Significance
RNA viruses often use RNA structures to hijack the required factors from the host cell to complete their life cycle. One example includes uncapped viral RNA interactions with the mRNA 5′ cap-binding protein eIF4E, an essential component of the mRNA translation through canonical mechanisms. Here, we performed RNA crystallography, eIF4E binding, and molecular docking studies using an RNA domain from the saguaro cactus virus and demonstrated that this uncapped RNA mimics the 5′ cap for interactions with the eIF4E. While several studies have shown how the 5′ cap interacts with eIF4E, our study represents crystallographic structures of an eIF4E-binding uncapped RNA, highlighting how the viral RNA structures exploit host factors by mimicking their binding partners to promote viral genome translation.
Keywords: RNA crystallography, crystallization chaperones, viral RNAs, 3′ cap-independent translation enhancers, RNA–eIF4E interactions
Abstract
The genomes of several plant viruses contain RNA structures at their 3′ ends called cap-independent translation enhancers (CITEs) that bind the host protein factors such as mRNA 5′ cap-binding protein eIF4E for promoting cap-independent genome translation. However, the structural basis of such 5′ cap-binding protein recognition by the uncapped RNA remains largely unknown. Here, we have determined the crystal structure of a 3′ CITE, panicum mosaic virus-like translation enhancer (PTE) from the saguaro cactus virus (SCV), using a Fab crystallization chaperone. The PTE RNA folds into a three-way junction architecture with a pseudoknot between the purine-rich R domain and pyrimidine-rich Y domain, which organizes the overall structure to protrude out a specific guanine nucleotide, G18, from the R domain that comprises a major interaction site for the eIF4E binding. The superimposable crystal structures of the wild-type, G18A, G18C, and G18U mutants suggest that the PTE scaffold is preorganized with the flipped-out G18 ready to dock into the eIF4E 5′ cap-binding pocket. The binding studies with wheat and human eIF4Es using gel electrophoresis and isothermal titration calorimetry, and molecular docking computation for the PTE–eIF4E complex demonstrated that the PTE structure essentially mimics the mRNA 5′ cap for eIF4E binding. Such 5′ cap mimicry by the uncapped and structured viral RNA highlights how viruses can exploit RNA structures to mimic the host protein-binding partners and bypass the canonical mechanisms for their genome translation, providing opportunities for a better understanding of virus-host interactions and non-canonical translation mechanisms found in many pathogenic RNA viruses.
The 5′ cap (m7GpppN, where N is any nucleotide) is an essential component of the mature eukaryotic mRNA, which promotes translation by recruiting the eukaryotic translation initiation factor 4E (eIF4E) to the mRNA during the translation initiation phase (1–4). However, genomes of many (+)-strand RNA viruses, which often directly serve as the mRNA for viral protein translation, lack this 5′ cap structure for initiating the genome translation through the canonical mechanisms. Instead of encoding the machinery for 5′ capping, these viruses seem to evolve to harbor structured RNA domains in their genomes that essentially bypass the requirement of the 5′ cap to interact with the host eIF4E, thus promoting the initiation of viral genome translation through cap-independent mechanisms (5–9). Nevertheless, the structural basis of how such a viral RNA domain mimics the mRNA 5′ cap for hijacking the host eIF4E remains unknown. Our study presents the crystal structure determination of an eIF4E-binding RNA domain from the saguaro cactus virus (SCV) genome and its interactions with human and wheat eIF4Es, revealing unique structural features that enable this RNA domain to mimic the 5′ cap for eIF4E binding during genome translation.
The cis-acting cap-independent translation enhancers (CITEs), including eIF4E-binding RNA domains, typically reside within the untranslated regions (UTRs) of viral genomes as a part of internal ribosome entry sites (IRESs) (10–12) at the 5′ or as the 3′ CITEs at the 3′ UTRs (13–16). The IRESs are frequently found in the 5′ UTRs of zoonotic virus genomes that recruit the 40S ribosome directly or through interactions with eukaryotic initiation factors (eIFs) such as eIF4E and eIF4G (10–12). More recently, some plant virus genomes have been shown to harbor CITEs at the 3′ UTRs, which essentially play similar roles as IRESs in recruiting eIFs or the ribosomal subunits and interact with the 5′ end to circularize the viral genome for priming the translation initiation (13–16). The RNA and ribonucleoprotein (RNP) complex structures within the 3′ CITEs generally contact with RNA domains of the 5′ UTR through long-range RNA–RNA and RNA–protein interactions to deliver the 3′ CITE-bound translation factors or the ribosomal subunits to the 5′ end, allowing the assembly of a translationally competent initiation complex at or near the start codon (17–19).
The 3′ CITEs, so far reported only for some plant viruses, have been categorized into seven classes based on the predicted secondary structures of their RNA domains and the types of the host translation initiation factors they bind (16). For example, a translational enhancer domain (TED) of satellite tobacco necrosis virus (STNV) binds the eIF4F complex (20), a four-way junction structure of barley yellow dwarf virus (BYDV)-like 3′ CITE (BTE) interacts with eIF4G subunit of eIF4F (21), and a T-shaped structure (TSS) of the turnip crinkle virus (TCV) directly binds the 60S ribosomal subunit (22). Similarly, among those seven classes, Y-shaped domains of panicum mosaic virus (PMV)-like translation element (PTE) found in the members of Panicoviruses, Carmoviruses, Aureviruses, and Umbraviruses, including SCV, are known to bind eIF4E with an affinity similar to that of 5′ cap-eIF4E binding (23, 24). While previous biochemical probing, phylogenetic analysis, and translation reporter assays have illustrated a strong correlation between the binding of initiation factors or the ribosomal subunits with 3′ CITE domains and the translation of the viral genomes (22–24), the lack of high-resolution three-dimensional structures of 3′ CITEs has constrained our understanding of how these viruses exploit RNA-based strategies to accomplish genome translation while competing host mRNAs for the same factors. Except for a few computational modeling studies of PMV and PEMV2 (Pea enation mosaic virus RNA2) PTEs (24), the NMR and SAXS-derived model of TCV TSS is the only three-dimensional structure of a viral 3′ CITE reported to date (25). Moreover, whereas RNA structures in the 5′ capped mRNAs have been shown to influence the cap-binding interactions with eIF4E compared to isolated cap analogs (26, 27), our work represents a high-resolution structural study of eIF4E binding with an uncapped RNA domain.
Previous biochemical probing and computational modeling studies have uncovered unique structural features of viral PTEs and their interactions with the eIF4E (24). The proposed secondary structure of a typical PTE consists of a three-way junction with a G-rich bulge (called G-domain) within the first helix, which interacts with the C-rich region (called C-domain) within the second and third helix junction (23, 24). Those studies also revealed that a specific guanine within the G-domain is hypersensitive to the SHAPE (selective 2′ hydroxyl acylation analyzed by primer extension) reagents but becomes protected in the presence of wheat eIF4E, suggesting its critical role in binding eIF4E protein (24). The loss of this hypersensitivity and eIF4E binding for C-domain mutants were also consistent with a pseudoknot formation between the G- and C-domain and the formation of a well-organized RNA scaffold for the protein binding. Further studies generated computational models for PEMV2 and PMV PTEs in which the hypersensitive guanine was flipped out from the rest of the RNA, allowing its docking with the eIF4E in an analogous way as the 5′ cap (24). The docking models predicted that two tryptophans of the wheat eIF4E binding pocket sandwiched the flipped guanine in the same way as the 5′ cap, and the peripheral residues clamped around the pseudoknot scaffold (24). These studies provided a model for an uncapped RNA structure that mimics the 5′ cap to bind eIF4E, but this model has not been demonstrated experimentally. Moreover, while the presence of hypersensitive guanine in the G-domain has been shown in other PTEs, such as TPAV and SCV, the C-rich domain seems variable, as observed in the SCV PTE, which contains a pyrimidine-rich sequence (18, 28). Although it is plausible to assume that these PTEs adopt a homologous structure mimicking the mRNA 5′ cap for binding the host eIF4E, this hypothesis remains to be tested due to the lack of any PTE three-dimensional structures.
Here, we used a Fab BL3-6 as a chaperone to determine the crystal structure of the PTE from SCV, a member of the carmovirus genus. The crystal structure adopts a λ-shaped three-way junction structure in which the first two helices are stacked coaxially, and the third helix protrudes from this coaxial helical axis. The long-range RNA interactions between the purine-rich and pyrimidine-rich domains form a pseudoknot architecture, consistent with previous in-solution biochemical results and phylogenetic analysis, including the PMV and PEMV2 PTEs (24). A specific guanine from the purine-rich domain, hypermodifiable by the SHAPE reagents (24), flips out from a pre-organized scaffold to comprise a major binding site for the eIF4E. The RNA–protein binding studies with the wild-type and mutant SCV PTEs and side-by-side comparisons with a 5′ cap analog (m7GTP) and an in vitro synthesized 5′ capped RNA oligo using gel electrophoresis and isothermal titration calorimetry (ITC) assays for both human and wheat eIF4Es suggest that this solvent exposed guanine within the SCV PTE essentially mimics the binding site of the mRNA 5′ cap. Consistent with these experimental results, the molecular docking of SCV PTE with both human and wheat eIF4E yielded models in which this flipped-out guanine fits well between two tryptophans of the eIF4E binding pocket in a similar fashion as the canonical 5′ cap, with the pseudoknot RNA scaffold complementing a claw-like shape of the eIF4E binding pocket to stabilize the PTE–eIF4E complex further. As observed in our in vitro translation reporter assay, the trans inhibitory effect of the SCV PTE on eIF4E-requiring canonical translation, but no significant effect on an IRES-dependent translation not involving eIF4E, further supports the competitive nature of this viral PTE and host mRNA 5′ cap for eIF4E binding. Overall, our wild-type SCV PTE and its G18A, G18C, and G18U mutant crystal structures, combined with phylogenetic, biochemical, and docking analyses, suggest that long-range interactions to form a pseudoknot between the purine-rich and pyrimidine-rich domains are perhaps the most conserved structural features among the viral PTEs. Our studies with SCV PTE presented here represent a starting point to pursue further structural studies with various PTEs and their eIF4E complexes and to search for potential PTE-like RNA structures in several other viral and cellular IRESs, which will help advance our understanding of different RNA-based cap-independent translation mechanisms observed in RNA viruses, providing opportunities for developing RNA-targeted antivirals.
