Abstract
The role of Pseudomonas aeruginosa quorum-sensing systems in the lung infections associated with cystic fibrosis (CF) has not been examined. The purpose of this study was to determine if genes regulated by the LasR-LasI quorum-sensing system were coordinately regulated by the P. aeruginosa populations during the lung infections associated with CF. We also wanted to ascertain if there was a relationship between the expression of lasR, a transcriptional regulator, and some P. aeruginosa virulence factors during these infections. We extracted RNAs from the bacterial populations of 131 sputa taken from 23 CF patients. These RNAs were blotted and hybridized with probes to P. aeruginosa lasA, lasB, and toxA. The hybridization signals from each probe were ranked, and the rankings were analyzed by a Spearman rank correlation to determine if there was an association between the population transcript accumulations for the three genes. The correlations between the transcript accumulation patterns of pairs of the genes suggested that lasA, lasB, and toxA might be coordinately regulated during CF lung infections. To determine if this coordinate regulation might be due to regulation by LasR, we probed RNAs, extracted from 84 sputa, with the lasR, lasA, lasB, toxA, and algD probes. Statistical analysis indicated that lasR transcript accumulation correlated to lasA, lasB, toxA, and algD transcript accumulations. These results indicated that lasR may at least partially regulate or be coordinately regulated with lasA, lasB, toxA, and algD during the lung infections associated with CF. These results also suggested that the LasR-LasI quorum-sensing system may control the expression of at least some virulence factors in the lungs of patients with CF.
A number of Pseudomonas aeruginosa virulence factors are regulated by two quorum-sensing systems (13, 29, 30, 33–36, 49). One of these sensing systems, the LasR-LasI system, controls the expression of lasA, lasB, apr, and toxA (14, 33, 48). The other system, RhlR-RhlI, controls lasB, lasA, rhlAB, rpoS, and potentially other genes (3, 22, 23, 29, 30). These two systems also interact to control the overall regulation of both systems and their target genes (36). The process of autoinduction in P. aeruginosa and the other bacterial systems that display this type of regulation was reviewed by Fuqua et al. (12). Their model suggests that when the bacterial density is low, the concentration of autoinducer is also low. As the bacterial numbers increase and if the bacteria are in a closed environment, the concentration of autoinducer will rise. At a critical level (the Kd for receptor binding) the autoinducer will bind the transcriptional activator and trigger the expression of a number of genes (12). In the case of the LasR-LasI system, LasR and autoinducer (PAI-1) increase the production of the autoinducer synthase (lasI product) and expression of the autoinducer (33, 42). Thus, once the critical concentration of autoinducer is reached, the level of induction remains high until the system is shut off in some manner.
A number of quorum-sensing regulatory systems are involved in or are speculated to be involved in interactions between bacteria and eucaryotic host organisms (12). Bacteria with quorum-sensing systems that may undergo pathogenic interactions with eucaryotic hosts are P. aeruginosa (13, 14, 33, 34), Erwinia spp. (1, 2, 4, 5, 20, 25, 37), and Agrobacterium tumefaciens (11, 18, 51). Bacteria with symbiotic relationships with eucaryotic hosts include Vibrio fischeri and Rhizobium leguminosarum (6–8, 16, 41). Potentially, the common environmental feature of these interactions is a space barrier that limits the growth of the bacteria and blocks diffusion of the autoinducer molecules.
The role of the P. aeruginosa quorum-sensing systems in infectious processes has not been extensively studied. One study revealed that a lasR mutant, PAOR1 (13), is significantly less virulent in a neonatal mouse model of infection (47). The regulatory role of the autoinducer was not specifically examined in this research, and the authors speculated that the lasR mutant was deficient in virulence because of a decrease in neuraminidase expression (47). To date no studies have been done to determine if P. aeruginosa quorum-sensing systems are active in human infections. The purpose of our research was to determine if the P. aeruginosa quorum-sensing systems are functional in the chronic lung infections associated with cystic fibrosis (CF).
The lung infections associated with CF should provide the perfect environment for control by a quorum-sensing system. The infections are localized to the airways of the lungs and so are spatially limited. The bacteria grow to a high density in the lungs (concentrations of 107 to 108 bacteria/ml in sputa are common) (43, 46), and the bacteria seem to grow in microcolonies or biofilms. These densities of bacteria should produce concentrations of autoinducer that would trigger the expression of the target genes. Furthermore, three genes that are regulated by the LasR–LasI–PAI-1 regulatory system, lasA, toxA, and lasB (13, 14, 33), are transcribed (38, 45, 46) and expressed (15, 17, 19, 21, 28) in the lungs of patients with CF. Thus, two questions arose which we wanted to address in this study. First, is there a correlation between the expressions of lasB, lasA, and toxA in the lungs of patients with CF? Second, is there a correlation between the transcriptions of lasR and the various virulence factors in the P. aeruginosa lung infections associated with CF?
In the present study we found a statistically significant correlation between the expressions of lasB, lasA, and toxA in the lungs of patients with CF. This correlation suggested that these three genes might be at least partially coordinately regulated in these lung infections. We also found a correlation between lasR transcript accumulation and lasA, lasB, and toxA transcript accumulations. Interestingly, there is also a statistically significant correlation between lasR and algD transcript accumulations. These results suggested that lasR may at least partially regulate or be coordinately regulated with lasA, lasB, toxA, and perhaps algD.
MATERIALS AND METHODS
Patient population.
