Abstract
Wenchang chicken, a prized local breed in Hainan Province of China renowned for its exceptional adaptability to tropical environments and good meat quality, is deeply favored by the public. However, an insufficient understanding of its population architecture and the unclear genetic basis that governs its typical attributes have posed challenges in the protection and breeding of this precious breed. To address these gaps, we conducted whole-genome resequencing on 200 Wenchang chicken samples derived from 10 distinct strains, and we gathered data on an array of 21 phenotype traits. Population genomics analysis unveiled distinctive population structures in Wenchang chickens, primarily attributed to strong artificial selection for different feather colors. Selection sweep analysis identified a group of candidate genes, including PCDH9, DPF3, CDIN1, and SUGCT, closely linked to adaptations that enhance resilience in tropical island habitats. Genome-wide association studies (GWAS) highlighted potential candidate genes associated with diverse feather color traits, encompassing TYR, RAB38, TRPM1, GABARAPL2, CDH1, ZMIZ1, LYST, MC1R, and SASH1. Through the comprehensive analysis of high-quality genomic and phenotypic data across diverse Wenchang chicken resource groups, this study unveils the intricate genetic backgrounds and population structures of Wenchang chickens. Additionally, it identifies multiple candidate genes linked to environmental adaptation, feather color variations, and production traits. These insights not only provide genetic reference for the purification and breeding of Wenchang chickens but also broaden our understanding of the genetic basis of phenotypic diversity in chickens.
Key words: Wenchang chicken, population structure, feather color, local adaptation, GWAS
INTRODUCTION
Chicken is one of the most economically valuable domesticated animals in the world (Lawal and Hanotte, 2021). After early domestication in southwestern China and Southeast Asia (Wang et al., 2020; Peng et al., 2022; Peters et al., 2022), chickens spread worldwide through multiple human migrations (Lawal and Hanotte, 2021). Consequently, various local breeds of chickens have been developed, adapted to diverse environmental conditions such as tropical climates (Tian et al., 2020; Xu et al., 2022; Shi et al., 2023), high altitudes (Wang et al., 2015), and cold tolerance (Xu et al., 2021; Zhao et al., 2022; Shi et al., 2023). These locally adapted characteristics make these breeds not only an economic foundation for smallholder farmers but also valuable genetic resources for poultry breeding and improvement (Jia et al., 2016; Bettridge et al., 2018). Understanding the genetic foundation of local adaptation in chickens is pivotal for sustainable poultry breeding and conservation, especially in preserving unique breeds like the Wenchang chickens. These endeavors not only help in preserving genetic diversity but also provide valuable insights for targeted breeding programs, ensuring the breed's resilience and productivity in diverse environments (Restoux et al., 2022; Nan et al., 2023).
Wenchang chicken is a local breed found on Hainan Island in southern China. Historical records indicate that during the Qing Dynasty, chickens were introduced to Hainan Island by immigrants from Fujian and Guangdong Provinces (Resources, 2011). The long-term breeding in tropical island environment contributed to the development of Wenchang chicken breed. Known for its extremely savory and nutritionally beneficial qualities, including thin skin and tender meat, Wenchang chicken has gained popularity across China and Southeast Asia (Resources, 2011). However, Wenchang chicken faces threats, including mixed genetic resources and challenges in stabilizing inherited high-quality traits due to multiple hybridizations with foreign breeds (Tian et al., 2023a).
Most recently, the high-throughput whole-genome resequencing (WGS) has been employed to investigate the genetic diversity of Wenchang chicken (Huang et al., 2020; Ren et al., 2023; Shi et al., 2023; Tian et al., 2023a). Generally, Wenchang chicken exhibits a close genetic affiliation with traditional Chinese yellow-feathered chicken breeds (Huang et al., 2020). However, Wenchang chickens also display certain genetic differentiation due to their isolation on Hainan Island (Tian et al., 2023a). Population genomics analyses have identified candidate genes potentially related to tropical adaptation and productive characteristics (Shi et al., 2023; Tian et al., 2023a,b). Nevertheless, because the sequenced Wenchang chickens were sourced from a limited number of local conservation farms (no more than 3) (Huang et al., 2020; Ren et al., 2023; Shi et al., 2023; Tian et al., 2023a,b), the population structure of Wenchang chicken remains unclear. Additionally, the breeding practices for Wenchang chicken have not been thoroughly evaluated.
In this study, in order to reveal the genetic background and explore the genetic mechanism of phenotypic diversity of Wenchang chickens, we generated WGS data of 200 Wenchang chickens from 10 different strains across Hainan Island. We collected data on a total of 21 traits and employed population genomic approaches to elucidate the population structure within Wenchang chickens. We detected selective genes associated with local adaptation to tropical island environments, including responses to ultraviolet radiation and pathogens. Our genome-wide selective scan identified candidate loci and genes linked to feather colors and agronomic traits in multiple Wenchang chicken strains. This study offers new insights into the genetic mechanisms of tropical island adaptation. Our findings have substantial practical implications, enhancing breeding strategies and improving Wenchang chicken breeds, thereby contributing to the conservation of local domestic chicken genetic resources. Furthermore, the identification of candidate genes associated with coat color and production traits expands our understanding of phenotypic diversity and evolutionary mechanisms within domestic chickens.
MATERIALS AND METHODS
Ethics Declarations
Ethical considerations regarding the welfare of the chickens were paramount throughout the study. The research protocol strictly adhered to established guidelines outlined by the Regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, revised in March 2017), and approved by the Institutional Animal Care and Use Committee at the Experimental Animal Center of Hainan Academy of Agricultural Science (HNXMSY-20210533). Additionally, protocols were in place to ensure appropriate housing conditions, adequate nutrition, and access to water. Veterinary care and oversight were also implemented to monitor the health and well-being of the chickens during the sampling and data collection processes.
Sample Collection and Genome Sequencing
Samples were collected from 10 Wenchang chicken populations that have been bred over the past 20 yr (Figure 1). These populations were retained according to different breeding strategies and selection criteria (Table S1) and are named according to the name of their breeding farms. A total of 200 samples were randomly selected, with 20 individuals (10 males and 10 females) from 10 populations. Breeding indexes included body weight at 120 d of age, egg production at 300 d of age, and feather color (Table S2). The assessment of phenotypic traits encompassed a range of characteristics crucial for understanding the breed's adaptation and production features. Traits such as body weight at 120 d of age, egg production at 300 d of age, and feather color were meticulously quantified using standardized methodologies. Body weight measurements were obtained using calibrated scales, ensuring accuracy and consistency in the assessment. Egg production was recorded using daily monitoring systems, with each egg traced back to the individual hen. Feather color was assessed through visual inspection, categorizing chickens into distinct groups based on color patterns and pigmentation, following established poultry breed standards. All procedures adhered to the Regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, revised in March 2017) and were approved by the Institutional Animal Care and Use Committee at the Experimental Animal Center of Hainan Academy of Agricultural Science (HNXMSY-20210533). Genomic DNA was extracted from blood samples collected on FTA cards. Whole genome resequencing was conducted for these 200 samples to an average depth of 15.10× using the MGI-2000/MGI-T7 platform at the Shenzhen BGI Technology Co. Ltd.
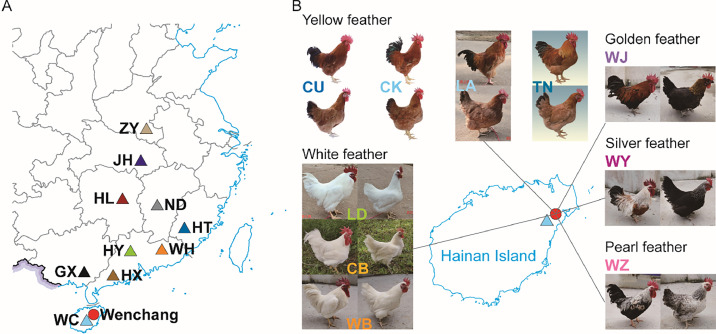
Figure 1.
Sampling map and appearance phenotypes of different strains of Wenchang chickens. (A) Geographical locations of all chicken breeds/populations. The triangles represent the previously published Yellow-feathered chickens, while the red circle represents the newly sequenced Wenchang chickens in this study. WC, Wenchang; GX, Guangxi Yellow; HX, Huaixiang; HY, Huiyang bearded; WH, Wuhua Yellow; HT, Hetian; ND, Ningdu Yellow; HL, Huanglang; JH, Jianghan; ZY, Zhengyang Yellow. (B) Appearance phenotypes of the newly sequenced Wenchang chicken strains. Wenchang chickens with yellow feathers include CU, CK, LA, and TN strains. Wenchang chicken with white feathers include LD, CB, and WB strains. The plumage feathers of the WJ strain are observed as golden in color. The plumage feathers of the WY strain are observed as silver in color, and the plumage feathers of the WZ strain are observed as pearl in color.
