Abstract
In this study, two virus concentration methods, namely Adsorption-Extraction (AE) and Nanotrap® Magnetic Virus Particles (NMVP) along with commercially available extraction kits were used to quantify endogenous pepper mild mottle virus (PMMoV) and severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in nucleic acid extracted from 48 wastewater samples collected over six events from eight wastewater treatment plants (WWTPs). The main aim was to determine which workflow (i.e., concentration and extraction methods) produces greater concentrations of endogenous PMMoV and SARS-CoV-2 gene copies (GC) in comparison with each other. Turbidity and total suspended solids (TSS) of wastewater samples within and among the eight WWTPs were highly variable (41–385 NTU and 77–668 mg/L TSS). In 58 % of individual wastewater samples, the log10 GC concentrations of PMMoV were greater by NMVP workflow compared to AE workflow. Paired measurements of PMMoV GC/10 mL from AE and NMVP across all 48 wastewater samples were weakly correlated (r = 0.455, p = 0.001) and demonstrated a poor linear relationship (r2 = 0.207). The log10 GC concentrations of SARS-CoV-2 in 69 % of individual samples were greater by AE workflow compared to NMVP workflow. In contrast to PMMoV, the AE and NMVP derived SARS-CoV-2 GC counts were strongly correlated (r = 0.859, p < 0.001) and demonstrated a strong linear relationship (r2 = 0.738). In general, the PMMoV GC achieved by the NMVP workflow decreased with increasing turbidity, but the PMMoV GC by the AE workflow did not appear to be as sensitive to either turbidity or TSS levels. These findings suggest that wastewater sample turbidity or suspended solids concentration, and the intended target for analysis should be considered when validating an optimal workflow for wastewater surveillance of viruses.
Keywords: Wastewater, SARS-CoV-2, PMMoV, Concentration methods, Health risks
GRAPHICAL ABSTRACT
1. Introduction
Infectious diseases caused by exposure to pathogens are rapidly increasing and responsible for many human deaths and significant economic losses around the world (Lindahl and Grace, 2015). Among these pathogens, enteric and respiratory viruses are the most important etiological agents due to mutations, greater transmissibility, and low infectious doses (Hall, 2012; Leung, 2021). The transmission of these viruses occurs via the fecal-oral, nasal mucosa, the conjunctiva, direct contact, fomite, droplet, and aerosol routes (Leung, 2021). Infected individuals may shed up to 1011 viral particles/g in feces (La Rosa et al., 2012), and 107–8 viral particles per unit volume in bodily fluids such as saliva and nasopharyngeal fluid (Kidd-Ljunggren et al., 2006).
Historically, wastewater surveillance of poliovirus has played an important role for the eradication of polio from Southeast Asia and Africa (El Bassioni et al., 2003; Chowdhary and Dhole, 2008). Recently, Sabin-like type 2 poliovirus has been detected in environmental samples in the USA and genetically linked to viruses detected in wastewater samples from United Kingdom and Israel (https://www.who.int/emergencies/diseaseoutbreak-news/item/2022-DON408). Wastewater surveillance has been conducted for enteric viruses (Chacón et al., 2021) and monitoring of bacterial antimicrobial resistance genes (Mtetwa et al., 2021). More recently, surveillance of wastewater for SARS-CoV-2 has gained much attention throughout the world to provide (i) an early warning system for the emergence or re-emergence of COVID-19 in the community (Medema et al., 2020; La Rosa et al., 2021a; Peccia et al., 2020; Hata et al., 2021), (ii) spatial and temporal trends that mirror the magnitude of COVID-19 in the community, (iii) information on the circulating and emerging variants (La Rosa et al., 2021b; Ahmed et al., 2022b; Reynolds et al., 2022). More recently, surveillance of wastewater has also proven useful to track other respiratory viruses circulating in communities including influenza and respiratory syncytial virus (Hughes et al., 2022; Wolfe et al., 2022).
In wastewater samples, the concentration of certain enteric (i.e., norovirus) and respiratory viruses (i.e., SARS-CoV-2) can be very low, depending on the variable shedding rates by the infected individuals and low disease prevalence in the community. Furthermore, other factors such as variability in wastewater composition, sampling approaches, flow and precipitation may influence the detection of low levels of viruses in wastewater (Ahmed et al., 2022c). Therefore, a concentration step is often required to obtain detectable amounts of virus nucleic acid (Ahmed et al., 2015). A wide range of virus concentration and extraction methods have been applied to concentrate viruses from wastewater samples (Ahmed et al., 2015; McMinn et al., 2021; Pecson et al., 2021; Sapula et al., 2021).
Recovery of viruses from wastewater using these workflows can be highly variable spanning several orders of magnitude (Ahmed et al., 2020a; McMinn et al., 2021; Pecson et al., 2021). Furthermore, recovery efficiency of viruses from wastewater samples can vary between and within WWTPs, even with the same concentration and extraction workflow, presumably due to variations in wastewater composition as well as types of viruses. Consequently, virus concentration efficiency may be a significant impediment to wastewater surveillance applications where virus levels are low, and the recovery efficiency is highly variable (Chik et al., 2021; Pecson et al., 2021).
Among these concentration methods, an Adsorption-Extraction (AE) protocol with a negatively charged membrane has been very effective for quantifying low concentrations of enteric and enveloped viruses from wastewater samples with minimal PCR inhibitory effects (Ahmed et al., 2015; Ahmed et al., 2020a; Jafferali et al., 2021). This workflow involves adding MgCl2 and/or HCl to a sample, adsorbing the viruses onto negatively charged membranes during filtration, followed by direct nucleic acid extraction from the membrane itself (Symonds et al., 2014; Ahmed et al., 2015; Babler et al., 2022). A recent study compared the efficiency of seven wastewater virus concentration protocols including AE, centrifugal filter devices, polyethylene glycol (PEG 8000) precipitation, and ultracentrifugation by seeding murine hepatis virus (MHV) in wastewater (Ahmed et al., 2020a). The mean MHV recoveries ranged from 26.7 to 65.7 % across all concentration methods. The most efficient method was AE with MgCl2 pre-treatment. There are several advantages of the AE concentration method including its speed (i.e., takes <15 min to process a typical 50 mL wastewater sample), ability to concentrate viruses from both the solid and liquid phases, use of filtration assemblies and vacuum pumps often readily available in microbiology laboratories, and multiple samples can be processed in parallel if filtration units and manifolds are available (Ahmed et al., 2020a).
Another concentration approach uses Nanotrap® Magnetic Virus Particles (NMVP) which utilize the NMVP materials high affinity for viruses to attract, capture and enrich low abundance viruses in solution. The NMVP concentration method has been demonstrated to capture and concentrate a wide range of viruses, including: influenza A, respiratory syncytial virus, coronavirus 229E (https://www.ceresnano.com/post/respiratory-pathogendetection), coronavirus OC43 (https://www.ceresnano.com/post/app-notecapture-and-concentration-of-coronavirus-oc43), SARS-CoV-2 (https://www.ceresnano.com/post/app-note-capture-and-concentration-of-sars-cov2-from-viral-transport-medium), Zika, chikungunya (Lin et al., 2020), and pepper mild mottle virus (https://www.ceresnano.com/post/app-notenanotrap-enhancement-reagent-1-improves-detection-of-viral-rna-fromwastewater-samples) from wastewater and clinical samples. The concentration method is inexpensive and wastewater processing workflows are available for manual or automated workflow. Karthikeyan and colleagues (2021) reported that when automated on the KingFisher Flex liquid-handling robot (Thermo Fisher Scientific, USA), using a 24-plex head, NMVP allowed the processing of 24 wastewater samples in single 40-min run. They also compared the performance of NMVP with electronegative membrane filtration and PEG workflows by seeding heat-inactivated SARS-CoV-2 into ninefold serial dilutions of a single 10-mL volumes of untreated wastewater sample. The average virus recovery efficiencies were 27, 15 and 8 %, for the NMVP, HA membrane filtration, and PEG methods, respectively. Furthermore, it has been reported that NMVP can improve sequencing workflows by enhancing current standard nucleic acid extraction methods (Andersen et al., 2021).
Little is known regarding the performance of NMVP (i.e., concentration and extraction workflow) in comparison with other concentration workflows. For the efficient surveillance of viruses, well-optimized, rapid, efficient (high recovery), and cost-effective virus concentration workflows are needed worldwide. The main aim of the present study was to compare the performance of two virus concentration workflows: (i) AE (i.e., direct nucleic acid extraction from a negatively charged membrane), and (ii) NMVP. The concentrations of endogenous PMMoV and SARS-CoV-2 were determined in nucleic acid extracted from wastewater samples from several wastewater treatment plants (WWTPs) to determine which workflow yields greater concentrations of PMMoV and SARS-CoV-2 gene copies (GC). PMMoV, which is an indicator virus for enteric viruses was selected because it has been used to normalize SARS-CoV-2 concentration data (Wu et al., 2021). The findings from this study can aid in selecting an optimal concentration workflow for wastewater surveillance studies.
2. Materials and methods
2.1. Sources of wastewater samples
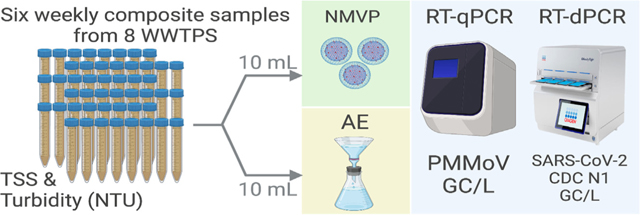
Wastewater samples used in this study were from the Queensland Health Wastewater Surveillance program. Wastewater samples were collected between 01/02/2022 and 09/03/2022 from eight wastewater treatment plants (de-identified as WWTPs A to H hereafter) in Queensland, Australia over six weeks yielding a total of 48 wastewater samples (i.e., six from each WWTP). The selected WWTPs employ activated sludge processes and treat mainly domestic wastewater from approximately ~15,000 to ~100,000 people. At each WWTP, 500 mL to 1 L of untreated wastewater samples were collected via time-based compositing using an automated sampler operating in time-proportional mode (taking 5–10 mL subsamples every 15 min for 24 h) at ambient temperature (Ahmed et al., 2022d).
