Abstract
Introduction
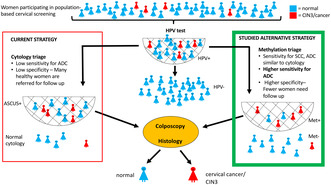
Methylation analysis of the promoter region of tumor‐suppressor genes has previously shown high sensitivity for detection of high‐grade cervical intraepithelial neoplasia (CIN) and cancer. HPV‐testing has a high sensitivity to identify women at risk to develop cancer, and has been implemented in cervical screening programs in several countries. But in most HPV‐positive women the infection will clear and they will not develop cancer. Testing for methylation could help to identify women who have potentially progressive cervical disease and need closer follow‐up. The goal of the present study was to investigate the potential use of methylation as a triage test of HPV‐positive women in the screening program.
Material and methods
A collection of liquid‐based cytology (LBC) samples from 106 women, collected between 4 months and 8 years before histologically confirmed cervical cancer or CIN3, was analyzed for hypermethylation of the human genes FAM19A4 and miR124‐2.
Results
Methylation was detected in 45% (33/73) of normal LBC samples from women who later developed CIN3+, compared with 10% (3/31) of normal LBC samples from women without subsequent dysplasia (P = 0.0006). Overall, methylation was detected in 39% (14/36), 51% (19/37), 61% (14/23) and 70% (7/10) of LBC samples from women who later developed CIN3, adenocarcinoma in situ (AIS), squamous cell carcinoma (SCC) and adenocarcinoma (ADC), respectively. Positive methylation analysis was not significantly more frequent than abnormal cytology of atypical squamous cells of unclear significance or worse (ASCUS+) in LBC samples collected 4 months to 8 years before SCC or AIS; however, prior to the development of ADC, methylation was observed in 7/10 LBC samples, despite normal cytology. Overall, LBC samples collected before invasive cancer (ADC and SCC) were more frequently positive in the methylation analysis than in cytological analysis of ASCUS+ (P = 0.048). For LBC samples collected more than 2 years before the development of AIS, SCC or ADC, methylation analysis showed a higher positivity rate than cytology did.
Conclusions
Testing for methylation of FAM19A4/miR124‐2 as a triage for HPV‐positive women would be useful to identify women at risk of cancer development, especially adenocarcinoma. Further studies are needed to estimate the cost‐effectiveness before introducing methylation testing in the screening program.
Keywords: cervical cancer, cervical dysplasia, cytology, HPV, methylation
We analyzed hypermethylation of the human genes FAM19A4/miR124‐2 in LBC samples from HPV‐positive women who later developed CIN3 or cancer. The methylation analysis was significantly better than cytology to detect future adenocarcinoma in these women.
Abbreviations
- ADC
adenocarcinoma
- AIS
adenocarcinoma in situ
- ASCUS
atypical squamous cells of unclear significance
- ASCUS+
ASCUS or more severe cytology
- CIN2
cervical intraepithelial neoplasia grade 2
- CIN3
cervical intraepithelial neoplasia grade 3
- CIN3+
CIN3 or worse (ie CIN3, cancer in situ or cancer)
- HPV
human papilloma virus
- HSIL
high‐grade squamous intraepithelial lesion (includes CIN2 and CIN3)
- LBC
liquid‐based cytology
- SCC
squamous cell carcinoma
Key message.
Analysis of methylation of the human genes FAM19A4/miR124‐2 in LBC samples from HPV‐positive women showed the same sensitivity as cytology to predict future squamous cell carcinoma and adenocarcinoma in situ, and a higher sensitivity for detection of adenocarcinoma.
1. INTRODUCTION
Cervical cancer screening has been successful in decreasing the number of cervical cancer cases. 1 Human papilloma virus (HPV) testing is more sensitive than cytology for finding women at risk of developing severe dysplasia or cancer. 2 , 3 For this reason, primary HPV testing has been implemented in the screening program in most parts of Sweden, as well as in other countries. However, in most of the HPV‐positive women, the infection will clear and they will not develop cancer. Only persisting HPV infections increase the cancer risk. 4 Therefore, to avoid follow‐up of healthy women, which could cause unnecessary anxiety among the women as well as inefficient use of resources, an additional test is needed. Cytology is often used for triage to help identify the fraction of HPV‐positive women in need of closer follow‐up. 5 According to Swedish guidelines, HPV‐positive women who have abnormal cytology of atypical squamous cells of undetermined significance or worse (ASCUS+) are referred for colposcopy, whereas HPV‐positive women with normal cytology are retested after 18 months when positive for HPV 16 or 18, and after 3 years when positive for other high‐risk HPV types. Continued evaluation of the screening program has shown that extremely few HPV‐negative women have severe histology‐confirmed dysplasia, even if the cytology is abnormal. 6 , 7 , 8
Cytology has a relatively low sensitivity and requires long training and continuous work to maintain high quality, which is a challenge for some laboratories. 9 Another problem with cytology triage is that cytotechnologists are hesitant to label the cytology as normal if they are aware that the woman is high‐risk HPV‐positive. 10 , 11 , 12 In addition, cytology has a low sensitivity to identify dysplasia in postmenopausal women. 12 , 13 , 14 Since the introduction of cytology screening in Sweden in the late 1960s, the incidence of adenocarcinoma has not decreased, unlike the incidence of squamous cell carcinoma, illustrating the difficulty of the cytology‐based screening program to prevent adenocarcinoma. 15
Genetic and epigenetic changes of the host cells are important parts in the oncogenic process, and DNA methylation of promoter regions can silence tumor suppressor genes. 16 Several human genes such as CADM1, EPB41L3, FAM19A4, MAL, miR‐124, PAX1 and SOX1 have been found to be hypermethylated in cervical cancer as well as in CIN2 and CIN3. 17 Methylation of certain nucleotides of the L1 and L2 genes of the HPV16 have also been associated with cancer development. The S5‐classifier gives a score by combination of methylation data from the human gene EPB41L3 and the HPV 18 and has been shown to identify women with CIN2/3 as well as to predict progression of untreated lesions. 19 Pyrosequencing is needed for the S5‐classifier, which can be a challenge for clinical laboratories with large volumes of screening samples. We have chosen to evaluate hypermethylation (in the following text referred to as “methylation”) of the genes FAM19A4 and miR124‐2, available as a commercial test, which has in several studies shown high sensitivity for detection of cervical intraepithelial neoplasia or worse (CIN3+). 20 , 21 , 22 , 23
The aim of this study was to determine the proportion of methylation in liquid‐based cytology (LBC) samples, collected prior to the development of histologically confirmed CIN3 and cervical cancer. We also wanted to evaluate how the methylation analysis performed in comparison with cytology, at different time points before the diagnosis of CIN3+.