Quality Control and Read Mapping
To ensure data quality, Fastp (version 0.20.0) (Chen et al., 2018) was used to filter raw reads. Data processing steps included: 1) remove adapter sequence; 2) eliminating paired reads with ≥10% unidentified nucleotides (N); 3) discarding paired reads with ≥40% low quality bases (Q ≤ 20). Clean reads were then aligned to the chicken reference genome (GRCg7b) using the BWA-MEM (version 0.7.17) (Li, 2014) alignment algorithm.
Variant Calling and Annotation
Variant calling utilized GATK's HaplotypeCaller (version 4.2.1.0) (McKenna et al., 2010). SNPs and INDELs were filtered by GATK's VariantFiltration with options “–filter-expression” "QD & lt; 2.0 || QUAL & lt; 30.0 || MQ & lt; 40.0 || FS & gt; 60.0 || SOR & gt; 3.0 || MQRankSum & lt; 12.5 || ReadPosRankSum & lt; -8.0” (Ma et al., 2020). ANNOVAR (version 2019Oct24) (Wang et al., 2010) was employed to annotate SNPs and INDELs. SNPs and INDELs were counted according to their different functional annotation on the genome. Only autosomal biallelic variants were retained for further analysis.
Population Structure and Genetic Diversity
To comprehend the genetic background and population structure of Wenchang chickens, the Wenchang chicken genome was integrated with published yellow chicken data: Guangxi Yellow (GX), Hetian (HT), Huaixiang (HX), Huanglang (HL), Huiyang bearded (HY), Jianghan (JH), Ningdu Yellow (ND), Wenchang (WC), Wuhua Yellow (WH), and Zhengyang Yellow (ZY), 10 samples for each breed. The PLINK (version 1.9) (Purcell et al., 2007) was used for principal component analysis (PCA) and to obtain pruned data with options “–indep-pairwise 50 10 0.1” for the ADMIXTURE (Patterson et al., 2012) analyses.
Various methods were employed to estimate of the genetic diversity of the tested populations, including nucleotide diversity (π), singleton count, heterozygosity, fixation statistics (Fst), linkage disequilibrium decay (LD decay), and runs of homozygosity (ROHs). Nucleotide diversity was calculated with the sliding windows of 50 Kb with the step size of 25 Kb across all tested populations using VCFtools (version 0.1.13) (Danecek et al., 2011). Singleton number in each chicken population were calculated using VCFtools with the command “—singletons.” Heterozygosity in each chicken population were calculated using VCFtools with the command “—het.” Fst was calculated using the sliding windows of 50 Kb with the step window of 25 Kb between pairwise tested populations. LD decay of all populations was computed using the PopLDdecay (version 3.4) (Zhang et al., 2019) with default parameters. The length and number of ROHs for each individual and mean size for each population were also calculated using PLINK (version 1.9) (Purcell et al., 2007).
Selection Sweep Analyses
To uncover the genetic basis for the exceptional phenotypical properties of Wenchang chicken, genome-wide scans for selection signals were conducted using composite likelihood ratio (CLR) (Pavlidis et al., 2013), Fst, Pi-ratio, the cross-population composite likelihood ratio test (XP-CLR) (Chen et al., 2010), and the cross population extended haplotype homozygosity test (XP-EHH) (Sabeti et al., 2007). CLR was calculated by SweeD (Pavlidis et al., 2013) to detect selection signals within a population. Fst values were calculated using VCFtools (Danecek et al., 2011) in a 50-kb sliding window with 25-kb stepwise increments. Pi-ratio was performed by first calculating the genetic diversity (π) for populations using VCFtools in 50-kb windows with 25-kb stepwise increments, then computing Pi-ratio (πA/πB). XP-CLR calculates cross-population composite likelihood ratio tests between populations with 50-kb windows with 25-kb stepwise increments. The data were first phased using Beagle (version 4.1) (Browning and Browning, 2007) and selscan (version 1.2.0a) (Szpiech and Hernandez, 2014) tool was used to calculate the XP-EHH. An empirical cutoff of the 99th percentile was used to identify candidate selective sweeps, which were then annotated using the online tools Variant Effect Predictor.
In the analysis of local environmental adaptation of Wenchang chickens, the Wenchang chickens (excluding LD strain) comprised the target group, while other Yellow-feathered chicken breeds were set as reference group. In the analysis of feather color, Wenchang chicken strains were categorized into 5 groups based on their feather color and population structure: white feathers (CB and WB), yellow feathers (CU, CK, LA, and TN), golden feathers (WJ), silver white feathers (WY) and pearl feathers (WZ). Each group was chosen as the target population, with the other 4 groups selected as the reference populations, respectively.
Genome Wide Association Studies
In this study, genome wide association studies (GWAS) utilized a univariate linear mixed model (ULMM) in GEMMA (version 0.98.5) (Zhou and Stephens, 2012). Data filtering steps were applied, including removing SNPs and individuals with missingness >0.01, low minor allele frequency (MAF < 0.01), deviations from Hardy Weinberg equilibrium (P < 1e-6), and heterozygosity (±3 standard deviations). PCA analysis and significance tests were conducted for each principal component. GEMMA was used to calculate a standardized kinship matrix.
The GWAS for feather color in Wenchang chickens were conducted between any 2 groups with different feather colors, and between each group and all other groups. GWAS for body weight and body size traits were performed in all populations except the LD strain, while GWAS for reproductive traits were conducted in all female individuals except LD strain. To mitigate the risk of increasing the false-positive error rate due to multiple tests, the Bonferroni correction (with a cutoff of −log10(0.05/number of variants)) or the FDR correction (implemented using the R language's p.adjust function with the parameter method set to “BH”) was employed to identify significant association sites.
Enrichment and Network Analysis
GO and KEGG enrichment analysis were conducted using R package clusterProfiler (version 3.18) (Yu et al., 2012) to gain insight into the biological functions of the candidate PSGs with significant selection signals in these genomic selection scans employed. The significance threshold was set at a Benjamini-Hochberg false discovery rate (FDR) of 0.05.
Data Availability
All newly generated whole genome sequencing (WGS) datasets in this study were deposited in the NCBI sequence read achieve (SRA) under accession number PRJNA1047735. The phenotypic data were listed in Table S2.
RESULTS
Variant Calling and Annotation
In this study, we conducted whole-genome resequencing on 200 Wenchang chickens from 10 specialized strains (Figure 1 and Table S2). The average sequencing depth was 15.1×. After quality control, we identified and annotated 24,011,680 SNPs and 5,165,113 INDELs (Figure S1 and Table S3). Approximately half of the variants were located in introns, with only 1.85% of SNPs and 0.54% of INDELs found in exons (Figure S1). Among these exonic variants, we identified 2,277 stopgain mutations, 261 stoploss mutations, and 173,460 other nonsynonymous SNPs (Table S4). For comparative analysis, we merged the data from the 200 newly sequenced Wenchang chickens with that of 100 previously published Yellow-feathered chickens (Figure 1) from NCBI under the accession ID PRJNA482210 (Huang et al., 2020).
Population Structure
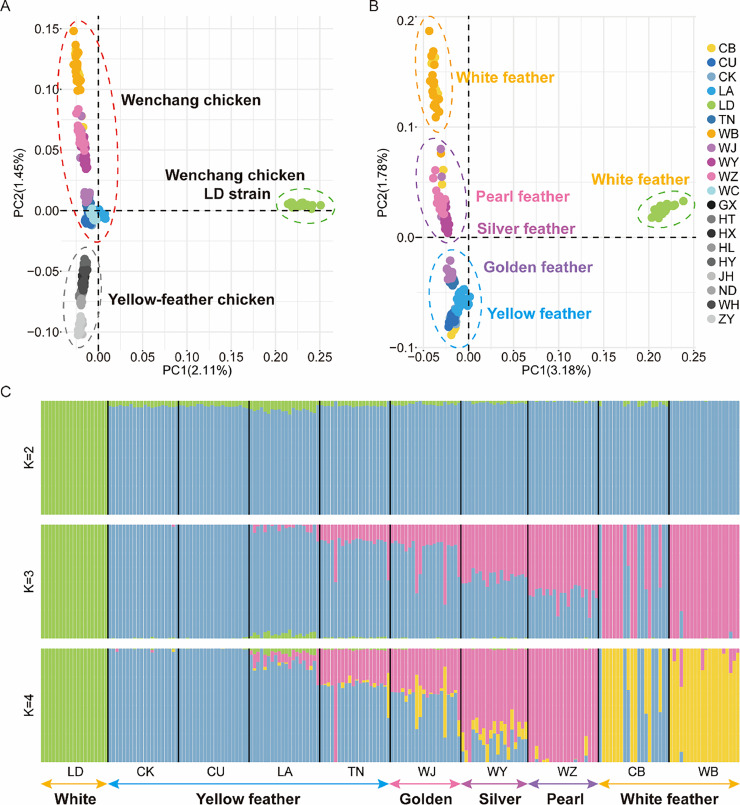
PCA results revealed that the LD strain of Wenchang chicken was separated from other populations along PC1, while the other Wenchang chickens were separated from the previously published Yellow-feathered chickens along PC2 (Figure 2A). The CB and WB strains of Wenchang chickens formed a cluster characterized by white feathers, the WZ and WY strains clustered together with variegated feathers, and the WJ, TN, LA, CU, and CK strains clustered as the yellow feather group (Figure 2B). ADMIXTURE analysis (Figure 2C) confirmed the PCA findings and further elucidated the clustering pattern of Wenchang chicken strains. These results were supported by genetic differentiation (Fst) analysis, where lower Fst values were observed among all Wenchang chickens, except for the LD strain (Figure S2). Additionally, the degree of genetic differentiation among populations with the same feather color was generally lower (Fst = 0.00065∼0.021057) than that among populations with different feather color (Fst = 0.012581∼0.033823) (Figure S2).
