Abstract
Real-world settings are necessary to improve the ecological validity of neuroscience research, and electroencephalography (EEG) facilitates mobile electrocortical recordings because of its easy portability and high temporal resolution. Table tennis is a whole-body, goal-directed sport that requires constant visuomotor feedback, anticipation, strategic decision-making, object interception, and performance monitoring – making it an interesting testbed for a variety of neuroscience studies. Although traditionally plagued by artifact contamination, recent advances in signal processing and hardware approaches, such as the dual-layer approach, have allowed high fidelity EEG recordings during whole-body maneuvers. Here, we present a dual-layer EEG dataset from 25 healthy human participants playing table tennis with a human opponent and a ball machine. Our dataset includes synchronized, multivariate time series recordings from 120 scalp electrodes, 120 noise electrodes, 8 neck electromyography electrodes, and inertial measurement units on the participant, paddles, and ball machine to record hit events. We also include de-identified T1 anatomical MR images and digitized electrode locations to create subject-specific head models for source localization. In addition, we provide anonymized video recordings and Adobe Premiere project files with hit events labeled (originally used to mark successful/missed hits). Researchers could use the videos to mark their own events of interest. We formatted our dataset in the Brain Imaging Data Structure (BIDS) format to facilitate data reuse and to adhere to the scientific community's new organization standard.
Keywords: Mobile Brain Body Imaging (MoBI), EEG, Independent component analysis, Sports neuroscience, Electrocortical dynamics
Specifications Table
| Subject | Neuroscience: General |
| Specific subject area | Mobile-Brain Body Imaging (MoBI) |
| Data format | Raw, Processed, Filtered |
| Type of data | Multivariate time-series data from electroencephalography (EEG), neck electromyography (EMG), inertial measurement units (IMU); T1 anatomical MR head images, video and Adobe Premiere project files; weight matrices from independent component analysis; metadata |
| Data collection | Twenty-five right-handed healthy participants played table tennis with a human player (cooperative or competitive play) and with a ball machine (stationary or moving conditions, replicating a rally shot or a serve). Data were acquired using dual-layer EEG with BrainVision ActiCap snap sensors and EeonTex LTT-PI-100 conductive fabric, data-logged on four BrainVision LiveAmp 64 amplifiers (500 Hz). We used the Cometas WaveTrack Inertial System in data-logging mode (2000 Hz), 3T Philips Ingenia Scanner for structural T1 MRI, GoPro Hero 7 video (30–240 fps), itSeez3D software and Structure Sensor from Occipital Inc. for electrode locations. |
| Data source location | Institution: University of Florida City/Town/Region: Gainesville, FL Country: United States |
| Data accessibility | Repository name: OpenNeuro Data identification number: ds004505 Direct URL to data: https://openneuro.org/datasets/ds004505/[1] |
| Related research article | A. Studnicki, R.D. Seidler, D.P. Ferris, A Table Tennis Serve versus Rally Hit Elicits Differential Hemispheric Electrocortical Power Fluctuations, Journal of Neurophysiology. 2023. 130(6):1444–1456. 10.1152/jn.00091.2023. |
1. Value of the Data
-
•
These data are useful because understanding the neural correlates of natural movement is important and these data advance the field of mobile brain imaging during a whole-body sporting task.
-
•
This multi-modal dataset can benefit researchers interested in sensorimotor integration, real-world object interception, performance monitoring, source-localized electrocortical connectivity, human-machine interaction, sports neurophysiology, and improving techniques for removing EEG artifacts during mobile tasks.
-
•
We formatted the dataset in the Brain Imaging Data Structure (BIDS) format to enable data reuse and interpretation.
-
•
We processed and selected participants that contained more than five source-localized brain components, which ensures that our data was high quality.
-
•
The subject-specific T1 anatomical MR head images and digitized electrode locations allow researchers to improve dipole source localization techniques.
-
•
The video data provides an opportunity to explore other events of interest.