2.2. Turbidity and total suspended solids measurements
The turbidity and total suspended solids (TSS) of wastewater samples can be highly variable within and between WWTPs. Furthermore, a recent study reported that sample turbidity can affect virus recovery from wastewater samples. Therefore, the turbidity of each wastewater sample was measured in nephelometric turbidity units (NTU) using a HACH turbidimeter (Model TU5200) following the method described in Standard Methods 2130 B (Standard Methods for the Examination of Water and Wastewater. http://standardmethods.org/). To measure total suspended solids (TSS), an aliquot of each wastewater sample was filtered through a pre-weighed standard glass-fibre filter, and the filter and resulting residue was dried at 103–105 °C (for 2 h). The measured increase in weight between the glass-fibre filter alone and the filter with dried residue was attributed to the retained solids (mg) and expressed as mg/L of TSS in proportion to the sample volume (L) tested. Owing to limited sample volume available, for turbidity and TSS, 40-mL and 500-mL wastewater samples were used, respectively. The samples were mixed well prior to subsampling to minimize subsampling variation.
2.3. Virus concentration
Endogenous viruses were concentrated from the wastewater samples using the AE and NMVP workflows. In this study, the AE workflow began with the addition of dissolved MgCl2 (Sigma-Aldrich, St. Louis, Missouri, USA) to a 10 mL wastewater sample to achieve a final concentration of 25 mM MgCl2. After amendment with MgCl2, wastewater samples were immediately filtered through a 0.45-μm pore-size, electronegative HA membrane (47-mm diameter HAWP04700; Merck Millipore Ltd., Sydney, Australia) via a magnetic filter funnel (Pall Corporation, Port Washington, New York, USA) and filter flask (Merck Millipore Ltd.) (Ahmed et al., 2020a). Following filtration, using aseptic technique, the membrane was immediately removed, rolled, and inserted into a 5-mL-bead-beating tube (Qiagen, Valencia, CA, USA) for nucleic acid extraction.
For the NMVP workflow, a 10 mL of wastewater sample was transferred into a 15 mL sterile conical falcon tube (Eppendorf, Hamburg, Germany). Each 10 mL wastewater sample was amended with 100 μL of Nanotrap® Enhancement Reagent 2 (ER2) (Ceres Nanosciences SKU 10112) and then vortexed thoroughly for 10 s. Next, 150 μL of NMVP (Ceres Nanosciences SKU 44202) were added to each sample. Samples were mixed thoroughly by inverting the tubes two to three times and then incubated at room temperature (24 ± 1 °C) for 10 min after mixing. The tubes were then placed on a magnetic rack (Dynal MPC™−6) to pelletize the NMVP at the bottom of the tube. The supernatant was carefully removed without disturbing the pellet via pipette and discarded, and then the pellet was resuspended by adding 1 mL of molecular grade water into each tube. The resulting virus particle suspension was transferred into a sterile 1.5 mL microcentrifuge tube using a pipette and placed on a magnetic rack (Invitrogen™ DynaMag™−2 Magnet) to create a final NMVP pellet, and then the supernatant was carefully discarded without disturbing the pellet for nucleic acid extraction. This concentration protocol was performed as described in Ceres Nanosciences APP-034 (https://www.ceresnano.com/_files/ugd/f7710c_215bb60d02c04572ae60153e24770650.pdf).
2.4. Nucleic acid extraction
Immediately after AE virus concentration, nucleic acid was extracted directly from the HA membranes using the RNeasy PowerWater Kit (Cat. No. 14700–50-NF) (Qiagen, Valencia, CA, USA). Prior to homogenization, 990 μL of buffer PM1 and 10 μL of β-Mercaptoethanol (Cat. No. M6250–10 mL) (Sigma-Aldrich) were added into each 5 mL bead-beating tube. A known concentration (~1.5 × 104 gene count (GC)) of murine hepatitis virus (MHV) was added into each bead-beating tube as an inhibition process control. The bead-beating tubes were then homogenized using a Precellys 24 tissue homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France) set for 3 × 15 s at 10,000 rpm at a 10 s interval. After homogenization, the tubes were centrifuged at 4000g for 4 min to pellet the filter debris and beads. Sample lysate supernatant ranging from 600 to 800 μL in volume was then passed into the extraction to yield purified nucleic acids following the manufacturer’s specified protocol. One modification was made: the use of DNase I solution was omitted from the protocol to isolate nucleic acid (i.e., both RNA and DNA). This was done to reduce the sample processing time in the laboratory. Omission of DNase I solution does not affect RNA quantification (data not shown).
Immediately after separating the NMVP from the sample as described above, nucleic acids were extracted from the resulting pellet using the QIAamp Viral RNA Mini Kit (Qiagen) and the procedure described in Ceres Nanosciences APP-034. Briefly, 150 μL of 1× PBS, 560 μL of Buffer AVL and 5.6 μL of carrier RNA were added to each pellet. A known concentration (~1.5 × 104 GC of murine hepatitis virus (MHV) was added into each 1.5 mL microcentrifuge tube as an inhibition process control. The NMVP pellet was homogenized by vortexing the tube for 30 s followed by incubation at room temperature for 10 min. The Invitrogen™ DynaMag™−2 Magnet rack (Invitrogen™, Waltham, Massachusetts, USA) was used to separate the NMVP from the resulting lysate suspension. The supernatant was transferred into a new sterile 1.5 mL microcentrifuge tube using a pipette and the pellet was discarded. Nucleic acid was then extracted from each lysate according to the manufacturer’s instructions.
The Agilent TapeStation 4200 was used to determine the quality score of the purified nucleic acid resulting from the extraction of each AE and NMVP sample. The integrity was assessed via the RNA integrity number (RIN). The instrument makes use of the entire electrophoretic trace of the purified nucleic acid to assign a RIN between 0 and 10, with 10 indicating maximum integrity. Nucleic acid purity (as measured by the A260/280 ratio) and concentration of nucleic acid (ng/μL) were measured using a DeNovix Spectrophotometer & Fluorometer (DeNovix, Wilmington, DE, USA).
2.5. Inhibition assessment
The presence of PCR inhibition in nucleic acid samples extracted from wastewater was assessed using an MHV RT-PCR assay (Besselsen et al., 2002). Known quantities (~1.5 × 104 GC of MHV were seeded into each bead tube containing membrane and lysis buffer (for AE workflow) and each pellet (for NMVP workflow) (described in ‘Nucleic acid extraction’ above) and added to a distilled water extraction control and subjected to extraction. The reference PCR quantification cycle (Cq) values obtained for MHV seeded into distilled water were compared with the Cq values of the MHV seeded into wastewater lysate to obtain information on potential RT-qPCR inhibition for nucleic acid samples extracted by both the AE and NMVP workflows. If the Cq value resulting from the sample was >2 different from the reference Cq value for the distilled water control, the sample was considered inhibited (Ahmed et al., 2022a). In addition to the extraction control, all samples were analyzed alongside three PCR negative controls.
2.6. MHV RT-PCR, PMMoV RT-qPCR and SARS-CoV-2 RT-dPCR analysis
A previously published RT-PCR assay was used for MHV detection (Besselsen et al., 2002), US CDC N1 RT-digital PCR (dPCR) assay was used for SARS-CoV-2 quantification (CDC, 2019), and RT-qPCR assay was used for PMMoV quantification (Rosario et al., 2009; Haramoto et al., 2013) (Supplementary Table ST1). For the MHV RT-PCR and PMMoV RT-qPCR assays, positive control materials in the form of gBlocks gene fragments were purchased from Integrated DNA Technologies (Integrated DNA Technology Coralville, IA, US). PMMoV standard curves were prepared from gBlocks with dilutions ranging from 6 × 105 to 0.6 GC/reaction. Gamma-irradiated SARS-CoV-2 was used as an RT-dPCR positive control for the CDC N1 dPCR assay (Ahmed et al., 2022a–d). Primer and probe sequences, reaction concentrations, and thermal cycling conditions are listed in Supplementary Table ST1. MHV RT-PCR and PMMoV RT-qPCR analyses were performed in 20-μL reactions using TaqMan™ Fast Virus 1-Step Master Mix (Applied Biosystem, California, USA). The RT-PCR (for MHV) and RT-qPCR (for PMMoV) experiments were performed using a Bio-Rad CFX96 thermal cycler (Bio-Rad Laboratories, Richmond, CA, USA) using manual settings for threshold and baseline.
The US CDC N1 RT-dPCR assay was performed in 40-μL reaction mixtures using the QIAcuity One-Step Viral RT-PCR Kit (Cat No. 1123145; Qiagen) and 26 K 24-well Nanoplates (Cat No. 250001; Qiagen). Two RT-dPCR replicates were analyzed for each sample. Each RT-dPCR nanoplate contained duplicate RT-dPCR no-template and positive (gamma-irradiated SARS-CoV-2 RNA) controls. Data were analyzed using the QIAcuity Suite Software V1.1.3193 (Qiagen, Germany) and quantities exported as GC/μL of reaction. The RT-dPCR assays were performed using automatic settings for threshold and baseline.