2. MATERIAL AND METHODS
2.1. Large LBC collection from women who developed CIN3+
We have a collection of 1225 LBC samples, stored at −80°C, from women who developed cervical intraepithelial neoplasia (CIN3) or cancer (CIN3+) diagnosed by histology at Clinical Pathology Malmö, between November 2007 and December 2015 (Table 1). At sampling, the women were between 15 and 85 years of age, average 33 years, and 94% within the screening age, defined as 23–70 years. The LBC samples were taken 4 months to 8 years prior to the CIN3+ diagnosis and were collected between May 2007 and January 2012. Information on cytological and histological diagnoses, date of sampling and age of the women, was available in the pathology patient registry. The samples (n = 1225) had previously been used to evaluate HPV‐mRNA and HPV‐DNA tests and 85% were positive for HPV‐mRNA, HPV‐DNA or both. 24 The histological diagnoses of the women with stored LBC samples were as follows: CIN3 n = 1094, squamous cell carcinoma (SCC) n = 32, adenocarcinoma in situ (AIS) n = 45, adenocarcinoma (ADC) n = 18 (Table 1).
TABLE 1.
Characteristics of the biobanked LBC samples, a n = 1225.
| Histological diagnosis | Time between LBC sample and histologic diagnosis | |||||||
|---|---|---|---|---|---|---|---|---|
| 4 months‐2 years | >2 to 5 years | >5 to 8 years | 4 months to 8 years | |||||
| ASCUS+/total | % ASCUS+ | ASCUS+/total | % ASCUS+ | ASCUS+/total | % ASCUS+ | ASCUS+/total | % ASCUS+ | |
| CIN3 | 471/507 | 93 | 97/472 | 21 | 21/115 c | 18 | 589/1094 c | 54 |
| AIS | 17/17 | 100 | 4/17 | 24 | 1/11 | 9 | 22/45 | 49 |
| SCC | 18/20 | 90 | 0/11 | 0 | 0/1 | 0 | 18/32 | 56 |
| ADC | 2/5 | 40 | 0/8 | 0 | 0/5 | 0 | 2/18 | 11 |
| Other cancers b | 4/15 | 27 | 2/16 | 13 | 0/5 | 0 | 6/36 | 17 |
| Total | 512/564 | 91 | 103/524 | 20 | 22/137 | 16 | 637/1225 | 52 |
Abbreviations: ADC, adenocarcinoma; AIS, adenocarcinoma in situ; ASCUS+, atypical squamous cells of unclear significance or worse; CIN3, cervical intraepithelial neoplasia grade 3; SCC, squamous cell carcinoma.
Number of samples in different time intervals between histological samples (differentiated by histological diagnosis) that defined the cohort (collected between November 2007 and December 2015) and the preceding LBC sample (collected between May 2007 and January 2012), and the share of LBC samples with ASCUS+ cytology.
Other cancers than cervical carcinoma, eg metastases, lymphoma, transitional epithelium, endometrioid adenocarcinoma and sarcoma.
One sample was collected 8 years and 4 months before histological diagnosis of CIN3+, with ASCUS+ cytology.
2.2. Selection of LBC samples for methylation assay
Some of the LBC samples from women with invasive cancer (n = 50) and AIS had been used in other experiments and did not contain enough volume for methylation testing. All available LBC samples from women who developed AIS (n = 37), SCC (n = 23) or ADC (n = 10), were analyzed for methylation, together with a fraction of samples from women who developed CIN3 (n = 36). The mean age of the tested women was 38.0 years at the time of the LBC sample. The ages of the women with different histological diagnoses are presented in Table S1, along with the mean time between the cytology sample and the histological diagnosis CIN3+. To test whether normal cytology samples collected before histological CIN3+ would be methylation‐positive, the majority of the selected samples had normal cytology (73/106; 69%). Analysis of LBC samples collected before histological diagnoses of AIS, SCC, ADC or CIN3 found benign cytology in 54% (20/37), 48% (11/23), 100% (10/10) and 89% (32/36) of the cases, respectively.