2. Data Description
Our dataset contains high-density, dual-layer electroencephalography (EEG), neck electromyography (EMG), inertial measurement unit (IMU) acceleration, T1 structural MR images, and video data for 25 healthy, cognitively-intact participants playing real-world table tennis. Participants played 60 min of table tennis (in total) with a ball machine and a human player (i.e., the researcher, A.S.), with an additional 10 min of standing baseline. All data files are separated for each participant and follow the Brain Imaging Data Structure (BIDS) format [4] (Fig. 1). A high-level overview of the dataset can be found in the dataset_description.json file.
Fig. 1.

Organization of the BIDS dataset on OpenNeuro.
2.1. Metadata
The participant.tsv and participant.json files hold the metadata information. In the participants.tsv file are the participant's age, sex, head circumference size, and answers to a nine-question survey to gauge the participant's table tennis and racquet sport experience level. Accompanying the TSV file is the participants.json file that describes each column in detail.
2.2. Raw data
The sourcedata folder contains the raw subject data, separated by subject into different folders. For example, “sub-01” corresponds to “Subject 1”. Within each subject folder inside the sourcedata folder are “.eeg”, “.vhdr”, and “.vmrk” files that come from the original BrainVision amplifier recordings. The “.eeg” files contain the raw EEG data. The “.vhdr” files contain the recording parameters and other meta information (sampling rate, channel resolution, amplifier battery levels, number of data points, etc.). The “.vmrk” files contain the raw event data. Four individual amplifiers were used to record the data (left/right hemisphere and scalp/noise layer of electrodes), and each file is labeled accordingly. An “_Impedance.txt” file contains the impedance values at the start of the experiment. The “_chanlocs.txt” file contain the digitized electrode locations from the get_chanlocs function in EEGLAB [5]. Electrode locations are in millimeters units and the fiducials (nasion and left/right preauricular points) are found at the bottom rows of the file. Multiple “_Cometas_##.txt” files correspond to the raw data from the inertial measurement units. Different participants had a different subset of sensors, and the “_Cometas_KEY.txt” file outlines which sensors correspond to which data stream. The rising edge of the square wave from the Cometas “sync” data stream and the “M 1” events from the BrainVision amplifier can be used to align the data.
The sourcedata folder also contains a directory titled “Merged”. Inside the “Merged” folder, the data are separated again by individual subject folders. Within the individual subject folders, the data from all the sensors have been synchronized and merged into a single subject's dataset (i.e., the “.fdt” and “.set” files) and high-pass filtered at 1 Hz. The “.fdt” and “.set” files can be read using EEGLAB. After loading the data, a field in the data structure (EEG.etc.ImportEvents.sensor_ch) explains what each of the inertial measurement units were for that particular subject. The subject paddle corresponds to “sub”, “res” is the researcher paddle, “mach” is the ball machine, “net” is the net, “table_BM” is the table on the side opposite the subject, “table_sub” is the table on the same side of the subject, “head” is on the subject's forehead, and “torso” is within the backpack.
2.3. Processed data
Processed data are found in subject folders (sub-01, sub-02, …) outside of the sourcedata directory. The subfolders within each subject folder are “anat”, “eeg”, and “video”. The “anat” subfolder contains the T1 structural MR scans. The “eeg” subfolder contains all the electrophysiological data. The “video” subfolder, present for participants who agreed to release them, hold videos of individual trials. Video data is missing from sub-01 to sub-05, sub-08, sub-18, and sub-22.
Within the “eeg” subfolder of the subject's processed data are time series recordings saved as EEGLAB “.set” and “.fdt” files. Using EEGLAB, both of the “.set” and “.fdt” files can be loaded. After loading, the time series data is in the EEG structure “data” field. Each row corresponds to one of the scalp channels (120 rows), noise channels (120 rows), neck electromyography (8 rows), 3-axis accelerometer channels that were built-in each LiveAmp 64 amplifier (12 rows), and Cometas IMU channels. The name, data type, and units of each data are found in the _channels.tsv file. The “_electrodes.tsv” file shows the digitized electrode location of the scalp channels in millimeters. The “_events.tsv” and “_events.json” contain the synchronization pulse events (“M 1”) and the hit events from the participant, human player, and ball machine. The timing of “M 1” events from the EEG line up with the rising edge of the square wave pulse from the IMU channel labeled “SyncPulse(uV)”. The timings of the hit events line up with peaks in the acceleration of the inertial measurement units.