2.7. PMMoV RT-qPCR and SARS-CoV-2 RT-dPCR assay limit of detection
To determine the PMMoV RT-qPCR assay limit of detection (ALOD), gBlocks were diluted (6 × 105 to 0.6 GC/reaction) and analyzed using RT-qPCR. At each dilution, 15 replicates were analyzed. The 95 % ALOD was defined by fitting an exponential survival model to the proportion of PCR replicates positive at each step along the gradient (Verbyla et al., 2016). The ALOD for SARS-CoV-2 RT-dPCR was previously determined in our study (Ahmed et al., 2022d).
2.8. Quality control
To minimize RT-qPCR and RT-dPCR contamination, nucleic acid extraction and RT-qPCR/dPCR set up were performed in separate laboratories. A sample negative control was included during the concentration process. An extraction negative control was also included during nucleic acid extraction to account for any contamination during extraction. All sample and extraction negative controls were negative for the analyzed targets.
2.9. Data analysis
For PMMoV RT-qPCR, samples were considered positive (SARS-CoV-2 detected) if amplification was observed in all three replicates within 45 cycles. Samples were considered quantifiable if amplification was observed in all three replicates with concentrations above the ALOD. For SARS-CoV-2 RT-dPCR, samples were considered positive if amplification was observed in both replicates within 45 cycles. Samples were considered quantifiable by RT-dPCR if the concentrations were above the ALOD, and the average number of partitions was >18,000 per sample well. Statistical differences in the extraction yields and RIN resulting from each concentration/extraction workflow combination were assessed using unpaired t-tests with Welch’s correction for unequal variances with a false discovery rate of 1 %. Inhibition control Cq values for samples from each WWTP were compared using the Kruskal-Wallis test with Dunn’s correction for multiple comparisons. Differences between paired PMMoV and SARS-CoV-2 GC derived from AE and NMVP performed on single samples were assessed by paired t-tests stratified by WWTP. Owing to the limited sample size from each WWTP tests of normality were not performed. However, since the basis of comparison was the difference in the log10 mean value of independent replicate RT-qPCR/RT-dPCR measurements and the standard curves displayed linearity, an assumption of normality of replicate measurements is not unreasonable. Correlations between PMMoV and SARS-CoV-2 GC/10 mL by each workflow across all samples were assessed by means of Pearson’s coefficient (r) and linear regression, according to the assumption that perfect agreement between the two would yield a line with slope equal to one and intercept equal to zero. Correlation between turbidity (NTU) and TSS was also assessed by means of Pearson’s coefficient (r) and linear regression.
3. Results
3.1. Turbidity and total suspended solids
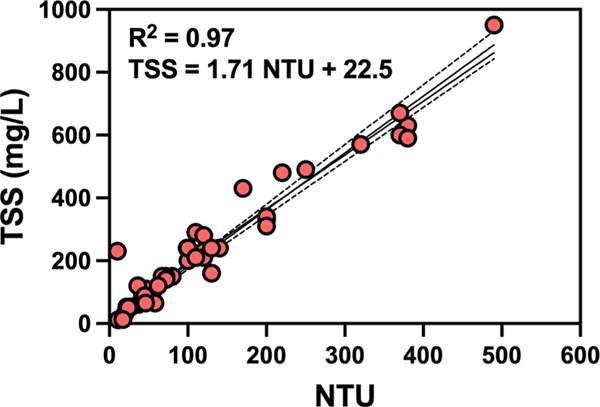
Summary statistics for measured turbidity and TSS are tabulated in Table 1. For 7 of the 8 WWTPs the mean NTU was in the typical range for domestic wastewater with means from 40 to 130 NTU, while WWTP H had mean of 385 NTU (i.e. far greater than other WWTP samples). Similar trends were observed for TSS with WWTP H having much greater mean TSS than the other WWTPs. Large variabilities of NTU and TSS were observed in wastewater samples collected from the same WWTP over time. For example, WWTP B showed 88 % variability in NTU over time, followed by 83 and 73 % variability in NTU for WWTP G and F. Similarly, high levels of variability were observed for TSS measurements, where WWTP B showed 98 % variability in TSS over time, followed by WWTP F (82 %) and WWTP D (76 %). The observed turbidity and total suspended solids concentration in the 48 wastewater samples were strongly and significantly correlated (r = 0.97, p < 0.0001). A linear regression fit to the paired NTU (x) and TSS (y) data points (Fig. 1) indicated a good linear fit (TSS = 1.713 NTU + 22.53, r2 = 0.94).
Table 1.
Summary statistics for turbidity (NTU) and total suspended solids (TSS)as observed in six untreated waste water samples from eight WWTPs including minimum (Min.),maximum (Max.), mean and standard deviation, and coefficient of variation (CV).
| WWTPs (No. of samples | Turbidity (NTU) | TSS (mg/L) | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Min. | Max. | Mean ± SD | CV (%) | Min. | Max. | Mean ± SD | CV (%) | |
|
| ||||||||
| WWTP A (8) | 11 | 54 | 41 ± 160 | 29 | 12 | 110 | 77 ± 36 | 47 |
| WWTP B (8) | 20 | 110 | 40 ± 350 | 88 | 33 | 290 | 100 ± 98 | 98 |
| WWTP C (8) | 72 | 140 | 105 ± 35 | 25 | 150 | 280 | 215 ± 53 | 25 |
| WWTP D (8) | 26 | 170 | 93 ± 54 | 59 | 53 | 430 | 179 ± 136 | 76 |
| WWTP E (8) | 24 | 200 | 130 ± 80 | 61 | 51 | 289 | 216 ± 132 | 61 |
| WWTP F (8) | 45 | 220 | 91.0 ± 66 | 73 | 65 | 480 | 189 ± 155 | 82 |
| WWTP G (8) | 10 | 250 | 106 ± 88 | 83 | 13 | 490 | 232 ± 152 | 65 |
| WWTP H (8) | 320 | 490 | 385 ± 56 | 15 | 570 | 950 | 668 ± 142 | 21 |
Fig. 1.
Linear relationship between turbidity (NTU) and total suspended solids (TSS) (mg/L) as observed in 48 untreated wastewater samples from eight WWTPs (8 samples per WWTP).
3.2. Nucleic acid purity and yield
Summary statistics for measured RIN, yield and A260/280 are shown in Table 2 for nucleic acid extracted through both AE and NMVP workflows. Purified nucleic acid yields from the extracts of both concentration and extraction workflows were statistically equivalent except for extractions from WWTP H samples. For WWTP H, AE yielded significantly higher nucleic acid yields than NMVP (p < 0.0001). For nucleic acid integrity as assessed by the RIN, NMVP yielded significantly greater RIN for extractions from WWTP B, C, and F while AE yielded significantly greater RIN from WWTP H. Extractions of concentrates from both AE and NMVP produced A260/280 ratios within or above the acceptable the range (2.0 to 2.2) for “pure” nucleic acid.
Table 2.
Mean, standard deviation (SD) and 95 % confidence interval (CI) of RNA integrity number (RIN), extraction yield (ng/μL), A260/280 ratio as observed for extractions of six untreated wastewater samples from eight WWTPs.