Results
SCV PTE Contains R and Y Domains.
The proposed RNA secondary structure of the SCV 3′ UTR based on the SHAPE analysis (Fig. 1A) consists of two distinct modular regions (18). The domain near the extreme 3′ end, composed of three stem-loops and a potential pseudoknot, is predicted to adopt a T-shaped structure (TSS), which may bind to the 60S ribosome as reported previously for other carmoviruses such as TCV (22). The region preceding the TSS assumes a Y-shaped secondary structure, termed PTE, similar to those observed in several other carmoviruses and umbraviruses (23). All known PTEs bind eIF4E with high affinity, and this protein–RNA interaction correlates with the genome translation, as shown previously for some selected viruses in the reporter gene assays (16, 24, 28). The secondary structures of most PTEs (SI Appendix, Fig. S1) consist of a three-way junction (3WJ) with an asymmetric G-rich bulge within the first stem (P1) and a C-rich sequence within the second stem (P2) and third stem (P3) intersection (21, 23, 24). These G- and C-rich domains are proposed to form an RNA pseudoknot through the base-pairing interactions (23, 24). However, despite being functionally analogous in binding eIF4E, the SCV PTE harbors a U-rich unpaired sequence between the second and third stems while maintaining a similar G-rich bulge as in other PTEs (Fig. 1B). As non-canonical G•U pairs are frequently observed in RNA structures (29, 30), it is reasonable to assume that the SCV PTE adopts a similar fold as other PTEs, perhaps still forming a pseudoknot through such non-canonical interactions. Given this diversity of sequences among these known PTEs, we have proposed a generalized designation for the previous G-domain and C-domain as R domain (purine-rich, R = G and A nucleotides) and Y domain (pyrimidine-rich, Y = C and U nucleotides), respectively, which describe secondary structures and nucleotide identities of all known viral PTEs more inclusively. Nevertheless, how these features define the PTE three-dimensional structures and promote the interactions of a PTE with the eIF4E remains largely undetermined without the high-resolution structures of these RNAs and their eIF4E complexes.
Fig. 1.
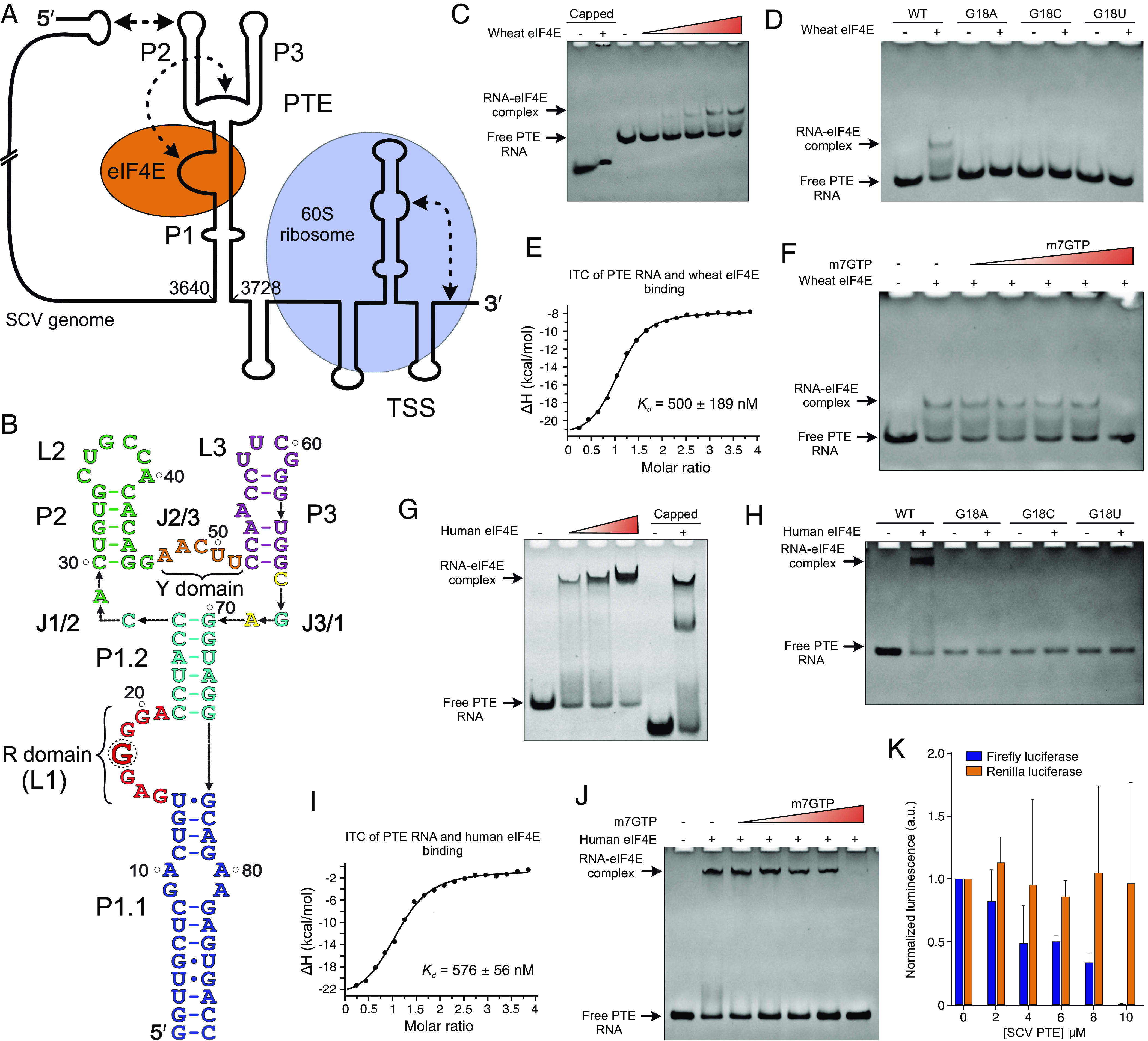
SCV genome organization and the PTE–eIF4E interactions. (A) Schematic representation of the SCV (+)-sense RNA genome with the predicted RNA structures at the 5′ and 3′ ends. Proposed binding sites for the 60S ribosome (TSS) and eIF4E (PTE) are indicated. The dotted arrows depict long-range RNA–RNA interactions. (B) The 89-nt SCV PTE construct (wild-type) was used in this study. Key features of the PTE elements are labeled, and a dotted circle indicates the SHAPE-hypermodifiable G18 within the R domain. The native polyacrylamide gel electrophoresis (nPAGE) assays showing the binding of the (C) wild-type and (D) G18 mutant PTEs with wheat eIF4E. (E) The ITC measurements for the PTE binding with wheat eIF4E. (F) The nPAGE of wheat eIF4E-PTE interactions in the presence of the m7GTP. Similar experiments were performed with human eIF4E. The nPAGE showing the binding of the (G) wild-type and (H) G18 mutant PTEs with human eIF4E. (I) The ITC measurements for the PTE binding with human eIF4E and (J) the nPAGE of the eIF4E-PTE interactions in the presence of m7GTP. (K) The expression levels (average ± SD, n = 6) of firefly (cap-dependent) and renilla (HCV IRES-dependent) luciferases in the presence of the PTE. The ability of the SCV PTE RNA to bind wheat and human eIF4Es effectively, the obvious displacement of the PTE RNA from the eIF4E-RNA complex by the m7GTP, and inhibition of the eIF4E-requiring cap-dependent translation in vitro strongly suggest similar binding interactions of the PTE and mRNA 5′ cap.
SCV PTE Interacts with Wheat eIF4E with Nanomolar Affinity.
As SCV is a plant virus not known to infect animals, we first sought to measure the binding interactions of its PTE RNA with the wheat eIF4E. Although previous studies have provided a semi-quantitative analysis of the PTE binding with eIF4E and shown that uncapped PTE RNAs could also interact effectively with this 5′ cap-binding protein (28), no study so far has reported thorough and quantitative measurements of such PTE–eIF4E interactions. To investigate the binding interactions of the SCV PTE with wheat eIF4E, we purified the recombinant protein with residues 39 to 215 as described previously (23) (see SI Appendix, Table S1 for the sequences of all constructs used in this study) and performed native electrophoretic mobility shift assays (EMSAs) and ITC measurements. The SCV PTE construct for these studies included the 89 nts (nucleotides) from the SCV genome position 3640 to 3728 (18) with the P1 helix closing-pair mutated to G-C (Fig. 1B), which would help stabilize the helix and enhance the in vitro transcription. We referred to this construct as the wild-type (WT) construct. We also used a 5′ cap analog m7GTP and a 31-nt RNA oligomer with an in vitro installed authentic 5′ cap (called 5′ capped RNA hereafter) as the positive controls for our binding studies. As observed in our native gel electrophoresis (Fig. 1C), the disappearance of the RNA bands and the appearance of slower migrating bands in the presence of wheat eIF4E, similar to that observed for the 5′ capped RNA, suggested a stable PTE–eIF4E complex formation, consistent with previous studies for SCV, PMV, and PEMV2 PTEs (28). Moreover, the mutations to the SHAPE hypersensitive guanine (G18) reduced the PTE binding to wheat eIF4E (Fig. 1D), implying that G18 comprises the major interaction site for the PTE–eIF4E binding, which agrees well with previous protection assays (24).