Two overlapping groups of patients were used in this study. The first group was identical to the patients selected for a previous study described by Storey et al. (45). This was a group of 23 patients with CF. Of these patients, 13 were female and 10 were male. The median age for these patients was 15.5 years at the beginning of the study. A subset of 16 patients was also used in the analysis for this study. These patients were all from the Alberta Children’s Hospital Cystic Fibrosis Clinic. Of these patients, seven were female and nine were male. The age range of the patients at the beginning of the study was 10 to 19 years, with a median age of 13 years. All patients and their guardians gave their voluntary consent to participate in this study. The study design was given ethical review and approval by the Conjoint Research Ethics Board at the University of Calgary. Selection of the patients was carried out to ensure that equal numbers of patients would have mild, moderate, or severe pulmonary disease. Patients with a forced expiratory volume in 1 s (FEV1) of <40% of predicted values were considered to be in the severe category of disease. A patient FEV1 in the range of >40% but <70% of predicted values was considered to be in the moderate range of pulmonary disease. The mild category of disease severity consisted of those patients with an FEV1 of >70% of predicted values.
Sputum collection and storage and extraction of total RNA.
At each clinic visit sputum was collected from each patient in the study. The samples were diluted with Sputalysin (Calbiochem-Behring, Horescht)-H2O and divided into two parts. One part was frozen at −70°C for later extraction of the total RNA. The second part was used to determine the number and types of bacteria present in the sputum. Extraction of total RNA from the sputa, slot blots of the RNA, hybridization, autoradiography, and measurement of the sample intensity were all performed as previously described (46).
DNA probes.
A 550-bp PstI fragment from pMJG1.7 was used to hybridize to lasR-specific mRNAs. Plasmid pMJG1.7 was a generous gift from B. H. Iglewski (13). The probes used for lasA, lasB, toxA, and algD mRNAs have been described previously (45).
Growth conditions and extraction of control RNA.
Control RNAs (algD, lasA, lasB, lasR, and toxA) were included on each blot to normalize the results from different blots and hybridizations. Preparation of the control RNAs for algD, lasA, lasB, and toxA has been previously described (45). Control RNA for lasR was extracted from P. aeruginosa PAO1 grown in peptone Trypticase soy broth (32) to an optical density at 540 nm of 4.0. RNA was extracted from 2 × 108 cells, as previously described (9, 10).
Determination of lasR mRNA half-life.
The half-life of lasR mRNA was determined by using a culture of P. aeruginosa PAO1 grown in peptone Trypticase soy broth (32) to an optical density at 540 nm of 4.0. Rifampin was added to the culture at a concentration of 200 μg/ml, and an aliquot of the culture was removed. RNA was extracted from the aliquot as previously described (9). Additional aliquots were removed from the culture at 2.5-min intervals. As each aliquot was removed, the RNA was immediately extracted. The RNA from each time point was then electrophoresed on a glyoxal gel, blotted onto Nytran (Schleicher and Schuell), probed, autoradiographed, and scanned for signal intensity (46).
Statistical analysis.
Correlations between the groups of hybridizations were analyzed by using the Spearman rank correlation (50). For comparison, the results from each sample were ranked on the level of hybridization with each probe. The subprogram Rank from Minitab was used to generate these ranks (40). To generate a Spearman rank correlation, the ranks from two probes were subtracted from each other and the difference was squared. The squares were then summed and used to calculate a Spearman rank correlation coefficient (rs) (50). As in the study described by Storey et al. (45), we initially analyzed 151 sputa that contained RNA based on a measurement of optical density at 260 nm. In 20 of these samples we were unable to detect any hybridization with our probes. Thus, we excluded these 20 samples from the analysis presented in this paper.
RESULTS
Correlation between toxA, lasB, and lasA transcript accumulations.
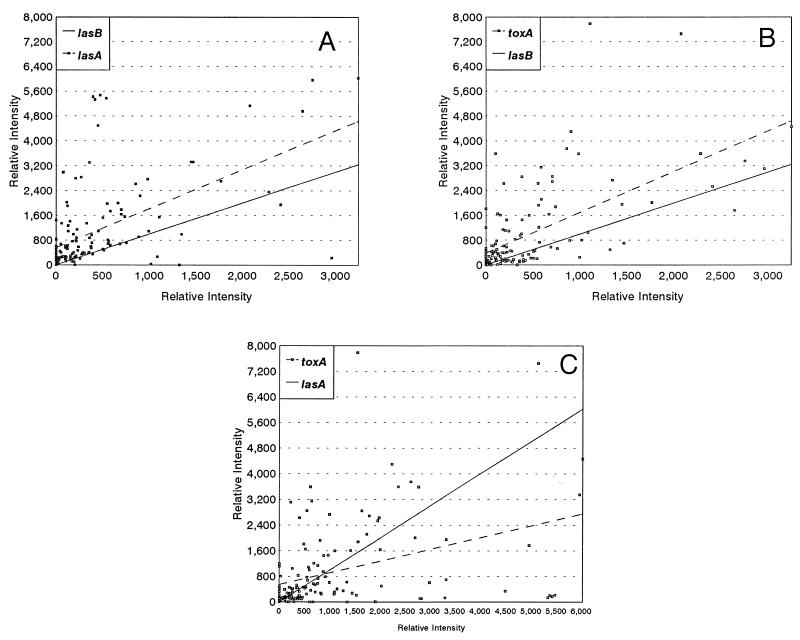
Recently our laboratory has shown correlations between the population transcript accumulation for algD and that for either lasA or lasB. In the present study we wanted to address the possibility of an association between lasA, lasB, and toxA transcript accumulations. To determine if there were correlations between the transcript accumulations for the three virulence determinants, we probed 131 RNA samples extracted directly from the sputa of 23 patients. Table 1 shows the Spearman rank correlation coefficients for these comparisons, and Fig. 1 shows the direct comparisons. Interestingly, there were statistically significant correlations between lasA and lasB, lasB and toxA, and lasA and toxA. The highest correlation was between lasA and lasB (Table 1; Fig. 1A), followed closely by the correlation between lasB and toxA (Table 1; Fig. 1B). The lowest correlation was between lasA and toxA (Table 1; Fig. 1C). These results suggested that during the lung infections associated with CF, P. aeruginosa populations coordinated the regulation of these three virulence factors. The two most likely explanations for these correlations are that the genes are responding to a common environmental cue or to a common regulatory protein. In a clonal situation all three genes have been shown to be regulated by LasR (14, 48). Thus, a common regulatory gene that might coordinately regulate these three genes in the lungs of CF patients is lasR. It therefore was of interest to determine if lasR population transcript accumulation corresponded to the population transcript accumulations for lasA, lasB, and toxA.