| WWTPs (No. of samples | AE (mean ± SD; 95 % CI) | NMVP (mean ± SD; 95 % CI) | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| RIN | Yield (ng/μL) | A260/280 | RIN | Yield (ng/μL) | A260/280 | |
|
| ||||||
| WWTP A (8) | 4.50 ± 0.50 (4.0–5.0) | 99.0 ± 49.0 (48.0–150) | 2.00 ± 0.04 (2.0–2.1) | 5.10 ± 0.14 (4.9–5.3) | 40.0 ± 36.0 (3.10–78.0) | 2.80 ± 0.37 (2.4–3.2) |
| WWTP B (8) | 5.00 ± 0.27 (4.7–5.3) | 78.0 ± 53.0 (23.0–133) | 2.10 ± 0.04 (2.0–2.1) | 6.00 ± 0.73 (5.2–6.7) | 69.0 ± 15.0 (53.0–84.0) | 2.70 ± 0.19 (2.5–2.9) |
| WWTP C (8) | 5.50 ± 0.23 (5.2–5.7) | 201 ± 24.0 (177–226) | 2.10 ± 0.02 (2.0–2.1) | 6.40 ± 0.45 (5.6–7.1) | 91.0 ± 88.0 (82.0–101) | 2.50 ± 0.08 (2.4–2.6) |
| WWTP D (8) | 5.50 ± 0.53 (4.9–6.0) | 119 ± 64.0 (52.0–186) | 2.00 ± 0.02 (2.2–2.1) | 5.80 ± 0.29 (5.5–6.1) | 71.0 ± 10.0 (60.0–82.0) | 2.60 ± 0.11 (2.5–2.7) |
| WWTP E (8) | 5.00 ± 0.52 (4.4–5.5) | 187 ± 117 (65.0–310) | 2.10 ± 0.03 (2.0–2.1) | 6.40 ± 1.20 (5.0–7.9) | 93.0 ± 38.0 (53.0–134) | 2.50 ± 0.16 (2.3–2.7) |
| WWTP F (8) | 4.90 ± 0.69 (4.1–5.6) | 130 ± 100 (25.0–235) | 2.00 ± 0.02 (2.0–2.1) | 6.10 ± 0.49 (5.6–6.6) | 64.0 ± 29.0 (34.0–94.0) | 2.50 ± 0.12 (2.4–2.6) |
| WWTP G (8) | 5.50 ± 0.31 (5.2–5.8) | 177 ± 121 (50.0–304) | 2.10 ± 0.02 (2.0–2.1) | 6.20 ± 0.70 (5.3–7.1) | 70.0 ± 19.0 (49.0–90.0) | 2.40 ± 0.19 (2.2–2.6) |
| WWTP H (8) | 4.70 ± 0.10 (4.6–4.8) | 682 ± 131 (545–820) | 2.00 ± 0.03 (2.0–2.1) | 3.60 ± 0.30 (3.3–5.9) | 154 ± 40.0 (112–196) | 2.20 ± 0.09 (2.1–2.3) |
3.3. PCR inhibition
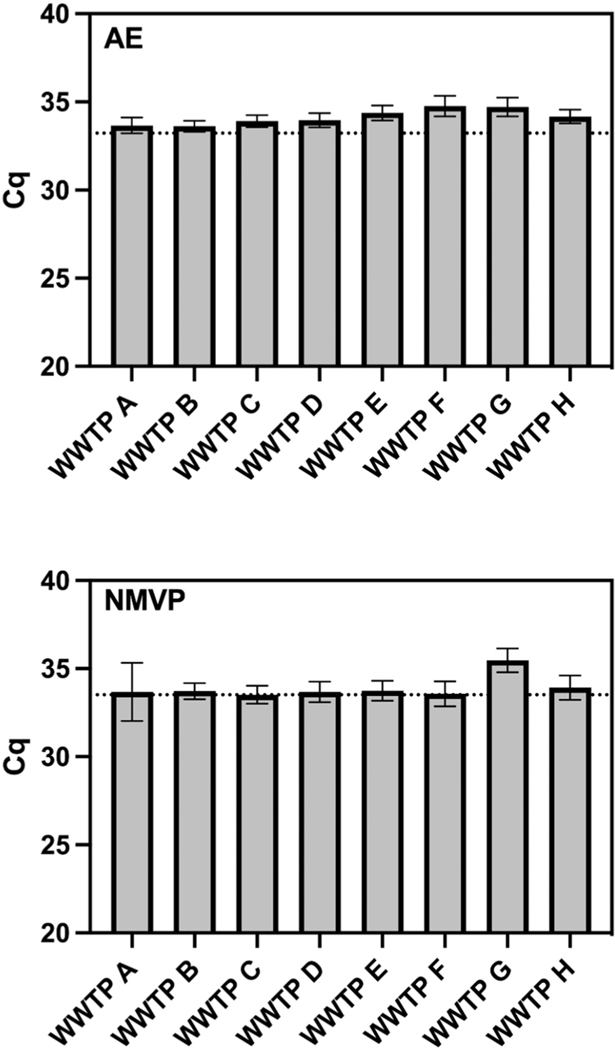
Median Cq values of the MHV inhibition control were significantly different between WWTPs for both the AE and NMVP workflows (Fig. 2). For AE, samples from WWTPs F and G yielded significantly greater Cq values compared to results for all other WWTPs, while WWTP E was significantly greater than WWTP A, and WWTP E and H were significantly greater than WWTP B. NMVP, on the other hand, yielded significantly greater median Cq values only for samples from WWTP G. Of the 48 wastewater samples extracted using the AE workflow, nucleic acid extracts of 46 samples were within the 2-Cq values of the reference Cq value, whereas Cq values of two samples from (WWTP F and G) were > 2-Cq values of the reference sample. Of the 48 wastewater samples extracted using the NMVP workflow, nucleic acid extracts of 43 samples were within the 2-Cq values of the reference Cq value, whereas Cq values of five samples from (one sample from WWTP A and four samples from WWTP G) were > 2-Cq values of the reference sample.
Fig. 2.
Observed Cq values from RT-qPCR of the inhibition control (MHV) seeded into purified nucleic acids derived from six untreated wastewater samples per WWTP shown.
3.4. RT-qPCR and RT-dPCR performance characteristics
The PMMoV RT-qPCR standard curves had a linear dynamic range of quantification from 6 × 105 to 0.6 GC/reaction. The slope of the standard curve was −3.36. The amplification efficiency and y-intercept values were 98.4 % and − 40.2, respectively within the prescribed range of MIQE guidelines (Bustin et al., 2009). The correlation coefficient (r2) value was 0.99. All positive controls or standard curves amplified in each PCR run and negative controls were negative. For US CDC N1 RT-dPCR, the number of partitions ranged from 19,212 to 25,320 with a mean ± standard deviation (SD) of 20,904 ± 2765. The ALOD for the PMMoV RT-qPCR assay was 6.80 GC/reaction.
3.5. Concentrations (log10 GC) of PMMoV and SARS-CoV-2 by AE and NMVP methods
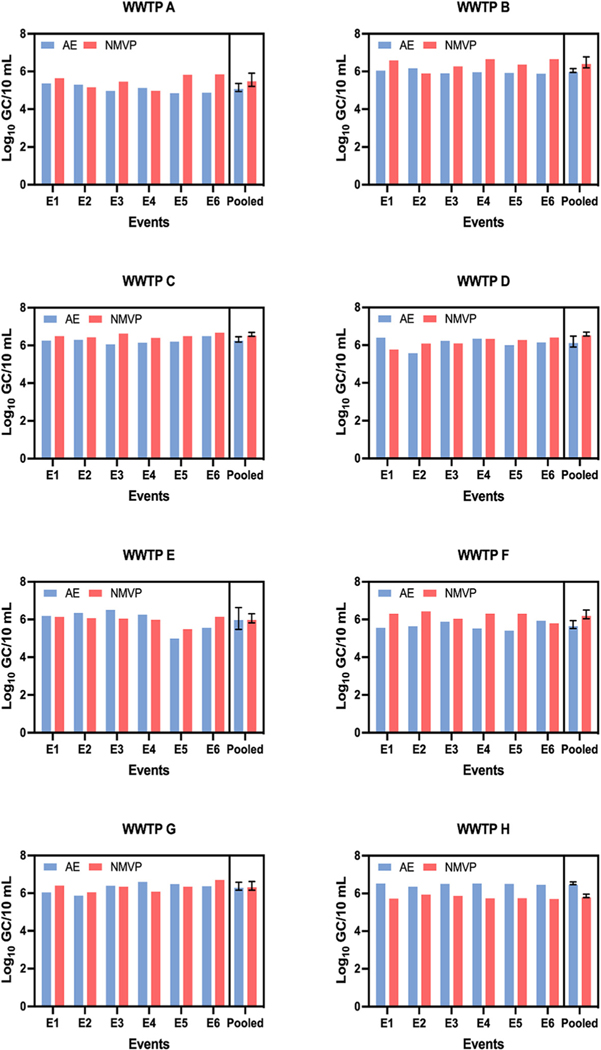
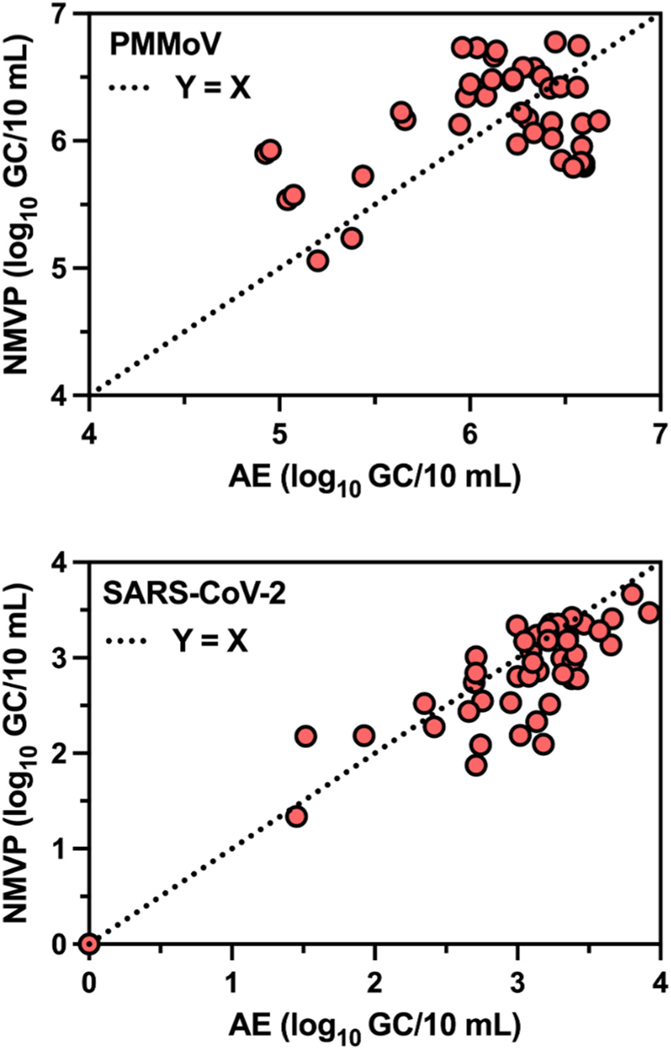
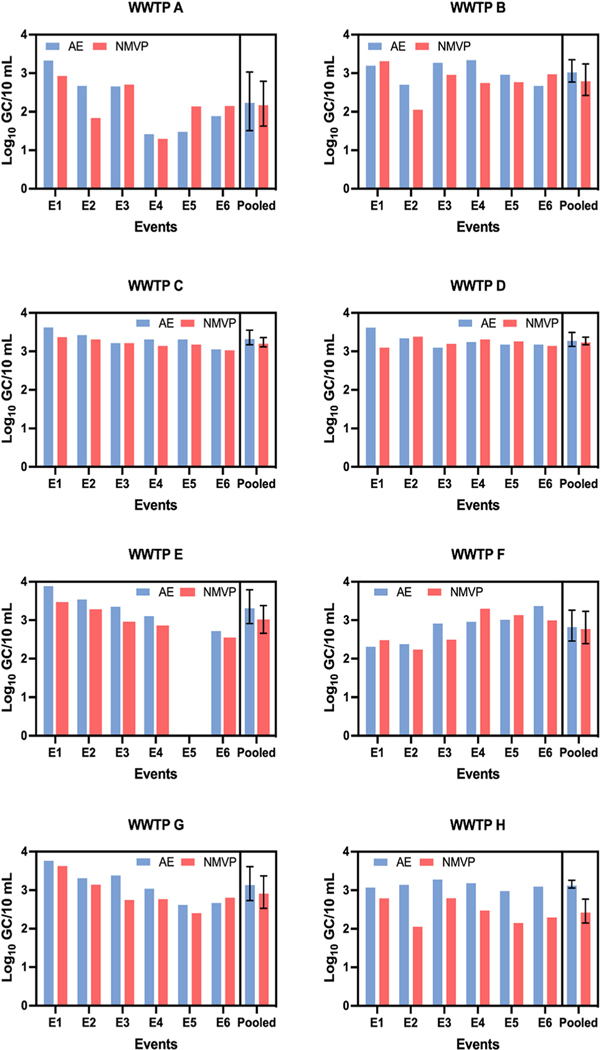
The log10 GC concentrations of PMMoV in 28 (58.3 %) of 48 individual samples were greater by NMVP workflow compared to AE workflow (the other 20 had greater concentrations by AE workflow). As shown in Fig. 3, across all six sampling events NMVP produced greater mean PMMoV log10 GC/10 mL than AE for WWTPs B (p = 0.039), C (p = 0.007), and F (p = 0.027) while AE yielded greater PMMoV GC concentrations for WWTP H (p < 0.001). For the remaining four WWTPs, AE and NMVP produced PMMoV log10 GC concentrations that were statistically similar across the six sampling events. Paired measurements of PMMoV log10 GC/10 mL from AE and NMVP across all 48 sampling events (Fig. 5) were weakly correlated (r = 0.455, p = 0.001) and demonstrated a poor linear relationship (r2 = 0.207).