Next, we performed ITC measurements to measure the SCV PTE and eIF4E binding affinity quantitatively. The N-terminally his-tagged wheat eIF4E bound to our positive controls the m7GTP and 5′ capped RNA construct with similar affinity (apparent dissociation constant, Kd = 1.8 ± 0.8 µM, and Kd = 900 ± 100 nM, respectively; see Table 1 and SI Appendix, Fig. S2), consistent with previous binding analysis for similar analogs using NMR, fluorescence quenching and stop-flow approaches (31). The SCV PTE bound more tightly (Kd = 500 ± 189 nM) than the m7GTP or 5′ capped RNA (Fig. 1E and SI Appendix, Fig. S2 and Table S2), which agrees with the inhibitory effect of the SCV PTE on cap-dependent translation observed previously (28). Interestingly, excess m7GTP prevented the SCV PTE–eIF4E complex formation in our native EMSA, indicating that the PTE RNA likely competes for the same binding site within the eIF4E as the 5′ cap (Fig. 1F). Consistent with these observations, the SCV PTE G18A, G18U, and G18C mutants bound with ~29, ~24, and ~22 times lower affinity (Kd = 14.5 ± 2.5 µM, 11.8 ± 3.2 µM, and 10.8 ± 1.5 µM, respectively) compared to the WT PTE (Table 1 and SI Appendix, Fig. S2 and Table S2). However, the measurable binding affinities of these mutants also suggested additional contacts between the PTE RNA and eIF4E beyond major interactions with the G18, which explains a tighter binding of the PTE compared to the m7GTP and 5′ capped-RNA construct. Taken together, our EMSAs and ITC measurements strongly support site-specific interactions of the SCV PTE with wheat eIF4E analogous to the mRNA 5′ cap, perhaps using the same cap-binding pocket of the eIF4E as the major interaction site.
Table 1.
ITC measurements for the binding interactions of SCV PTE with wheat and human eIF4E
| RNA constructs | Wheat eIF4E | Human eIF4E | ||
|---|---|---|---|---|
| Kd (mean ± SD) | N (mean ± SD) | Kd (mean ± SD) | N (mean ± SD) | |
| m7GTP | 1.8 ± 0.8 µM | 1.8 ± 0.2 | 126 ± 5.0 nM | 1.5 ± 0.5 |
| 5′ capped RNA | 900 ± 100 nM | 1.0 ± 0.1 | 76 ± 3.0 nM | 1.2 ± 0.1 |
| SCV PTE G18 wild-type | 500 ± 189 nM | 1.0 ± 0.1 | 576 ± 56 nM | 1.0 ± 0.1 |
| SCV PTE G18A mutant | 14.5 ± 2.5 µM | 1.9 ± 0.7 | 6.2 ± 0.9 µM | 0.8 ± 0.3 |
| SCV PTE G18C mutant | 10.8 ± 1.5 µM | 2.0 ± 0.3 | N.B. | N.B. |
| SCV PTE G18U mutant | 11.8 ± 3.2 µM | 2.3 ± 0.8 | N.B. | N.B. |
The Kd represents mean ± SD values from three independent measurements (n = 3). N.B. depicts no detectable binding within the tested conditions. Details of data collection and statistics are shown in SI Appendix, Figs. S2, S7, and S8 and Tables S2 and S3.
Human eIF4E Binds the SCV PTE with a Similar Affinity As m7GTP and 5′ Capped-RNA.
The overall three-dimensional structure of eIF4E protein is highly conserved across plants and animals (RMSD for the all-atoms superposition of human and wheat eIF4E crystal structures = 0.677 Å; see SI Appendix, Fig. S3 for details), including the residues integrally involved in binding the 5′ cap. However, several residues highly conserved in plant eIF4Es differ from the human and yeast eIF4Es, with the most notable C113 and C151 residues close to the cap-binding site forming the disulfide bond in wheat eIF4E (31). Such plant-specific residues are accessible on the protein surface, possibly accounting for the differential recognition of mammalian and plant eIF4Es observed in previous reporter gene translation assays (28). While relative affinities of eIF4E and PTE interactions might be specific to the host and infecting virus species, both wheat and human eIF4Es have previously been shown to interact with the same plant virus PTE in the semi-quantitative gel electrophoresis assays (28).
To compare the binding interactions of the SCV PTE between wheat and human eIF4Es, we also expressed and purified a recombinant human eIF4E. The disappearance of the RNA bands in an eIF4E dose-dependent manner in our native EMSAs indicated the PTE and eIF4E binding interactions (SI Appendix, Fig. S4). However, unlike wheat eIF4E, the human eIF4E-PTE complexes did not show defined bands in the gel. Therefore, we expressed and purified GB1-eIF4E fusion protein as the GB1 domain has been used to solubilize and stabilize other proteins (32). Excitingly, we observed defined PTE–eIF4E complex bands with this fusion protein, similar to that observed for the control 5′ capped-RNA and consistent with the analogous results for the wheat eIF4E (Fig. 1G). While multiple factors, such as the protein’s isoelectric point, thermal stability, and kinetics of RNA–protein complex formation, can contribute to the band-smearing in gel electrophoresis, higher thermal stability (Tm ~ 92 °C) and slightly slower dissociation (koff = 2.1 × 10−3 ± 1.34 × 10−5 s−1) for the GB1-eIF4E compared to that for eIF4E only (Tm ~ 82 °C and koff = 1.7 × 10−3 ± 1.05 × 10−5 s−1) are consistent with the band-smearing for the eIF4E-RNA complex (see SI Appendix, Fig. S5 for the details of kinetics and SI Appendix, Fig. S6 for thermal melting measurements). Notably, the multiple bands for eIF4E complexes with the 5′ capped RNA (Fig. 1G) perhaps reflect the conformational heterogeneity associated with the RNA and GB1-eIF4E fusion (see SI Appendix, Fig. S4 for the details). Expectedly, the human eIF4E with the G18A, G18C, and G18U mutants also showed a similar binding trend observed for the wheat eIF4E (Fig. 1H), suggesting that the PTE organizes an RNA structure with the G18 as a major binding site for wheat and human eIF4Es. For human eIF4E, the G18C and G18U mutations were more disruptive than the G18A mutation, indicating that purines at this position may have more favorable hydrogen bonding and stacking interactions within the cap-binding site. To further support this hypothesis, we also performed ITC assays with the human eIF4E without the GB1 fusion. The human eIF4E bound to the m7GTP (Kd = 126 ± 5.0 nM) and 5′ capped-RNA (Kd = 76 ± 3.0 nM), consistent with that for the wheat eIF4E as discussed above (Fig. 1I and SI Appendix, Fig. S7). Moreover, the wild-type PTE and G18A bound to human eIF4E with Kd = 576 ± 56 nM, 6.2 ± 0.9 µM, respectively, while G18C and G18U mutants showed no detectable binding in our ITC measurements (Table 1 and SI Appendix, Fig. S7 and Table S3). Notably, the GB1-eIF4E did not significantly affect the binding with the PTE RNA (Kds = 900 ± 236 nM and 576 ± 56 nM with and without the GB1, SI Appendix, Fig. S8). Like the wheat eIF4E-PTE binding, excess m7GTP abrogated the human eIF4E binding to the PTE, supporting an analogous mode of the SCV PTE recognition by wheat and human eIF4Es (Fig. 1J). Although the human eIF4E binding with SCV PTE followed a similar overall trend as wheat eIF4E, the human eIF4E, in general, seems to bind to the m7GTP, 5′ capped-RNA with slightly higher and the SCV PTE with similar affinities compared to the wheat eIF4E. Due to subtle structural differences, other minor interactions from the PTE structure beyond the G18 may be important to recognize the human and wheat eIF4Es differently. Additionally, these differences may reflect the protein stability and buffer conditions, as wheat eIF4E tends to precipitate quickly in the buffer used for human eIF4E. Although we used freshly purified proteins for our experiments, the instability and precipitation of recombinant eIF4Es have been well documented (31, 33).
SCV PTE Likely Competes with mRNA 5′ Cap for eIF4E Binding in Host Cells.
The ability of the SCV PTE to interact with both wheat and human eIF4Es in our binding studies is consistent with previous biochemical analyses (24, 28, 31) and agrees well with the eIF4E’s 5′ cap-binding pocket being the major interaction site for the uncapped PTE, strongly supporting our hypothesis that the viral PTEs assume a common structural fold to mimic the mRNA 5′ cap that recruits the eIF4E for viral genome translation. To further understand the nature of the SCV PTE interactions with animal eIF4E during viral genome translation, we performed an in vitro translation assay in a mammalian cell lysate using a bicistronic reporter construct that encodes for the firefly and renilla luciferases. The T7 promotor controls the transcription of the bicistronic mRNA, and the canonical (cap-dependent) and HCV IRES (cap-independent) mechanisms direct the firefly and renilla luciferase translation, respectively (34). The expression level measurements for each protein using dual luciferase assays revealed that the presence of the SCV PTE in trans significantly decreases the firefly luciferase expression dose dependently. In contrast, the expression levels of the corresponding HCV IRES-controlled renilla luciferase remained mostly unaffected (Fig. 1K). These observations are consistent with the sequestering of eIF4E by the SCV PTE, inhibiting the eIF4E-requiring cap-dependent translation. However, such reduction in the eIF4E availability is less likely to influence the renilla luciferase expression as HCV IRES is known to recruit the intact 40S ribosome for translation initiation directly without requiring eIF4E (35). Whereas other translation initiation factors, such as eIF4G, are likely to influence the affinity of eIF4E-PTE interactions in cells, taking together with the inhibitory effects of m7GTP on the SCV PTE binding for both wheat and human eIF4Es (Fig. 1 F and J), these in vitro translation results support for a direct competition of the viral PTEs with the host mRNA 5′ cap for binding eIF4E.
SCV PTE Cocrystallizes with a Fab Chaperone.
To understand how the SCV PTE architecture promotes its interactions with eIF4E, we sought to determine the high-resolution structure of the SCV PTE by X-ray crystallography. As our attempts to crystallize the wild-type SCV PTE were unsuccessful, we turned our efforts to the Fab-assisted RNA crystallography (36–39) with Fab BL3-6 as a crystallization chaperone, which has proven helpful in determining the crystal structure of several other RNAs (39–44). To create the Fab binding site within the SCV PTE, we replaced the L2 loop sequence, 5′ CUGCCA 3′, with the Fab BL3-6 binding hairpin motif, 5′ GAAACAC 3′. Although the L2 loop has been proposed to engage in long-range interactions with the 5′ end of the SCV genome, mutational and biochemical probing studies have shown that eIF4E binding activity is independent of this highly solvent-exposed loop, indicating that this loop has minimal influence on the overall structure of the SCV PTE (18, 24). Consistent with those results, this mutant crystallization construct, designated PTE-BL3-6 (SI Appendix, Fig. S9), binds with the Fab BL3-6 (SI Appendix, Fig. S9) as expected for the RNAs grafted with this motif (41, 43). Moreover, this crystallization construct binds both human and wheat eIF4Es in the presence and absence of the Fab BL3-6, as illustrated by our EMSAs (SI Appendix, Fig. S10) and ITC measurements (SI Appendix, Figs. S2 and S7 and Tables S2 and S3), suggesting that the grafted motif and subsequent binding of the Fab had no significant effect on the overall structure of the SCV PTE.