TABLE 1.
Statistical analysis of the associations between the population transcript accumulations for lasA, lasB, toxA, lasR, and algD during the chronic lung infections associated with CF
| Comparison |
n = 131
|
n = 84
|
n = 35a
|
|||
|---|---|---|---|---|---|---|
| rsb | P | rs | P | rs | P | |
| lasA-lasB | 0.595 | <0.001 | 0.560 | <0.001 | ||
| lasB-toxA | 0.570 | <0.001 | 0.557 | <0.001 | ||
| lasA-toxA | 0.423 | <0.001 | 0.455 | <0.001 | ||
| lasR-lasA | 0.759 | <0.001 | 0.731 | <0.001 | ||
| lasR-lasB | 0.680 | <0.001 | 0.708 | <0.001 | ||
| lasR-toxA | 0.569 | <0.001 | 0.626 | <0.001 | ||
| lasR-algD | 0.309 | <0.01 | 0.350 | <0.05 | ||
Thirty-five samples which contain P. aeruginosa and no other pathogenic strains.
rs, Spearman rank correlation coefficient.
FIG. 1.
Scatter plots for the population transcript accumulations for lasA, lasB, and toxA. In these plots the results for 131 sputum samples are presented. For each sample, 10 μg of the total RNA extracted from the pelleted bacterial population was blotted onto a Nytran membrane (Schleicher and Schuell) and hybridized with the toxA probe. The blots were then washed to remove the probe, and the membrane was rehybridized with the lasB probe. The process was repeated with the lasA probe. The relative intensity for the sample was measured from the autoradiograph with a soft laser scanning densitometer. The results represent 10 μg of RNA from equivalent numbers of cells. (A and B) Comparison of lasB population transcript accumulation to lasA (A) and toxA (B) population transcript accumulations. The results were sorted to generate a linear plot for the lasB hybridizations. The solid lines represent the sorted results. The dashed lines represent the trend line for lasA or toxA. (C) Comparison of lasA population transcript accumulation to toxA population transcript accumulation. The results were sorted to generate a linear plot for the lasA. The solid line represents the sorted results. The dashed line represents the trend line for toxA.
Determination of the half-life of lasR mRNA.
To determine if we could compare the transcript accumulations for the genes of interest, we determined the half-life of lasR. For P. aeruginosa PAO1 we found that the half-life of lasR mRNA was 2 min (44a). Thus, it appears that lasR mRNA has a considerably shorter half-life than toxA (10 min [24]), lasA (8.5 min [45]), and lasB (11 min [46]) mRNAs. The difference in mRNA half-lives between the transcripts suggested that we might have difficulty detecting a loose association between the genes because of the rapid degradation of the transcripts.
Comparison of transcript accumulations in an individual patient with CF.
To determine if there was a correlation between the various virulence factors and lasR transcript accumulation, we monitored a number of patients over a 3- to 5-year period. We have also included in our study the algD probe, because transcript accumulation for this gene has recently been shown to correlate with lasA and lasB transcript accumulations in the lung infections associated with CF (45).
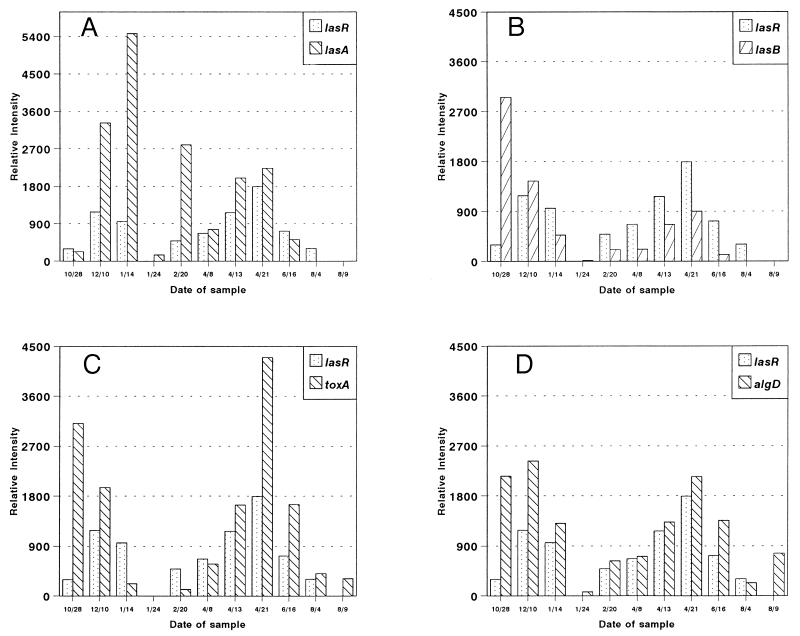
Figure 2 shows the results from one such patient (patient 15) over a period of about 1 year. In all but one sample the concentration of P. aeruginosa was ≥108 bacteria/ml of sputum. The one exception was the sample taken at the beginning of the study, on 28 October (5 × 106 bacteria/ml of sputum). These concentrations should be high enough to trigger the quorum-sensing regulation in P. aeruginosa. Figure 2 shows that lasR transcript accumulation seems to correspond to the transcript accumulations for all four other genes. In particular, in the last eight samples lasR transcript accumulation corresponded fairly tightly to toxA, lasB, lasA, and algD population transcript accumulations, whereas the first three samples showed less correlation between lasR population transcript accumulation and those for four other genes (Fig. 2). Overall, the results from this patient suggested that a correlation might exist between lasR and some of virulence factors produced by P. aeruginosa in the lungs of patients with CF. Thus, we wanted to analyze a larger number of samples from a number of CF patients at various stages of their disease.