Figure 2.
Population structure of Wenchang chickens. (A) Principal component analysis (PCA) was conducted on 200 Wenchang chickens from this study and 100 samples of previously published Yellow-feathered chickens. The analysis revealed 2 primary clusters among all Wenchang chickens. The LD strain of Wenchang chickens formed a distinctive single cluster, indicated by a green circle. All other Yellow-feathered chickens formed a separate distinct cluster, denoted by a gray circle. (B) The PCA plot of the 200 Wenchang chickens displays different strains represented by distinct colors. (C) The ADMIXTURE analysis conducted on 200 Wenchang chickens for K = 2, K = 3, and K = 4 highlighted significant genetic differences between the Wenchang chicken LD strain and other populations. The Wenchang chicken appears genetically separate from other Yellow-feathered chicken populations. Additionally, except for the LD strain, the Wenchang chicken populations clustered according to their feather colors.
Genetic Diversity
Nucleotide diversity, heterozygosity levels, and the number of singletons were lower in the LD strain compared to the other strains (Figures S3–S5). The level of linkage disequilibrium decay (LD decay) and runs of homozygosity (ROHs) were higher in the LD strain compared to others (Figures S6 and S7). In comparison with Yellow-feathered chicken breeds, Wenchang chickens exhibited lower nucleotide diversity, fewer singletons, and a lower level of linkage disequilibrium decay (Figures S3–S6), along with a higher level of heterozygosity and ROHs (Figures S4 and S7). These patterns align with the isolation environments of Wenchang chickens on Hainan Island (Resources, 2011).
Local Adaptation
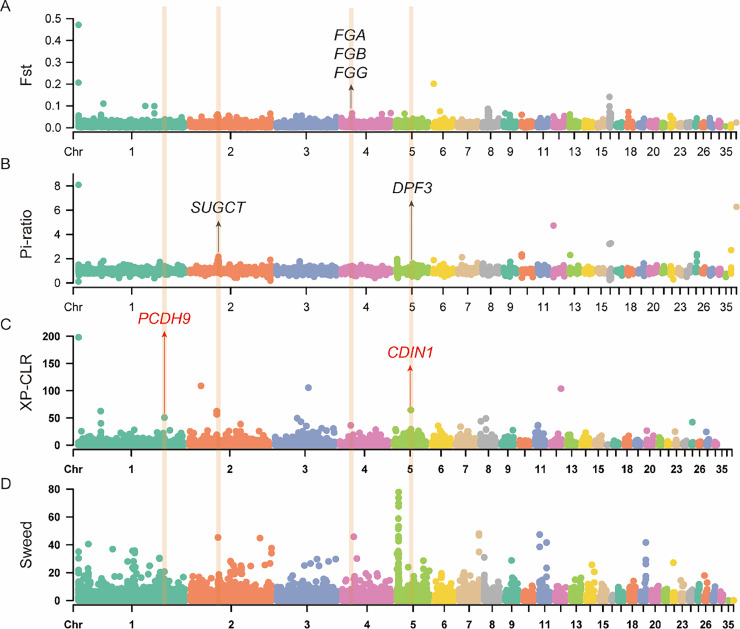
Over centuries of environmental adaptation, the genome of Wenchang chickens has undergone significant changes to thrive in the hot and humid island environment. To explore the genetic mechanisms behind Wenchang chickens’ adaptation to these conditions, we conducted selection sweep analyses, using continental environment breeds with Yellow-feathered chickens as a control. The results revealed 30 positively selected genes (PSGs) identified by at least 3 methods (Table S5 and Figure S8). Among these PSGs, CDH12, CDIN1, and PCDH9 genes were consistently detected by all 4 methods (Figure 3).
Figure 3.
Selection sweep of environmental adaptation in Wenchang chickens. (A) Fst, (B) Pi-ratio, (C) XP-CLR, and (D) SweeD analyses were conducted with the Wenchang chickens (excluding LD, including WC) as the target population and previously published Yellow-feathered chickens (excluding WC) as the reference population. The genes labeled in the figure are candidate genes for tropical island environment adaptation in Wenchang chickens. The genes labeled in red color were detected in all 4 methods, and the genes in black color were detected in 3 methods. Among them, PCDH9 and DPF3 may be related to the adaptation of Wenchang chickens to a strong ultraviolet environment. CDIN1 may be beneficial for Wenchang chickens to resist avian malaria. SUGCT gene is related to adaptation to hot environment.
CDH12 encodes a type II classical cadherin of the cadherin superfamily, playing a vital role in neurite outgrowth (Guo et al., 2021). CDIN1 encodes a novel type of restriction endonuclease belonging to the Holliday junction resolvase family, contributing to erythrocyte differentiation (King et al., 2022). PCDH9 encodes a calcium-dependent cell-cell adhesion molecule, belonging to the cadherin superfamily of calcium-binding proteins (Kim et al., 2011; Izycka et al., 2019; Zhang et al., 2022). Additionally, 2 PSGs (FGA and FGB) are involved in “hemostasis,” “coagulation,” and “regulation of body fluid levels.” Two PSGs (CDIN1 and INHBA) are significantly enriched in “erythrocyte differentiation,” “erythrocyte homeostasis,” and “myeloid cell homeostasis.” Two PSGs (PCLO and SEMA3A) play a role in “synapse organization” (Figure S8). Furthermore, DPF3, OCA2, SASH1, ZMIZ1, SUGCT, FGG, CNTN5, SEMA3A, and GRID2 may also contribute to Wenchang chickens’ ability to thrive in the tropical island environment (Figure 3).
Feather Colors
Feather color is one of the most important genetic markers in breeding and presents an obvious phenotype. To uncover the genetic mechanisms regulating feather color diversity in Wenchang chickens, we conducted GWAS (Figure 4A and B, Figures S9–S24, and Tables S6 and S7) and selection sweep analysis (Figure 4C–F, Figures S25–S29, and Table S8) for different feather color populations. Significant loci identified by GWAS are presented in Table S9.
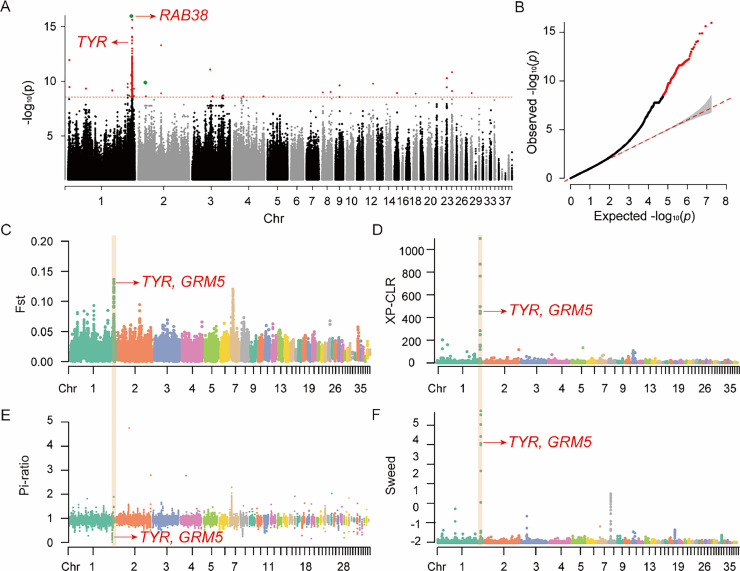
Figure 4.
Genome-wide association analysis and selection sweep of white feather traits in Wenchang chickens. (A) Manhattan plot and (B) quantile-quantile plot (QQ plot) showing the observed P values of the genome-wide association analysis for white feather and nonwhite feather traits in Wenchang chickens. The Manhattan plot indicates the red dashed line depicts the genome-wide significant threshold (2.88 × 10−9), and the green loci represent 2 missense mutations. In the QQ plot (B), the red loci represent the genome-wide significant associations. (C) Fst, (D) XP-CLR, and (E) Pi-ratio analyses were conducted with the white feather strain as the target population and yellow feather strain as the reference population. (F) SweeD analysis was performed based on the white feather strain. The orange shadows denote regions of candidate genes. The TYR gene is a candidate gene for white plumage, and GRM5 may be linked to the TYR gene.
