesis in mice and compared our findings with past research on how this function works in primates. Given that Sirt6 is an aging-related gene that is significantly involved in the human nervous system, studying its influence on embryonic NPCs furthers our understanding of how it controls adult NSC behavior in mice and the origins and future moderation of neuroaging and neurodegenerative disorders in our worldwide aging human population.
Limitations of the study
The major limitation of the present study is that we have not evaluated the compensatory effect of other sirtuins after Sirt6 cKO. What is more, we were unable to perform ChIP-qPCR of H3K9ac on the Notch1 promoter due to limitations in the availability of experimental reagents. The possibility that Sirt6 regulates the transcription of Notch1 through H3K9ac could not be excluded. The current research shows that the Sirtuin family regulates Notch signaling in a complex manner. At this point, we lack sufficient evidence to offer a comprehensive description. We hope our work can shed new light on this issue.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Sirt6 | Cell Signaling Technology | Cat#12486; RRID: AB_2636969 |
| Anti-Sirt1 | Abcam | Cat#ab12193; RRID: AB_298923 |
| Anti-Sirt2 | Proteintech | Cat#19655-1-AP; RRID: AB_2878592 |
| Anti-Sirt3 | Proteintech | Cat#10099-1-AP; RRID: AB_2239240 |
| Anti-Sirt4 | Proteintech | Cat#66543-1-lg; RRID: AB_2881905 |
| Anti-Sirt5 | Proteintech | Cat#15122-1-AP; RRID: AB_2188778 |
| Anti-Sirt7 | Proteintech | Cat#12994-1-AP; RRID: AB_10644276 |
| Anti-H3K56ac | Invitrogen | Cat#PA5-40101; RRID: AB_2608322 |
| Anti-H3K9ac | Abcam | Cat#ab10812; RRID: AB_297491 |
| Anti-Sox2 | Abcam | Cat#ab97959; RRID: AB_2341193 |
| Anti-β-actin | Sigma | Cat#A5441; RRID: AB_476744 |
| Anti-GFAP | Abcam | Cat#ab7260; RRID: AB_305808 |
| Anti-CNP | Cell Signaling Technology | Cat#5664; RRID: AB_10705455 |
| Anti-H3 | Abcam | Cat#ab1791; RRID: AB_302613 |
| Anti-mycn | Abcam | Cat#ab227822 |
| Anti-myc | Abcam | Cat#ab32072; RRID: AB_731658 |
| Anti-EGFR | Abcam | Cat#ab32077; RRID: AB_732101 |
| Anti-Olig2 | Abcam | Cat#ab109186 |
| Anti-NICD | Cell Signaling Technology | Cat#2421; RRID: AB_2314204 |
| Anti-Notch1 | Cell Signaling Technology | Cat#4380: RRID: AB_10691684 |
| Anti-MAP2 | Abcam | Cat#ab11267: RRID: AB_297885 |
| Anti-Ki67 | Abcam | Cat#ab16667: RRID: AB_302459 |
| Anti-Brn2 | Santa Cruz | Cat#sc-6029: RRID: AB_2167385 |
| Anti-Tbr2 | Abcam | Cat#ab23345; RRID: AB_778267 |
| Anti-Tbr2 | Oasis Biofarm | Cat#OB-PGP022; RRID: AB_2934262 |
| Anti-p27 | Cell Signaling Technology | Cat#3686; RRID: AB_2077850 |
| Chemicals, peptides, and recombinant proteins | ||
| DAPT γ-secretase inhibitor | Sigma-Aldrich | Cat#D5942 |
| Critical commercial assays | ||
| Click-iT plus EdU Alexa Fluor Imaging Kit | Life Technologies | Cat#C10640 |
| ChIP Kit | Cell Signaling Technology | Cat#9004S |
| Deposited data | ||
| RNA-seq data | This paper | GEO: GSE236460 |
| Original western blot | This paper | https://doi.org/10.17632/vkbc59rmsx.1 |
| Experimental models: Cell lines | ||
| Mouse: N1E-115 cell line | ATCC | Cat# CRL-2263, RRID: CVCL_0451 |
| Mouse: primary culture neural stem cells | This paper | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: Sirt6 fl/fl C57BL/6 | Yongsheng Chang’s Laboratory46 | N/A |