FIG. 2.
Population transcript accumulations for P. aeruginosa lasR, lasA, lasB, toxA, and algD in sputum from a pediatric CF patient (patient 15). For each sample, 10 μg of the total RNA extracted from the pelleted bacterial population was blotted onto a Nytran membrane (Schleicher and Schuell) and hybridized with the toxA probe. The blots were then washed to remove the probe, and the membrane was rehybridized with the lasB probe. The process was repeated with the lasA, algD, and lasR probes. The relative intensity for the sample was measured from the autoradiograph with a soft laser scanning densitometer. The results represent 10 μg of RNA from equivalent numbers of cells. lasR population transcript accumulation was compared to lasA population transcript accumulation (A), lasB population transcript accumulation (B), toxA population transcript accumulation (C), and algD population transcript accumulation (D). Patient 15 is a male pediatric patient with severe lung disease. Patient 15 was receiving ciprofloxacin and tobramycin on 12/10, ceftazidime on 1/14, ciprofloxacin and tobramycin on 1/24 and 4/8, and ceftazidime again on 4/13 and 4/21.
Comparison of P. aeruginosa lasR population transcript accumulation to lasA, lasB, toxA, and algD population transcript accumulations.
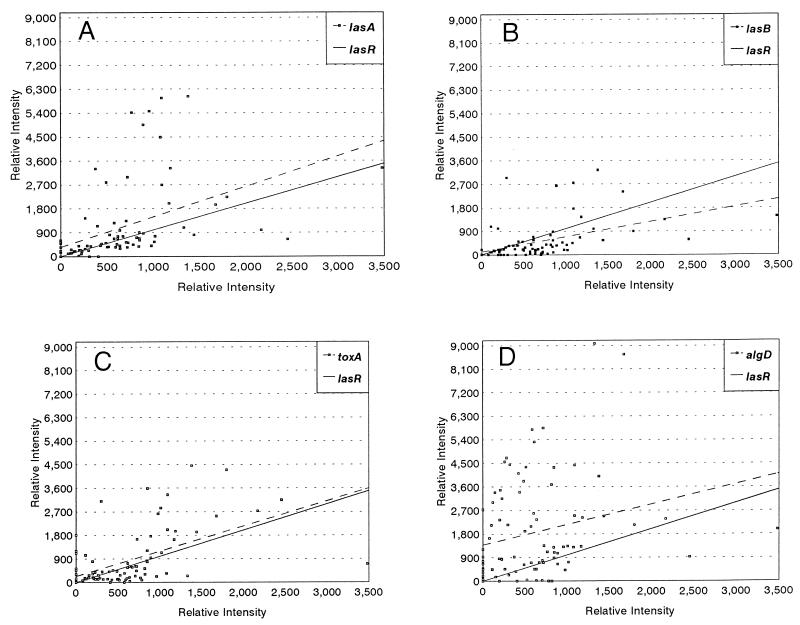
To take a systematic approach to the question of whether lasR transcript accumulation correlates to those for lasA, lasB, toxA, and algD, we probed 84 RNA samples extracted from the sputa from 16 pediatric CF patients. Table 1 shows the Spearman rank correlation coefficients for these comparisons. The highest correlation was between lasR and lasA (Table 1; Fig. 3A), followed by lasR and lasB (Table 1; Fig. 3B). The third best correlation was between lasR and toxA (Table 1; Fig. 3C). The weakest correlation was between lasR and algD (Table 1; Fig. 3). These results suggested that lasR might regulate or be coordinately regulated with lasA, lasB, and toxA. Furthermore, the relationship between lasR transcript accumulation and algD transcript accumulation might indicate a partial regulation of algD by lasR or a common environmental cue triggering the two genes.
FIG. 3.
Scatter plots for the population transcript accumulations for lasR, lasA, lasB, toxA, and algD. In these plots the results for 84 sputum samples are presented. The samples were prepared as described in the legend for Fig. 2. For all four panels the results were sorted to generate a linear plot for the lasR hybridizations. The solid lines represent the sorted results. The dashed lines represent the trend line for lasA, lasB, toxA, or algD. lasR population transcript accumulation was compared to lasA population transcript accumulation (A), lasB population transcript accumulation (B), toxA population transcript accumulation (C), and algD population transcript accumulation (D).
Potential cross-reaction between lasR and lasR-like genes from other organisms in the sputa.