Nearly all significant SNPs associated with white feathers were found in a region spanning from 187.6 Mb to 188.1 Mb of chromosome 1 (Figure 4A and Figures S9–S12; Tables S7 and S9). TYR, RAB38, and TRPM1 genes were identified as candidate genes for white feathers. TYR encodes tyrosinase, the rate-limiting enzyme in melanin synthesis (Sturm, 2009; Yang et al., 2019; Seruggia et al., 2021; Varghese et al., 2021). A retroviral insertion into the fourth intron of the TYR gene in chickens leads to impaired tyrosinase expression and the production of recessive white feathers (Chang et al., 2006; Cho et al., 2021). One significant SNP (Chr1:188181166 G>C) located in the first exon of RAB38, which is a missense mutation. RAB38 gene participates in the transport of melanogenic enzymes (such as TYR and TYRP1) from the Golgi apparatus to immature melanosomes, promoting melanosome maturation, and melanin production (Marubashi et al., 2016). TRPM1 gene is a member of the TRPM family that encodes transient receptor potential ion channels and is associated with pigmentation in human melanocytes (Oancea et al., 2009).
Additionally, we identified pigment-related genes, including LYST, GABARAPL2 and CDH1, in the association analysis. These GWAS results were supported by selection sweep analyses (Table S8). TYR, RAB38, and TRPM1 genes were among the PSGs in Wenchang chicken strains with white feathers. GABARAPL2 gene was among the PSGs in strains with yellow feathers. LYST gene was under selection in strain with golden feathers. CDH1 gene may play a role in the formation of both golden and silver feathers. Furthermore, we identified other candidate genes: ZMIZ in strain with golden feathers, MC1R in strains with golden and silver feathers, and SASH1 in strains with pearl feathers. GO enrichment and KEGG cluster analysis showed that candidate genes of various feather colors were significantly enriched in cell adhesion-related pathways (Figure S30).
Production Traits
This study examined the differences in all measured production traits (Table S10) among Wenchang chickens with different feather colors. The results indicated that Wenchang chickens with yellow feathers had the highest body weight, body size, and egg production performance, while those with white feathers exhibited the lowest (Figure S31). The production abilities of Wenchang chickens with other feather colors fell between those of the 2 mentioned strains, and there were no significant differences (Figure S31).
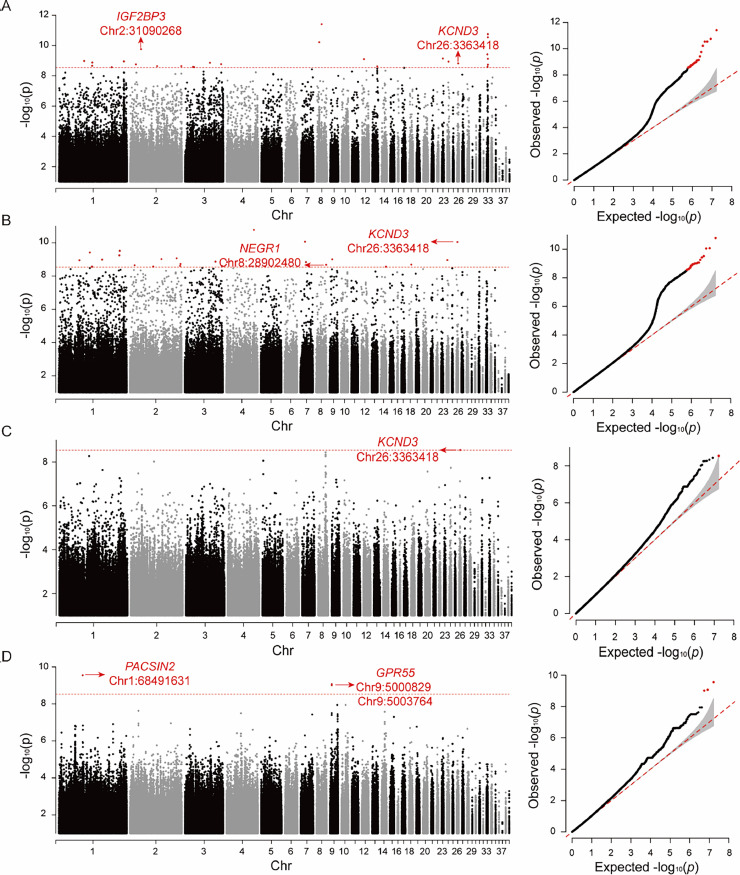
Furthermore, we conducted a preliminary exploration of candidate genes related to production traits in Wenchang chickens, albeit with a small sample size. GWAS for some production traits did not yield significant results (Figures S32–S33), partly due to the limited sample size and the restricted range of phenotypic variation. Significant loci identified in other GWAS are presented in Table S11. In the association analysis of body oblique length, IGF2BP3 and KCND3 genes were identified as candidate genes (Figure 5A). The IGF2BP3 gene plays a crucial role in the growth and development of chicken skeletal muscle (Lin et al., 2017; Shen et al., 2021). IGF2BP3 can bind to insulin-like growth factor 2 (IGF2), regulating cell proliferation and migration, thereby promoting cell growth and development (Wang et al., 2022). The KCND3 gene encodes the potassium ion channel Kv4.3, responsible for generating an instantaneous outgoing potassium current, expressed in both the heart and brain (Pollini et al., 2020). NEGR1 and KCND3 genes were associated with chest depth (Figure 5B). Several studies have indicated that reduced expression of NEGR1 in the hypothalamus may increase body weight by elevating food intake (Lee et al., 2012; Boender et al., 2014; Joo et al., 2019). Additionally, this gene is linked to physiological conditions like obesity (Dennis et al., 2014; Kim et al., 2017; Rana and Mobin, 2020) and Alzheimer's disease (Singh et al., 2019). KCND3 gene was also associated with body weight at 8 wk of age (Figure 5C). PACSIN2 and GPR55 genes were linked to egg weight (Figure 5D).
Figure 5.
Genome-wide association analysis of agronomic traits in Wenchang chickens. Manhattan plot (left) and QQ plot (right) displaying the observed P values of the genome-wide association analysis for body oblique length (A), chest depth (B), 8 wk old weight (C), and 66 wk old egg weight (D). The Manhattan plot indicates that the red dashed line represents the genome-wide significance threshold (A, B, and C: 2.93 × 10−9; D: 3.00 × 10−9). In the association analysis of body oblique length, significant locus Chr2:31090268 was annotated to IGF2BP3. In the association analysis of chest depth, Chr8:28902480 was annotated to NEGR1. In the association analysis of body oblique length, chest depth, and 8 wk old weight, Chr26:3363418 was annotated to KCND3. These genes may be related to the growth and development of Wenchang chickens. Additionally, PACSIN2 and GPR55 may play a role in egg weight.
DISCUSSION
Local Adaptation to Tropical Island
Understanding the genetic adaptation of domestic animals to tropical island environments can help address global climate change and develop appropriate breeding programs (Razgour et al., 2019; Aguirre-Liguori et al., 2021). In this study, we focused on Wenchang chickens and identified multiple candidate genes related to the local adaptation to tropical island environments. We found that PSGs were associated with the nervous system, kidney function, lipid metabolism, and the melanogenesis pathway. The nervous system plays an important role in thermoregulatory responses (Tan and Knight, 2018; Nakamura et al., 2022). CDH12 has an effect on the axonal extension and has been associated with various neuropsychiatric diseases (Guo et al., 2021). CNTN5 is an immunoglobulin cell adhesion molecule that is exclusively expressed in the central nervous system (Kleijer et al., 2018). Metabotropic glutamate receptor (mGluR), encoded by GRM1 gene, is one of the most abundant mGluRs in the mammalian central nervous system (Niswender and Conn, 2010).
Kidney development and function are essential for heat acclimation (Chapman et al., 2021). SEMA3A encodes a secreted guidance protein that functions as an extracellular negative regulator of the integrity and function of the glomerular filtration barrier (Tufro, 2014). Heat stress affects feed intake and adipogenesis (Belhadj Slimen et al., 2016; Xiao et al., 2020). SUGCT encodes a mitochondrial enzyme that synthesizes glutaryl-CoA from glutarate in tryptophan and lysine catabolism (Niska-Blakie et al., 2020). SUGCT-null mice show imbalanced lipid and acylcarnitine metabolism (Niska-Blakie et al., 2020). This gene may be involved in adaptation to hot environments in the study of chickens (Guo et al., 2020). The cutaneous melanin pigment plays a critical role in protecting animals against the harmful effects of solar radiation (Slominski et al., 2004; Lin and Fisher, 2007). OCA2, SASH1, and ZMIZ1 play roles in pigmentation (Lin and Fisher, 2007). PCDH9 may increase the adhesion of melanoma cells by binding to calcium ions, thereby inhibiting the proliferation and migration of melanoma cells (Zhang et al., 2022). Another related gene is DPF3, which is an important candidate gene that causes gray horse melanoma (Druml et al., 2022).