| Mouse: C57BL/6Smoc-emx1em1(IRES-Cre) Smoc | Shanghai Model Organisms, Inc. | NM-KI-200149 |
| Mouse: ICR pregnant mouse | Department of Laboratory Animal Science, Peking University Health Science Center | N/A |
| Oligonucleotides | ||
| Primers for genotyping, see Table S1 | This paper | N/A |
| Primers for qPCR, see Table S2 | This paper | N/A |
| siRNA sequence: si-488: GCUGCACGGAAA CAUGUUUTTAAACAUGUUUCCGUGCAGCTT |
This paper | N/A |
| siRNA sequence: si-781: GGUCAUUGUCAA CCUGCAATTUUGCAGGUUGACAAUGACCTT |
This paper | N/A |
| siRNA sequence: si-987: GCAGUGCAUGUUU CGUAUATTUAUACGAAACAUGCACUGCTT |
This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pCIG | Weimin Zhong’s Laboratory47 | N/A |
| Plasmid: pCIG-Sirt6 | This paper | N/A |
| Plasmid: pGEM-3zf (+) | Promega | Cat#P2271 |
| Plasmid: pGEM-3zf-Sirt6 | This paper | N/A |
| Software and algorithms | ||
| ImageJ | Schindelin J et al.48 | https://imagej.nih.gov/ij/ |
| IGV software | Robinson JT et al.49,50 | https://www.igv.org/ |
| DAVID bioinformatics database | Huang DW et al.26,27 | https://david.ncifcrf.gov/ |
| the OmicShare tools | GENE DENOVO | https://www.omicshare.com/tools |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xiaozhong Peng (pengxiaozhong@pumc.edu.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Data reported in this paper will be shared by the lead contact upon request. RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Animals
Animal studies were conducted according to protocols approved by the Institutional Animal Care and Use Committee of the Academy of Medical Sciences and Peking Union Medical College (PUMC). Floxed Sirt6 C57BL/6 mice were a gift from Dr Yongsheng Chang. Adult Emx1-Cre mice were obtained from Shanghai Model Organisms Center, Inc. Sirt6 conditional knockout mice were produced by crossing Sirt6fl/fl mice with Emx1-Cre mice. The development stages were described in figures or figure legends. The sex of the mice was not identified because it was difficult to detect in embryo mice and not relevant to dorsal cortex development. The sequences of primer used for genotyping by PCR are shown in Table S1. Three-month-old pregnant ICR mice were used to conduct in utero electroporation. All mice had free access to food and water.
NSC culture
NSCs were isolated from E14.5 or P7 mice by dissection of the cerebral cortex and digested into a single-cell suspension with Accutase (Sigma, A6964). Cells were cultured in DMEM/F12 proliferation medium (Gibco, 11330032) supplemented with 2% B27 supplement (Gibco, 17504-044), 20 ng/ml EGF (peprotech, 100-15-100), 20 ng/ml bFGF (peprotech, 100-1813-100) and 0.2% BSA for proliferation. The differentiation medium was the same except for growth factors. Cells were cultured at 37°C in 5% carbon dioxide. For γ-secretase inhibition, 75 μM DAPT (Sigma-Aldrich, D5942) was added to the culture medium to reduce NICD generation.
Method details
RNA in situ hybridization
The Sirt6 probe was synthesized according to the primer sequences of Allen Brain in situ hybridization. Procedures for ISH were the same as previously described.51 The digoxigenin-labeled Sirt6 probe was synthesized using T7 or SP6 RNA polymerase (New England Biolabs, M0251, M0207), pGEM-3zf plasmids and the DIG RNA Labeling Kit (Roche Diagnostics, 11277073910) according to the manufacturer’s instructions.