Quorum-sensing mechanisms are found in numerous microorganisms. In particular, gram-negative bacteria utilize these systems to control numerous processes (12). In our study, 58% of the sputa collected contained at least one other pathogenic organism (Table 2). However, Table 2 also shows that in the majority of samples with other pathogenic organisms, P. aeruginosa was the predominant organism. Overall, in 75% of all the sputa analyzed, P. aeruginosa was the predominant organism by at least 100-fold. Nonetheless, a possible interfering factor in our analysis might be cross-reactivity of our probes with the mRNAs from other organisms. This was particularly a problem with lasR, because lasR or luxR homologs have been found in numerous organisms (12). To specifically address this problem, we segregated the 35 sputum samples that contained P. aeruginosa and no other pathogenic organisms and reanalyzed the results. Table 1 shows that for lasR-lasA, lasR-lasB, and lasR-toxA comparisons we still achieved strong, statistically significant correlations between the transcript accumulation patterns of these genes with the 35 sputum samples. For the comparison of lasR to algD, we also achieved a statistically significant correlation between the transcript accumulations for these genes, but the correlation was not as strong. Thus, in our analysis it did not seem to matter whether we used the total number of samples with P. aeruginosa as the predominant organism or samples with P. aeruginosa as the only pathogenic organism. Overall, this comparison suggested that cross-reactivity with our probes and the mRNAs from other microbes was not a significant factor in our analysis.
TABLE 2.
Sputum samples containing pathogenic organisms in addition to P. aeruginosaa
| Organism isolated from sputa | No. of sputa containing the microorganism | No. of sputa in which P. aeruginosa was the predominant pathogen |
|---|---|---|
| Candida albicans | 20 | 18 |
| Staphylococcus aureus | 13 | 11 |
| Streptococcus group A | 8 | 4 |
| Burkholderia cepacia | 8 | 2 |
| Stenotrophomonas maltophilia | 7 | 4 |
| Aspergillus sp. | 5 | 5 |
| Haemophilus influenzae | 2 | 0 |
| Serratia marcescens | 2 | 1 |
| Klebsiella oxytoca | 1 | 1 |
| Streptococcus group G | 1 | 1 |
| Citrobacter freundii | 1 | 0 |
| Enterobacter cloacae | 1 | 0 |
| Escherichia coli | 1 | 0 |
Samples were from the 84 sputum samples that were probed with the lasR probe.
DISCUSSION
Traditionally, analysis of coordinate regulation involves the deletion of the regulatory gene and an analysis of the mutant bacterium growing under laboratory conditions. Commonly, a single environmental cue is used to trigger the coordinate expression (26, 27). To establish the role of coordinate regulation in the pathogenicity of an organism, a regulatory mutant is usually tested in comparison to a wild-type strain in an inbred animal model. The situation with the chronic P. aeruginosa lung infections in CF patients is more complex. CF affects a wide range of individuals with different genetic backgrounds, who present with different clinical manifestations of their disease (52). The infections tend to involve numerous strains of P. aeruginosa with potentially different phenotypes (31, 39, 44). Finally, over time the patients have exacerbations of their respiratory symptoms. Our approach to the analysis of regulation in the lung infections associated with CF has been to look for correlations between the transcript accumulations for various genes in the bacterial populations found in the sputum taken directly from the patient. Thus, we are examining the organisms actually causing the infection rather than a single isolate introduced into an animal model. One difficulty with our approach is that different populations of bacteria in the sputum may be expressing different sets of genes at the same time. This may bias the results, because independently expressed genes may appear to correlate with each other or genes which are coordinately regulated may not appear to correlate. To partially minimize this problem, we have taken a large number of samples from a diverse set of patients over a fairly lengthy period of time. This ensured that we were sampling a diverse set of organisms exposed to a variety of environmental cues. Once the transcript accumulation has been measured for each sample, we then rank the results for each probe. The rankings are then compared for two probes with the Spearman rank correlation (50). For a statistically significant correlation to result, there has to be close association of the rankings across all the samples. For this approach to result in a false correlation, various subpopulations of bacteria would have to be expressing different genes at various levels across a wide range of patients and environmental conditions. A more likely scenario is that of a false null correlation in which the various subpopulations confuse the results by expressing different genes at different times. Thus, one might expect that we would have difficulties finding correlations between expression of genes even if they were regulated by the same transcriptional activator. However, in the current study and in past studies (38, 45) we have found statistically significant correlations between the transcript accumulations for various genes. These correlations occurred even with large sample sizes from a diverse set of patients (Table 1) (45). While these correlations do not exclude the possibility that subpopulations of bacteria may be expressing different genes, they do suggest that some process is coordinating the expression across the bacterial populations found in sputum.
A number of the quorum-sensing systems regulate genes involved with interactions between bacterial populations and their eucaryotic hosts (12). However, it has not been established that the quorum-sensing systems of P. aeruginosa are important in interactions with eucaryotic hosts. In particular, it has not been established that either of the quorum-sensing systems (LasR-LasI or RhlR-RhlI) regulates virulence factors during human infections. As a first step in determining the importance of the P. aeruginosa LasR-LasI quorum-sensing system in the lung infections associated with CF, we wanted to determine if there was a coordinated expression of some of the virulence factors that might be controlled by the LasR-LasI regulatory system. Additionally, we wanted to determine if there was a correlation of those virulence factors with lasR expression.