Fig. 3.
Comparative log10 GC/10 mL of pepper mild mottle virus (PMMoV) in 48 wastewater samples from eight WWTPs using Adsorption Extraction (AE) and Nanotrap® Magnetic Virus Particles (NMVP) methods.
Fig. 5.
Correlation (log10 GC/10 mL) between Adsorption Extraction (AE) and Nanotrap® Magnetic Virus Particles (NMVP) as observed in 48 untreated wastewater samples from eight WWTPs (8 samples per WWTP).
The log10 GC concentrations of SARS-CoV-2 in 33 (68.8 %) of 48 individual samples were greater by AE workflow compared to NMVP workflow (the other 15 had greater concentrations by NMVP workflow). Mean SARS-CoV-2 concentrations (log10 GC/10 mL) for each sampling event and pooled across all sampling events at each WWTP are shown in Fig. 4. For samples from WWTPs C, E, and H, AE concentration yielded greater SARCoV-2 log10 GC/10 mL than NMVP with p-values of 0.027, 0.001, and 0.002, respectively. For the remaining WWTPs AE and NMVP yielded statistically similar SARS-CoV-2 GC/10 mL across the six sampling events. As shown in Fig. 5 across all 48 samples, AE and NMVP derived SARS-CoV-2 GC counts were strongly correlated (r = 0.859, p < 0.001) and demonstrated a strong linear relationship (r2 = 0.738). For PMMoV-normalized SARS-CoV-2 counts (SARS-CoV-2 log10 GC/PMMoV log10 GC), when the workflows produced significantly different counts of PMMoV or SARSCoV-2 GC, the resulting PMMoV-normalized counts were also significantly different from one another.
Fig. 4.
Comparative log10GC/10 mL ofSARS-CoV-2 in 48 wastewater samples from eight WWTPs using Adsorption Extraction (AE) and Nanotrap®Magnetic Virus Particles (NMVP) methods.
3.6. Trends between NTU/TSS and viruses
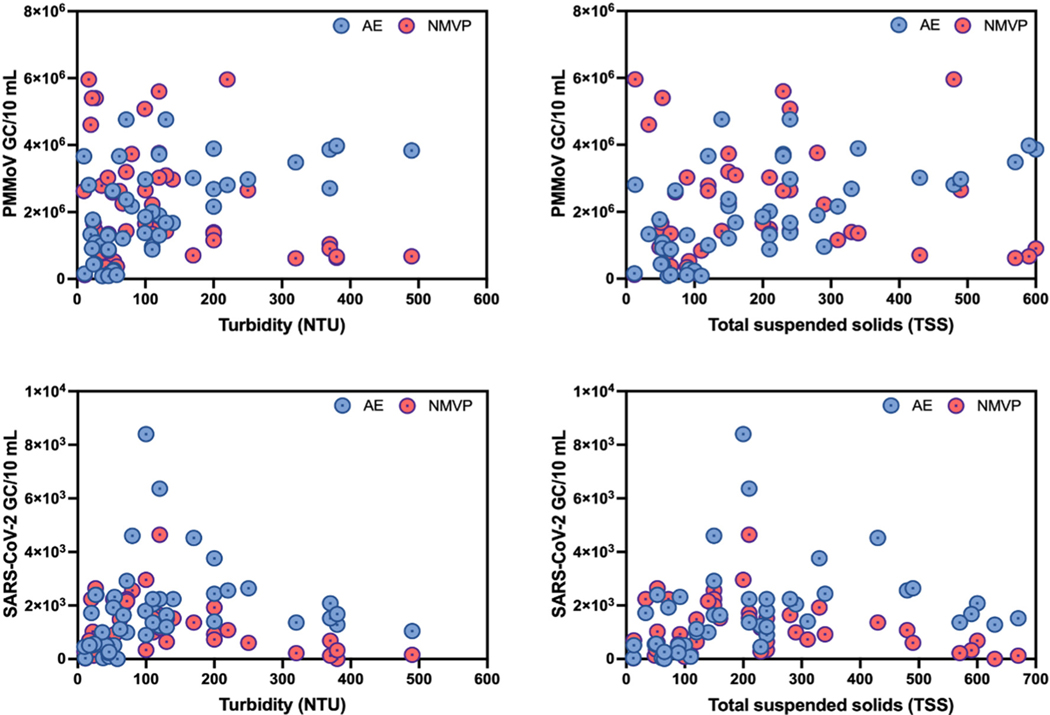
PMMoV and SARS-CoV-2 GC/10 mL versus turbidity and TSS are displayed in Fig. 6. In general, NMVP-derived PMMoV counts decreased with increasing turbidity. Meanwhile, AE-derived PMMoV GC counts did not appear to be sensitive to turbidity or TSS levels. For SARS-CoV-2, however, both NMVP and AE workflows yielded the greatest GC/10 mL in the middle ranges of turbidity (100–200 NTU) and TSS (150–450 mg/L). For low turbidity and TSS samples, SARS-CoV-2 GC/10 mL increased for both workflows up to the mid-range and then decreased with further increased turbidity and TSS.
Fig. 6.
Trends between viruses (PMMoV and SARS-CoV-2) and wastewater turbidity (NTU) and total suspended solids (TSS).
4. Discussion
Viral pathogens, including SARS-CoV-2 and enteric viruses, are typically present in low concentrations in wastewater and thus require a concentration step to attain levels of viral nucleic acids detectable using molecular methods (Ahmed et al., 2015). Considering the multitude of concentration methods available, ranging from AE to different types of ultrafiltration, chemical precipitation, and centrifugation (Ahmed et al., 2020a), each with its own caveats (recently reviewed in Rusiñol et al., 2020; Cervantes-Avilés et al., 2021; Pecson et al., 2021; Ahmed et al., 2022c), and often with highly variable recovery efficiencies (Ahmed et al., 2020a; McMinn et al., 2021; Pecson et al., 2021), selecting the optimal concentration and extraction workflow is paramount for wastewater surveillance or environmental studies. Additionally, adsorption dependent methods, such as AE and NMVP, rely on electrostatic interactions that are dependent on the isoelectric points of the viruses being adsorbed (Armanious et al., 2016). In this study, we compared two rapid and inexpensive workflows (i.e., AE and NMVP) for their ability to concentrate endogenous SARS-CoV-2 and PMMoV nucleic acid from wastewater samples. We did not seed exogenous viruses as seeding exogenous stock viruses may result in inaccurate recovery assessment due to differential structural form and partitioning behaviour of viruses in the stock compared to those shed by infected humans (Kantor et al., 2020). Ultimately, the observations in the current study are empirical in nature as an investigation of the fundamental electrostatic interactions for each of the workflows and viruses was beyond the scope of the investigation. It is important to note that the inherent methodological differences between the two concentration methods necessitated use of two different nucleic acid extraction kits, which precludes a direct comparison between the two workflows. We acknowledge that the extractions kit efficiency used in the AE workflow may be significantly greater than that of the NMVP workflow, or vice versa, and require further investigation. We have compensated for this limitation through a carefully executed experimental design by using the same wastewater sample volume (i.e., 10 mL) to ensure comparability and by challenging both concentration techniques with multiple wastewater samples from eight different WWTPs with variable physico-chemical parameters.
Across the 48 wastewater samples analyzed in this study, turbidity and total suspended solids were highly variable and ranged from 10 to 490 NTU and 12–950 mg/L of TSS, respectively, and were the lowest for WWTP A and the highest for WWTP H. The composition of the wastewater affected the performance of the workflows regarding both nucleic acid yield and purity, as well as estimated concentrations of PMMoV and SARS-CoV-2. Measured nucleic acid mass was comparable across all WWTPs and both workflows, except for WWTP H where turbidity and TSS were notably greater. However, NMVP generated higher purity nucleic acid for samples from three WWTP, while AE produced higher purity nucleic acid for only one WWTP (H). As previously mentioned, different nucleic acid extraction kits were used with each concentration method, so the result must be interpreted considering the concentration/extraction workflow as a whole method. Nonetheless, this finding has important implications for concentration and extraction method selection for applications other than quantitative PCR, such as next generation sequencing, which require higher concentrations and greater purity of the nucleic acids for high resolution of SARS-CoV-2 variants detection (Karthikeyan et al., 2022).