Western blot analysis
Forebrains were quickly dissected out and homogenized in TNTE buffer (50 mM Tris-HCl, 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, 1 mM Na3VO4, 25 mM NaF, 10 mM Na4P2O7 and protease inhibitors [5 mg/ml PMSF, 0.5 mg/ml leupeptin, 0.7 mg/ml pepstatin, and 0.5 mg/ml aprotinin]). Lysates were incubated for 15 min on ice and centrifuged at 12,000 rpm for 20 min at 4°C. Protein electrophoresis was performed using 8%–12% SDS-PAGE gels, and protein were transferred to nitrocellulose membranes. The antibodies used in this study are listed in the key resources table.
RNA isolation and quantitative RT–PCR
Total RNA from control and Sirt6 cKO brains was extracted using TRIzol reagent (Invitrogen, 15596026) according to the manufacturer’s instructions. Complementary DNA synthesis was performed with a Reverse Transcriptase Kit (TransGene, AH341-01) following the manufacturer’s instructions. Quantitative RT–PCR was performed using a SYBR Green-containing PCR kit (TaKaRa, RR820A), and signals were detected with a Bio-Rad CFX96 Touch Real-Time PCR detection system. Relative mRNA amounts were assayed by comparing PCR cycles to GAPDH using normalization to control samples. The sequences of primers used in this study are given in Table S2.
EdU staining and immunofluorescence staining of brain sections
Pregnant mice were injected intraperitoneally with 75 mg/kg body weight of EdU (Thermo, E10187). EdU staining was conducted on cryosections with a Click-iT plus EdU Alexa Fluor Imaging Kit (Thermo, C10640) according to the manufacturer’s instructions. Immunofluorescence analyses were conducted as previously described.52 Brains were fixed with 4% paraformaldehyde in PBS overnight at 4°C, cryoprotected in 25% sucrose in PBS, embedded in OCT compound (ZSGB-Bio, Zil-9302), and sectioned coronally (14 mm thickness). Tissue sections were blocked and permeabilized with blocking solution (5% normal sheep serum and 0.1% Triton X-100 in PBS) for 1 hr at room temperature, followed by incubation with primary antibodies at 4°C overnight. Secondary antibodies were applied to the sections for 2 hr at room temperature. Nuclei were visualized by incubation for 5 min with DAPI (Sigma-Aldrich, F6057). Images were acquired using a confocal laser scanning microscope (FV1000MPE-BX61WI, Olympus) and analyzed using FluoView (Olympus) and Adobe Illustrator (Adobe Systems). The antibodies used in this study are listed in the key resources table.
Plasmid constructs and transfection
Sirt6 siRNA was synthesized by Gemma. Sirt6 sequences (termed mSirt6) were cloned and inserted into the pCIG vector using the following primers:
Sirt6-F: 5'- CGTGTGACCGGCGGCTCTAGACGATGTCGGTGAATTATGC-3', Sirt6-R: 5'- CCCCCGGGCTGCAGGAATTCTGTGTGGTTCCTTCAAGTTC-3'.
All plasmids were purified using a Plasmid Mini Kit (QIAGEN, DP103), and then N1E-115 cells were transfected using LipofectamineTM 2000 (Invitrogen, 11668030).
Luciferase assay
N1E-115 cells at a density of 1×105 cells per well were plated into 24-well plates without antibiotics. Cells were transfected with 0.8 mg of shRNA plasmid along with 20 ng of pRL-TK. The culture media were replaced with medium containing antibiotics 6 hr after transfection. After 48 hr, the cells were collected for the luciferase assay.
MTS assay
The proliferation ability of NSCs was assessed using MTS reagent. The MTS assay was performed on Days 0,1,3,5,7 and 9; cell viability was measured using Cell Titer 96 AQueous One Solution Reagent (Promega, G3582). Cells were cultured in 96-well plates with 100 μl of proliferation medium per well; 20 μl of MTS reagent was added to each well for two hours of incubation. Absorbance was detected at 490 nm and 630 nm with a microplate reader.