More information on events can be found in the EEG structure from the “.set” and “.fdt” files in EEG.event. The “EEG.event.condlabel” field specifies the trial type (cooperative, competitive, stationary_hit, stationary_serve, moving_hit, moving_serve, and standing_baseline). Each “cometas_checked” type of event corresponds to the hit event (timing comes from peaks in the IMU acceleration) that was double-checked with the video data. Other event fields include performance (1=in bounds, 2=error/out of bounds, 3=error/in the net, 4=other) and bounces (2Bounces_Human = human player serve and participant return of serve, 1Bounce_Human = rally hit from the human player and participant, Serve_Human = participant serve, 2Bounces_BM = hits with the ball machine that simulate a return of serve with two bounces, and 1Bounce_BM = hits with the ball machine that simulate a rally hit with one bounce). Segmented datasets were concatenated together from each trial type, and the “event.video” field specifies which GoPro video was associated with that trial. The GoProFrame event field corresponds to the hit's timestamp from the video data in the Adobe project file.
The “etc.” field in the EEG structure contains the weight and sphering matrices that resulted from adaptive mixture independent component analysis (AMICA) [6]. The “etc.” field also contains dipole modeling information that was estimated with DIPFIT3.3 in “EEG.etc.dipfit”. These dipoles were fitted with subject-specific head models, and the coorindates of the dipoles were transformed to MNI space. The components retained in analyses of the brain data (selected with ICLabel, residual variance, and manual inspection) are specified in “EEG.etc.KeepComponents” [7], [8].
Within the “anat” subfolder of the subject's processed data are de-identified T1 anatomical head images. The “_T1w.nii” files were de-identified with FieldTrip functions to remove the participant's face [9]. These files were used to create subject-specific head models for dipole source localization.
Within the “video” subfolder of the subject's processed data are videos from all the trials. The timing of hit events were manually labeled in Adobe Premiere Pro software. The Adobe project file can be loaded with Premiere Pro software and the relative timing between hit events may be exported. Chapter markers indicate the time when the participant hit the ball. Flash Cue points mark when the human player hit the ball. Comment markers labeled the time of the first hits (i.e., serves) in the human player trials.
3. Experimental Design, Materials and Methods
3.1. Participants
Thirty-seven healthy, cognitively-intact participants were recruited to participate in our study (ages 23.5 +/- 6.7 years, mean +/- SD; 13 females). We only present here the data from twenty-five participants who yielded more than five brain source-localized components according to ICLabel and manual inspection. All participants self-identified as right-hand dominant, had normal or corrected-to-normal vision, and were free from any musculoskeletal or neurological injuries. Participants had a wide range of table tennis and racquet sport experience. The University of Florida Institutional Review Board approved our protocol, and all participants gave written informed consent.
3.2. Materials and equipment
The experiment took place in an indoor lab on the University of Florida campus. The table was a Joola, 15 mm thick. We provided participants with the same wooden paddles, which we put an inertial measurement unit on the bottom of the handle and secured with self-adherent tape. The ball machine was a Robo-Pong 2040+ Ping Pong Robot (Newgy Industries).
