Abstract
We previously isolated from a 1994 isolate of Vibrio cholerae O139 a filamentous lysogenic bacteriophage, choleraphage 493, which inhibits pre-O139 but not post-O139 El Tor biotype V. cholerae strains in plaque assays. We investigated the role of the mannose-sensitive hemagglutinin (MSHA) type IV pilus as a receptor in phage 493 infection. Spontaneous, Tn5 insertion, and mshA deletion mutants are resistant to 493 infection. Susceptibility is restored by mshA complementation of deletion mutants. Additionally, the 493 phage titer is reduced by adsorption with MSHA-positive strains but not with a ΔmshA1 strain. Monoclonal antibody against MSHA inhibits plaque formation. We conclude that MSHA is the receptor for phage 493. The emergence and decline of O139 in India and Bangladesh are correlated with the susceptibility and resistance of El Tor strains to 493. However, mshA gene sequences of post-O139 strains are identical to those of susceptible pre-O139 isolates, indicating that phage resistance of El Tor is not due to a change in mshA. Classical biotype strains are (with rare exceptions) hemagglutinin negative and resistant to 493 in plaque assays. Nevertheless, they express the mshA pilin gene. They can be infected with 493 and produce low levels of phage DNA, like post-O139 El Tor strains. Resistance to 493 in plaque assays is thus not equivalent to resistance to infection. The ability of filamentous phages, such as 493, to transfer large amounts of DNA provides them, additionally, with the potential for quantum leaps in both identity and pathogenicity, such as the conversion of El Tor to O139.
Prior to 1992, two biotypes, classical and El Tor, of Vibrio cholerae serogroup O1 were responsible for all epidemic cholera. In 1992, V. cholerae O139 (synonym Bengal), a newly recognized serogroup, emerged explosively in India and Bangladesh—soon outnumbering the previously resident El Tor biotype. However, in 1994, the El Tor biotype again began to predominate (7, 27). The O139 serogroup is closely related to the El Tor biotype (1, 20) but lacks the O1-specific antigen and expresses a polysaccharide capsule. These characteristics are the result of the replacement of the O1 rfb locus by exogenous DNA encoding the O139 antigen and capsular polysaccharide synthesis (3, 28) as a consequence of horizontal transmission of genetic information (3, 24).
The El Tor biotype closely resembles the classical biotype but differs in several phenotypic properties, including the ability to agglutinate chicken erythrocytes in a slide test with agar-grown vibrios (8). This activity is dependent on a mannose-sensitive hemagglutinin (MSHA) type IV pilus (14, 18, 22). With only rare exceptions, the MSHA test is a reliable method for differentiating El Tor and classical biotypes (8). Like El Tor, O139 expresses the MSHA (1).
We recently isolated and partially characterized a filamentous lysogenic bacteriophage, choleraphage 493, from a 1994 isolate of a V. cholerae O139 strain, AJ27-493, from Bangladesh (19). This phage can assume a pseudolysogenic plasmid form or can randomly integrate into the bacterial chromosome. Depending on the host, the phage can form turbid plaques or can inhibit bacterial growth without forming defined plaques. In plaque assays, choleraphage 493 was inhibitory at a high titer for all pre-O139 El Tor biotype strains tested but not for classical biotype strains (19). Interestingly, however, one of five isolates of El Tor in 1994 and all 1995 and 1996 El Tor isolates tested were resistant to 493 in the plaque assay. These results correlate with our hypothesis that choleraphage may play a significant role in the population dynamics or territoriality of choleragenic vibrios (19) by affecting susceptible strains in the environment.
Another recently discovered filamentous bacteriophage, CTXφ, which carries the structural genes for the cholera enterotoxin pathogenicity island (including ctxAB, zot, cep, ace, and orfU), uses a type IV pilus, toxin-coregulated pilus protein (TcpA), as its receptor (30). We had previously excluded the possibility that TcpA could also be the receptor for choleraphage 493 (19): a ΔtcpA mutant was as sensitive to 493 as its wild-type parent. To rule out immunity, we also demonstrated that the resistant new El Tor strains were not lysogenized by 493.