In utero electroporation
The Institutional Animal Care and Use Committee of Medical Sciences Academy and Peking Union Medical College approved all experiments. Pentobarbital sodium (0.7 mg/g) was used to anesthetize pregnant mice. Five pulses of 30 V (50 ms on/950 ms interval) were delivered across the head of the E13.5 embryos. pCIG-GFP and pCIG-Sirt6 (2.5 ng/μl) were electroporated.
RNA-seq
RNA-seq was performed by Annoroad, China. For neurosphere RNA-seq, neural stem cells were isolated from three P7 mice per group and cultured for seven days until they formed neuroshpheres. RNA was isolated from the collected neuroshere with TRIzol reagent (Invitrogen, 15596026) following the manufacturer’s instructions.
ChIP-qPCR
ChIP-qPCR was performed using the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Cell Signaling Technology, 9004S) according to the protocol. Three pairs of P7 neural stem cells were collected for the experiment. 37% formaldehyde was used for cross-linking. Micrococcal Nuclease was added and DNA was digested for 17 min at 37°C. Anti-H3K56ac (Proteintech, 12994-1-AP) was used at the recommended dose (2 μg per IP sample). 00.
Quantification and statistical analysis
Data are presented as mean±s.d. from at least three independent experiments or three individual animals, unless otherwise stated in the figure legend. An unpaired two-tailed Student’s t-test was used to compare two groups, unless otherwise stated in the figure legend. The statistically significant P-values are shown as ns (P > 0.05), ∗ (P < 0.05), and ∗∗ (P < 0.01). Statistical tests were performed using GraphPad Prism 8. RNA-seq data analysis was performed by Active Motif, Novogene, and OmicShare tools.
Acknowledgments
The authors gratefully acknowledge the research grants that supported this study, including CAMS Innovation Fund for Medical Sciences (CIFMS, 2021-I2M-1-024, 2021-I2M-1-019, 2019-RC-HL-007) and The National Key Research and Development Program of China (2021ZD0200902).
Author contributions
X. P. supervised the research. W. L. with help from P. S., B. Y. and L. H. designed and supervised the study. Y. W. and X. W. performed the majority of experiments and gathered data. Z. M. performed plasmid construction. P. X. helped perform EdU and immunofluorescence staining of brain sections. W. L. performed in utero electroporation and NPCs culture. Y. W. prepared figures and wrote the article with contributions from X. W. and G. L.
Declaration of interests
The authors declare no competing interests.
Published: December 9, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108706.
Contributor Information
Wei Liu, Email: liuwei@ibms.cams.cn.
Xiaozhong Peng, Email: pengxiaozhong@pumc.edu.cn.
Supplemental information
References
- 1.Noctor S.C., Flint A.C., Weissman T.A., Wong W.S., Clinton B.K., Kriegstein A.R. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J. Neurosci. 2002;22:3161–3173. doi: 10.1523/JNEUROSCI.22-08-03161.2002. 20026299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shibata M., Gulden F.O., Sestan N. From trans to cis: transcriptional regulatory networks in neocortical development. Trends Genet. 2015;31:77–87. doi: 10.1016/j.tig.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye F., Chen Y., Hoang T., Montgomery R.L., Zhao X.H., Bu H., Hu T., Taketo M.M., van Es J.H., Clevers H., et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat. Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montgomery R.L., Hsieh J., Barbosa A.C., Richardson J.A., Olson E.N. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc. Natl. Acad. Sci. USA. 2009;106:7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castelo-Branco G., Lilja T., Wallenborg K., Falcão A.M., Marques S.C., Gracias A., Solum D., Paap R., Walfridsson J., Teixeira A.I., et al. Neural Stem Cell Differentiation Is Dictated by Distinct Actions of Nuclear Receptor Corepressors and Histone Deacetylases. Stem Cell Rep. 2014;3:502–515. doi: 10.1016/j.stemcr.