Despite the higher purity nucleic acid (sometimes observed with the NMVP workflow), RT-qPCR inhibition was more frequently detected in more wastewater samples that were concentrated using the NMVP workflow when compared to results using AE workflow. Again, this could be an artifact of the different extraction kits used with each concentration method. The QIAamp Viral RNA Mini Kit employed with the NMVP concentration method does not have PCR inhibitor removal technology; whereas, the RNeasy PowerWater Kit used with the AE concentration method does. The manufacturer does provide a protocol for using the NMVP with an extraction kit that features inhibitor removal technology (Qiagen AllPrep PowerViral DNA/RNA Kit); however, the performance of the NMVP concentration method in conjunction with this kit has not yet been characterized in the literature and merits further investigation.
The ability of the AE concentration and extraction methods to successfully concentrate a variety of viruses, including PMMoV and SARS-CoV-2, from wastewater has been previously documented (Ahmed et al., 2020a; Ahmed et al., 2021; Ahmed et al., 2022a), along with its improved performance compared to other concentration and extraction protocols (Ahmed et al., 2020a). Additionally, prior to the COVID-19 pandemic and widespread application for wastewater surveillance, the AE concentration method was explicitly developed and widely used to concentrate low abundance targets from aqueous matrices in the environment (Ahmed et al., 2020b). The NMVP concentration method, on the other hand, was initially developed to concentrate multiple viruses from urine, cells, viral supernatants, and a variety of clinical samples (e.g., saliva, nasal fluid, nasal aspirate) with improved sensitivity and detection rates (Jaworski et al., 2014; Shafagati et al., 2013; Shafagati et al., 2015; Shafagati et al., 2016; Barclay et al., 2020; Lin et al., 2020). However, there are now several published studies reporting the application of NMVP to concentrate SARS-CoV-2 and indicator viruses from wastewater for both RT-qPCR and sequencing (Karthikeyan et al., 2021a; Karthikeyan et al., 2021b; Karthikeyan et al., 2021b; Babler et al., 2022; Hall et al., 2022; Lawal et al., 2022; Safford et al., 2022). For samples derived from clinical matrices, NMVP has improved both the analytical sensitivity of RT-qPCR and sequencing coverage for SARS-CoV-2 (Andersen et al., 2021; Barclay et al., 2020). But much less is known regarding the performance of the NMVP workflow compared to other concentration and extraction workflows for wastewater. In a four-sample comparison, Safford et al. (2022) found NMVP appeared to be more effective than centrifugal ultrafiltration for the detection of SARS-CoV-2 in wastewater. While Karthikeyan et al. (2021a) reported comparable results for NMVP and electronegative membrane filtration (n = 24), critical methodological details surrounding the processing of the membranes (e.g., homogenization and extraction) were not described. The largest electronegative membrane filtration and NMVP comparison yet published featured 65 samples, but variations between the method workflows (e.g., different sample volumes processed) and from previously published protocols (e.g., pipette mixing homogenization of membranes) make an “apples to apples” comparison more difficult (Babler et al., 2022). While the current study also has methodological limitations (e.g., different extraction kits), we sought to execute a robust comparison utilizing the manufacturer’s recommendations, especially concerning volumetric equivalency for both concentration methods. We have prioritized volumetric equivalency in our comparison, because the assumption that “more is more” regarding the volume of wastewater concentrated only holds if the recovery efficiency is not itself dependent on the volume concentrated – a hypothesis which as of yet has not been systematically tested.
Across all samples in the current study, we found the NMVP workflow yielded greater PMMoV concentrations in 58.3 % of samples, while the AE workflow generated greater concentrations of SARS-CoV-2 in 68.8 % of samples. Interestingly, the estimated concentration of SARS-CoV-2 was more strongly correlated (r = 0.859) between two workflows than PMMoV (r = 0.455). Safford et al. (2022) also reported mixed results with a centrifugal ultrafiltration workflow yielding greater concentrations of PMMoV, and NMVP yielding greater concentrations of SARS-CoV-2. These results suggest the concentration and extraction workflow performance is likely influenced by characteristics of both the wastewater and the morphology and structural characteristics of the target virus. Thus, concentration and extraction workflow performance should be validated for individual virus in wastewater.
When considering concentration workflow performance on a per-WWTP basis, comparable concentrations of PMMoV were derived at four WWTPs, irrespective of the concentration workflow employed. The NMVP workflow yielded greater PMMoV concentrations at three WWTPs (compared to yields by AE), and the AE workflow performed better than NMVP for the remaining one WWTP. For SARS-CoV-2, both workflows yielded comparable concentrations at five WWTPs, but the AE workflow performed better at the remaining three WWTPs. On the whole, these results suggest comparable performance for many WWTPs, but there was a non-negligible proportion of WWTP samples where the AE workflow was better for SARS-CoV-2 and the NMVP workflow was better for PMMoV. In their comparison, Babler et al. (2022) concluded that NMVP and electronegative membrane concentration and extraction methods provide equivalent results during RT-qPCR analysis. Although, they also noted variability for individual measurements which is consistent with our observations. Concentration methods combining liquid and solid fractions of the wastewater (such as AE and NMVP) generally outperform those that concentrate only liquid portions (Ahmed et al., 2020a), which could explain similar concentrations observed at five out of eight WWTPs.
While it is convenient to accept the concentration and extraction methods as equivalent, further development of wastewater surveillance methodology, especially for trace detection of biological analytes, must concern itself with the fundamental parameters driving method performance. In this regard, our study also yields some suggestive results. For the WWTPs with divergent results, the NMVP workflow yielded greater PMMoV concentrations at three WWTPs with low to moderate turbidity and TSS while the AE workflow only performed better at the WWTP with the greatest observed turbidity and TSS (WWTP H). For SARS-CoV-2 concentration, turbidity and TSS at the three WWTPs where the AE workflow performed better were above the 50th percentile including the two WWTPs with the highest turbidity and TSS (H, E). These results suggest that suspended solids loading, especially at the extremes, may affect the overall workflow performance, which is consistent with observations that substantial portions of enveloped viruses may be bound to particulate matter (Gundy et al., 2009; Ye et al., 2016).
Others have also noted decreased recovery efficiency with increasing solids loading for electronegative membrane concentration (Feng et al., 2021). In general, we found both concentration and extraction workflows yielded the greatest SARS-CoV-2 concentrations in the middle range of suspended solids loading and then decreased with further increases beyond this range. However, for SARS-CoV-2 the AE workflow appeared to be more resilient against extremely high solids loading than the NMVP workflow. PMMoV enumerations, meanwhile, seemed less affected by solids loading for both workflows. The effect of wastewater turbidity on chemical precipitation and ultrafiltration methods has been demonstrated before (Kevill et al., 2022), but the finding that the AE workflow appears to be less influenced by turbidity is novel. Owing to the constraints of our experimental design, it was outside of the scope of the current study to provide possible explanations for this observation, but nonetheless our results indicate that characteristics of the wastewater should be taken into consideration when selecting the concentration and extraction method. Our results, also suggest opportunities to further optimize virus concentration methods by controlling suspended solids loading.
If equivalent in performance, AE and NMVP workflows may provide additional advantages over other concentration and extraction workflows including demonstrated effectiveness for diverse groups of viruses, cost-effectiveness, and rapid concentration. However, one primary drawback of either approach presented in this study is the relatively limited sample volume (10-mL) processed for workflow comparison purpose. We acknowledge this volume may not be sufficient for trace detections and may represent a major limitation when virus loads in the wastewater are very low due to low infections in the community especially in the early stage of a pandemic. However, sample volume can be increased for both workflows if more reagents and larger pore membranes are used. Some additional considerations specific to each approach are propensity for clogging, more frequently observed with AE (Ahmed et al., 2020a), and requirements for expensive and specialized equipment (e.g., magnetic racks for NMVP and homogenizers for AE) that may not be readily available in many microbiology laboratories. AE clogging issues may be alleviated through initial centrifugation or prefiltration step or using a larger pore size membrane, this could also result in target loss and should be carefully considered when selecting pretreatment steps. To circumvent the need for expensive equipment, vortexing the filter paper using less-expensive vortex adaptors can be a substitute for a homogenzier and some laboratories have reported fabricating their own magnetic racks with plans available on the NIH 3D Print Exchange (https://3dprint.nih.gov/discover/3dpx-009008). One advantage that several wastewater surveillance studies have noted for NMVP is the ability to automate the wastewater concentration and extraction steps on liquid handling platforms such as the KingFisher Flex for high throughput processing (Karthikeyan et al., 2021a; Karthikeyan et al., 2021b; Safford et al., 2022). While possible, such high throughput efforts will require additional high-cost specialized equipment; whereas membrane filtration apparatus are likely to be found in many microbiology laboratories.
In this study, we compared two rapid and inexpensive concentration and extraction workflows (i.e., AE and NMVP) for their ability to concentrate endogenous PMMoV and SARS-CoV-2 from wastewater. Our findings indicate that the performance of each workflow was affected by the wastewater characteristics, namely suspended solids, as well as morphology of the viral target. The influence of turbidity and TSS on the recovery of enveloped and non-enveloped viruses requires further investigations using studies design for such a priori. The NMVP workflow performed better than the AE workflow for PMMoV at four of eight WWTPs, while the AE workflow performed better than the NMVP workflow for SARS-CoV-2 at three of eight WWTPs. For the remaining WWTPs, the performance of the two concentration and extraction workflows were comparable. These findings suggest that wastewater characteristics, such as suspended solids concentration, and the intended target for analysis should be considered when validating an optimal concentration and extraction workflow for wastewater surveillance.
Supplementary Material
HIGHLIGHTS.