2.3. Healthy controls
LBC samples from women in the Malmö area who took part in the screening program between 2007 and 2012 were used as controls (n = 31). These women had normal cytology in the methylation‐analyzed sample, as well as normal cytology and/or a negative HPV result in the subsequent round of screening 3–5 years later. The pathology patient registry was checked to ensure that there were no subsequent (abnormal) histological results for these women until 2022. The samples were anonymized prior to the testing. The healthy controls were of similar age as the women with CIN3+ (41.4 years on average; Table S1).
2.4. Dynamics of methylation and cytology in LBC samples collected prior to CIN3+
To investigate the dynamics of methylation and cytology, the time from sampling of LBC to histological diagnosis of CIN3+ was calculated, and stratified in periods of 4 months to 2 years, 2–5 years and 5–8 years (Table 3, Table S2). The cytology diagnoses for all samples in the large collection (n = 1225) at these time intervals before histology CIN3+, are presented in Table 1.
TABLE 3.
Proportions of methylated LBC (liquid‐based cytology) samples, collected at different time points prior to histological diagnosis of CIN3+.
| Histology | 4 months to 2 years before histological diagnosis | >2 to 5 years before histological diagnosis | >5 to 8 years before histological diagnosis | 4 months to 8 years before histological diagnosis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All cytologies | Normal cytology | Cytology ASCUS + | All cytologies | Normal cytology | Cytology ASCUS+ | All cytologies | Normal cytology | Cytology ASCUS+ | All cytologies | Normal cytology | Cytology ASCUS+ | |
| CIN3 | 46% (12/26) | 41% (9/22) | 75% (3/4) | NA | NA | NA | 20% (2/10) | 20% (2/10) | NA | 39% (14/36) | 34% (11/32) | 75% (3/4) |
| AIS | 50% (7/14) | NA | 50% (7/14) | 67% (8/12) | 60% (6/10) | 100% (2/2) | 36% (4/11) | 40% (4/10) | 0% (0/1) | 51% (19/37) | 50% (10/20) | 53% (9/17) |
| SCC | 77% (10/13) | 100% (1/1) | 75% (9/12) | 44% (4/9) | 44% (4/9) | NA | 0% (0/1) | 0% (0/1) | NA | 61% (14/23) | 45% (5/11) | 75% (9/12) |
| ADC | 50% (1/2) | 50% (1/2) | NA | 86% (6/7) | 86% (6/7) | NA | 0% (0/1) | 0% (0/1) | NA | 70% (7/10) | 70% (7/10) | NA |
| All | 55% (30/55) | 44% (11/25) | 63% (19/30) | 64% (18/28) | 62% (16/26) | 100% (2/2) | 26% (6/23) | 27% (6/22) | 0% (0/1) | 51% (54/106) | 45% (33/73) | 64% (21/33) |
Abbreviations: ADC, adenocarcinoma; AIS, adenocarcinoma in situ; ASCUS+, atypical squamous cells of unclear significance or worse; CIN3, cervical intraepithelial neoplasia grade 3; SCC, squamous cell carcinoma.
2.5. DNA extraction
The LBC samples were originally collected in SurePath medium, pelleted and stored at −80°C. Before DNA extraction, the samples were resuspended in 420 μL of Digene Specimen Transport Medium (STM, Qiagen) and 4 μL Proteinase K (50 mg/mL, Qiagen) was added to the solution following incubation overnight at 37°C. The pretreatment with Proteinase K is said to reduce protein cross‐linking which occurs in samples stored in SurePath medium. 25 A portion of this study, 41 samples, did not undergo Proteinase K treatment before DNA extraction. Low DNA concentrations (<10 ng/μL) were observed for these samples, whereas for samples with Proteinase K treatment we achieved DNA concentrations >200 ng/μL. DNA was extracted from a 400‐μL sample, diluted 1:4 in H2O, and eluted in 50 μL, using a routine standard operating DNA extraction protocol, based on the magLEAD 12cG (PSS bio system net). DNA concentrations were measured in Qubit® 2.0 (Invitrogen).
2.6. Bisulfite conversion
Bisulfite conversion was performed with the EZ DNA Methylation kit (Zymo Research) according to the manufacturer's instructions.
2.7. Methylation analysis
Bisulfite‐converted DNA was used as input for QIAsure Methylation test (Qiagen). The standard DNA input was 2000 ng, but for samples with reduced DNA concentrations, as low as 100 ng was used. The methylation test was performed according to the manufacturer's instructions, where 2.5 μL bisulfite‐converted DNA was used as input for the PCR, conducted on the Rotor‐Gene Q MDx 5plex HRM instrument (Qiagen). The ASSAYMANAGER software enables automated interpretation of the results and uses the housekeeping gene β‐actin (ACTB) as a quality control and reference for successful bisulfite‐conversion. The software calculates the ddCt values for both FAM19A4 and hsa‐miR124‐2, and a sample is positive for hypermethylation if either FAM19A4 or hsa‐miR124‐2 or both are above the cutoff point. When ACTB was not detected, the analysis was invalid according to the manufacturer's instructions.
LBC samples (n = 113) from women who developed CIN3+ were analyzed, along with 31 healthy controls (described above).
2.8. Statistical analyses
Associations between proportions of methylated samples and development of CIN3+, and between proportions of methylated samples and abnormal cytology, were calculated using Fisher's exact test, GRAPHPAD Software (https://www.graphpad.com). P < 0.05 was considered statistically significant. Confidence intervals were calculated by the modified Wald method, using the GRAPHPAD Software. The Chi‐square test for trend was performed to assess the increased proportion of methylated samples with increasing severity of histological diagnosis, using Epitools (https://epitools.ausvet.com.au).