3.3. Dual-layer EEG system
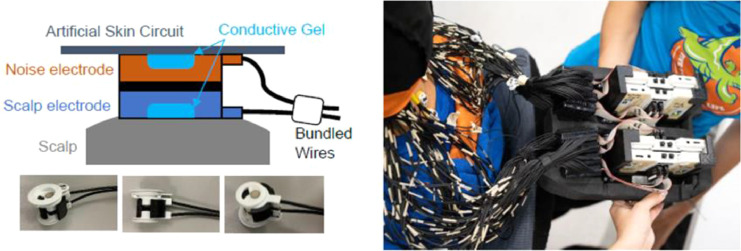
The EEG data was recorded from a custom-made dual-layer EEG system [10] that consisted of 120 scalp electrodes and 120 noise electrodes (BrainVision ActiCAP snap sensors). Scalp electrodes were mechanically joined and electrically isolated from inverted noise electrodes (Fig. 2). Wires from individual scalp and noise pairs of electrodes were secured using tape so that both cables experienced similar motion artifacts. Conductive fabric (EeonTex elastic piezoresistive fabric, LTT-PI-100) stretched over the noise layer of electrodes and acted as an artificial skin circuit. We re-purposed eight of the original 128 scalp electrodes to measure neck electromyography from the left/right and upper/lower sternocleidomastoid and trapezius muscles. The channels we repurposed were at the back of the head (TP9, P9, PO9, O9, O10, PO10, P10, and TP10). Four BrainVision LiveAmp 64 amplifiers data-logged and recorded data at 500 Hz. Each LiveAmp recorded data from one hemisphere in one layer of electrodes. The online reference (CPz) and online ground (Fpz) were kept separate for the scalp and noise layers to keep them electrically isolated. BrainVision provided us with cables to share the ground and reference between hemispheres from each layer. We shortened the stock BrainVision ribbon cables to 14 cm and housed the amplifiers in 3D-printed cases inside a small backpack (35 × 25 × 11 cm) (Fig. 2). We adjusted the straps of the backpack so that it rested on the upper back of each participant. In total, the dual-layer EEG system weighed 2.7 kg (6 lbs).
Fig. 2.
Dual-Layer EEG and backpack to hold the BrainVision LiveAmp 64 amplifiers. Figure adopted from a previous article [2].
3.4. Inertial measurement units and video
Inertial measurement units (Cometas WaveTrack Inertial System, 2000 Hz) and video (GoPro Hero 7, range of 30–240 frames per second) were used to record timing of hit events. We placed the inertial measurement units (IMUs) on the handles of the two wooden paddles and the ball machine for all participants. We also placed IMUs on a subset of the participant's foreheads, lower back underneath the backpack, inside the backpack, underneath both sides of the table, and on the net to measure peaks in acceleration for timing of events. We placed the IMU on the participant's forehead since head motion is thought to be a common source of noise in EEG data [11,12]. The IMUs on the participant's lower back and inside the backpack allowed us to check the time synchronization between the EEG and IMUs since we could compare the acceleration to the EEG amplifier's built-in accelerometer. Each IMU sensor's memory logged the IMU data and was offloaded after the experiment. The video data was used in post processing to filter out any mislabeled events and to derive other events of interest.
3.5. Data Synchronization Approach
For the processed data and the data in the “Merged” folder inside the sourcedata folder, we synchronized the data that came from different recording systems. The BrainVision EEG system and Cometas IMU system were synchronized with pulses from an Arduino timer module every five seconds. An analog pulse was sent to a Cometas EMG sensor at the same time as a TTL pulse was sent to the BrainVision EEG system [13]. The Cometas EMG sensor was a purpose-built sensor for synchronizing the Cometas system to external devices. The GoPro video data was not synchronized to the other two systems in real time. Rather, we manually tagged timing of hit events in Adobe Premiere Pro software using markers. We exported the hit timing relative to the first hit event and then aligned these GoPro hits to the resultant acceleration of the paddle IMU data using a cross-correlation. Any hit events from the IMUs that were outside of the 200 ms from the video markers were filtered out as mislabeled events (e.g., if the participant accidentally hit the table with their paddle). The video data also allowed us to mark other parameters of the event, such as whether the ball was in-bounds or out-of-bounds.
3.6. Participant prep
After consenting, we prepared the participants for the experiment. We measured the distance between preauricular points and between the nasion and inion to place the Cz electrode according to the 10–20 system. We gelled the scalp layer of electrodes with Abralyt gel and ensured that impedance values were below 20 kOhms. We put adhesive washers on the repurposed neck electromyography electrodes, prepared the skin with alcohol and light abrasion, and gelled the neck electrodes like the scalp. Next, we digitized the electrode locations with itSeez3D software and a Structure Sensor (Occipital Inc.) attached to an iPad. Then, we put a small amount of SuperVisc gel on the tops of each of the noise electrodes. Two people stretched the conductive fabric over the noise layer of electrodes, taking care not to smear the gel and bridge the electrodes. The noise layer of electrodes were also kept below 20 kOhms. We put the amplifiers in the backpack, connected EEG electrodes to the amplifiers, and adjusted the backpack so it rested on the participant's upper back. We also used a chest strap to better distribute the backpack's weight.