AE and NMVP methods were used to quantify endogenous PMMoV and SARS-CoV-2.
The concentrations of PMMoV were greater by NMVP method than AE method.
The concentrations of SARS-CoV-2 were greater by AE method than NMVP method.
Suspended solids concentration and the viral target should be considered for WBE.
Acknowledgements
We thank CSIRO Land and Water for strategic funding to complete this research project. We thank Urban Utilities for providing untreated wastewater samples.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.160072.
Disclaimer
The views expressed in this article are those of the author(s) and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. The U.S. Environmental Protection Agency through the Office of Research and Development provided technical direction but did not collect, generate, evaluate, or use the environmental data described herein.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Data will be made available on request.
References
- Ahmed W, Bivins A, Wendy JM, Metcalfe S, Stephens M, Jennison AV, Moore FAJ, Bourke J, Schlebusch S, McMahon, Hewitson JG, Nguyen S, Barcelon J, Jackson G, Mueller JF, Ehret J, Hosegood I, Tian W, Wang H, Yang L, Bertsch PM, Tynan J, Thomas KV, Bibby K, Graber TE, Ziels R, Simpson SL, 2022b. Detection of the Omicron (B.1.1.529) variant of SARS-CoV-2 in aircraft wastewater. Sci. Total Environ 820, 153171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Harwood VJ, Gyawali P, Sidhu JP, Toze S, 2015. Comparison of concentration methods for quantitative detection of sewage-associated viral markers in environmental waters. Appl. Environ. Microbiol 81, 2042–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Bertsch PM, Bivins A, Bibby K, Farkas K, Gathercole A, Haramoto E, Gyawali P, Korajkic A, McMinn BR, Mueller JF, Simpson SL, Smith W, Symonds EM, Thomas KV, Verhagen R, Kitajima M, 2020. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ 739, 139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Payyappat S, Cassidy M, Harrison N, Besley C, 2020b. Sewage-associated marker genes illustrate the impact of wet weather overflows and dry weather leakage in urban estuarine waters of Sydney, Australia. Sci. Total Environ 705, 135390. [DOI] [PubMed] [Google Scholar]
- Ahmed W, Bivins A, Metcalfe S, Smith W, Verbyla ME, Symonds EM, Simpson SL, 2022a. Evaluation of process limit of detection and quantification variation of SARS-CoV-2 RT-qPCR and RT-dPCR assays for wastewater surveillance. Water Res. 213, 118132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Bivins A, Simpson SL, Smith W, Metcalfe S, McMinn B, Symonds EM, Korajkic A, 2021. Comparative analysis of rapid concentration methods for the recovery of SARS-CoV-2 and quantification of human enteric viruses and a sewage-associated marker gene in untreated wastewater. Sci. Total Environ 799, 149386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Simpson SL, Bertsch PM, Bibby K, Bivins A, Blackall LL, Bofill-Mas S, Bosch A, Brandão J, Choi PM, Ciesielski M, Donner E, D’Souza N, Farnleitner AH, Gerrity D, Gonzalez R, Griffith JF, Gyawali P, Haas CN, Hamilton KA, Hapuarachchi HC, Harwood VJ, Haque R, Jackson G, Khan SJ, Khan W, Kitajima M, Korajkic A, La Rosa G, Layton BA, Lipp E, McLellan SL, McMinn B, Medema G, Metcalfe S, Meijer WG, Mueller JF, Murphy H, Naughton CC, Noble RT, Payyappat S, Petterson S, Pitkänen T, Rajal VB, Reyneke B, Roman FA Jr., Rose JB, Rusiñol M, Sadowsky MJ, Sala-Comorera L, Setoh YX, Sherchan SP, Sirikanchana K, Smith W, Steele JA, Sabburg R, Symonds EM, Thai P, Thomas KV, Tynan J, Toze S, Thompson J, Whiteley AS, Wong JCC, Sano D, Wuertz S, Xagoraraki I, Zhang Q, Zimmer-Faust AG, Shanks OC, 2022c. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ 805, 149877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Smith WJM, Metcalfe S, Jackson G, Choi PM, Morrison M, Field D, Gyawali P, Bivins A, Bibby K, Simpson SL, 2022d. Comparison of RT-qPCR and RT-dPCR platforms for the trace detection of SARS-CoV-2 RNA in wastewater. ACS ES&T Water 10.1021/acsestwater.1c00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen P, Barksdale S, Barclay RA, Smith N, Fernandes J, Besse K, Goldfarb D, Barbero R, Dunlap R, Jones-Roe T, Kelly R, Miao S, Ruhunusiri C, Munns A, Mosavi S, Sanson L, Munns D, Sahoo S, Swahn O, Hull K, White D, Kolb K, Noroozu F, Seelam J, Patnaik A, Lepene B, 2021. Nanotrap Particles Improve Nanopore Sequencing of SARS-CoV-2 and Other Respiratory Viruses. bioRxiv, p. 471814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanious A, Aeppli M, Jacak R, Refardt D, Sigstam T, Kohn T, Sander M, 2016. Viruses at solid-water interfaces: a systematic assessment of interactions driving adsorption. Environ. Sci. Technol 50, 732–743. [DOI] [PubMed] [Google Scholar]
- Babler KM, Amirali A, Sharkey ME, Williams SL, Boone MM, Cosculluela GA, Currall BB, Grills GS, Laine J, Mason CE, Redings BD, Schurer SC, Stevenson M, Vidovic D, Solo-Gabriele HM, 2022. Comparison of electronegative filtration to magnetic bead-based concentration and V2G-qPCR to RT-qPCR for quantifying viral SARS-CoV-2 RNA from wastewater. ACS EST Water 10.1021/acsestwater.2c00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay RA, Akhrymuk I, Patnaik A, Callahan V, Lehman C, Andersen P, Barbero R, Barksdale S, Dunlap R, Goldfarb D, Jones-Roe T, Kelly R, Kim B, Miao S, Munns A, Munns D, Patel S, Porter E, Ramsey R, Sahoo S, Swahn O, Warsh J, Kehn-Hall K, Lepene B, 2020. Hydrogel particles improve detection of SARS-CoV-2 RNA from multiple sample types. Sci. Rep 10, 22425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besselsen DG, Wagner AM, Loganbill JK, 2002. Detection of rodent coronaviruses by use of fluorogenic reverse transcriptase-polymerase chain reaction analysis. Comp. Med 52, 111–116. [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT, 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem 55, 611–622. [DOI] [PubMed] [Google Scholar]
- CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel, n.d.CDC 2019novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel (n.d.). https://www.fda.gov/media/134922/download. [DOI] [PMC free article] [PubMed]
- Cervantes-Avilés P, Moreno-Andrade I, Carrillo-Reyes J, 2021. Approaches applied to detect SARS-CoV-2 in wastewater and perspectives post-COVID-19. J. Water Process Eng. 40, 101947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón L, Morales E, Valiente C, Reyes L, Barrantes K, 2021. Wastewater-based epidemiology of enteric viruses and surveillance of acute gastrointestinal illness outbreaks in a resource-limited region. Am. J. Trop. Med. Hyg 105, 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chik AHS, Glier MB, Servos M, Mangat CS, Pang XL, Qiu Y, D’Aoust PM, Burnet JB, Delatolla R, Dorner S, Geng Q, Giesy JP Jr., McKay RM, Mulvey MR, Prystajecky N, Srikanthan N, Xie Y, Conant B, Hrudey SE, 2021. Canadian SARSCoV-2 inter-laboratory consortium. Comparison of approaches to quantify SARS-CoV-2 in wastewater using RT-qPCR: results and implications from a collaborative interlaboratory study in Canada. J. Environ. Sci. (China) 107, 218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary R, Dhole TN, 2008. Interrupting wild poliovirus transmission using oral poliovirus vaccine: environmental surveillance in high-risks area of India. J. Med. Virol 80, 1477–1488. [DOI] [PubMed] [Google Scholar]
- El Bassioni L, Barakat I, Nasr E, de Gourville EM, Hovi T, Blomqvist S, Burns C, Stenvik M, Gary H, Kew OM, Pallansch MA, Wahdan MH, 2003. Prolonged detection of indigenous wild polioviruses in sewage from communities in Egypt. Am. J. Epidemiol 158, 807–815. [DOI] [PubMed] [Google Scholar]
- Feng S, Roguet A, McClary-Gutierrez JS, Newton RJ, Kloczko N, Meiman JG, McLellan SL, 2021. Evaluation of sampling, analysis, and normalization methods for SARS-CoV-2 concentrations in wastewater to assess COVID-19 burdens in Wisconsin communities. ACS EST Water. 1, 1955–1965. [Google Scholar]
- Gundy PM, Gerba CP, Pepper IL, 2009. Survival of coronaviruses in water and wastewater. Food Environ. Virol 1, 10. [Google Scholar]
- Hall AJ, 2012. Noroviruses: the perfect human pathogens? J. Infect. Dis 205, 1622–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall GJ, Page EJ, Rhee M, Hay C, Krause A, Langenbacher E, Ruth A, Grenier S, Duran AP, Kamara I, Iskander JK, Thomas DL, Bock E, Porta N, Pharo J, Osterink BA, Zelmanowitz S, Fleischmann CM, Liyanage D, Gray JP, 2022. Wastewater Surveillance of U.S., Coast Guard Installations and Seagoing Military Vessels to Mitigate the Risk of COVID-19 Outbreaks. medrXiv. 10.1101/2022.02.05.22269021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E, Kitajima M, Kishida N, Konno Y, Katayama H, Asami M, Akiba M, 2013. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl. Environ. Microbiol 79, 7413–7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A, Hara-Yamamura H, Meuchi Y, Imai S, Honda R, 2021. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ 758, 143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes B, Duong D, White BJ, Wiggington KR, Chan EMG, Wolfe MK, Boehm AB, 2022. Respiratory syncytial virus (ESV) RNA is wastewater settled solids reflects RSV clinical positivity rates. Environ. Sci. Technol. Lett 9, 173–178. [Google Scholar]