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanfi Y., Naiman S., Amir G., Peshti V., Zinman G., Nahum L., Bar-Joseph Z., Cohen H.Y. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 7.Satoh A., Brace C.S., Rensing N., Cliften P., Wozniak D.F., Herzog E.D., Yamada K.A., Imai S.I. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18:416–430. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Venzor A., Toiber D. SIRT6 Through the Brain Evolution, Development, and Aging. Front. Aging Neurosci. 2021;13 doi: 10.3389/fnagi.2021.747989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imai S., Armstrong C.M., Kaeberlein M., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 10.Ferrer C.M., Alders M., Postma A.V., Park S., Klein M.A., Cetinbas M., Pajkrt E., Glas A., van Koningsbruggen S., Christoffels V.M., et al. An inactivating mutation in the histone deacetylase SIRT6 causes human perinatal lethality. Genes Dev. 2018;32:373–388. doi: 10.1101/gad.307330.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McBurney M.W., Yang X., Jardine K., Hixon M., Boekelheide K., Webb J.R., Lansdorp P.M., Lemieux M. The mammalian SIR2 alpha protein has a role in embryogenesis and gametogenesis. Mol. Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prozorovski T., Schulze-Topphoff U., Glumm R., Baumgart J., Schröter F., Ninnemann O., Siegert E., Bendix I., Brüstle O., Nitsch R., et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat. Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 13.Mostoslavsky R., Chua K.F., Lombard D.B., Pang W.W., Fischer M.R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M.M., et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 14.Geng A., Tang H., Huang J., Qian Z., Qin N., Yao Y., Xu Z., Chen H., Lan L., Xie H., et al. The deacetylase SIRT6 promotes the repair of UV-induced DNA damage by targeting DDB2. Nucleic Acids Res. 2020;48:9181–9194. doi: 10.1093/nar/gkaa661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawahara T.L.A., Michishita E., Adler A.S., Damian M., Berber E., Lin M., McCord R.A., Ongaigui K.C.L., Boxer L.D., Chang H.Y., Chua K.F. SIRT6 Links Histone H3 Lysine 9 Deacetylation to NF-κB-Dependent Gene Expression and Organismal Life Span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smirnov D., Eremenko E., Stein D., Kaluski S., Jasinska W., Cosentino C., Martinez-Pastor B., Brotman Y., Mostoslavsky R., Khrameeva E., Toiber D. SIRT6 is a key regulator of mitochondrial function in the brain. Cell Death Dis. 2023;14:35. doi: 10.1038/s41419-022-05542-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haigis M.C., Sinclair D.A. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raj S., Dsouza L.A., Singh S.P., Kanwal A. Sirt6 Deacetylase: A Potential Key Regulator in the Prevention of Obesity, Diabetes and Neurodegenerative Disease. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.598326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W., Wan H., Feng G., Qu J., Wang J., Jing Y., Ren R., Liu Z., Zhang L., Chen Z., et al. SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature. 2018;560:661–665. doi: 10.1038/s41586-018-0437-z. [DOI] [PubMed] [Google Scholar]