In addition, candidate genes FGA and FGG, which are consistent with the adaptations to tropical climates in Sri Lankan indigenous chickens, may be involved in the regulation of blood vessel diameter to cope with the excessive oxygen demand caused by heat stress (Tian et al., 2020). We also pay attention to CDIN1, which is associated with diseases including congenital dyserythropoietic anemia type Ia and Ib (King et al., 2022). Malaria is prevalent in the central and southern parts of Hainan, and it is often accompanied by various blood diseases (Sun et al., 2023). Therefore, the CDIN1 gene may be advantageous for Wenchang chickens to resist avian malaria.
Candidate Genes for Feather Color
Compared with previous reports based on limited strains (Huang et al., 2020; Shi et al., 2023; Tian et al., 2023a,b), this study provides a broader and more comprehensive understanding of Wenchang chickens based on 10 strains with different phenotypes. Our results suggest that feather colors are critical markers in the breeding management of Wenchang chickens. Additionally, we have identified some candidate genes that likely control the feather colors of Wenchang chickens. For instance, we found a candidate gene for yellow feathers, GABARAPL2, which contributes to the formation of autophagosomes, thus forming melanosomes (Schaaf et al., 2016; Waku et al., 2023). In mice, mutations in LYST (a yellow feather candidate gene in Wenchang chickens) cause melanosomes to become larger, leading to reduced melanosis (Barbosa et al., 1996; Perou et al., 1996; Runkel et al., 2006; Trantow et al., 2011). The white feather candidate gene, TRPM1, regulates Ca2+ homeostasis, which may affect the proliferation and differentiation of melanocytes, plays a role in regulating melanin production (Devi et al., 2009; Guo et al., 2012). Research has found that the insertion of a 1,378-bp endogenous retroviral long terminal repeat in the first intron of TRPM1 disrupts its transcription process, resulting in the leopard complex spotting phenotype in horses (Bellone et al., 2010; Bellone et al., 2013).
CDH1, encoding E-cadherin, may be an important candidate gene for golden and silver feathers of Wenchang chickens. Research has found that before clinical lesions appear in vitiligo patients, E-cadherin between melanocytes and keratinocytes appears to have discontinuous distribution or disappears, resulting in the loss of melanocytes and the appearance of white spots on the skin (Wagner et al., 2015; Kubelis-Lopez et al., 2023). Knockdown of ZMIZ1 (a golden feather candidate gene in Wenchang chickens) inhibits the proliferation and migration of melanocytes (Li et al., 2020). A mutation in SASH1 leads to Dyschromatosis Symmetrica Hereditaria (DSH), a hereditary disorder characterized by skin macules with both hyper- and hypo-pigmentation (Liu et al., 2021). Additionally, there are genes that affect the feather color of Wenchang chickens by affecting the proliferation, migration, and localization of melanocytes.
Breeding Management and Improvement
This study unveils critical insights that could significantly impact the breeding management and improvement of Wenchang chickens. The genetic background analysis revealed the population structure of Wenchang chickens and its relationship with other Yellow-feathered chickens (Figures 1 and 2). Notably, our results showed that the LD strain of Wenchang chicken exhibits reduced genetic diversity and elevated homozygosity (Figures S3–S7), suggestive of potential inbreeding or genetic assimilation by commercial white-feathered chickens. As for other strains, strong artificial selection for different feather colors drove the population structure of Wenchang chickens. It is important to note that singularly focusing on 1 trait may overlook other crucial phenotypic aspects like adaptability (Darwin, 1868). Further investigations into the varying adaptability of different strains to their environment are warranted.
Additionally, the detailed exploration of candidate genes associated with environmental adaptation, feather color, and production traits offers a valuable resource for refining breeding strategies and promoting the preservation of desirable traits. Leveraging this genetic knowledge could facilitate targeted breeding programs geared toward fortifying resilience, productivity, and adaptability in domestic chickens (Bettridge et al., 2018; Lawal and Hanotte, 2021). These findings provide a roadmap for breeders and researchers to implement precise breeding methodologies, thereby safeguarding the unique genetic resources within this local breed and advancing the overall efficiency and success of poultry breeding practices.
Limitations of the Study
While this study provides valuable insights into the population structure, genetic diversity, and traits associated with Wenchang chickens, several limitations should be acknowledged. Firstly, despite the comprehensive analysis conducted on 200 Wenchang chicken samples across 10 distinct strains, the sample size might not fully encapsulate the entire genetic diversity of the breed. Expanding the sample scope to include a more extensive range of populations or integrating data from diverse geographic locations could offer a more comprehensive understanding of their genetic architecture. Additionally, while the GWAS and selection sweep analyses shed light on candidate genes associated with specific traits, a more extensive functional validation of these genes and their precise roles in governing the observed phenotypic traits would be beneficial.
CONCLUSIONS
In summary, this study has enhanced our understanding of the genetic background of Wenchang chickens and provided essential genomic and phenotypic resources for further research. The loci related to environmental adaptation, feather color, and production traits have potential practical value and lay the foundation for the protection and utilization of Wenchang chickens and other local domestic chicken breeds.
ACKNOWLEDGMENTS
We thank the breeding farm and its staff for assistance in sampling. This work was supported by the National Key R&D Program of China (Grant No. 2021YFD1300100), the Key R&D projects in Hainan Province (ZDYF2023XDNY036), the Project of Wenchang city Wenchang Chickens Research Institute (WWXM20230301), and the Animal Branch of the Germplasm Bank of Wild Species, Chinese Academy of Sciences.
Author Contributions: L.-H. G., M.-S. P. and C. M. conceived and supervised this project. R.-R. W., C. M., and T.-S. X analyzed the data. X.-L. Z., A. F., Z.-Y. X, Y.-Y. C., Z.-C. H., L.-Z. L., Y.-T. Q., and A.-H. C. conducted the sample and phenotype data. C. M. and R.-R. W. prepared for the manuscript. L.-H. G., M.-S. P., and Y.-P. Z. revised the manuscript. All authors revised and approved the final manuscript.
DISCLOSURES
All authors declare that they have no financial or nonfinancial conflicts of interest that could be perceived as influencing the research presented in this manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2023.103376.
Appendix. Supplementary materials
REFERENCES
- Aguirre-Liguori J.A., Ramirez-Barahona S., Gaut B.S. The evolutionary genomics of species' responses to climate change. Nat. Ecol. Evol. 2021;5:1350–1360. doi: 10.1038/s41559-021-01526-9. [DOI] [PubMed] [Google Scholar]
- Barbosa M.D., Nguyen Q.A., Tchernev V.T., Ashley J.A., Detter J.C., Blaydes S.M., Brandt S.J., Chotai D., Hodgman C., Solari R.C., Lovett M., Kingsmore S.F. Identification of the homologous beige and Chediak-Higashi syndrome genes. Nature. 1996;382:262–265. doi: 10.1038/382262a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhadj Slimen I., Najar T., Ghram A., Abdrrabba M. Heat stress effects on livestock: molecular, cellular and metabolic aspects, a review. J. Anim. Physiol. Anim. Nutr. (Berl.) 2016;100:401–412. doi: 10.1111/jpn.12379. [DOI] [PubMed] [Google Scholar]
- Bellone R.R., Forsyth G., Leeb T., Archer S., Sigurdsson S., Imsland F., Mauceli E., Engensteiner M., Bailey E., Sandmeyer L., Grahn B., Lindblad-Toh K., Wade C.M. Fine-mapping and mutation analysis of TRPM1: a candidate gene for leopard complex (LP) spotting and congenital stationary night blindness in horses. Brief Funct. Genom. 2010;9:193–207. doi: 10.1093/bfgp/elq002. [DOI] [PubMed] [Google Scholar]