3. RESULTS
Valid methylation results were obtained from 94% (106/113) of the samples. Overall, 51% (54/106) of the tested samples, 45% (33/73) of samples with normal cytology and 64% (21/33) of samples with ASCUS+, showed hypermethylation of either FAM19A4 (n = 28) or miR124‐2 (n = 3) or both genes (n = 23) (Table 2).
TABLE 2.
Methylation results for normal and abnormal LBC samples collected 4 months to 8 years before histologically confirmed CIN3+, with results for the subset with cancer shown separately.
| Cytology | Women with histology CIN3+ 4 months to 8 years later | Women with histologically verified cancer (SCC, ADC) 4 months to 8 years later | ||||
|---|---|---|---|---|---|---|
| Methylation‐ positive | Methylation‐ negative | Total | Methylation‐positive | Methylation‐negative | Total | |
| ASCUS+ | 21 | 12 | 33 (31%) | 9 | 3 | 12 (36%) |
| Normal | 33 | 40 | 73 (69%) | 12 | 9 | 21 (64%) |
| Total | 54 (51%) | 52 (49%) | 106 (100%) | 21 (64%) | 12 (36%) | 33 (100%) |
Abbreviations: ADC, adenocarcinoma; ASCUS+, atypical squamous cells of unclear significance or worse; CIN3+, CIN3, cancer in situ or cancer; SCC, squamous cell carcinoma.
Ten percent (3/31) of the healthy controls were positive for methylation, giving a specificity of approximately 90% (95% CI 74%–98%). Methylation was significantly more frequent in samples from women who developed CIN3+ than in samples from the healthy controls (P < 0.0001). This also applied to samples with normal cytology from women with later CIN3+ (P < 0.001).
LBC samples from women with subsequent CIN3, AIS, SCC and ADC histology were positive for methylation in 39% (14/36), 51% (19/37), 61% (14/23) and 70% (7/10) of the cases, respectively (Table 3). The proportion of methylated LBC samples increased significantly along with the severity of the dysplasia (Chi‐square for linear trend P < 0.0001).
3.1. Correlation between cytology and methylation
Overall, we observed low agreement (57%, Kappa 0.157) between ASCUS+ cytology and methylation among the women with future CIN3+ lesions (n = 106) (Table 2). The proportion of methylation was similar for samples with ASCUS+ cytology and samples with normal cytology (64% vs 45%). However, 89% (95% CI 55%–100%, 8/9) of samples with more severe cytology, CIN3, were methylated.
Regarding only LBC samples with normal cytology, methylation was detected in one‐third to 70% of women with subsequent CIN3+, depending on the histological diagnosis (Table 3).
3.2. Methylation and cytology prior to CIN3
Of the samples selected for methylation analysis, from women with subsequent histology CIN3, 11% (4/36) had ASCUS+ cytology. This is a lower share than for the total number of LBC samples collected before CIN3 in the large collection, where 54% (589/1094) had ASCUS+ cytology (Table 1). Despite mainly normal cytology, 39% of the tested samples collected before CIN3 were methylation‐positive, 46% within 24 months (Table 3). The women with positive methylation tests were of similar age to the women with negative methylation tests prior to CIN3, on average 35.6 and 33.2 years, respectively.
3.3. Methylation and cytology prior to cancer
Regarding only invasive cancer (SCC, ADC), 64% (21/33) and 36% (12/33) of the previous LBC samples were methylated and had ASCUS+, respectively (Table 2). The methylation assay identified more cases compared with cytology (P = 0.048). Among SCC, 61% (14/23) of the samples were methylation‐positive and 52% (12/23) had ASCUS+ cytology. For ADC, 70% (7/10) of the analyzed samples were methylation‐positive, despite all having normal cytology (Table 3). Within 12 months prior to SCC, 90% (9/10) LBC samples were methylation‐positive. The only methylation‐negative sample came from a woman with a microinvasive squamous cell carcinoma.
ASCUS+ cytology was seen in 56% (18/32) of LBC samples collected 4–95 months before SCC (histological diagnosis) and in 11% (2/18) of samples collected before ADC (Table 1). Figure 1 shows the percentage of samples with ASCUS+ cytology and the percentage positive for methylation 4–60 months before CIN3, AIS, SCC and ADC.
FIGURE 1.
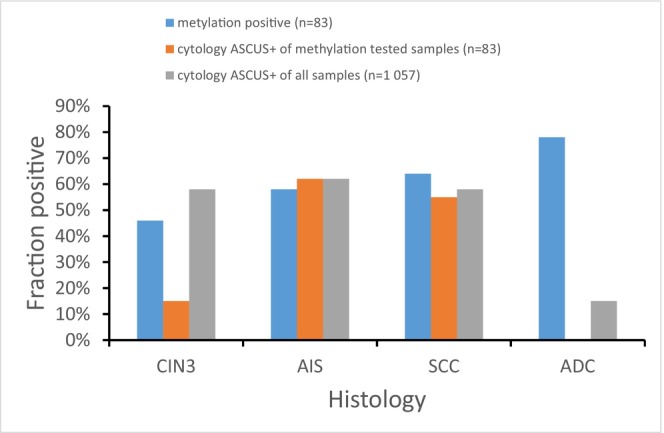
Percentage of LBC samples methylation‐positive, positive for ASCUS+ cytology among samples tested for methylation, and positive for ASCUS+ cytology among all samples in the large collection, respectively, from women with histological diagnosis of CIN3+, 4–60 months later. ADC, adenocarcinoma; AIS, adenocarcinoma in situ; ASCUS+, atypical squamous cells of unclear significance or worse; CIN3, cervical intraepithelial neoplasia grade 3; SCC, squamous cell carcinoma.