- 20.Schwer B., Schumacher B., Lombard D.B., Xiao C., Kurtev M.V., Gao J., Schneider J.I., Chai H., Bronson R.T., Tsai L.H., et al. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc. Natl. Acad. Sci. USA. 2010;107:21790–21794. doi: 10.1073/pnas.1016306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaluski S., Portillo M., Besnard A., Stein D., Einav M., Zhong L., Ueberham U., Arendt T., Mostoslavsky R., Sahay A., Toiber D. Neuroprotective Functions for the Histone Deacetylase SIRT6. Cell Rep. 2017;18:3052–3062. doi: 10.1016/j.celrep.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simeone A., Gulisano M., Acampora D., Stornaiuolo A., Rambaldi M., Boncinelli E. Two vertebrate homeobox genes related to the Drosophila empty spiracles gene are expressed in the embryonic cerebral cortex. EMBO J. 1992;11:2541–2550. doi: 10.1002/j.1460-2075.1992.tb05319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molnár Z., Clowry G.J., Šestan N., Alzu'bi A., Bakken T., Hevner R.F., Hüppi P.S., Kostović I., Rakic P., Anton E.S., et al. New insights into the development of the human cerebral cortex. J. Anat. 2019;235:432–451. doi: 10.1111/joa.13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molyneaux B.J., Arlotta P., Menezes J.R.L., Macklis J.D. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 25.Etchegaray J.P., Zhong L., Li C., Henriques T., Ablondi E., Nakadai T., Van Rechem C., Ferrer C., Ross K.N., Choi J.E., et al. The Histone Deacetylase SIRT6 Restrains Transcription Elongation via Promoter-Proximal Pausing. Mol. Cell. 2019;75:683–699.e7. doi: 10.1016/j.molcel.2019.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu M., Liang K., Zhen J., Zhou M., Wang X., Wang Z., Wei X., Zhang Y., Sun Y., Zhou Z., et al. Sirt6 deficiency exacerbates podocyte injury and proteinuria through targeting Notch signaling. Nat. Commun. 2017;8:413. doi: 10.1038/s41467-017-00498-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bray S.J., Gomez-Lamarca M. Notch after cleavage. Curr. Opin. Cell Biol. 2018;51:103–109. doi: 10.1016/j.ceb.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Pierfelice T., Alberi L., Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron. 2011;69:840–855. doi: 10.1016/j.neuron.2011.02.031. [DOI] [PubMed] [Google Scholar]
- 31.Gao F., Zhang Y.F., Zhang Z.P., Fu L.A., Cao X.L., Zhang Y.Z., Guo C.J., Yan X.C., Yang Q.C., Hu Y.Y., et al. miR-342-5p Regulates Neural Stem Cell Proliferation and Differentiation Downstream to Notch Signaling in Mice. Stem Cell Rep. 2017;8:1032–1045. doi: 10.1016/j.stemcr.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lui J.H., Hansen D.V., Kriegstein A.R. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo H., Mao C., Jin X.L., Wang H., Tu Y.T., Avasthi P.P., Li Y. Cre-mediated cerebellum- and hippocampus-restricted gene mutation in mouse brain. Biochem. Biophys. Res. Commun. 2000;269:149–154. doi: 10.1006/bbrc.2000.2263. [DOI] [PubMed] [Google Scholar]
- 34.Koh K.P., Yabuuchi A., Rao S., Huang Y., Cunniff K., Nardone J., Laiho A., Tahiliani M., Sommer C.A., Mostoslavsky G., et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Etchegaray J.P., Chavez L., Huang Y., Ross K.N., Choi J., Martinez-Pastor B., Walsh R.M., Sommer C.A., Lienhard M., Gladden A., et al. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat. Cell Biol. 2015;17:545–557. doi: 10.1038/ncb3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H., Diao D., Shi Z., Zhu X., Gao Y., Gao S., Liu X., Wu Y., Rudolph K.L., Liu G., et al. SIRT6 Controls Hematopoietic Stem Cell Homeostasis through Epigenetic Regulation of Wnt Signaling. Cell Stem Cell. 2016;18:495–507. doi: 10.1016/j.stem.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 37.Pan H., Guan D., Liu X., Li J., Wang L., Wu J., Zhou J., Zhang W., Ren R., Zhang W., et al. SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res. 2016;26:190–205. doi: 10.1038/cr.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhai X.Y., Yan P., Zhang J., Song H.F., Yin W.J., Gong H., Li H., Wu J., Xie J., Li R.K. Knockdown of SIRT6 Enables Human Bone Marrow Mesenchymal Stem Cell Senescence. Rejuvenation Res. 2016;19:373–384. doi: 10.1089/rej.2015.1770. [DOI] [PubMed] [Google Scholar]