3.7. Experimental protocol
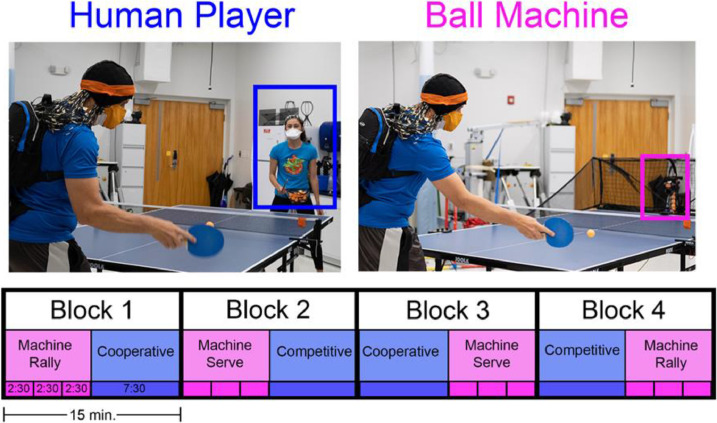
Participants played table tennis with a human player and a ball machine (Fig. 3). We aimed to collect around 70 min of total data, excluding breaks, which included 10 min of standing baseline and 60 min of table tennis play. We separated trials into four 15-minute blocks of play. In a single block, participants played a continuous 7.5 min with a human player (either cooperatively or competitively) and also played three back-to-back 2.5 min trials with a ball machine.
Fig. 3.
Organization of trials with a human player and ball machine. Figure adopted from a previous article [3].
For the ball machine trials, we used a feed rate of approximately one ball every two seconds (0.5 Hz) on the slowest speed setting. The ball machine trials alternated between stationary and moving conditions. In the stationary condition, the rate of oscillation was set to 0, the ball's trajectory was predictable (always landing in the middle of the table), and the participant did not have to move their feet to intercept the ball. In the moving condition, we put the rate of oscillation on the machine's slowest setting, and the ball machine fed balls towards all directions on the table. The mechanism for feed rate and oscillation were decoupled, so the feed rate stayed at a constant 0.5 Hz while the landing positions varied in the moving condition. Half of the ball machine trials replicated a rally hit (1 bounce) and the other half replicated a serve (2 bounces). The angle and speed of the feed were adjusted so that the ball landed halfway between the net and the end of the table that the participant was standing on. The order of trials across and within blocks was randomized.
For the human player trials, the researcher hitting with participants was experienced enough to scale their play to match the level of each participant. In the cooperative trials, participants were instructed to keep as many balls in play with the human player as possible. In the competitive trials, participants were instructed to try winning a 21-point game (switching serves every 5 points) against the human player. In all of the trials, we encouraged participants to stay as relaxed as possible and avoid clenching their jaws. We did not give participants specific targets to hit nor did we instruct them to hit any particular type of shot. Breaks were given between blocks and between trials, as needed.
On a different day and time, participants came in to get a T1 anatomical MR head scan. All metal was removed from the participant, and they were scanned with a 3T Philips Ingenia Scanner (32-channel head coil), magnetization-prepared rapid-acquisition gradient-echo (MP-RAGE) sequence. Eight of the twenty-five participants were scanned with 7.00 ms repetition time (TR), 3.17 ms echo time (TE), 8-degree flip angle, 240 mm x 240 mm x 176 mm field of view, and 1 mm3 recon voxel size. The remaining participants were scanned with 11.13 ms repetition time (TR), 5.10 ms echo time (TE), 8-degree flip angle, 256 mm x 240 mm x 179.9 mm field of view, and 0.67 × 0.67 × 0.70 mm3 recon voxel size.
We followed up the study with an email survey to gauge the participant's table tennis and racquet sport experience.