Following the binding characterization of the crystallization construct with Fab and eIF4Es, we set up the crystallization screening for the PTE-BL3-6 RNA in a 1:1 complex with the Fab BL3-6. Out of 576 conditions screened using commercial kits, the complex crystallized in two conditions. After further optimization of these conditions, we obtained robust crystals within a week in 0.2 M LiSO4.H2O, 0.1 M Tris (pH 8.6), and 20% (w/v) polyethylene glycol at 22 °C that diffracted to 3.13 Å resolution. The analysis of crystallographic data (see SI Appendix, Table S4 for crystallographic statistics) suggested that the SCV PTE-BL3-6 complex crystallized in a lattice space group I 2 2 2 and contained a single Fab-RNA complex within the crystallographic asymmetric unit (a = 77.44 Å, b = 81.20 Å, c = 321.20 Å, α = 90°, β = 90°, and γ = 90°). The analogous screening trials for the same construct without Fab and the wild-type RNA did not produce any crystals, suggesting a substantial role of Fab BL3-6 in facilitating the SCV PTE crystallization.
The Crystal Structure of the SCV PTE Adopts a Three-Way Junction Fold.
To solve the crystal structure of the SCV PTE complex with Fab BL3-6, we used a previous crystal structure of the Fab BL3-6 (PDB code: 6B14) as a molecular replacement search model (38). The initial phasing by this process generated a robust electron density map, allowing us to model the RNA nucleotides unambiguously. The structure of the Fab-RNA complex reported here represents the model solved at 3.13 Å resolution through the iterative rounds of model building and refinement (see SI Appendix, Table S4 for the details of data collection and refinement statistics). Expectedly, while most of the crystal contacts, including the Fab-RNA binding interface, are mediated by the Fab (86.5%), we also observed RNA–RNA interactions within the crystal lattice through coaxial helical stacking of symmetry-related helices that account for 13.5% of the crystal contacts (SI Appendix, Fig. S11).
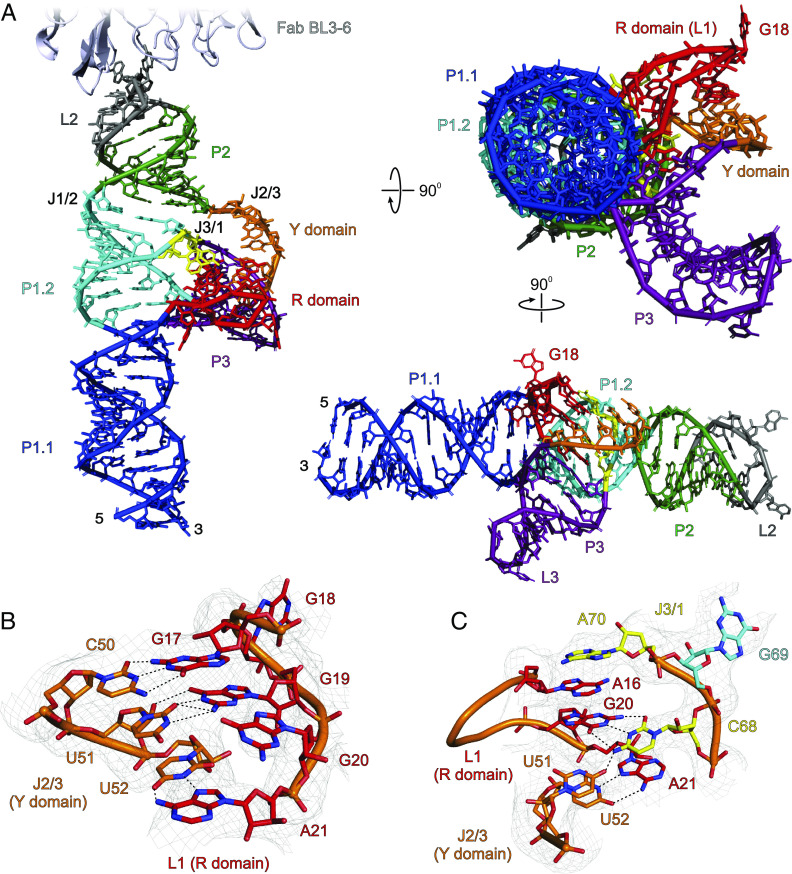
The observed crystal structure of SCV PTE adopts an A-type 3WJ fold (45) that consists of a P1 helix formed by base-pairing of the 5′ and 3′ ends, and two helices, P2 and P3, closed by loops L2 and L3, respectively (Fig. 2A and SI Appendix, Fig. S12). These three helices (P1, P2, and P3) organize into a λ-shaped architecture where the P1 represents the base of the λ and the P2 and P3 the bifurcated arms. Within this three-way junction architecture, the P1 and P2 helices are coaxially stacked, whereas the P3 protrudes from the P1–P2 helical axis, orienting toward the P1 helix. The entire P1 helix consists of two distinct helical regions designated as the sub-helices P1.1 and P1.2. While the P1.1 helix consists of two sets of non-canonical base pairs, U4•G87 and G5•U86, and G9•A82 and A10•A81, the L1 nucleotides between P1.1 and P1.2 with a G-rich sequence (G15 to A21, R domain) adopts a unique structure that interacts extensively with both P2-P3 (J2/3) and P3-P1 (J3/1) junction nucleotides through tertiary contacts to constitute a pseudoknot (Fig. 2C). Following the R domain, with no unpaired nucleotides within the junction, the coaxial stacking of the P1.1 with P1.2 forms almost a continuous A-form helix of the entire P1. The P2 helix and junction J1/2 also have no unpaired nucleotides. The loop L2 that closes the P2 helix constitutes the Fab BL3-6 epitope, which remains bound with the Fab in the crystal structure as expected. The junction J2/3 harbors five unpaired nucleotides (A48 to U52, Y domain) that heavily interact with the R domain nucleotides to stabilize the overall 3WJ architecture. Moving forward, the P3 helix with five canonical base pairs is closed by the L3 loop, which adopts a GNRA-type tetraloop structure. The J3/1 consists of three nucleotides, C68, G69, and A70, where C68 and A70 interact with L2 nucleotides, while the G69 base pairs with the C28 within junction J1/2. Because our crystallized construct with grafted Fab binding loop binds to eIF4E with similar affinity compared to the wild-type SCV PTE (SI Appendix, Figs. S2, S7, and S10), and key structural features such as the G-domain and three-way junction in the crystal structure do not involve any Fab interactions, the position, and structure of the Fab-binding loop are unlikely to influence the overall folding of the SCV PTE. Notably, although SCV PTE crystal structure appeared topologically similar to previously proposed models for PMV and PEMV2 PTEs (24), some key structural features, such as the relative orientation of the helices around the three-way junction and pseudoknot structure, seem pretty different. However, the unavailability of those models in public repositories hampered detailed comparisons with our crystal structures.
Fig. 2.
The crystal structure of SCV PTE. (A) The overall structure of the SCV PTE RNA in complex with a crystallization chaperone Fab BL3-6 solved at 3.13 Å resolution. For clarity, Fab is obscured in the rotated views of the PTE RNA (see SI Appendix, Fig. S12 for the complete structure of the Fab-RNA complex). (B) Specific interactions between the R (L1 nucleotides) and Y domain (J2/3 nucleotides). (C) The J3/1 nucleotides interact with the R and Y domains through hydrogen bonding and base-stacking. Gray mesh in (B) and (C) represents the composite simulated anneal-omit 2|Fo|-|Fc| electron density map at contour level 1σ and carve radius 2.5 Å. The dotted black lines depict the heteroatoms within the hydrogen bonding distances (≤3.0 Å). Figure panels and labels are colored analogously for facile comparison.
SCV PTE Crystal Structure Revealed Unique Features.