- Jafferali MH, Khatami K, Atasoy M, Birgersson M, Williams C, Cetecioglu Z, 2021. Benchmarking virus concentration methods for quantification of SARS-CoV-2 in raw wastewater. Sci. Total Environ 755, 142939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski E, Saifuddin M, Sampey G, Shafagati N, Van Duyne R, Iordanskiy S, KehnHall K, Liotta L, Petricoin III E, Young M, Lepene B, Kashanchi F, 2014. The use of nanotrap particles technology in capturing HIV-1 virions and viral proteins from infected cells. PLoS One 9, e96778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor RS, Nelson KL, Greenwald HD, Kennedy LC, 2020. Challenges in measuring the recovery of SARS-CoV-2 from wastewater. Environ. Sci. Technol 55, 3514–3519. [DOI] [PubMed] [Google Scholar]
- Karthikeyan S, Ronquillo N, Belda-Ferre P, Alvarado D, Javidi T, Longhurst CA, Knight R, 2021. High-throughput wastewater SARS-CoV-2 detection enables forecasting of community infection dynamics in San Diego County. mSystems 6 (2), e00045–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan S, Levy JI, De Hoff P, et al. , 2021b. Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission. Nature 609, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan S, Nguyen A, McDonald D, Zong Y, Ronquillo N, Ren J, Zoou J, Farmer S, Humphrey G, Henderson D, Javidi T, Messer K, Anderson C, Schooley R, Martin NK, Night R, 2022. Rapid, large-scale wastewater surveillance and automated reporting system enable early detection of nearly 85% of COVID-19 cases on a university campus. mSystems. 6 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevill JL, Pellett C, Farkas K, Brown MR, Bassano I, Denise H, McDonald JE, Malham SK, Porter J, Warren J, Evens NP, Paterson S, Singer AC, Jones DL, 2022. A comparison of precipitation and filtration-based SARS-CoV-2 recovery methods and the influence of temperature, turbidity, and surfactant load in urban wastewater. Sci. Total Environ 808, 151916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd-Ljunggren K, Holmberg A, Bläckberg J, Lindqvist B, 2006. High levels of hepatitis B virus DNA in body fluids from chronic carriers. J. Hosp. Infect 64 (4), 352–357. [DOI] [PubMed] [Google Scholar]
- La Rosa G, Fratini M, della Libera S, Iaconelli M, Muscillo M, 2012. Emerging and potentially emerging viruses in water environments. Ann. Ist. Super Sanità 4, 397–406. [DOI] [PubMed] [Google Scholar]
- La Rosa G, Mancini P, Bonanno Ferraro G, Veneri C, Iaconelli M, Bonadonna L, Lucentini L, Suffredini E, 2021a. SARS-CoV-2 has been circulating in northern Italy since December 2019: evidence from environmental monitoring. Sci. Total Environ 750, 141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G, Mancini P, Bonanno Ferraro G, Veneri C, Iaconelli M, Lucentini L, Bonadonna L, Brusaferro S, Brandtner D, Fasanella A, Pace L, Parisi A, Galante D, Suffredini E, 2021b. Rapid screening for SARS-CoV-2 variants of concern in clinical and environmental samples using nested RT-PCR assays targeting key mutations of the spike protein. Water Res. 197, 117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawal OU, Zhang L, Parreira VR, Brown RS, Chettleburgh C, Dannah N, Delatolla R, Gilbride KA, Graber TE, Islam G, Knockleby J, Ma S, McDougall H, McKay RM, Mloszewska A, Oswald C, Servos M, Swinwood-Sky M, Ybazeta G, Habash M, Goodridge L, 2022. Metagenomics of wastewater influent from wastewater treatment facilities across Ontario in the era of emerging SARS-CoV-2 variants of concern. Microbiol. Resour. Announc 11, e0036222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung NHL, 2021. Transmissibility and transmission of respiratory viruses. Nat. Rev. Microbiol 19, 528–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Carey BD, Callahan V, Lee JH, Bracci N, Patnaik A, Smith AK, Narayanan A, Lepene B, Kehn-Hall K, 2020. Use of nanotrap particles for the capture and enrichment of zika, chikungunya and dengue viruses in urine. PLoS One 15 (1), e0227058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl JF, Grace D, 2015. The consequences of human actions on risks for infectious diseases: a review. Infect. Ecol. Epidemiol 5, 30048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMinn BR, Korajkic A, Kelleher J, Herrmann MP, Pemberton AC, Ahmed W, Villegas EN, Oshima K, 2021. Development of a large volume concentration method for recovery of coronavirus from wastewater. Sci. Total Environ 774, 145727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A, 2020. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett 7, 511–516. [DOI] [PubMed] [Google Scholar]
- Mtetwa HN, Amoah ID, Kumari S, Bux F, Reddy P, 2021. Wastewater-based surveillance of antibiotic resistance genes associated with tuberculosis treatment regimen in KwaZulu Natal, South Africa. Antibiotics (Basel, Switzerland). 10, 1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J, Zulli A, Brackney DE, Grubaugh ND, Kaplan EH, Casanovas-Massana A, Ko AI, Malik AA, Wang D, Wanh M, Warren JL, Weinberger DM, Arnold W, Omer SB, 2020. Measurements of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol 38, 1164–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson BM, Darby E, Haas CN, Amha YM, Bartolo M, Danielson R, Dearborn Y, Giovanni GD, Ferguson C, Fevig S, Gaddis E, Gray D, Lukasik G, Bull M, Olivas L, Olivieri A, Qu Y, 2021. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the US. Environ. Sci.: Water Res. Technol 7, 504–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds LJ, Gonzalez G, Sala-Comorera L, Martin NA, Byrne A, Fennema S, Holohan N, Kuntamukkula SR, Sarwar N, Nolan TM, Stephens JS, Whitty M, Bennett C, Luu Q, Morley U, Yandle Z, Dean J, Joyce E, O’Sullivan JJ, Cuddihy JM, McIntyre AM, Robinson EP, Dahly D, Fletcher NF, Carr M, Gascun CD, Meijer WM, 2022. SARS-CoV-2 variant trends in Ireland: wastewater-based epidemiology and clinical surveillance. Sci. Total Environ 838, 155828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K, Symonds EM, Sinigalliano C, Stewart J, Breitbart M, 2009. Pepper mild mottle virus as an indicator of fecal pollution. Appl. Environ. Microbiol 75, 7261–7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol M, Martínez-Puchol S, Forés E, Itarte M, Girones R, Bofill-Mas S, 2020. Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater. Curr. Opin. Environ. Sci. Health 17, 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safford H, Zuniga-Montanez RE, Kim M, Wu X, Wei L, Sharpnack J, Shapiro K, Bischel H, 2022. Wastewater Surveillance for COVID-19 Response at Multiple Geographic Scales: Aligning Wastewater and Clinical Results at the Census-block Level and Addressing Pervasiveness of qPCR Non-detects. medRxiv. 10.1101/2022.01.28.22269911. [DOI] [Google Scholar]
- Sapula SA, Whittall JJ, Pandopulos AJ, Gerber C, Venter H, 2021. An optimized and robust PEG precipitation method for detection of SARS-CoV-2 in wastewater. Sci. Total Environ 785, 147270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafagati N, Narayanan A, Baer A, Fite K, Pinkham C, Bailey C, Kashanchi F, Lepene B, Kehn-Hall K, 2013. The use of NanoTrap particles as a sample enrichment method to enhance the detection of Rift Valley fever virus. PLoS Negl. Trop. Dis 7, e2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafagati N, Lundberg L, Baer A, Patanarut A, Fite K, Lepene B, Kehn-Hall K, 2015. The use of nanotrap particles in the enhanced detection of Rift Valley fever virus nucleoprotein. PLoS One 10, e0128215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafagati N, Fite K, Patanarut A, Baer A, Pinkham C, An S, Foote B, Lepene B, KehnHall K, 2016. Enhanced detection of respiratory pathogens with nanotrap particles. Virulence. 7, 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds EM, Verbyla ME, Lukasik JO, Kafle RC, Breitbart M, Mihelcic JR, 2014. A case study of enteric virus removal and insights into the associated risk of water reuse for two wastewater treatment pond systems in Bolivia. Water Res. 65, 257–270. [DOI] [PubMed] [Google Scholar]
- Verbyla ME, Symonds EM, Kafle RC, Cairns MR, Iriarte M, Mercado Guzmán A, Coronado A, Breitbart M, Ledo C, Mihelcic JR, 2016. Managing microbial risks from indirect wastewater reuse for irrigation in urbanizing watersheds. Environ. Sci. Technol 50, 6803–6813. [DOI] [PubMed] [Google Scholar]
- Wolfe MK, Duong D, Bakker KM, Ammermann M, Mortenson L, Hughes B, Arts P, Lauring AS, Fitzsimmons WJ, Bendall E, Hwang CE, Martin ET, White BJ, Boehm AB, Wiggington KR, 2022. Wastewater-based detection of two influenza out-breaks. Environ. Sci. Technol. Lett 9 (8), 687–692. [Google Scholar]
- Wu F, Zhang J, Xiao A, Gu X, Lee WL, Armas F, Kauffman K, Hanage W, Matus M, Ghaeli N, Endo N, Duvallet C, Poyet M, Moniz K, Washburne AD, Ericson TB, Chai PR, Thompson J, Alm EJ, 2021. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems 5, e00614–e00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Ellenberg RM, Graham KE, Wigginton KR, 2016. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol 50, 5077–5085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.