3.8. Data processing
For the processed data, we used custom MATLAB (R2020A), EEGLAB (v2021.0) [5], and SPM/CAT12 (CAT12.8) functions. The data were 1 Hz high-pass filtered to remove drift. The individual datasets from each LiveAmp 64 amplifier and from the Cometas IMUs were merged using the sync pulses from the Arduino. We marked hit events when the first derivative of the resultant acceleration of the IMUs exceeded 0.75 gravity. We visually inspected and manually separated trials into individual EEG datasets by looking for continuous blocks of hit events. Then, we used Cleanline to remove 60 Hz line noise [14]. We downsampled each dataset to 250 Hz, rejected bad channels (any channel more than three standard deviations in voltage from the median of the other channels), averaged re-referenced the data with full rank, and interpolated the rejected channels with spherical interpolation. iCanClean removed motion and muscle artifacts from the scalp data [15]. We used a two-second sliding window, an r2 threshold of 0.85 for the scalp x noise iCanClean, and an r2 threshold of 0.40 for the scalp × neck EMG iCanClean. Then, we rejected time windows using the clean_artifacts function with a standard deviation threshold of 30 and a window criterion of 0.3.
We ran adaptive mixture independent component analysis (AMICA) on just the scalp data to separate it into maximally independent sources of activity [6]. We first used principal component analysis (PCA) to ensure rank by reducing the principal components by the maximum number of rejected/interpolated scalp channels across trials for each participant. Then, we applied the weight matrix to the data before time window rejection.
Subject-specific, custom head models were created with the FieldTrip-SimBio pipeline (finite element method) [9]. The participant's T1 anatomical head images were segmented into five tissue layers using CAT12.8 (tissue conductivity values: skin 430, skull 10, cerebrospinal fluid 1790, gray matter 330, and white matter 140 mS/m) [16]. Digitized electrode locations were found by manually selecting electrode locations on the 3D head scan .obj file with EEGLAB get_chanlocs function. We specified an inward shift of 10 mm to account for the dual-layer height of electrodes (default for one layer is 5 mm). We aligned the head model from the segmented T1 scan with the electrode locations using fiducials (nasion and left/right preauricular points). We meshed the tissues into hexahedrons, and computed the source model, transfer matrix, and leadfield matrix. We source-localized and fit each independent component to an equivalent dipole model with DIPFIT3.3. The dipole locations were converted to MNI space.
Limitations
For someone pursuing a similar table tennis experiment, I recommend a few improvements to the methods and approach. First, I would use motion capture to track the kinematics of the participant's body and arm. The information on body and arm kinematics would give timing information on phases of the stroke and quality or type of hit. Secondly, I recommend recording electromyography from the participant's arm to record movement onset for a table tennis hit. The electroencephalography could then be response-locked to movement onset. Lastly, I recommend using Lab Streaming Layer to record all of these multi-modal data streams. Lab Streaming Layer simplifies the process for synchronizing multi-modal data streams – handling data merging, and providing access to view the data streams in near real-time.
Ethics Statement
All participants provided written informed consent before participating, and the University of Florida Institutional Review Board approved our protocol (IRB201801727). The research was carried out in accordance with the Declaration of Helsinki.
CRediT authorship contribution statement
Amanda Studnicki: Conceptualization, Methodology, Software, Validation, Formal analysis, Data curation, Writing – original draft, Writing – review & editing. Daniel P. Ferris: Conceptualization, Writing – review & editing, Supervision, Funding acquisition.