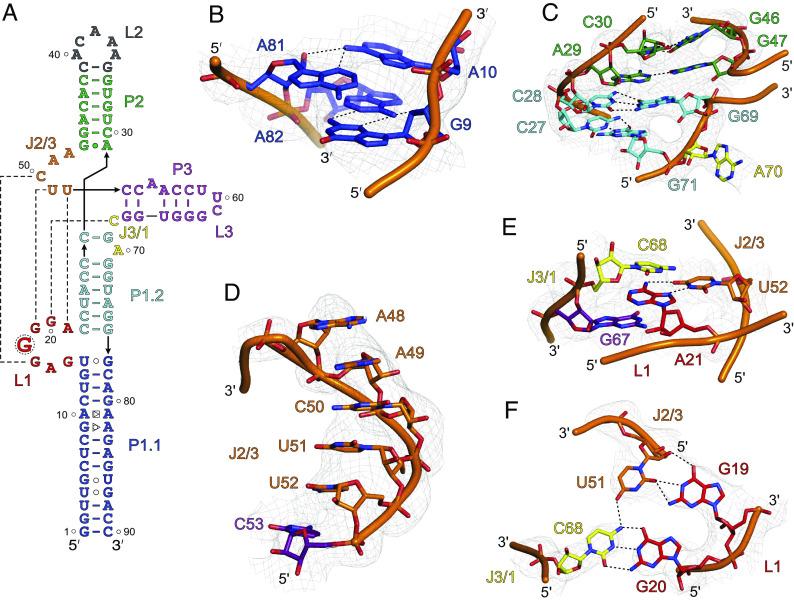
The crystal-derived secondary structure of SCV PTE (Fig. 3A) agrees with the previous biochemical data and a proposed model based on phylogenetic comparisons with analogous PTEs in terms of the paired helices, loops, and bulged regions (14). However, our crystal structure differs from the proposed model in several respects, particularly with non-canonical base pairs, the location of the three-way junction, and the nucleotides involved in the pseudoknot formation. First, unlike the previous prediction of a symmetric bulge, four nucleotides (G9, A10, A81, and A82) within the P1.1 helix form two non-canonical base pairs (G9•A82 and A10•A81). In these pairs, the G9 sugar-edge face interacts with the Hoogsteen face of A82, whereas A10 and A81 interact through the Hoogsteen and sugar-edge faces, respectively (Fig. 3B). Despite these non-canonical purine–purine base pairs, we observed no significant distortion or bending of the P1 helix from a standard A-form within this region (see SI Appendix, Fig. S13 for comparison). Second, within the J1/2, J2/3, and J3/1 junction nucleotides in the crystal structure are involved in complex secondary and tertiary interactions, which were predicted to be two (C28, A29), six (G47 to U52), and three (C68 to A70) unpaired nucleotides, respectively, in the predicted models. In the crystal structure, the C28 within the J1/2 forms a canonical base pair with G69 within the J3/1, G47 within the J2/3 interacts with A29 through Hoogsteen–Watson–Crick hydrogen bonds, and G47 remains stacked with G69, creating a near-ideal coaxial stacking of the overall P1 and P2 helices in the three-way junction (Fig. 3C). Third, the R domain, predicted to form an asymmetric bulge, adopts a well-structured fold further stabilized by extensive interactions with nucleotides within or near the J2/3 and J3/1 junctions. These long-range interactions result in a stable pseudoknot formation, consistent with previous predictions based on the biochemical probing and SHAPE results for other analogous PTEs (24). For a typical PTE, the J2/3 consists of a C-rich sequence (so-called C-domain), which mostly interacts with the so-called G-domain (here, R domain) through expected canonical G-C base pairs. Nevertheless, specific nucleotides within the SCV PTE pseudoknot and their interactions are quite different from those described previously for typical G-domain-C-domain interactions. As discussed above, the SCV crystal structure supported a more consensus architecture of the pseudoknot formation between the R and Y domains.
Fig. 3.
The unique features of SCV PTE crystal structure. (A) The crystal-derived secondary structure of the PTE RNA. The dotted lines represent tertiary interactions between the nucleotides (B) The non-canonical G9•A82 (sugar-edge-Hoogsteen) and A10•A81 (Hoogsteen-sugar-edge) pairs within the P1.1 helix. (C) The structure and helical stacking interactions within the J1/2. (D) Helical base-stacking of the J2/3 nucleotides that interact with L1 nucleotides to form a pseudoknot structure. (E) Additional interactions between the J2/3. The U52 base pairs with the A21, which sandwiches between the cross-strand stacking of the G67 and C68. (F) The J3/1 C68 nucleotide forms a canonical base pair with G20 and contacts U51 through a hydrogen bond that creates a planer tetrad structure involving G19 and G20, U51, and C68. Gray mesh represents the composite simulated anneal-omit 2|Fo|-|Fc| electron density map at contour level 1σ and carve radius 2.5 Å, and dotted black lines depict the heteroatoms within hydrogen bonding distances (≤3.0 Å).
R and Y Domain Interactions Stabilize the SCV PTE 3WJ.
The observed structural features in our crystal structure of SCV PTE, such as the number of junction nucleotides and the overall orientations of the P1, P2, and P3 helices, resemble a type A 3WJ. Consistent with typical features of type A 3WJs (45), the SCV PTE structure contains no nucleotides within the J1/2 between coaxially stacked P1 and P2 helices, five nucleotides (A48-U52) within the J2/3, and one nucleotide (C68) within the J3/1. Consistent with typical features of type A 3WJs (45), the SCV PTE structure contains no nucleotides within the J1/2 between coaxially stacked P1 and P2 helices, five nucleotides (A48-U52) within the J2/3, and one nucleotide (C68) within the J3/1. The P3 is slightly oriented toward the P1, which creates a λ-shaped topology for the overall PTE fold. Although this λ-shape matches the general topology with type B three-way junctions, all other structural features agree with the type A junctions. Nevertheless, unlike observed previously in typical 3WJs, the J2/3 and J3/1 in the SCV PTE make long-range tertiary interactions with the R domain. Beginning from the 5′ end of the J2/3 nucleotides, a dinucleotide stack of A48 and A49 does not make any direct contacts yet maintains the helical stacking with the remaining nucleotides within the junction (Fig. 3D). Whereas C50 forms a canonical base pair with G17, U51 contacts G19 through the sugar-edge/Watson–Crick hydrogen bonding interactions (Fig. 3D). The last nucleotide of the J2/3, U52 base pairs with the A21 (the last nucleotide of the R domain) that remains sandwiched between the cross-strand stacking of the G67 and C68 (Fig. 3E). The C68 nucleotide in the J3/1 junction forms a canonical base pair with G20 and contacts with U51 through a hydrogen bond that creates a planner tetrad structure involving G19 and G20 from the R domain, U51 (J2/3) and C68 (J3/1) (Fig. 3F). These long-range interactions between the R domain, Y domain (J2/3), and J3/1 nucleotides essentially leave no unpaired nucleotides within the SCV PTE 3WJ except A48 and A49 of the J2/3, which stabilize the overall fold of the junction. These interactions also stabilize the R domain’s 7-nt loop structure (G15 to A21), providing a unique arrangement of parallel-strand base pairing to organize a platform for the eIF4E binding. Consistent with the prominent role of the pseudoknot-forming nucleotides for the PTE stability, mutations to the R and Y domains have been shown previously to disrupt the overall folding and the binding of the PTE RNAs with eIF4E (24, 28).
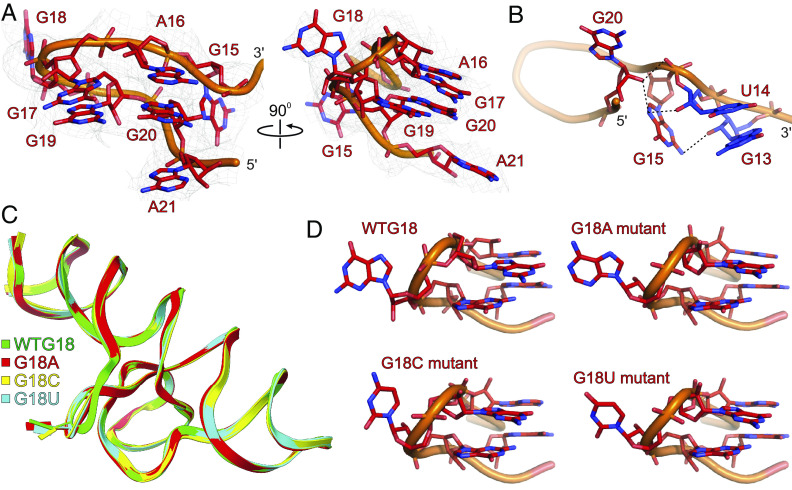
The G18 Structure Correlates Well with eIF4E Binding.
With a G-rich 5′ G15A16G17G18G19G20A21 sequence, the R domain forms a unique structure at the junction of P1.1 and P1.2 helices, which has been hypothesized to be the major binding site for the eIF4E (24). Within this loop structure (Fig. 4A), G15 interacts extensively with G13, U14, and the back of G20 through a network of hydrogen bonding interactions (Fig. 4B). Specifically, whereas 2′ OH of U14 and G13 interact with the G15 amino group and N7, respectively, the G20 2′ OH contacts the G15 phosphodiester backbone and N7. Moving upward, the A16 sandwiches between the flipped-out A70 of the P1.2 helix near the junction J1/2 from the top and G20 from the bottom, stabilizing the R domain’s structure through cross-strand stacking (Fig. 2C). The strand sharply turns between the G17 and G19, causing G18 to flip out from the helix. Apart from the G17 and G19 base-pairing interactions with the C50 and U51 of the Y domain, respectively (Figs. 2 B and C and 4B), base-stacking interactions between G17 and G19, as well as A16 and G20, stabilize this configuration. The highly solvent-exposed conformation of the G18 in our crystal structure correlates well with the higher SHAPE reactivity as G18 2′ OH is easily accessible to the SHAPE reagents. However, a strong electron density map and a lower B-factor (SI Appendix, Fig. S14) for G18 compared to some base-paired nucleotides within the same structure (P1.1 nucleotides, for example) contradicts its SHAPE reactivity profile, which implies a high degree of flexibility. Although we did not observe direct crystal contacts of the G18, it appears near the neighboring Fab molecule in the crystal lattice, which may contribute to its crystallographic stability and stronger density map (SI Appendix, Fig. S15). Nevertheless, consistent with the G18 being a major binding site for eIF4E, as evident from previous protection assays and our binding experiments, the unique configuration of the R domain, further stabilized by interactions with the Y domain, strongly holds the G18 in this configuration away from the core of the PTE RNA that likely provides a ready-made docking platform for the eIF4E similar to the 5′ cap-eIF4E binding.
Fig. 4.
The structural features of the R domain and G18 mutant SCV PTE. (A) The overall structure of the SCV PTE’s R domain showing the flipped-out G18 (major binding site for the 5′ cap-binding protein, eIF4E). (B) Specific interactions of the G13, U14, G15, and G20 nucleotides within the R domain. (C) Superposition of the G18A (red), G18C (yellow), and G18U (cyan) mutant PTE crystal structures with the wild-type PTE. (D) Comparisons of the R domain structures for wild-type and G18 mutants showing that SCV PTE adopts a preorganized structure with flipped-out G18 that constitutes a ready-made platform for the eIF4E binding. Gray mesh represents the composite simulated anneal-omit 2|Fo|-|Fc| electron density map at contour level 1σ and carve radius 2.5 Å, and dotted black lines depict the heteroatoms within hydrogen bonding distances (≤3.0 Å).
R Domain Scaffold Is Preorganized to Create a Mimic of the mRNA 5′ Cap.