3.4. Dynamics of methylation and ASCUS+ cytology prior to CIN3+
The proportion of methylated samples, stratified for different time spans before the histological diagnosis, is shown in Table 3. Among women who developed CIN3+, methylation was detected in 57% (27/51) and 27% (6/22) of LBC samples with normal cytology, collected 4–60 months and 61–95 months before CIN3+, respectively. The proportion of methylated normal cytology samples, collected 4 months to 5 years before CIN3+, was higher than the proportion of methylation in samples from the healthy controls (57% vs 10%, P < 0.0001), whereas no statistical difference was observed between normal cytology samples taken more than 5 years before CIN3+ and samples from the controls (27% vs 10%).
Generally, samples collected up to 5 years prior to the histological diagnosis were methylation‐positive slightly more often than samples collected up to 8 years earlier. LBC samples were positive for methylation in 46% (12/26), 58% (15/26), 64% (14/22) and 78% (7/9) of the cases up to 5 years before a histological diagnosis of CIN3, AIS, SCC and ADC, respectively (Table 3).
A comparison of the proportion of samples with ASCUS+ cytology and the proportion of methylation‐positive samples is shown in Figure 2 and Table S2.
FIGURE 2.
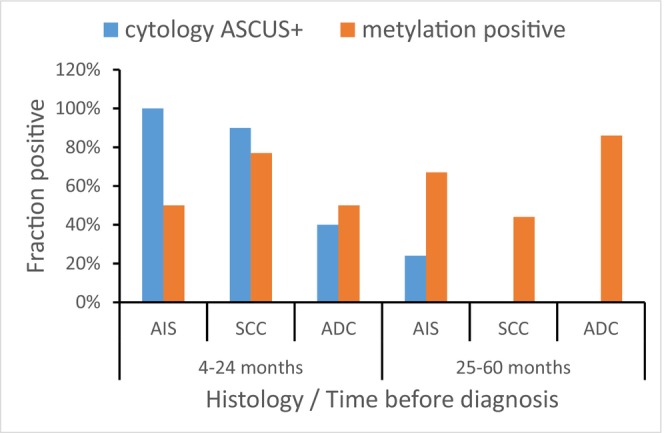
Dynamics of ASCUS+ and methylation in women who developed AIS and cancer. The proportions of ASCUS+ cytology were derived from a large LBC collection of samples (n = 1225), whereas corresponding proportions of methylated samples were calculated from the samples analyzed for methylation (n = 106). ADC, adenocarcinoma; AIS, adenocarcinoma in situ; ASCUS+, atypical squamous cells of unclear significance or worse; CIN3, cervical intraepithelial neoplasia grade 3; SCC, squamous cell carcinoma.
Between 2 and 5 years before the diagnosis of AIS, methylation was more frequent than ASCUS+ cytology (67% vs 24%; P = 0.03). No samples had abnormal cytology 2–5 years before the diagnosis of cancer, but 44% (4/9) were methylation‐positive before the diagnosis of SCC and 86% (6/7) before ADC (Figure 2). Also for samples collected up to 8 years before ADC, a positive methylation test was more common than ASCUS+ cytology (70% vs 11%; P = 0.002). For AIS and SCC, up to 8 years before the diagnosis, the proportion of methylation‐ and cytology‐positive samples was similar, but more than 2 years before the diagnosis, methylation was more frequent than ASCUS+ cytology (AIS, P = 0.0016; SCC, P = 0.029) whereas up to 2 years before diagnosis of AIS, ASCUS+ cytology was more frequent (P = 0.0013). No significant difference was seen between methylation and cytology for samples collected up to 2 years prior to SCC (Figure 2, Table S2).
For samples taken >5 years before histological diagnosis of CIN3+, there was no statistical difference between the proportion of methylation (27%, 6/22) and ASCUS+ (16%, 22/127).
3.5. Sensitivity for CIN3+ for cytology combined with methylation
In all, 31% (33/106) of the methylation‐tested samples showed ASCUS+ cytology, 51% (54/106) were positive for methylation (Table 2), and 61% (66/106) were positive for either cytology (defined as ASCUS+) or methylation.
4. DISCUSSION
LBC samples collected 4 months to 8 years before histological diagnosis CIN3+ were methylation‐positive and positive for ASCUS+ cytology in similar proportions. The methylation assay identified as many cases of future SCC and a higher number of future ADC than did cytology. Testing methylation of FAM19A4/miR124‐2 for triage of HPV‐positive women could therefore be considered, especially in settings without access to cytology, for instance in low‐income countries. 26
The proportion of methylated LBC samples increased with the severity of the developed lesions, which is in agreement with previous studies. 27 , 28 The elevated methylation levels are suggestive of progressive CIN disease and may be related to a higher number of methylated cells, and larger and more genetically aberrant lesions. 29 Concerning our cancer cases (SCC and ADC), 73% (11/15) had methylated LBC samples up to 2 years prior to the cancer, which is similar to that in a large European multicenter study where 95% (19 of 20) of cervical cancers were methylated. 30
Only about half of the LBC samples collected within 24 months before CIN3 histological diagnosis were methylation‐positive. This is in agreement with previous studies which have shown that only about two‐thirds of CIN3 lesions are methylated, and that methylation is more common in CIN3 lesions that have persisted for a long time, and in those that are likely to progress. 27 , 28 , 29 If CIN3 lesions that are likely to regress are not detected in the methylation test, this could actually be an advantage. Treatment of precancerous lesions can be associated with an increased risk for premature birth 31 and it is therefore crucial to avoid overtreatment. In this study, we were not able to investigate whether CIN 3 would have regressed more often in women with a negative methylation test, since all women with CIN3 all underwent conization, according to routine practice.