- Bellone R.R., Holl H., Setaluri V., Devi S., Maddodi N., Archer S., Sandmeyer L., Ludwig A., Foerster D., Pruvost M., Reissmann M., Bortfeldt R., Adelson D.L., Lim S.L., Nelson J., Haase B., Engensteiner M., Leeb T., Forsyth G., Mienaltowski M.J., Mahadevan P., Hofreiter M., Paijmans J.L., Gonzalez-Fortes G., Grahn B., Brooks S.A. Evidence for a retroviral insertion in TRPM1 as the cause of congenital stationary night blindness and leopard complex spotting in the horse. PLoS One. 2013;8:e78280. doi: 10.1371/journal.pone.0078280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettridge J.M., Psifidi A., Terfa Z.G., Desta T.T., Lozano-Jaramillo M., Dessie T., Kaiser P., Wigley P., Hanotte O., Christley R.M. The role of local adaptation in sustainable village chicken production. Nat. Sustain. 2018;1:574–582. doi: 10.1038/s41893-018-0150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boender A.J., van Gestel M.A., Garner K.M., Luijendijk M.C., Adan R.A. The obesity-associated gene Negr1 regulates aspects of energy balance in rat hypothalamic areas. Physiol. Rep. 2014;2 doi: 10.14814/phy2.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.M., Coville J.L., Coquerelle G., Gourichon D., Oulmouden A., Tixier-Boichard M. Complete association between a retroviral insertion in the tyrosinase gene and the recessive white mutation in chickens. BMC Genom. 2006;7:19. doi: 10.1186/1471-2164-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman C.L., Johnson B.D., Parker M.D., Hostler D., Pryor R.R., Schlader Z. Kidney physiology and pathophysiology during heat stress and the modification by exercise, dehydration, heat acclimation and aging. Temperature (Austin) 2021;8:108–159. doi: 10.1080/23328940.2020.1826841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Patterson N., Reich D. Population differentiation as a test for selective sweeps. Genome. Res. 2010;20:393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E., Kim M., Manjula P., Cho S.H., Seo D., Lee S.S., Lee J.H. A retroviral insertion in the tyrosinase (TYR) gene is associated with the recessive white plumage color in the Yeonsan Ogye chicken. J. Anim. Sci. Technol. 2021;63:751–758. doi: 10.5187/jast.2021.e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., McVean G., Durbin R. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. Cambridge University Press; Cambridge, UK: 1868. The Variation of Animals and Plants under Domestication. [Google Scholar]
- Dennis E.L., Jahanshad N., Braskie M.N., Warstadt N.M., Hibar D.P., Kohannim O., Nir T.M., McMahon K.L., de Zubicaray G.I., Montgomery G.W., Martin N.G., Toga A.W., Wright M.J., Thompson P.M. Obesity gene NEGR1 associated with white matter integrity in healthy young adults. Neuroimage. 2014;102(Pt. 2):548–557. doi: 10.1016/j.neuroimage.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi S., Kedlaya R., Maddodi N., Bhat K.M., Weber C.S., Valdivia H., Setaluri V. Calcium homeostasis in human melanocytes: role of transient receptor potential melastatin 1 (TRPM1) and its regulation by ultraviolet light. Am. J. Physiol. Cell Physiol. 2009;297:C679–C687. doi: 10.1152/ajpcell.00092.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druml T., Brem G., Horna M., Ricard A., Grilz-Seger G. DPF3, A putative candidate gene for melanoma etiopathogenesis in gray horses. J. Equine Vet. Sci. 2022;108 doi: 10.1016/j.jevs.2021.103797. [DOI] [PubMed] [Google Scholar]
- Guo H., Carlson J.A., Slominski A. Role of TRPM in melanocytes and melanoma. Exp. Dermatol. 2012;21:650–654. doi: 10.1111/j.1600-0625.2012.01565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Qu L., Dou T.C., Shen M.M., Hu Y.P., Ma M., Wang K.H. Genome-wide association study provides insights into the genetic architecture of bone size and mass in chickens. Genome. 2020;63:133–143. doi: 10.1139/gen-2019-0022. [DOI] [PubMed] [Google Scholar]
- Guo B., Qi M., Huang S., Zhuo R., Zhang W., Zhang Y., Xu M., Liu M., Guan T., Liu Y. Cadherin-12 regulates neurite outgrowth through the PKA/Rac1/Cdc42 pathway in cortical neurons. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.768970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Otecko N.O., Peng M., Weng Z., Li W., Chen J., Zhong M., Zhong F., Jin S., Geng Z., Luo W., He D., Ma C., Han J., Ommeh S.C., Zhang Y., Zhang X., Du B. Genome-wide genetic structure and selection signatures for color in 10 traditional Chinese yellow-feathered chicken breeds. BMC Genomics [Electronic Resource] 2020;21:316. doi: 10.1186/s12864-020-6736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izycka N., Sterzynska K., Januchowski R., Nowak-Markwitz E. Semaphorin 3A (SEMA3A), protocadherin 9 (PCdh9), and S100 calcium binding protein A3 (S100A3) as potential biomarkers of carcinogenesis and chemoresistance of different neoplasms, including ovarian cancer - review of literature. Ginekol. Pol. 2019;90:223–227. doi: 10.5603/GP.2019.0040. [DOI] [PubMed] [Google Scholar]
- Jia C.L., He L.J., Li P.C., Liu H.Y., Wei Z.H. Effect of egg composition and oxidoreductase on adaptation of Tibetan chicken to high altitude. Poult. Sci. 2016;95:1660–1665. doi: 10.3382/ps/pew048. [DOI] [PubMed] [Google Scholar]
- Joo Y., Kim H., Lee S., Lee S. Neuronal growth regulator 1-deficient mice show increased adiposity and decreased muscle mass. Int. J. Obes. (Lond.) 2019;43:1769–1782. doi: 10.1038/s41366-019-0376-2. [DOI] [PubMed] [Google Scholar]
- Kim H., Chun Y., Che L., Kim J., Lee S., Lee S. The new obesity-associated protein, neuronal growth regulator 1 (NEGR1), is implicated in Niemann-Pick disease Type C (NPC2)-mediated cholesterol trafficking. Biochem. Biophys. Res. Commun. 2017;482:1367–1374. doi: 10.1016/j.bbrc.2016.12.043. [DOI] [PubMed] [Google Scholar]
- Kim S.Y., Yasuda S., Tanaka H., Yamagata K., Kim H. Non-clustered protocadherin. Cell Adh. Migr. 2011;5:97–105. doi: 10.4161/cam.5.2.14374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R., Gallagher P.J., Khoriaty R. The congenital dyserythropoieitic anemias: genetics and pathophysiology. Curr. Opin. Hematol. 2022;29:126–136. doi: 10.1097/MOH.0000000000000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijer K.T.E., van Nieuwenhuize D., Spierenburg H.A., Gregorio-Jordan S., Kas M.J.H., Burbach J.P.H. Structural abnormalities in the primary somatosensory cortex and a normal behavioral profile in Contactin-5 deficient mice. Cell Adh. Migr. 2018;12:5–18. doi: 10.1080/19336918.2017.1288788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubelis-Lopez D.E., Zapata-Salazar N.A., Salinas-Santander M.A., Sanchez-Dominguez C.N., Morlett-Chavez J.A., Ocampo-Candiani J. Association of e-cadherin gene CDH1 polymorphism -160 C/A with susceptibility to develop vitiligo. An. Bras. Dermatol. 2023;98:376–378. doi: 10.1016/j.abd.2022.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawal R.A., Hanotte O. Domestic chicken diversity: origin, distribution, and adaptation. Anim. Genet. 2021;52:385–394. doi: 10.1111/age.13091. [DOI] [PubMed] [Google Scholar]
- Lee A.W., Hengstler H., Schwald K., Berriel-Diaz M., Loreth D., Kirsch M., Kretz O., Haas C.A., de Angelis M.H., Herzig S., Brummendorf T., Klingenspor M., Rathjen F.G., Rozman J., Nicholson G., Cox R.D., Schafer M.K. Functional inactivation of the genome-wide association study obesity gene neuronal growth regulator 1 in mice causes a body mass phenotype. PLoS One. 2012;7:e41537. doi: 10.1371/journal.pone.0041537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Fan Y., Wang Y., Xu J., Xu H. ZMIZ1 promotes the proliferation and migration of melanocytes in vitiligo. Exp. Ther. Med. 2020;20:1371–1378. doi: 10.3892/etm.2020.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.Y., Fisher D.E. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–850. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- Lin S., Luo W., Ye Y., Bekele E.J., Nie Q., Li Y., Zhang X. Let-7b regulates myoblast proliferation by inhibiting IGF2BP3 expression in dwarf and normal chicken. Front. Physiol. 2017;8:477. doi: 10.3389/fphys.2017.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.W., Habulieti X., Wang R.R., Ma D.L., Zhang X. Two novel SASH1 mutations in Chinese families with dyschromatosis universalis hereditaria. J. Clin. Lab. Anal. 2021;35:e23803. doi: 10.1002/jcla.23803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Khederzadeh S., Adeola A.C., Han X.M., Xie H.B., Zhang Y.P. Whole genome resequencing reveals an association of ABCC4 variants with preaxial polydactyly in pigs. BMC Genomics [Electronic Resource] 2020;21:268. doi: 10.1186/s12864-020-6690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marubashi S., Shimada H., Fukuda M., Ohbayashi N. RUTBC1 functions as a GTPase-activating protein for Rab32/38 and regulates melanogenic enzyme trafficking in melanocytes. J. Biol. Chem. 2016;291:1427–1440. doi: 10.1074/jbc.M115.684043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome. Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Nakamura Y., Kataoka N. A hypothalamomedullary network for physiological responses to environmental stresses. Nat. Rev. Neurosci. 2022;23:35–52. doi: 10.1038/s41583-021-00532-x. [DOI] [PubMed] [Google Scholar]