To test whether this R domain G18 results from a pre-organized scaffold, we determined the crystal structures of the mutant SCV PTE constructs using the Fab chaperone discussed above. All three mutant constructs, G18A, G18C, and G18U, crystallized in the same conditions and the space group as the corresponding non-mutant construct, suggesting a similar overall fold of the RNA with no significant role of this nucleotide position in crystal packing. Remarkably, the crystal structures of the G18A (solved at 3.17 Å resolution), G18C (solved at 3.18 Å resolution), and G18U (solved at 3.29 Å resolution) were very similar (RMSDs for the all-atoms superposition of G18A, G18C, and G18U with the WT PTE = 0.521 Å, 0.428 Å, and 0.323 Å, respectively) with the 18th nucleotide being projecting out in all structures (Fig. 4C). The flipping out of the 18th nucleotide within the R domain from the rest of the RNA (Fig. 4D), regardless of its identity (G, U, or A), supports that the R domain is pre-organized to project the G18 into the binding pocket of the eIF4E as proposed previously and also consistent with our binding results for both wheat and human eIF4E. Moreover, the titration of the m7GTP against the SCV PTE–eIF4E complex suggests that the SCV PTE RNA competes for the same binding site within the eIF4E as the 5′ cap (Fig. 1 F and J). It means the G18 is most likely sandwiched between the two tryptophans (W62 and W108 for wheat and W56 and W102 for human eIF4E) of the eIF4E binding pocket similar to the 5′ cap (31, 46, 47), which also agrees well with the observed binding affinities for the G18 mutants in our ITC and EMSA assays. The stronger binding of eIF4E with the G18A is likely to favor the efficient stacking and hydrogen bonding interactions compared to G18C and G18U mutants. Overall, given the high conservation of the wheat and human eIF4E structures, similar binding of the SCV PTE with both eIF4Es and the likelihood of the G18 fitting into the 5′ cap-binding pocket strongly support our hypothesis that SCV PTE structure essentially mimics the mRNA 5′ cap for binding eIF4E.
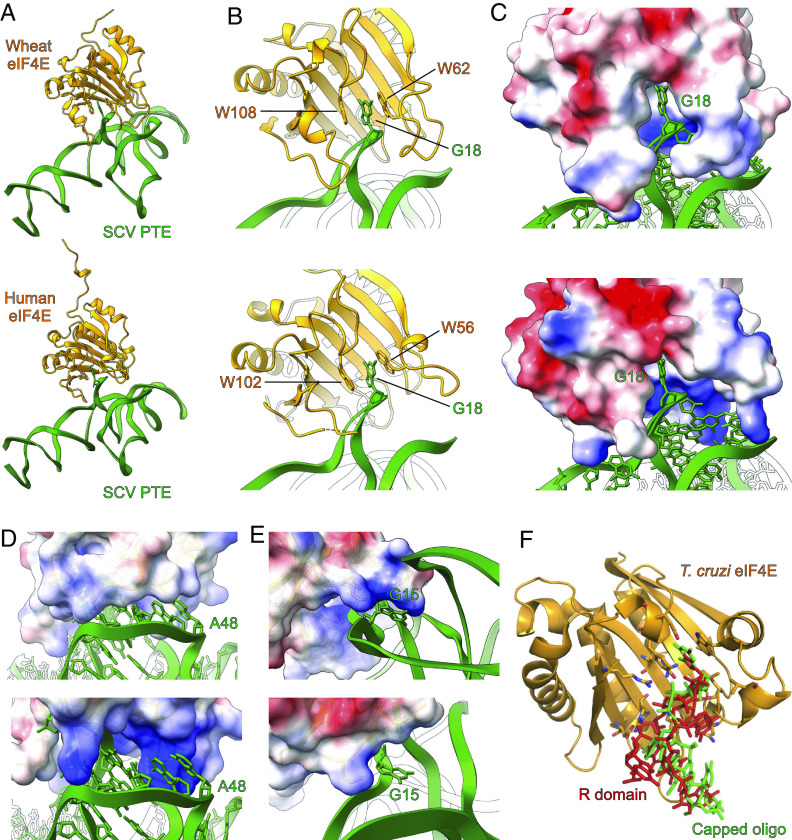
Molecular Docking Suggests Similar Modes of eIF4E Recognition by the SCV PTE and 5′ Cap.
To further understand the structural basis of eIF4E recognition by the viral PTE at the atomic level, we sought to determine the crystal structures of SCV PTE in complex with human and wheat eIF4Es. As our attempts to crystallize these complexes were unsuccessful, we performed computational docking studies. In silico docking of the SCV PTE crystal structure with eIF4E was performed using human and wheat m7GTP-bound eIF4E structures as a template (PDB IDs 1IPC and 2IDV, respectively). Initial docking positions were generated by aligning the R domain G18 from the crystal PTE structure with the m7GTP in the bound eIF4E structures to determine the feasibility of cap-pocket binding mimicry of the PTE. From this starting point, an iterative rigid-body rotation of the PTE structure was performed in three axes centered on the R domain G18, minimizing steric clashes between the structures while maintaining the stacking between R domain G18 and two tryptophan residues present in the eIF4E binding site (i.e., wheat eIF4E, 2IDV: W62 and W108; human eIF4E, 1IPC: W56 and W102).
The results of these docking simulations support the likelihood of this π–π stacking interaction as the basis for RNA–protein docking, with the PTE R domain fitting neatly within the binding pocket of eIF4E (Fig. 5 A–C). In the docking models of the PTE and wheat eIF4E complex, eIF4E loops β1-β2, α2-β3, and β5-β6 form a channel that fits neatly with the Y domain of the SCV PTE (Fig. 5C). In the homologous regions of human eIF4E (Fig. 5C), the pocket is not as shape complementary as the wheat eIF4E to the RNA structure interface, but the respective loops contain three additional positively charged lysine residues (K52, K119, and K159), which likely contribute to the electrostatic affinity between the PTE Y domain and the protein (Fig. 5D). These potential specific interactions are consistent with a higher affinity of the PTE observed for human eIF4E compared to wheat eIF4E. Additionally, the α6-β8 loop near the C-terminus of both human and wheat eIF4E structures contains two lysine residues, interacting with G15 of the PTE’s R domain (Fig. 5E). Although five of the α6-β8 loop residues reside closer to the PTE’s pseudoknot region, these residues in the reported crystal structure of human eIF4E are of indeterminate configuration to compare the interactions with the PTE (46, 47). Interestingly, the overall direction of the PTE R domain emerging out from the cap-binding pocket in our docking models is similar to the crystal structure (PDB ID: 6O7Y) of a 5-nt capped RNA complexed with the Trypanosoma cruzi eIF4E homolog (Fig. 5F), suggesting that the peripheral nucleotides of the SCV PTE beyond G18 have important roles in maintaining the shape-complementarity and specific interactions with the eIF4E, consistent with previous observations on the effect of sequence and secondary structures in binding eIF4E to the 5′ capped RNAs (48, 49).
Fig. 5.
Molecular docking studies of SCV PTE with eIF4E. (A) The overall model with wheat eIF4E (Top) and human eIF4E (Bottom). (B) The docking models for both wheat (Top) and human (Bottom) eIF4E showed that the R domain G18 occupies the same binding pocket as the 5′ cap and emulates similar interactions with the protein. (C) The electrostatic model of eIF4E shows that the PTE R domain fits neatly within the binding pocket of wheat (Top) and human (Bottom) eIF4E. The human eIF4E binding pocket (Bottom) appears more open compared to that of wheat, with three additional positively charged lysine residues around the binding pocket. (D) The Y domain of the SCV PTE also fits in a groove formed by the eIF4E loops β1-β2, α2-β3, and β5-β6. Compared to the wheat eIF4E (Top), the homologous channel of the human eIF4E (Bottom) is more open and more positively charged. (E) The α6-β8 loop near the wheat eIF4E C-terminal interacts with the G15 R domain (Top). However, five residues of the loop are omitted as indeterminate in the published structure of human eIF4E (Bottom, PDB: 1IPC) that might contribute to this binding interaction. (F) The alignment of the PTE G18 with the 5′ cap (m7GTP) of an oligonucleotide (green) within the cocrystal structure with T. cruzi eIF4E (PDB: 6O7Y) resulted in PTE’s R domain (red) exiting the binding pocket in the same orientation as the oligonucleotide. The key nucleotides and residues involved in the PTE–eIF4E interactions are labeled.
Discussion
The structural basis of the mRNA 5′ cap binding with eIF4E has been well established by using the 5′ cap analogs such as m7GTP. However, the effect of RNA secondary and tertiary structures in the 5′ cap-eIF4E binding for larger mRNA constructs remains largely unknown. The 5-nt capped RNA oligomer is the longest construct to be crystallized in a complex with eIF4E. On the other hand, although several RNA domains, especially from viruses, are known to interact with this protein in a cap-independent manner, no high-resolution structural data existed before this study to understand the molecular basis of such uncapped RNA–eIF4E interactions. Our studies with SCV PTE provide high-resolution structural information and a structural basis for this uncapped class of viral RNA interactions with wheat and human eIF4Es. Moreover, the resemblance of the computationally predicted structures for PMV and PEMV2 PTEs (24) with our SCV PTE structure provides validation for such prediction and strong evidence for the modular nature and robust folding of this viral RNA class.
The structural and eIF4E binding studies for the SCV PTE presented here underscore how a viral RNA domain can mimic the host 5′ cap to bypass the host cap-dependent mechanism and promote viral genome translation via cap-independent mechanisms. We have determined the high-resolution crystal structure of the SCV PTE and revealed long-range tertiary interactions between the R and Y domains (so-called G and C domains, respectively, in other PTEs) that constitute an unusual pseudoknot architecture. Within this unique architecture, the R domain flips out a guanine nucleobase (G18) from the rest of the PTE structure to comprise the major eIF4E binding site. While computed PTE models for PMV and PEMV2 accurately predicted such flipping out of this guanine to interact with the cap-binding pocket (24), its structural role and specificity were not defined. We have shown that the G18A, G18C, and G18U mutations in SCV PTE structure reduce or abrogate binding with both wheat and human eIF4E without perturbing the overall PTE structure, demonstrating that the R domain is preorganized to cause the nucleotide at this position to be flipped out regardless of the nucleotide identity at this position.