Within 2 years before the histological diagnosis of CIN3+, the vast majority (91%) of LBC samples showed ASCUS+ cytology, whereas the percentage of methylation‐positive samples was lower (55%). But the fact that many of the women with dysplasia were diagnosed thanks to an abnormal cytology sample, means that there will be a selection towards a higher percentage of abnormal cytology than if a different test, for example methylation, had been used primarily. No abnormal cytology was seen in samples taken 2–5 years before the diagnosis of SCC (n = 11) or ADC (n = 8). The probable explanation for this is that if abnormal cytology had been detected, the women would have been referred for prompt follow‐up and would not have waited until the next cytology sample.
Another explanation could be that the total number of ASCUS+ diagnoses is high, about 50% of HPV‐positive women participating in the screening in 2017 had ASCUS+ cytology, 32 but the specificity for severe dysplasia is low. The majority (77%) of cytology samples with ASCUS+ have the diagnoses ASCUS and LSIL. 32 Data from the Swedish National Cervical Screening Registry (https://nkcx.se/index_e.htm) shows that only 4% of women with ASCUS cytology in 2020 had a histological diagnosis of HSIL, AIS or cancer within 12 months. The corresponding number for LSIL was 10%.
Methylation was seen in LBC samples collected 2–5 years before the diagnosis, to a greater extent than abnormal cytology (ASCUS+). For samples taken more than 5 years before histological diagnosis of CIN3+, there was no statistical difference between the proportions of methylation‐positive and ASCUS+ cytology, or between women who developed CIN3+ and healthy controls. Thus, it seems this methylation assay can detect dysplasia earlier than cytology, but not >5 years ahead. Since only one LBC sample of each SCC and ADC was taken more than 5 years before cancer, it was not possible to draw any conclusions on the possibilities to detect methylation >5 years prior to invasive cancer. However, De Strooper et al. reported that substantial proportions of cancer cases had methylated cytology samples 5–9 years (64% methylated) and 10–14 (33% methylated) years prior to cancer, indicating that methylation can be detected early during cancer development, when the cytology is frequently classified as normal. 22 Identification of women several years before development of cancer or severe dysplasia may not be purely beneficial, since it would create the need for long‐term follow‐up of these women and could cause unnecessary anxiety.
Even though methylation was frequently seen in LBC samples collected 2–5 years before CIN3+, the proportion of methylation was generally higher among samples collected a short time before the diagnosis. There was a higher proportion of methylated samples collected within 5 years before the CIN3+ diagnosis, than among samples taken a longer time before the diagnosis, consistent with the accumulation of methylated promoter tumor suppressor genes during cancer and pre‐cancer development. 28
Methylation was detected in the majority (7/10) of LBC samples from women with normal cytology who subsequently developed ADC. Only 11% (2/18) of all cases of ADC had abnormal cytology in LBC samples collected 4 months to 5 years before the diagnosis. Even within 2 years from the diagnosis, only two of five ADC cases showed abnormal cytology, illustrating the difficulty of finding these cases through cytology. The prevalence of adenocarcinoma has not decreased since the introduction of cytological cervical cancer screening in Sweden in the 1960s. 15 In fact, the incidence of adenocarcinoma was 31% higher during 2014–2015 than during 2002–2013. 33 A probable reason for this is the difficulty of identifying adenocarcinoma and its precursors through cytology, even when searching carefully. In a nationwide Swedish review of cytology samples taken before CIN3+, the diagnosis was changed from normal to atypical glandular cells or adenocarcinoma in only 6% of the cases. 9
Overall, we observed low agreement (Kappa 0.157) between ASCUS+ cytology and methylation among the women with future CIN3+ lesions. This could probably be explained by moderate sensitivity and low specificity for cytology. 9 As mentioned previously, for the samples tested in this study, the proportion of methylation was similar for samples with ASCUS+ and samples with normal cytology (64% vs 45%). Since methylation was detected in nearly half of the samples with normal cytology, our study indicates that methylation could identify a substantial proportion of cases that will develop CIN3+ but which are missed by cytology. However, approximately 10% of the LBC samples from the healthy controls were also found to be methylated (for a more exact estimation of the specificity of the assay, a larger number of samples from healthy controls would need to be analyzed). This is in line with previous data with methylation of 9.3% among HPV‐negative cervical scrapes with no evidence of CIN2+. 28 A substantially lower methylation rate (1.5% by GynTect) among normal cytology screening samples has also been described. 27 Overall, this may reflect that a small subset of normal cytology samples harbor methylation that is not clinically relevant. It is possible that older women more often would test positive in the FAM19A4/miR124 methylation assay, since the degree of methylation tends to increase with age. 34 This could either mean that the test is less specific for older women or that cancer in general is more common at older ages. We have not studied methylation in older women particularly, but that would be an interesting future project.
Testing HPV‐positive women with normal cytology for methylation would increase the sensitivity to find cancer, especially adenocarcinoma. Extensive testing of normal cytology samples would increase the cost of the screening, so the cost‐effectiveness has to be considered before implementation, given that many samples would probably need to be analyzed to prevent one case of adenocarcinoma.