- Nan J., Yang S., Zhang X., Leng T., Zhuoma J., Zhuoma R., Yuan J., Pi J., Sheng Z., Li S. Identification of candidate genes related to highland adaptation from multiple Chinese local chicken breeds by whole genome sequencing analysis. Anim. Genet. 2023;54:55–67. doi: 10.1111/age.13268. [DOI] [PubMed] [Google Scholar]
- Niska-Blakie J., Gopinathan L., Low K.N., Kien Y.L., Goh C.M.F., Caldez M.J., Pfeiffenberger E., Jones O.S., Ong C.B., Kurochkin I.V., Coppola V., Tessarollo L., Choi H., Kanagasundaram Y., Eisenhaber F., Maurer-Stroh S., Kaldis P. Knockout of the non-essential gene SUGCT creates diet-linked, age-related microbiome disbalance with a diabetes-like metabolic syndrome phenotype. Cell Mol. Life Sci. 2020;77:3423–3439. doi: 10.1007/s00018-019-03359-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender C.M., Conn P.J. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu. Rev. Pharmacol. Toxicol. 2010;50:295–322. doi: 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oancea E., Vriens J., Brauchi S., Jun J., Splawski I., Clapham D.E. TRPM1 forms ion channels associated with melanin content in melanocytes. Sci. Signal. 2009;2:ra21. doi: 10.1126/scisignal.2000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P., Živkovic D., Stamatakis A., Alachiotis N. SweeD: likelihood-based detection of selective sweeps in thousands of genomes. Mol. Biol. Evol. 2013;30:2224–2234. doi: 10.1093/molbev/mst112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M.S., Han J.L., Zhang Y.P. Missing puzzle piece for the origins of domestic chickens. Proc. Nat. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2210996119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou C.M., Moore K.J., Nagle D.L., Misumi D.J., Woolf E.A., McGrail S.H., Holmgren L., Brody T.H., Dussault B.J., Jr., Monroe C.A., Duyk G.M., Pryor R.J., Li L., Justice M.J., Kaplan J. Identification of the murine beige gene by YAC complementation and positional cloning. Nat. Genet. 1996;13:303–308. doi: 10.1038/ng0796-303. [DOI] [PubMed] [Google Scholar]
- Peters J., Lebrasseur O., Irving-Pease E.K., Paxinos P.D., Best J., Smallman R., Callou C., Gardeisen A., Trixl S., Frantz L., Sykes N., Fuller D.Q., Larson G. The biocultural origins and dispersal of domestic chickens. Proc. Nat. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2121978119. e2121978119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollini L., Galosi S., Tolve M., Caputi C., Carducci C., Angeloni A., Leuzzi V. KCND3-related neurological disorders: from old to emerging clinical phenotypes. Int. J. Mol. Sci. 2020;21:5802. doi: 10.3390/ijms21165802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana S., Mobin M. Association of the NEGR1 rs2815752 with obesity and related traits in Pakistani females. Ups J. Med. Sci. 2020;125:226–234. doi: 10.1080/03009734.2020.1756996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razgour O., Forester B., Taggart J.B., Bekaert M., Juste J., Ibanez C., Puechmaille S.J., Novella-Fernandez R., Alberdi A., Manel S. Considering adaptive genetic variation in climate change vulnerability assessment reduces species range loss projections. Proc. Nat. Acad. Sci. USA. 2019;116:10418–10423. doi: 10.1073/pnas.1820663116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Guan Z., Li H., Wen J., Zhao X., Wang G., Zhang X., Wang H., Zhang L., Yu F., Qu L. Extensive intra- and inter-genetic admixture of Chinese gamecock and other indigenous chicken breeds revealed by genomic data. Poult. Sci. 2023;102 doi: 10.1016/j.psj.2023.102766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- o.A.G. Resources C.N.C. PoultryChina Agriculture Press; Beijing, China: 2011. Animal Genetic Resources in China. [Google Scholar]
- Restoux G., Rognon X., Vieaud A., Guemene D., Petitjean F., Rouger R., Brard-Fudulea S., Lubac-Paye S., Chiron G., Tixier-Boichard M. Managing genetic diversity in breeding programs of small populations: the case of French local chicken breeds. Gene. Sel. Evol. 2022;54:56. doi: 10.1186/s12711-022-00746-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runkel F., Bussow H., Seburn K.L., Cox G.A., Ward D.M., Kaplan J., Franz T. Grey, a novel mutation in the murine Lyst gene, causes the beige phenotype by skipping of exon 25. Mamm. Genome. 2006;17:203–210. doi: 10.1007/s00335-005-0015-1. [DOI] [PubMed] [Google Scholar]
- Sabeti P.C., Varilly P., Fry B., Lohmueller J., Hostetter E., Cotsapas C., Xie X., Byrne E.H., McCarroll S.A., Gaudet R., Schaffner S.F., Lander E.S., International HapMap C., Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., Pasternak S., Wheeler D.A., Willis T.D., Yu F., Yang H., Zeng C., Gao Y., Hu H., Hu W., Li C., Lin W., Liu S., Pan H., Tang X., Wang J., Wang W., Yu J., Zhang B., Zhang Q., Zhao H., Zhao H., Zhou J., Gabriel S.B., Barry R., Blumenstiel B., Camargo A., Defelice M., Faggart M., Goyette M., Gupta S., Moore J., Nguyen H., Onofrio R.C., Parkin M., Roy J., Stahl E., Winchester E., Ziaugra L., Altshuler D., Shen Y., Yao Z., Huang W., Chu X., He Y., Jin L., Liu Y., Shen Y., Sun W., Wang H., Wang Y., Wang Y., Xiong X., Xu L., Waye M.M., Tsui S.K., Xue H., Wong J.T., Galver L.M., Fan J.B., Gunderson K., Murray S.S., Oliphant A.R., Chee M.S., Montpetit A., Chagnon F., Ferretti V., Leboeuf M., Olivier J.F., Phillips M.S., Roumy S., Sallee C., Verner A., Hudson T.J., Kwok P.Y., Cai D., Koboldt D.C., Miller R.D., Pawlikowska L., Taillon-Miller P., Xiao M., Tsui L.C., Mak W., Song Y.Q., Tam P.K., Nakamura Y., Kawaguchi T., Kitamoto T., Morizono T., Nagashima A., Ohnishi Y., Sekine A., Tanaka T., Tsunoda T., Deloukas P., Bird C.P., Delgado M., Dermitzakis E.T., Gwilliam R., Hunt S., Morrison J., Powell D., Stranger B.E., Whittaker P., Bentley D.R., Daly M.J., de Bakker P.I., Barrett J., Chretien Y.R., Maller J., McCarroll S., Patterson N., Pe'er I., Price A., Purcell S., Richter D.J., Sabeti P., Saxena R., Schaffner S.F., Sham P.C., Varilly P., Altshuler D., Stein L.D., Krishnan L., Smith A.V., Tello-Ruiz M.K., Thorisson G.A., Chakravarti A., Chen P.E., Cutler D.J., Kashuk C.S., Lin S., Abecasis G.R., Guan W., Li Y., Munro H.M., Qin Z.S., Thomas D.J., McVean G., Auton A., Bottolo L., Cardin N., Eyheramendy S., Freeman C., Marchini J., Myers S., Spencer C., Stephens M., Donnelly P., Cardon L.R., Clarke G., Evans D.M., Morris A.P., Weir B.S., Tsunoda T., Johnson T.A., Mullikin J.C., Sherry S.T., Feolo M., Skol A., Zhang H., Zeng C., Zhao H., Matsuda I., Fukushima Y., Macer D.R., Suda E., Rotimi C.N., Adebamowo C.A., Ajayi I., Aniagwu T., Marshall P.A., Nkwodimmah C., Royal C.D., Leppert M.F., Dixon M., Peiffer A., Qiu R., Kent A., Kato K., Niikawa N., Adewole I.F., Knoppers B.M., Foster M.W., Clayton E.W., Watkin J., Gibbs R.A., Belmont J.W., Muzny D., Nazareth L., Sodergren E., Weinstock G.M., Wheeler D.A., Yakub I., Gabriel S.B., Onofrio R.C., Richter D.J., Ziaugra L., Birren B.W., Daly M.J., Altshuler D., Wilson R.K., Fulton L.L., Rogers J., Burton J., Carter N.P., Clee C.M., Griffiths M., Jones M.C., McLay K., Plumb R.W., Ross M.T., Sims S.K., Willey D.L., Chen Z., Han H., Kang L., Godbout M., Wallenburg J.C., L'Archeveque P., Bellemare G., Saeki K., Wang H., An D., Fu H., Li Q., Wang Z., Wang R., Holden A.L., Brooks L.D., McEwen J.E., Guyer M.S., Wang V.O., Peterson J.L., Shi M., Spiegel J., Sung L.M., Zacharia L.F., Collins F.S., Kennedy K., Jamieson R., Stewart J. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf M.B., Keulers T.G., Vooijs M.A., Rouschop K.M. LC3/GABARAP family proteins: autophagy-(un)related functions. FASEB J. 2016;30:3961–3978. doi: 10.1096/fj.201600698R. [DOI] [PubMed] [Google Scholar]