Despite being a cactus virus, our EMSA and ITC binding studies showed that both human and wheat eIF4Es effectively interact with the SCV PTE within the 5′ cap-binding site, supporting that this PTE scaffold represents a structured RNA mimic of the mRNA 5′ cap. Moreover, our docking studies with human and wheat eIF4Es also agree that the G18 is a major binding site within the SCV PTE for eIF4E recognition. Nevertheless, while the m7 modification of the G18 within the SCV genome is unknown, the interactions around the rim of the cap-binding pocket perhaps compensate for the higher affinity of the 5′ cap to eIF4E due to such modification. The ability of uncapped RNAs to bind cap-binding protein tightly also suggests that these viral PTEs might have evolved to exploit multiple surfaces of eIF4E, including the cap-binding site, to directly compete with the host mRNA 5′ cap for the eIF4E binding during viral genome translation. Given the topological conservation and modular nature of these PTEs in plant viruses with the ability to bind human eIF4E further support the likelihood of similar RNA structures in several other viral and cellular IRESs that require eIF4E for their genome translation in a cap-independent manner.
Given that human and wheat eIF4E structures are highly conserved and SCV only infects plants, it is interesting that the human eIF4E binds the SCV PTE with a different affinity than wheat eIF4E. Whereas both wheat and human eIF4Es have previously been shown to bind with similar low nanomolar affinities to the synthetically 5′ capped TPAV PTE (estimated Kd = 160 nM and 31 nM for human and wheat eIF4E) and PEMV2 PTE (estimated Kd = 285 nM and 41 nM for human and wheat eIF4E) in the semi-quantitative gel electrophoresis assays (28), the binding affinities for the corresponding uncapped PTEs were slightly different for wheat eIF4E (estimated Kd = 110 nM and 65 nM for PEMV2 and TPAV PTE) and human eIF4E (estimated Kd = 2,524 nM and 838 nM for PEMV2 and TPAV PTE). Comparatively, both TPAV and PEMV2 PTEs bound human eIF4E with a lower affinity compared to the corresponding wheat eIF4E, but the TPAV PTE showed over threefold higher affinity for human eIF4E (estimated Kd = 838 nM) compared to PEMV2 PTE (estimated Kd = 2,524 nM), which correlated positively with a stronger ability of the TPAV to promote PTE-mediated translation in mammalian extracts compared to that in the wheat-germ extracts (28). The SCV PTE in our EMSA and ITC measurements showed a comparable nanomolar binding affinity with human eIF4E and wheat, consistent with the TPAV PTE-like behavior of the SCV PTE in promoting PTE-mediated translation in mammalian and wheat-germ extracts (28). Nevertheless, these observations for various PTE–eIF4E binding suggest that both eIF4Es and PTEs contain unique additional structural features beyond the major interaction sites that perhaps are responsible for the slightly different binding behavior among viral PTEs and eIF4Es.
Recent studies on target-specific gene expression regulation achieved through eIF-binding RNA domains delivered in trans to the mRNA 3′ UTR (50) not only underline the tremendous potential of the viral PTEs in biotechnology and therapeutics as efficient translation activators or inhibitors but also suggest similar function of the viral PTEs for delivering the translation initiation factors to the 5′ end for translation through genome circularization. While our SCV PTE crystal structures and previous phylogenetic, biochemical, and computational modeling studies have revealed that long-range interactions to form a pseudoknot between the R and Y domains are conserved structural features among viral PTEs, the chimeric PTEs with these domains swapped from different PTEs did not retain their function (24), suggesting that eIF4E binding is specific to the structural context of the particular PTE. Therefore, although this structural study with SCV PTE represents the very first steps, future studies on the determination of high-resolution structures of other PTEs and PTE–eIF4E complexes and quantitative measurements of their interactions with the host and viral proteins will help uncover the unique structural features of this particular class of viral RNAs, provide mechanistic insights into how they facilitate virus-host interactions and decipher more precise mechanisms of the RNA structure-based cap-independent translation in plant viruses and beyond.
Materials and Methods
RNA Synthesis and Purification.
The RNA constructs were synthesized by in vitro transcription and purified by denaturing polyacrylamide gel electrophoresis (dPAGE), as described elsewhere (43). See SI Appendix for further details.
Human and Wheat eIF4E Expression and Purification.
The human and wheat eIF4E genes were cloned into a pET-16b (+) vector (GenScript, https://www.genscript.com/) with a 6×-His tag at the N-terminal. The expression plasmid for the GB1 tag fused human eIF4E was a generous gift from Michael Summers. The proteins were expressed in BL21 (DE3) Escherichia coli and purified by standard chromatographic techniques using affinity and size-exclusion columns. See SI Appendix for further details.
Fab Expression and Purification.
The Fab BL3-6 expression plasmid was a kind gift from Joseph Piccirilli, the University of Chicago. The Fab was expressed and purified according to published protocols (36, 37, 51). See SI Appendix for further details.
5′ Capped RNA Synthesis and Purification.
The 5′ capped RNA oligomer was synthesized and purified according to the previously reported protocols (52, 53). See SI Appendix for further details.
Native Gel Electrophoresis Assay.
About 200 ng RNA was refolded and then incubated for 30 min at room temperature with different Fab or eIF4E protein equivalents. The gels were stained with ethidium bromide and imaged using the Azure 200 gel documentation system (Azure Biosystems). See SI Appendix for further details.
Isothermal Titration Calorimetry.
The isothermal titration calorimetry (ITC) experiments were carried out in the MicroCal PEAQ-ITC automated equipment (Malvern Panalytical) using freshly prepared RNA and protein samples. The injection syringe contained 150 μL of 100 µM of human or 100 to 200 µM of wheat eIF4E protein, and the calorimetry cell was loaded with 500 μL of 5 µM RNA. After thermal equilibration at 25 °C and an initial 60-s delay, a single injection of 0.2 μL followed by the 19 serial injections of 2 μL of the protein was made into the calorimetry cell. See SI Appendix for further details.
In Vitro Translation Assay.
The in vitro translation assays were performed using a similar protocol described elsewhere (34). Briefly, the bicistronic gene construct (DNA template) was prepared via PCR amplification of the pFR_HCV_xb plasmid (Addgene plasmid # 11510), a gift from Phil Sharp. The translation reactions were performed using a rabbit reticulocyte lysate-based system (Promega) in a final volume of 50 μL for 90 min at 30 °C with about 4 μg DNA template in the presence or absence of the PTE. After the reaction, expression levels of the firefly and renilla luciferases were measured as the luminescence signals generated through a dual luciferase reporter assay (Promega) using a plate reader (Molecular Devices).
Crystallization.
The Xtal3 Mosquito liquid handling robot (TTP Labtech, https://www.ttplabtech.com) was used to set up hanging-drop vapor-diffusion crystallization screens at room temperature (22 °C) within a humidity-controlled (70%) chamber using commercially available screening kits from Hampton Research. The initial hit conditions were further optimized for pH, precipitant, and salt concentration to grow larger crystals. Drops containing suitable crystals were brought up to 40% glycerol for cryoprotection without changing the other compositions. The crystals were flash-frozen in liquid nitrogen after being fished in the loops from the drops and sent to Argonne National Laboratory for X-ray diffraction screening and data collection. See SI Appendix for further details.
Crystallographic Data Collection, Processing, and Analysis.
The X-ray diffraction datasets were collected at the Advanced Photon Source NE-CAT section beamlines 24-ID-C and 24-ID-E. All the datasets were then integrated and scaled using its on-site RAPD automated programs (https://rapd.nec.aps.anl.gov/). The initial phases were obtained by molecular replacement with the previously reported structure of Fab BL3-6 (PDB code: 8DP3) (43) as the search model using Phaser on Phenix (54). Iterative model building and refinement were performed using the COOT (55) and the Phenix package (54). The solvent-accessible surface area and buried surface area for the crystal lattice interactions were calculated using PDBePISA (https://www.ebi.ac.uk/pdbe/pisa/) (56). See SI Appendix for further details.
Molecular Docking.
The wheat and human eIF4E structures bound with m7GTP were used as docking modeling templates [PDB IDs 1IPC (46) and 2IDV (31), respectively]. The PTE crystal structure was aligned to these templates using C2, C4, and C6 atoms of guanine G18 with the corresponding atoms in the m7GTP of the complex. The m7GTP was subsequently removed, and an iterative rigid-body rotation of the PTE structure was performed in all three axes centered on the R domain’s G18 to minimize the steric clashes between the eIF4E and PTE structures. Each model was scored based on steric clashes, determined by a Van der Waals radius overlap ≥0.60 Å. From these initial models, 1,000 decoy models were generated by applying a random rotation of ±5° in three axes centered on the C2, C4, and C6 atoms of guanine G18. These decoys were again scored by Van der Waals radius overlap, and the lowest-scoring results were then iterated through another round of random rotations centered on G18. The process was repeated three times, with subsequent iterations producing no significant improvements in the modeling scores. The script used for the molecular docking has been provided in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by the NIH (MIRA 1R35GM150869-01) and University of Maryland Baltimore County (start-up funds, START grant, and SURFF fellowship) to D.K. and partly by the NIH T32 grant (GM066706) to M.O., H.A.B., and N.K.D. The crystallographic work is based on research conducted at the NE-CAT beamlines, 24-ID-C, and 24-ID-E, funded by the National Institute of General Medical Sciences from the NIH (P30 GM124165). The Eiger 16 M detector on 24-ID-E is funded by an NIH-ORIP HEI grant (S10OD021527). This research used the Advanced Photon Source resources—a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. We thank Prof. Michael F. Summers, the University of Maryland Baltimore County, for kindly providing the ITC facility and critical feedback on the manuscript.
Author contributions
M.O. and D.K. designed research; M.O., J.V., N.K.D., E.R., K.S., H.A.B., T.S., and D.K. performed research; K.S. contributed new reagents/analytic tools; M.O., J.V., and D.K. analyzed data; and M.O., J.V., and D.K. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
The atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under the accession codes 8T29 (57), 8T2A (58), 8T2B (59), and 8T2O (60) for the WT SCV PTE and its G18A, G18C, and G18U mutants, respectively. All other data, associated protocols, codes, and additional information, if any, have been provided in the Materials and Methods or SI Appendix.