We speculated that the quorum-sensing systems of P. aeruginosa may play a role in the chronic lung infections associated with CF for a number of reasons. In these infections P. aeruginosa grows to a high bacterial density (43, 46) and is spatially limited by the lung environment. Both of these factors are critical to the triggering of quorum sensing in many diverse systems (12). Another reason we suspect that quorum sensing might play a role in these infections is the detection of lasA, lasB, and toxA transcript accumulation in the bacterial populations found in CF sputa (45). When P. aeruginosa is grown in the laboratory, it appears that lasA and lasB are tightly regulated by the LasR-LasI quorum-sensing system (14, 33) and that toxA transcription is modulated by the system (48). Therefore, if toxA, lasB, and lasA are coordinately regulated in the lung infections associated with CF, then a correlation might exist for the transcript accumulation patterns of these three genes. Indeed, Table 1 and Fig. 1 revealed a correlation of the transcript accumulations for the three genes during the chronic lung infections associated with CF. The strongest correlation was between lasA and lasB (Spearman rank correlation, n = 131, rs = 0.595, P < 0.001), followed by lasB and toxA (n = 131, rs = 0.570, P < 0.001) and lasA and toxA (n = 131, rs = 0.423, P < 0.001) (Table 1). The most likely explanations for these correlations are that in the lungs of CF patients, these genes might be coordinately regulated or might be responding to a common environmental cue.
In light of previous work showing that in a clonal situation lasA, lasB, and to some extent toxA were controlled by a common regulatory element, lasR (14, 33, 48), we decided to probe our RNA samples with a lasR-specific probe. The results from an individual patient (Fig. 2), as well as the results from numerous patients (Table 1; Fig. 3), revealed that lasR transcript accumulation correlated with lasA, lasB, and toxA transcript accumulations. A potential problem with our probes could be cross-reactivity with the mRNAs from other organisms found in the sputum. This is particularly a problem with the lasR probe, as lasR and luxR homologs have been found in a number of pathogenic organisms (12). To alleviate this problem, we segregated 35 of our sputum samples that contained only P. aeruginosa and no other pathogenic organism and reevaluated our data. Table 1 shows that the same correlations existed for the 35 samples with only P. aeruginosa as the pathogenic organism as for the sputa containing P. aeruginosa and at least one other pathogen. These results suggested that our probes were specific for the targeted genes.
Our results are the first to implicate LasR as a transcriptional activator in a human infection and to suggest that it might control multiple virulence factors in these infections. Furthermore, since lasR must be activated by PAI in order to transcribe lasA and lasB (14, 33), our results suggested that the LasR–LasI–PAI-1 autoinduction system was active in the lung infections associated with CF.
The significant but not perfect correlation between lasR, lasA, and lasB population transcript accumulations suggested that other regulatory systems might also be active in the lung environment. One possibility is the RhlR–RhlI–PAI-2 system, which has also been shown to influence the regulation of all three of these genes (3, 36, 49) and to interact with the LasR–LasI–PAI-1 system. Thus, one might speculate that a combination of control by the RhlR-RhlI and the LasR-LasI systems could account for our results. We are currently studying the role that the RhlR-RhlI system has in the regulation of P. aeruginosa virulence factors in the chronic lung infections associated with CF.
A curious finding of this study is the statistically significant correlation (n = 84, rs = 0.309, P < 0.01; n = 35, rs = 0.350, P < 0.05) of lasR population transcript accumulation to algD population transcript accumulation. While this association may not be particularly strong, it fits with a previous finding that in the lung infections associated with CF, algD population transcript accumulation correlates with lasA and lasB transcript accumulations (45). The correlation between lasR and algD could be explained in a number of ways. First, lasR may directly modulate expression of algD. Notably, there is a putative lasR box upstream of the algD promoter that matches the lasR box upstream of lasB at 9 of 20 nucleotides, which could possibly serve as a binding site. Second, one of the regulators of algD could be regulated by lasR, thus accounting for the partial correlation between lasR and algD. A third possibility is that a common environmental cue may control both systems and that this accounts for the correlation of population transcript accumulation between the two genes. A final explanation is that algD mRNA has the longest half-life of the transcripts and so by its longevity (45) and circumstances correlates to lasR transcript accumulation. This last explanation is not likely the case, because one would then expect to see a low level of correlation between algD and all the other transcripts which have longer half-lives than lasR. However, we previously reported that there was not a statistically significant association between algD and toxA (45). Thus, it is not likely that the correlation between lasR and algD population transcript accumulations occurs by chance.
The correlation of lasR population transcript accumulation to toxA transcript accumulation (Table 1) is also curious in light of the previous finding that there is a strong association between expression of the regAB operon and toxA in the lungs of patients with CF (38). In P. aeruginosa PAO1, LasR has been shown to enhance toxA transcription (48). However, this evidently does not occur by enhancement of regA transcription (48). At least some isolates from CF patients appear to regulate toxA in a manner different from that of strain PAO1 and similar to that of strain PA103 (38). Strain PA103 has a gene, regB, that appears to enhance activity from the P1 promoter of the regAB operon. Strain PAO1 lacks regB; thus, it is possible that LasR and RegB interact to promote expression of the P1 promoter and cause a high level of expression of toxA in strains from CF patients. Another possibility is that in isolates from CF patients, LasR may control toxA regulation by interacting with the regAB operon. A third possibility is that LasR may only partially regulate toxA expression and that regA is needed for full control of toxA expression. Further work will be needed to test these possibilities.
In summary, we revealed a correlation between lasA, lasB, and toxA population transcript accumulations that might suggest a coordinated regulation of these genes during the chronic lung infections associated with CF. We also detected lasR population transcript accumulation in the bacterial populations found in sputa. Furthermore, we found a correlation between lasR population transcript accumulation and those for four P. aeruginosa virulence genes. Thus, in the chronic lung infections associated with CF, lasR may at least partially regulate the virulence genes lasA, lasB, and toxA. This finding may indicate that the LasR-LasI quorum-sensing system may be active in these infections. Our results also suggested that lasR may either partially regulate algD or respond to the same environmental cues as algD.