Acknowledgments
We would like to thank all the undergraduate research assistants who helped with data collections: Christina Collings, Corey Orlando, Sebastian Huerta, and Juan De La Espriella. This work was supported by the National Science Foundation (Division of Behavior and Cognitive Sciences, grant number BCS-1835317).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data Availability
Real World Table Tennis (Original data) (OpenNeuro)
References
- 1.Studnicki A., Ferris D.P. Real World Table Tennis. OpenNeuro. 2023 doi: 10.18112/openneuro.ds004505.v1.0.1. [DOI] [Google Scholar]
- 2.Studnicki A., Downey R.J., Ferris D.P. Characterizing and removing artifacts using dual-layer EEG during table tennis. Sensors. 2022;22:5867. doi: 10.3390/S22155867/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Studnicki A., Ferris D.P. Parieto-occipital electrocortical dynamics during real-world table tennis. eNeuro. 2023;10 doi: 10.1523/ENEURO.0463-22.2023. 0463-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorgolewski K.J., Auer T., Calhoun V.D., Craddock R.C., Das S., Duff E.P., Flandin G., Ghosh S.S., Glatard T., Halchenko Y.O., Handwerker D.A., Hanke M., Keator D., Li X., Michael Z., Maumet C., Nichols B.N., Nichols T.E., Pellman J., Poline J.-B., Rokem A., Schaefer G., Sochat V., Triplett W., Turner J.A., Varoquaux G., Poldrack R.A. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data. 2016;3 doi: 10.1038/sdata.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delorme A., Makeig S. EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods. 2004;134:9–21. doi: 10.1016/J.JNEUMETH.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Palmer J.A., Makeig S., Kreutz-Delgado K., Rao B.D. IEEE International Conference on Acoustics, Speech and Signal Processing - Proceedings. 2008. Newton method for the ica mixture model, ICASSP; pp. 1805–1808. [DOI] [Google Scholar]
- 7.Pion-Tonachini L., Kreutz-Delgado K., Makeig S. ICLabel: an automated electroencephalographic independent component classifier, dataset, and website. Neuroimage. 2019;198:181. doi: 10.1016/J.NEUROIMAGE.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klug M., Gramann K. Identifying key factors for improving ICA-based decomposition of EEG data in mobile and stationary experiments. Eur. J. Neurosci. 2020;00:1–15. doi: 10.1111/EJN.14992. [DOI] [PubMed] [Google Scholar]
- 9.Vorwerk J., Oostenveld R., Piastra M.C., Magyari L., Wolters C.H. The FieldTrip-SimBio pipeline for EEG forward solutions. Biomed. Eng. Online. 2018;17:1–17. doi: 10.1186/S12938-018-0463-Y/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordin A.D., Hairston W.D., Ferris D.P. Dual-electrode motion artifact cancellation for mobile electroencephalography. J. Neural Eng. 2018;15 doi: 10.1088/1741-2552/AAD7D7. [DOI] [PubMed] [Google Scholar]
- 11.Daly I., Billinger M., Scherer R., Müller-Putz G. On the automated removal of artifacts related to head movement from the EEG. IEEE Trans. Neural Syst. Rehabilit. Eng. 2013;21:427–434. doi: 10.1109/TNSRE.2013.2254724. [DOI] [PubMed] [Google Scholar]
- 12.O'Regan S., Faul S., Marnane W. Automatic detection of EEG artefacts arising from head movements using EEG and gyroscope signals. Med. Eng. Phys. 2013;35:867–874. doi: 10.1016/J.MEDENGPHY.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 13.Artoni F., Barsotti A., Guanziroli E., Micera S., Landi A., Molteni F. Effective synchronization of EEG and EMG for mobile brain/body imaging in clinical settings. Front. Hum. Neurosci. 2018;11 doi: 10.3389/fnhum.2017.00652. https://www.frontiersin.org/articles/10.3389/fnhum.2017.00652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mullen: CleanLine EEGLAB plugin - Google Scholar, (n.d.). https://scholar.google.com/scholar_lookup?title=CleanLine%20EEGLAB%20Plugin&publication_year=2012&author=T.%20Mullen (Accessed 21 September 2022).
- 15.R.J. Downey, D.P. Ferris, The iCanClean algorithm: how to remove artifacts using reference noise recordings, (2022). https://arxiv.org/abs/2201.11798v1.
- 16.Vorwerk J., Aydin Ü., Wolters C.H., Butson C.R. Influence of head tissue conductivity uncertainties on EEG dipole reconstruction. Front. Neurosci. 2019;13:531. doi: 10.3389/FNINS.2019.00531/. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Real World Table Tennis (Original data) (OpenNeuro)