For our methylation‐tested samples, the proportion of samples with abnormal cytology was approximately 30%, whereas the proportion of samples positive for either cytology or methylation was about 60%. However, since we only analyzed relatively few samples, and since the analyzed samples had a higher percentage of normal cytology than the entire group of women who developed CIN3+ (69% vs 46%), it is not possible to estimate how much the sensitivity would increase.
Even though addition of methylation would increase the cost for testing, a higher specificity could lower the cost for colposcopies, for example, if women with ASCUS or LSIL would not need follow‐up immediately if they are methylation‐negative. But this strategy could not be implemented unless the methylation assay has a high sensitivity to predict CIN3+.
A limitation of our study is the relative low number of samples analyzed, which makes it difficult to make any robust conclusions. We are currently planning a larger prospective study analyzing LBC samples from 2017, to estimate the sensitivity and specificity for methylation, to identify high‐grade dysplasia and cancer within 5 years, in HPV‐positive women participating in the screening program.
5. CONCLUSION
Our results indicate that almost half of normal cytology samples collected prior to development of severe cervical dysplasia, manifest methylation. In addition, our data indicate that methylation can be detected earlier than abnormal cytology among women who develop severe cervical dysplasia. We propose testing for methylation of FAM19A4/miR124‐2 among HPV‐positive women with normal cytology to attempt to improve detection of cancer, especially adenocarcinoma.
AUTHOR CONTRIBUTIONS
OF had the idea for the study design and contributed to the writing the draft. YL planned the study, analyzed the data and wrote the draft. LP, JA and TB performed the laboratory work and contributed with their practical expertise. AS and CB contributed to the writing of the paper.
FUNDING INFORMATION
This work was supported by grants from Region Skåne Research Fund and Royal Physiographic Society, Lund.
CONFLICT OF INTEREST STATEMENT
The authors have stated explicitly that there are no conflicts of interest in connection with this article.
6. Ethics statement
The study was approved by the Swedish ethics review authority, reference number DNR 2019–01464, on February 14, 2019. When the samples were collected, the women gave consent for storage in the biobank.
Supporting information
Table S1.
Table S2.
Lindroth Y, Pedersen L, Alssamaray J, et al. Cervix cytology samples revealed increased methylation of the human markers FAM19A4/ miR124‐2 up to 8 years before adenocarcinoma. Acta Obstet Gynecol Scand. 2024;103:378‐386. doi: 10.1111/aogs.14707
REFERENCES
- 1. Gustafsson L, Adami HO. Cytologic screening for cancer of the uterine cervix in Sweden evaluated by identification and simulation. Br J Cancer. 1990;616:903‐908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bulkmans NW, Berkhof J, Rozendaal L, et al. Human papillomavirus DNA testing for the detection of cervical intraepithelial neoplasia grade 3 and cancer: 5‐year follow‐up of a randomised controlled implementation trial. Lancet. 2007;3709601:1764‐1772. [DOI] [PubMed] [Google Scholar]
- 3. Ronco G, Dillner J, Elfstrom KM, et al. Efficacy of HPV‐based screening for prevention of invasive cervical cancer: follow‐up of four European randomised controlled trials. Lancet. 2014;3839916:524‐532. [DOI] [PubMed] [Google Scholar]
- 4. Soderlund‐Strand A, Kjellberg L, Dillner J. Human papillomavirus type‐specific persistence and recurrence after treatment for cervical dysplasia. J Med Virol. 2014;864:634‐641. [DOI] [PubMed] [Google Scholar]
- 5. Maver PJ, Poljak M. Primary HPV‐based cervical cancer screening in Europe: implementation status, challenges, and future plans. Clin Microbiol Infect. 2020;265:579‐583. [DOI] [PubMed] [Google Scholar]
- 6. Borgfeldt C, Leksell A, Forslund O. Co‐testing in cervical screening among 40‐ to 42‐year‐old women is unreasonable. Acta Obstet Gynecol Scand. 2022;1013:374‐378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kares S, Kholova I, Tirkkonen M, Vuento R, Kujala P. Cytohistological findings in HPV‐negative cases from HPV primary screening programme: quality assurance study. APMIS. 2022;13010:599‐604. [DOI] [PubMed] [Google Scholar]
- 8. Kleppe SN, Andersson H, Elfstrom KM, Dillner J. Evaluation of co‐testing with cytology and human papillomavirus testing in cervical screening. Prev Med. 2023;166:107364. [DOI] [PubMed] [Google Scholar]
- 9. Edvardsson H, Wang J, Andrae B, Sparen P, Strander B, Dillner J. Nationwide rereview of Normal cervical Cytologies before high‐grade cervical lesions or before invasive cervical cancer. Acta Cytol. 2021;655:377‐384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doxtader EE, Brainard JA, Underwood D, Chute DJ. Knowledge of the HPV status biases cytotechnologists’ interpretation of Pap tests originally diagnosed as negative for intraepithelial lesion or malignancy. Cancer Cytopathol. 2017;1251:60‐69. [DOI] [PubMed] [Google Scholar]