ACKNOWLEDGMENTS
We thank B. H. Iglewski for the generous gift of plasmids pMJG1.7, containing the lasR clone, and pPS1816, containing the lasA clone. We also thank D. E. Ohman for plasmid pCC27, containing the algD clone. Thanks are due to K. Jamieson for her organization of the patient results and K. E. Sanderson and H. Ceri for critically reading the manuscript. We are indebted to the patients who participated in this study.
This study was supported by the RDP II of the Canadian Cystic Fibrosis Foundation and a grant to D.G.S. from the Canadian Cystic Fibrosis Foundation.
REFERENCES
- 1.Bainton N J, Bycroft B W, Chabra S R, Stead P, Geldhill L, Hill P J, Rees C E, Winson M K, Salmond G P, Stewart G S A B, Williams P. A general role for the lux autoinducer in bacterial cell signalling: control of antibiotic synthesis in Erwinia. Gene. 1992;116:87–91. doi: 10.1016/0378-1119(92)90633-z. [DOI] [PubMed] [Google Scholar]
- 2.Beck von Bodman S, Farrand S. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acylhomoserine lactone autoinducer. J Bacteriol. 1995;177:5000–5008. doi: 10.1128/jb.177.17.5000-5008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RHLR-RHLI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1996;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee A, Cui Y, Lui Y, Dumenyo C K, Chatterjee A K. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Appl Environ Microbiol. 1995;61:1959–1967. doi: 10.1128/aem.61.5.1959-1967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui Y, Chatterjee A, Liu Y, Dumenyo C K, Chatterjee A K. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J Bacteriol. 1995;177:5108–5115. doi: 10.1128/jb.177.17.5108-5115.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eberhard A, Burlingame A L, Eberhard C, Kenyon G K, Nealson K H, Oppenheimer N J. Structural identification of autoinducer of Photobacterium fischeri. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 7.Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 8.Engebrecht J, Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci USA. 1984;81:4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank D W, Iglewski B H. Kinetics of toxA and regA mRNA accumulation in Pseudomonas aeruginosa. J Bacteriol. 1988;170:4477–4483. doi: 10.1128/jb.170.10.4477-4483.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank D W, Storey D G, Hindahl M S, Iglewski B H. Differential regulation by iron of regA and toxA transcript accumulation in Pseudomonas aeruginosa. J Bacteriol. 1989;171:5304–5313. doi: 10.1128/jb.171.10.5304-5313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuqua W C, Winans S C. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuqua W C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 13.Gambello M J, Iglewski B H. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gambello M J, Kaye S, Iglewski B H. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granstrom M, Ericsson A, Strandvik B, Wretlind B, Pavlovskis O R, Berka R, Vasil M L. Relation between antibody response to Pseudomonas aeruginosa exoproteins and colonization/infection in patients with cystic fibrosis. Acta Paediatr Scand. 1984;73:772–777. doi: 10.1111/j.1651-2227.1984.tb17774.x. [DOI] [PubMed] [Google Scholar]
- 16.Gray K M, Pearson J P, Downie J A, Boboye B E A, Greenberg E P. Quorum sensing in the symbiotic nitrogen-fixing bacterium Rhizobium leguminosarum: autoinduction of a stationary phase and rhizosphere-expressed genes. J Bacteriol. 1996;178:372–376. doi: 10.1128/jb.178.2.372-376.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollsing A E, Granstrom M, Vasil M L, Wretlind B, Strandvik B. Prospective study of serum antibodies to Pseudomonas aeruginosa exoproteins in cystic fibrosis. J Clin Microbiol. 1987;25:1868–1874. doi: 10.1128/jcm.25.10.1868-1874.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang I, Li P, Zhang L, Piper K R, Cook D M, Tate M E, Farrand S K. TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc Natl Acad Sci USA. 1994;91:4639–4643. doi: 10.1073/pnas.91.11.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jagger K S, Robinson D L, Franz M N, Warren R L. Detection by enzyme-linked immunosorbent assays of antibody specific for Pseudomonas proteases and exotoxin A in sera from cystic fibrosis patients. J Clin Microbiol. 1982;15:1054–1058. doi: 10.1128/jcm.15.6.1054-1058.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones S, Yu B, Bainton N J, Birdsall M, Bycroft B W, Chhabra S R, Cox A J, Golby P, Reeves P J, Stephens S, Winson M K, Salmond G P C, Stewart G S A B, Williams P. The lux autoinducer regulates the production of exoenzyme determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 1993;12:2477–2482. doi: 10.1002/j.1460-2075.1993.tb05902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klinger J D, Straus D C, Hilton C B, Bass J A. Antibodies to proteases and exotoxin A of Pseudomonas aeruginosa in patients with cystic fibrosis: demonstration by radioimmunoassay. J Infect Dis. 1978;138:49–58. doi: 10.1093/infdis/138.1.49. [DOI] [PubMed] [Google Scholar]
- 22.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 23.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S A B, Lazdunski A, Williams P. Multiple homologues of LuxR and Luxl control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 24.Lory S. Effect of iron on accumulation of exotoxin A-specific mRNA in Pseudomonas aeruginosa. J Bacteriol. 1986;168:1451–1456. doi: 10.1128/jb.168.3.1451-1456.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGowan S, Sebaihia M, Jones S, Yu B, Bainton N, Chan, Bycroft B, Stewart G S, Williams P, Salmond G P. Carbapenem antibiotic production in Erwinia carotovora is regulated by CarR, a homologue of the LuxR transcriptional activator. Microbiology. 1995;141:541–550. doi: 10.1099/13500872-141-3-541. [DOI] [PubMed] [Google Scholar]