- 39.Cai Y., Xu L., Xu H., Fan X. SIRT1 and Neural Cell Fate Determination. Mol. Neurobiol. 2016;53:2815–2825. doi: 10.1007/s12035-015-9158-6. [DOI] [PubMed] [Google Scholar]
- 40.Ma X.R., Zhu X., Xiao Y., Gu H.M., Zheng S.S., Li L., Wang F., Dong Z.J., Wang D.X., Wu Y., et al. Restoring nuclear entry of Sirtuin 2 in oligodendrocyte progenitor cells promotes remyelination during ageing. Nat. Commun. 2022;13:1225. doi: 10.1038/s41467-022-28844-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang C.L., Ohkubo R., Mu W.C., Chen W., Fan J.L., Song Z., Maruichi A., Sudmant P.H., Pisco A.O., Dubal D.B., et al. The mitochondrial unfolded protein response regulates hippocampal neural stem cell aging. Cell Metab. 2023;35:996–1008.e7. doi: 10.1016/j.cmet.2023.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guarani V., Deflorian G., Franco C.A., Krüger M., Phng L.K., Bentley K., Toussaint L., Dequiedt F., Mostoslavsky R., Schmidt M.H.H., et al. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature. 2011;473:234–238. doi: 10.1038/nature09917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collesi C., Felician G., Secco I., Gutierrez M.I., Martelletti E., Ali H., Zentilin L., Myers M.P., Giacca M. Reversible Notch1 acetylation tunes proliferative signalling in cardiomyocytes. Cardiovasc. Res. 2018;114:103–122. doi: 10.1093/cvr/cvx228. [DOI] [PubMed] [Google Scholar]
- 44.Hisahara S., Chiba S., Matsumoto H., Tanno M., Yagi H., Shimohama S., Sato M., Horio Y. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc. Natl. Acad. Sci. USA. 2008;105:15599–15604. doi: 10.1073/pnas.0800612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han Q., Xie Q.R., Li F., Cheng Y., Wu T., Zhang Y., Lu X., Wong A.S.T., Sha J., Xia W. Targeted inhibition of SIRT6 via engineered exosomes impairs tumorigenesis and metastasis in prostate cancer. Theranostics. 2021;11:6526–6541. doi: 10.7150/thno.53886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao L., Cui X., Chen Q., Yang X., Fang F., Zhang J., Liu G., Jin W., Chang Y. Cold-Inducible SIRT6 Regulates Thermogenesis of Brown and Beige Fat. Cell Rep. 2017;20:641–654. doi: 10.1016/j.celrep.2017.06.069. [DOI] [PubMed] [Google Scholar]
- 47.Shu P., Wu C., Ruan X., Liu W., Hou L., Fu H., Wang M., Liu C., Zeng Y., Chen P., et al. Opposing Gradients of MicroRNA Expression Temporally Pattern Layer Formation in the Developing Neocortex. Dev. Cell. 2019;49:764–785.e4. doi: 10.1016/j.devcel.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 48.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson J.T., Thorvaldsdóttir H., Wenger A.M., Zehir A., Mesirov J.P. Variant Review with the Integrative Genomics Viewer. Cancer Res. 2017;77:E31–E34. doi: 10.1158/0008-5472.CAN-17-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson J.T., Thorvaldsdottir H., Turner D., Mesirov J.P. igv.js: an embeddable JavaScript implementation of the Integrative Genomics Viewer (IGV) Bioinformatics. 2023;39 doi: 10.1093/bioinformatics/btac830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou J., Wang R., Zhang J., Zhu L., Liu W., Lu S., Chen P., Li H., Yin B., Yuan J., et al. Conserved expression of ultra-conserved noncoding RNA in mammalian nervous system. Biochim. Biophys. Acta. Gene Regul. Mech. 2017;1860:1159–1168. doi: 10.1016/j.bbagrm.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 52.Shu P., Fu H., Zhao X., Wu C., Ruan X., Zeng Y., Liu W., Wang M., Hou L., Chen P., et al. MicroRNA-214 modulates neural progenitor cell differentiation by targeting Quaking during cerebral cortex development. Sci. Rep. 2017;7:8014. doi: 10.1038/s41598-017-08450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data reported in this paper will be shared by the lead contact upon request. RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.