- 11. Aitken CA, Holtzer‐Goor KM, Uyterlinde A, et al. The impact of knowledge of HPV positivity on cytology triage in primary high‐risk HPV screening. J Med Screen. 2019;264:221‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moriarty AT, Nayar R, Arnold T, et al. The Tahoe study: bias in the interpretation of Papanicolaou test results when human papillomavirus status is known. Arch Pathol Lab Med. 2014;1389:1182‐1185. [DOI] [PubMed] [Google Scholar]
- 13. Aarnio R, Wikstrom I, Gustavsson I, Gyllensten U, Olovsson M. Diagnostic excision of the cervix in women over 40 years with human papilloma virus persistency and normal cytology. Eur J Obstet Gynecol Reprod Biol X. 2019;3:100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sahlgren H, Elfstrom KM, Lamin H, et al. Colposcopic and histopathologic evaluation of women with HPV persistence exiting an organized screening program. Am J Obstet Gynecol. 2020;2223:253.e1‐253.e8. [DOI] [PubMed] [Google Scholar]
- 15. Sundqvist A, Moberg L, Dickman PW, Hogberg T, Borgfeldt C. Time trends for incidence and net survival of cervical cancer in Sweden 1960‐2014—a nationwide population‐based study. Cancer Epidemiol Biomarkers Prev. 2022;31:1572‐1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chatterjee A, Rodger EJ, Eccles MR. Epigenetic drivers of tumourigenesis and cancer metastasis. Semin Cancer Biol. 2018;51:149‐159. [DOI] [PubMed] [Google Scholar]
- 17. Lorincz AT. Virtues and weaknesses of DNA methylation as a test for cervical cancer prevention. Acta Cytol. 2016;606:501‐512. [DOI] [PubMed] [Google Scholar]
- 18. Brentnall AR, Vasiljevic N, Scibior‐Bentkowska D, et al. A DNA methylation classifier of cervical precancer based on human papillomavirus and human genes. Int J Cancer. 2014;1356:1425‐1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Louvanto K, Aro K, Nedjai B, et al. Methylation in predicting progression of untreated high‐grade cervical intraepithelial neoplasia. Clin Infect Dis. 2020;7012:2582‐2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Strooper LM, Meijer CJ, Berkhof J, et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev Res. 2014;712:1251‐1257. [DOI] [PubMed] [Google Scholar]
- 21. Dick S, Kremer WW, De Strooper LMA, et al. Long‐term CIN3+ risk of HPV positive women after triage with FAM19A4/miR124‐2 methylation analysis. Gynecol Oncol. 2019;1542:368‐373. [DOI] [PubMed] [Google Scholar]
- 22. De Strooper LMA, Berkhof J, Steenbergen RDM, et al. Cervical cancer risk in HPV‐positive women after a negative FAM19A4/mir124‐2 methylation test: a post hoc analysis in the POBASCAM trial with 14 year follow‐up. Int J Cancer. 2018;1436:1541‐1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vink FJ, Meijer C, Clifford GM, et al. FAM19A4/miR124‐2 methylation in invasive cervical cancer: a retrospective cross‐sectional worldwide study. Int J Cancer. 2020;1474:1215‐1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Forslund O, Miriam Elfstrom K, Lamin H, Dillner J. HPV‐mRNA and HPV‐DNA detection in samples taken up to seven years before severe dysplasia of cervix uteri. Int J Cancer. 2019;1445:1073‐1081. [DOI] [PubMed] [Google Scholar]
- 25. Agreda PM, Beitman GH, Gutierrez EC, et al. Long‐term stability of human genomic and human papillomavirus DNA stored in BD SurePath and Hologic PreservCyt liquid‐based cytology media. J Clin Microbiol. 2013;518:2702‐2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. WHO . Global strategy to accelerate the elimination of cervical cancer as a public health problem. 2020. Available from https://www.who.int/publications/i/item/9789240014107
- 27. Schmitz M, Eichelkraut K, Schmidt D, et al. Performance of a DNA methylation marker panel using liquid‐based cervical scrapes to detect cervical cancer and its precancerous stages. BMC Cancer. 2018;181:1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. De Strooper LM, van Zummeren M, Steenbergen RD, et al. CADM1, MAL and miR124‐2 methylation analysis in cervical scrapes to detect cervical and endometrial cancer. J Clin Pathol. 2014;6712:1067‐1071. [DOI] [PubMed] [Google Scholar]
- 29. Bierkens M, Hesselink AT, Meijer CJ, et al. CADM1 and MAL promoter methylation levels in hrHPV‐positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer. 2013;1336:1293‐1299. [DOI] [PubMed] [Google Scholar]
- 30. Bonde J, Floore A, Ejegod D, et al. Methylation markers FAM19A4 and miR124‐2 as triage strategy for primary human papillomavirus screen positive women: a large European multicenter study. Int J Cancer. 2021;1482:396‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wiik J, Karrberg C, Nilsson S, Strander B, Jacobsson B, Sengpiel V. Associations between cervical intraepithelial neoplasia during pregnancy, previous excisional treatment, cone‐length and preterm delivery: a register‐based study from western Sweden. BMC Med. 2022;201:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lindroth Y, Borgfeldt C, Thorn G, Bodelsson G, Forslund O. Population‐based primary HPV mRNA cervical screening compared with cytology screening. Prev Med. 2019;124:61‐66. [DOI] [PubMed] [Google Scholar]
- 33. Aaltonen LM, Auvinen E, Dillner J, et al. Poor antibody response against human papillomavirus in adult‐onset laryngeal papillomatosis. J Med Microbiol. 2001;505:468‐471. [DOI] [PubMed] [Google Scholar]
- 34. Jones MJ, Goodman SJ, Kobor MS. DNA methylation and healthy human aging. Aging Cell. 2015;146:924‐932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Table S2.