- 26.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller J F. Coordinate regulation and sensory transduction in the control of bacterial virulence. Science. 1989;243:916–922. doi: 10.1126/science.2537530. [DOI] [PubMed] [Google Scholar]
- 28.Moss R B, Hsu Y, Lewiston N J, Curd J G, Milgrom H, Hart S, Dyer B, Larrick J W. Association of systemic immune complexes, complement activation and antibodies to Pseudomonas aeruginosa lipopolysaccharride and exotoxin A with mortality in cystic fibrosis. Am Rev Respir Dis. 1986;133:648–652. doi: 10.1164/arrd.1986.133.4.648. [DOI] [PubMed] [Google Scholar]
- 29.Ochsner U A, Fiechter A, Reiser J. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J Biol Chem. 1994;269:19787–19795. [PubMed] [Google Scholar]
- 30.Ochsner U A, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogle J W, Janda J M, Woods D E, Vasil M L. Characterization and use of a DNA probe as an epidemiological marker for Pseudomonas aeruginosa. J Infect Dis. 1987;155:119–126. doi: 10.1093/infdis/155.1.119. [DOI] [PubMed] [Google Scholar]
- 32.Ohman D E, Cryz S J, Iglewski B H. Isolation and characterization of a Pseudomonas aeruginosa PAO mutant that produces altered elastase. J Bacteriol. 1980;142:836–842. doi: 10.1128/jb.142.3.836-842.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 34.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson J P, Passador L, Iglewski B H, Greenberg E P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pesci E C, Pearson J P, Seed P C, Iglewski B H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3133. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pirhonen M, Flego D, Heikinheimo R, Palva E T. A small diffusible molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 1993;12:2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raivio T L, Ujack E E, Rabin H R, Storey D G. Association between transcript levels of the Pseudomonas aeruginosa regA, regB, and toxA genes in sputa of cystic fibrosis patients. Infect Immun. 1994;62:3506–3514. doi: 10.1128/iai.62.8.3506-3514.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Römling U, Fiedler B, Bosshammer J, Grothues D, Greipel J, von der Hardt H, Tümmler B. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J Infect Dis. 1994;170:1616–1621. doi: 10.1093/infdis/170.6.1616. [DOI] [PubMed] [Google Scholar]
- 40.Ryan B F, Joiner B L. Minitab handbook. Belmont, Calif: Duxbury Press; 1994. [Google Scholar]
- 41.Schripsema J, de Rudder K E E, van Vliet T B, Lankhorst P P, de Vroom E, Greenberg E P. Bacteriocin small of Rhizobium leguminosarum belongs to the class of N-acyl-homoserine lactone molecules, known as autoinducers and as quorum sensing co-transcription factors. J Bacteriol. 1996;178:366–371. doi: 10.1128/jb.178.2.366-371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seed P C, Passador L, Iglewski B H. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J Bacteriol. 1995;177:654–659. doi: 10.1128/jb.177.3.654-659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith A L, Redding G, Doershuk C, Goldmann D, Gore E, Hilman B, Marks M, Moss R, Ramsey B, Rubio T, Schwartz R H, Thomassen M J, Williams-Warren J, Weber A, Wilmott R W, Wilson H D, Yogev R. Sputum changes associated with therapy for endobronchial exacerbation in cystic fibrosis. J Pediatr. 1988;112:547–554. doi: 10.1016/s0022-3476(88)80165-3. [DOI] [PubMed] [Google Scholar]
- 44.Speert D P, Campbell M E, Farmer S W, Volpel K, Joffe A M, Paranchych W. Use of a pilin gene probe to study molecular epidemiology of Pseudomonas aeruginosa. J Clin Microbiol. 1989;27:2589–2593. doi: 10.1128/jcm.27.11.2589-2593.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Storey, D. G., and E. E. Ujack. Data not shown.
- 45.Storey D G, Ujack E E, Mitchell I, Rabin H R. Positive correlation of algD transcription to lasB and lasA transcription by populations of Pseudomonas aeruginosa in the lungs of patients with cystic fibrosis. Infect Immun. 1997;65:4061–4067. doi: 10.1128/iai.65.10.4061-4067.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Storey D G, Ujack E E, Rabin H R. Population transcript accumulation of Pseudomonas aeruginosa exotoxin A and elastase in sputa from patients with cystic fibrosis. Infect Immun. 1992;60:4687–4694. doi: 10.1128/iai.60.11.4687-4694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang H B, DiMango E, Bryan R, Gambello M, Iglewski B H, Goldberg J B, Prince A. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect Immun. 1996;64:37–43. doi: 10.1128/iai.64.1.37-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toder D S, Gambello M J, Iglewski B H. Pseudomonas aeruginosa LasA: a second elastase under the transcriptional control of lasR. Mol Microbiol. 1991;5:2003–2010. doi: 10.1111/j.1365-2958.1991.tb00822.x. [DOI] [PubMed] [Google Scholar]
- 49.Winson M K, Camara M, Latifi A, Foglino M, Chhabra S R, Daykin M, Bally M, Chapon V, Salmond G P, Bycroft B W, Lazdunski A, Stewart G S A B, Williams P. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zar J H. Rank correlation. In: McElroy W D, Swanson C P, editors. Biostatistical analysis. Englewood Cliffs, N.J: Prentice-Hall, Inc.; 1974. pp. 243–245. [Google Scholar]
- 51.Zhang L, Murphy P J, Kerr A, Tate M E. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature. 1993;362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]
- 52.Zielenski J, Tsui L C. Cystic fibrosis: genotypic and phenotypic variations. Annu Rev Genet. 1995;29:777–807. doi: 10.1146/annurev.ge.29.120195.004021. [DOI] [PubMed] [Google Scholar]