Supporting Information
References
- 1.Richter J. D., Sonenberg N., Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433, 477–480 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Jia Y., Polunovsky V., Bitterman P. B., Wagner C. R., Cap-dependent translation initiation factor eIF4E: An emerging anticancer drug target. Med. Res. Rev. 32, 786–814 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aitken C. E., Lorsch J. R., A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 19, 568–576 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Browning K. S., Bailey-Serres J., Mechanism of cytoplasmic mRNA translation. Arabidopsis Book 13, e0176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kneller E. L., Rakotondrafara A. M., Miller W. A., Cap-independent translation of plant viral RNAs. Virus Res. 119, 63–75 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez-Salas E., Francisco-Velilla R., Fernandez-Chamorro J., Embarek A. M., Insights into structural and mechanistic features of viral IRES elements. Front Microbiol. 8, 2629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du Z., Alekhina O. M., Vassilenko K. S., Simon A. E., Concerted action of two 3’ cap-independent translation enhancers increases the competitive strength of translated viral genomes. Nucleic Acids Res. 45, 9558–9572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lacerda R., Menezes J., Romão L., More than just scanning: The importance of cap-independent mRNA translation initiation for cellular stress response and cancer. Cell. Mol. Life Sci. 74, 1659–1680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaafar Z. A., Kieft J. S., Viral RNA structure-based strategies to manipulate translation. Nat. Rev. Microbiol. 17, 110–123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson R. J., Hellen C. U., Pestova T. V., The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jan E., Mohr I., Walsh D., A cap-to-tail guide to mRNA translation strategies in virus-infected cells. Annu. Rev. Virol. 3, 283–307 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Yang Y., Wang Z., IRES-mediated cap-independent translation, a path leading to hidden proteome. J. Mol. Cell Biol. 11, 911–919 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholson B. L., White K. A., 3′ Cap-independent translation enhancers of positive-strand RNA plant viruses. Curr. Opin. Virol. 1, 373–380 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Simon A. E., Miller W. A., 3′ cap-independent translation enhancers of plant viruses. Ann. Rev. Microbiol. 67, 21–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholson B. L., Zaslaver O., Mayberry L. K., Browning K. S., White K. A., Tombusvirus Y-shaped translational enhancer forms a complex with eIF4F and can be functionally replaced by heterologous translational enhancers. J. Virol. 87, 1872–1883 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Truniger V., Miras M., Aranda M. A., Structural and functional diversity of plant virus 3’-cap-independent translation enhancers (3’-CITEs). Front Plant Sci. 8, 2047 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo L., Allen E. M., Miller W. A., Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell 7, 1103–1109 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Chattopadhyay M., Shi K., Yuan X., Simon A. E., Long-distance kissing loop interactions between a 3’ proximal Y-shaped structure and apical loops of 5’ hairpins enhance translation of Saguaro cactus virus. Virology 417, 113–125 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miras M., et al. , A dual interaction between the 5’- and 3’-ends of the melon necrotic spot virus (MNSV) RNA genome is required for efficient cap-independent translation. Front Plant Sci. 9, 625 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gazo B. M., Murphy P., Gatchel J. R., Browning K. S., A novel interaction of Cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3’-untranslated region of satellite tobacco necrosis virus. J. Biol. Chem. 279, 13584–13592 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Wang Z., Kraft J. J., Hui A. Y., Miller W. A., Structural plasticity of Barley yellow dwarf virus-like cap-independent translation elements in four genera of plant viral RNAs. Virology 402, 177–186 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stupina V. A., et al. , The 3’ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA 14, 2379–2393 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z., Treder K., Miller W. A., Structure of a viral cap-independent translation element that functions via high affinity binding to the eIF4E subunit of eIF4F. J. Biol. Chem. 284, 14189–14202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z., Parisien M., Scheets K., Miller W. A., The cap-binding translation initiation factor, eIF4E, binds a pseudoknot in a viral cap-independent translation element. Structure 19, 868–880 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo X., et al. , Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3’ UTR of turnip crinkle virus. Proc. Natl. Acad. Sci. U.S.A. 107, 1385–1390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niedzwiecka A., et al. , Biophysical studies of eIF4E cap-binding protein: Recognition of mRNA 5’ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 319, 615–635 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Slepenkov S. V., Darzynkiewicz E., Rhoads R. E., Stopped-flow kinetic analysis of eIF4E and phosphorylated eIF4E binding to cap analogs and capped oligoribonucleotides: Evidence for a one-step binding mechanism. J. Biol. Chem. 281, 14927–14938 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Kraft J. J., et al. , The 3’ untranslated region of a plant viral RNA directs efficient cap-independent translation in plant and mammalian systems. Pathogens 8, 28 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varani G., McClain W. H., The G•U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 1, 18–23 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westhof E., Yusupov M., Yusupova G., The multiple flavors of GoU pairs in RNA. J. Mol. Recogn. 32, e2782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monzingo A. F., et al. , The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond. Plant Physiol. 143, 1504–1518 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song S. J., Diao H. P., Moon B., Yun A., Hwang I., The B1 domain of streptococcal protein G serves as a multi-functional tag for recombinant protein production in plants. Front Plant Sci. 13, 878677 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., Shirako Y., Association of VPg and eIF4E in the host tropism at the cellular level of Barley yellow mosaic virus and Wheat yellow mosaic virus in the genus Bymovirus. Virology 476, 159–167 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Koirala D., Lewicka A., Koldobskaya Y., Huang H., Piccirilli J. A., Synthetic antibody binding to a preorganized RNA domain of Hepatitis C virus internal ribosome entry site inhibits translation. ACS Chem. Biol. 15, 205–216 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraser C. S., Doudna J. A., Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 5, 29–38 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Ye J.-D., et al. , Synthetic antibodies for specific recognition and crystallization of structured RNA. Proc. Natl. Acad. Sci. U.S.A. 105, 82–87 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koldobskaya Y., et al. , A portable RNA sequence whose recognition by a synthetic antibody facilitates structural determination. Nat. Struct. Mol. Biol. 18, 100–106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koirala D., et al. , Affinity maturation of a portable Fab-RNA module for chaperone-assisted RNA crystallography. Nucleic Acids Res. 46, 2624–2635 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krochmal D., et al. , Structural basis for substrate binding and catalysis by a self-alkylating ribozyme. Nat. Chem. Biol. 18, 376–384 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Huang H., et al. , A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 10, 686–691 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roman C., Lewicka A., Koirala D., Li N. S., Piccirilli J. A., The SARS-CoV-2 programmed -1 ribosomal frameshifting element crystal structure solved to 2.09 Å using chaperone-assisted RNA crystallography. ACS Chem. Biol. 16, 1469–1481 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rees H. C., Gogacz W., Li N.-S., Koirala D., Piccirilli J. A., Structural basis for fluorescence activation by pepper RNA. ACS Chem. Biol. 17, 1866–1875 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das N. K., et al. , Crystal structure of a highly conserved enteroviral 5’ cloverleaf RNA replication element. Nat. Commun. 14, 1955 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shelke S. A., et al. , Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif. Nat. Commun. 9, 4542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lescoute A., Westhof E., The interaction networks of structured RNAs. Nucleic Acids Res. 34, 6587–6604 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomoo K., et al. , Crystal structures of 7-methylguanosine 5’-triphosphate (m(7)GTP)- and P(1)-7-methylguanosine-P(3)-adenosine-5’,5’-triphosphate (m(7)GpppA)-bound human full-length eukaryotic initiation factor 4E: Biological importance of the C-terminal flexible region. Biochem. J. 362, 539–544 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomoo K., et al. , Structural features of human initiation factor 4E, studied by X-ray crystal analyses and molecular dynamics simulations. J. Mol. Biol. 328, 365–383 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Carberry S. E., Friedland D. E., Rhoads R. E., Goss D. J., Binding of protein synthesis initiation factor 4E to oligoribonucleotides: Effects of cap accessibility and secondary structure. Biochemistry 31, 1427–1432 (1992). [DOI] [PubMed] [Google Scholar]
- 49.Uppala J. K., et al. , The cap-proximal RNA secondary structure inhibits preinitiation complex formation on HAC1 mRNA. J. Biol. Chem. 298, 101648 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao Y., et al. , RNA-based translation activators for targeted gene upregulation. Nat. Commun. 14, 6827 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paduch M., et al. , Generating conformation-specific synthetic antibodies to trap proteins in selected functional states. Methods 60, 3–14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown J. D., et al. , Structural basis for transcriptional start site control of HIV-1 RNA fate. Science 368, 413–417 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding P., et al. , 5’-Cap sequestration is an essential determinant of HIV-1 genome packaging. Proc. Natl. Acad. Sci. U.S.A. 118, e2112475118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Ojha M., Koirala D., Crystal structure of SCV PTE RNA in complex with Fab BL3-6. RCSB Protein Data Bank. https://www.rcsb.org/structure/8T29. Deposited 5 June 2023.
- 58.Ojha M., Koirala D., Crystal structure of SCV PTE G18A mutant RNA in complex with Fab BL3-6. RCSB Protein Data Bank. https://www.rcsb.org/structure/8T2A. Deposited 5 June 2023.
- 59.Ojha M., Koirala D., Crystal structure of SCV PTE G18C mutant RNA in complex with Fab BL3-6. RCSB Protein Data Bank. https://www.rcsb.org/structure/8T2B. Deposited 5 June 2023.
- 60.Ojha M., Koirala D., Crystal structure of SCV PTE G18U mutant RNA in complex with Fab BL3-6. RCSB Protein Data Bank. https://www.rcsb.org/structure/8T2O. Deposited 6 June 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under the accession codes 8T29 (57), 8T2A (58), 8T2B (59), and 8T2O (60) for the WT SCV PTE and its G18A, G18C, and G18U mutants, respectively. All other data, associated protocols, codes, and additional information, if any, have been provided in the Materials and Methods or SI Appendix.