- Seruggia D., Josa S., Fernandez A., Montoliu L. The structure and function of the mouse tyrosinase locus. Pigment. Cell Melanoma Res. 2021;34:212–221. doi: 10.1111/pcmr.12942. [DOI] [PubMed] [Google Scholar]
- Shen X., Wei Y., You G., Liu W., Amevor F.K., Zhang Y., He H., Ma M., Zhang Y., Li D., Zhu Q., Yin H. Circular PPP1R13B RNA Promotes Chicken Skeletal Muscle Satellite Cell Proliferation and Differentiation via Targeting miR-9-5p. Animals (Basel) 2021;11:2396. doi: 10.3390/ani11082396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Shao D., Yang L., Liang Q., Han W., Xue Q., Qu L., Leng L., Li Y., Zhao X., Dong P., Walugembe M., Kayang B.B., Muhairwa A.P., Zhou H., Tong H. Whole genome analyses reveal novel genes associated with chicken adaptation to tropical and frigid environments. J. Adv. Res. 2023;47:13–25. doi: 10.1016/j.jare.2022.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K., Jayaram M., Kaare M., Leidmaa E., Jagomae T., Heinla I., Hickey M.A., Kaasik A., Schafer M.K., Innos J., Lillevali K., Philips M.A., Vasar E. Neural cell adhesion molecule Negr1 deficiency in mouse results in structural brain endophenotypes and behavioral deviations related to psychiatric disorders. Sci. Rep. 2019;9:5457. doi: 10.1038/s41598-019-41991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski A., Tobin D.J., Shibahara S., Wortsman J. Melanin pigmentation in mammalian skin and its hormonal regulation. Physiol. Rev. 2004;84:1155–1228. doi: 10.1152/physrev.00044.2003. [DOI] [PubMed] [Google Scholar]
- Sturm R.A. Molecular genetics of human pigmentation diversity. Hum. Mol. Genet. 2009;18:R9–17. doi: 10.1093/hmg/ddp003. [DOI] [PubMed] [Google Scholar]
- Sun D., Chen Y., Wang L., Hu X., Wu Q., Liu Y., Liu P., Zeng X., Li S., Wang G., Zhang Y. Surveillance and control of malaria vectors in Hainan Province, China from 1950 to 2021: a retrospective review. Trop. Med. Infect. Dis. 2023;8:131. doi: 10.3390/tropicalmed8030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiech Z.A., Hernandez R.D. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol. Biol. Evol. 2014;31:2824–2827. doi: 10.1093/molbev/msu211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.L., Knight Z.A. Regulation of body temperature by the nervous system. Neuron. 2018;98:31–48. doi: 10.1016/j.neuron.2018.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S., Li W., Zhong Z., Wang F., Xiao Q. Genome-wide re-sequencing data reveals the genetic diversity and population structure of Wenchang chicken in China. Anim. Genet. 2023;54:328–337. doi: 10.1111/age.13293. [DOI] [PubMed] [Google Scholar]
- Tian S., Tang W., Zhong Z., Wang Z., Xie X., Liu H., Chen F., Liu J., Han Y., Qin Y., Tan Z., Xiao Q. Identification of runs of homozygosity islands and functional variants in Wenchang chicken. Animals (Basel) 2023;13 doi: 10.3390/ani13101645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S., Zhou X., Phuntsok T., Zhao N., Zhang D., Ning C., Li D., Zhao H. Genomic analyses reveal genetic adaptations to tropical climates in chickens. iScience. 2020;23 doi: 10.1016/j.isci.2020.101644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trantow C.M., Cuffy T.L., Fingert J.H., Kuehn M.H., Anderson M.G. Microarray analysis of iris gene expression in mice with mutations influencing pigmentation. Invest. Ophthalmol. Vis. Sci. 2011;52:237–248. doi: 10.1167/iovs.10-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufro A. Semaphorin3a signaling, podocyte shape, and glomerular disease. Pediatr. Nephrol. 2014;29:751–755. doi: 10.1007/s00467-013-2743-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese P.K., Abu-Asab M., Dimitriadis E.K., Dolinska M.B., Morcos G.P., Sergeev Y.V. Tyrosinase nanoparticles: understanding the melanogenesis pathway by isolating the products of tyrosinase enzymatic reaction. Int. J. Mol. Sci. 2021;22:734. doi: 10.3390/ijms22020734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R.Y., Luciani F., Cario-Andre M., Rubod A., Petit V., Benzekri L., Ezzedine K., Lepreux S., Steingrimsson E., Taieb A., Gauthier Y., Larue L., Delmas V. Altered E-cadherin levels and distribution in melanocytes precede clinical manifestations of vitiligo. J. Invest. Dermatol. 2015;135:1810–1819. doi: 10.1038/jid.2015.25. [DOI] [PubMed] [Google Scholar]
- Waku T., Nakada S., Masuda H., Sumi H., Wada A., Hirose S., Aketa I., Kobayashi A. The CNC-family transcription factor Nrf3 coordinates the melanogenesis cascade through macropinocytosis and autophagy regulation. Cell Rep. 2015;42 doi: 10.1016/j.celrep.2022.111906. [DOI] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic. Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.S., Li Y., Peng M.S., Zhong L., Wang Z.J., Li Q.Y., Tu X.L., Dong Y., Zhu C.L., Wang L., Yang M.M., Wu S.F., Miao Y.W., Liu J.P., Irwin D.M., Wang W., Wu D.D., Zhang Y.P. Genomic analyses reveal potential independent adaptation to high altitude in Tibetan chickens. Mol. Biol. Evol. 2015;32:1880–1889. doi: 10.1093/molbev/msv071. [DOI] [PubMed] [Google Scholar]
- Wang M.S., Thakur M., Peng M.S., Jiang Y., Frantz L.A.F., Li M., Zhang J.J., Wang S., Peters J., Otecko N.O., Suwannapoom C., Guo X., Zheng Z.Q., Esmailizadeh A., Hirimuthugoda N.Y., Ashari H., Suladari S., Zein M.S.A., Kusza S., Sohrabi S., Kharrati-Koopaee H., Shen Q.K., Zeng L., Yang M.M., Wu Y.J., Yang X.Y., Lu X.M., Jia X.Z., Nie Q.H., Lamont S.J., Lasagna E., Ceccobelli S., Gunwardana H., Senasige T.M., Feng S.H., Si J.F., Zhang H., Jin J.Q., Li M.L., Liu Y.H., Chen H.M., Ma C., Dai S.S., Bhuiyan A., Khan M.S., Silva G., Le T.T., Mwai O.A., Ibrahim M.N.M., Supple M., Shapiro B., Hanotte O., Zhang G., Larson G., Han J.L., Wu D.D., Zhang Y.P. 863 genomes reveal the origin and domestication of chicken. Cell Res. 2020;30:693–701. doi: 10.1038/s41422-020-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Yang Y.Z., Liu Y., Zhang Y.T., Song J.W., Wang H.Y., Li G.Q., Wang X.Z., Gong S.M., Chen S.F., He D.Q. Molecular characterization, expression pattern and genetic variant of insulin-like growth factor 2 mRNA-binding protein 3 gene in goose (Anser cygnoides) J. Appl. Anim. Res. 2022;50:574–581. [Google Scholar]
- Xiao Y., Kronenfeld J.M., Renquist B.J. Feed intake-dependent and -independent effects of heat stress on lactation and mammary gland development. J. Dairy Sci. 2020;103:12003–12014. doi: 10.3168/jds.2020-18675. [DOI] [PubMed] [Google Scholar]
- Xu N.Y., Liu Z.Y., Yang Q.M., Bian P.P., Li M., Zhao X. Genomic analyses for selective signatures and genes involved in hot adaptation among indigenous chickens from different tropical climate regions. Front. Genet. 2022;13:906447. doi: 10.3389/fgene.2022.906447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N.Y., Si W., Li M., Gong M., Lariviere J.M., Nanaei H.A., Bian P.P., Jiang Y., Zhao X. Genome-wide scan for selective footprints and genes related to cold tolerance in Chantecler chickens. Zool. Res. 2021;42:710–720. doi: 10.24272/j.issn.2095-8137.2021.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.W., Ran J.S., Yu C.L., Qiu M.H., Zhang Z.R., Du H.R., Li Q.Y., Xiong X., Song X.Y., Xia B., Hu C.M., Liu Y.P., Jiang X.S. Polymorphism in MC1R, TYR and ASIP genes in different colored feather chickens. 3 Biotech. 2019;9:203. doi: 10.1007/s13205-019-1710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Dong S.S., Xu J.Y., He W.M., Yang T.L. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics. 2019;35:1786–1788. doi: 10.1093/bioinformatics/bty875. [DOI] [PubMed] [Google Scholar]
- Zhang J., Yang H.Z., Liu S., Islam M.O., Zhu Y., Wang Z., Chen R. PCDH9 suppresses melanoma proliferation and cell migration. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.903554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Zhang J., Wang H., Li H., Qu C., Wen J., Zhang X., Zhu T., Nie C., Li X., Muhatai G., Wang L., Lv X., Yang W., Zhao C., Bao H., Li J., Zhu B., Cao G., Xiong W., Ning Z., Qu L. Genomic and transcriptomic analyses reveal genetic adaptation to cold conditions in the chickens. Genomics. 2022;114 doi: 10.1016/j.ygeno.2022.110485. [DOI] [PubMed] [Google Scholar]
- Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All newly generated whole genome sequencing (WGS) datasets in this study were deposited in the NCBI sequence read achieve (SRA) under accession number PRJNA1047735. The phenotypic data were listed in Table S2.