The present study was initiated to define the receptor for 493 and to examine the reason for the resistance of the new El Tor strains. As type IV pili are frequently associated with both bacterial pathogenesis and DNA uptake events (23), we chose to examine the MSHA.
(Portions of this work were presented at the 97th General Meeting of the American Society for Microbiology, 1997; at the 8th European Workshop Conference on Bacterial Protein Toxins, Staffelstein, Germany, 1997; at the Cold Spring Harbor Laboratory Conference on Microbial Pathogenesis and Host Defense, Cold Spring Harbor, N.Y., 1997; and at the U.S.-Japan Cooperative Medical Science Program, 33rd Joint Conference on Cholera and Related Diarrheal Diseases, St. Petersburg, Fla., 1997.)
MATERIALS AND METHODS
Bacterial strains and plasmids.
V. cholerae and Escherichia coli strains and plasmids used in this study are listed in Table 1. The source of phage 493, V. cholerae O139 strain AJ27-493, a 1994 isolate from Bangladesh, was provided by John Albert (1), who also provided the 1994 to 1996 El Tor biotype isolates used. Each of these relatively pristine strains was the lyophilized progeny of a colony from the primary isolation plate. Whereas these cultures are colonially homogeneous, other strains that we have received consist of mixtures of colony types (10). The mshA-phoA gene fusion strains, JM69 and JM191, were constructed by allelic exchange with pJM62 as previously described (22). Plasmid pTrc99A (Pharmacia) was used for the cloning and expression of the mshA gene from either C6706 or 150659 genomic DNA. The gene was isolated as a 450-bp mshA PCR product amplified with the following primer pair: MSH39, 5′-CTAGTCATGAAAAGACAAGGTGGTTTCACCC-3′, and MSH40, 5′-CATGAGCTCAATTATTGCGCTGGTTTACCACAAGC-3′. The product was inserted into pTrc99A as a BspHI-SacI fragment and recovered in E. coli JM109. The resulting plasmids, pJM232 (C6706) and pJM249 (150659), were introduced into the C6706 ΔmshA strain KHT46 by electroporation, yielding the isogenic strains JM137 and JM253, respectively. Plasmid pJM232 was transferred by electroporation into strain O395 to form Apr O395-mshA.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| V. cholerae | ||
| 17 | O1 El Tor, Ogawa | 13 |
| 17− | MSHA− spontaneous mutant of 17 | 13 |
| 26-3 | O1 El Tor, Ogawa, Philippines, 1961 | 13 |
| 26-3− | MSHA− spontaneous mutant of 26-3 | 13 |
| JBK70 | O1 El Tor, Inaba ΔctxAB | 21 |
| Tn5-1, -3, -9, and -10 | MSHA− Tn5 insertion mutants of JBK70 | 14 |
| C6706 | O1 El Tor, Inaba, Peru, spontaneous str-2, 1992 | 29 |
| KHT46 | C6706 ΔmshA1 | 29 |
| KHT52 | C6706 str-2 ΔtcpA10 | 29 |
| JM191 | C6706 str-2 mshA-phoA1 | This study |
| JM137 | KHT46(pJM232) | This study |
| JM253 | KHT46(pJM249) | This study |
| AJ17564 | O1 El Tor, Ogawa, Bangladesh, 1994 | J. Albert |
| AJ19290 | O1 El Tor, Ogawa, Bangladesh, 1994 | J. Albert |
| 150659 | O1 El Tor, Ogawa, Bangladesh, 1995 | J. Albert |
| 20049 | O1 El Tor, Ogawa, Bangladesh, 1996 | J. Albert |
| NIH41 (20-A-11) | O1 classical, Ogawa, MSHA+ or MSHA−, India, 1941 | 8 |
| 20-A-11+ | MSHA+ spontaneous mutant of 20-A-11 | 8 |
| O395 Sm | O1 classical, Ogawa, India, 1964, Smr | Laboratory collection |
| O395-mshA | O395(pJM232) | This study |
| JM69 | O395 mshA-phoA1 mshK::pGP704 | This study |
| AJ27-493 | O139, Bangladesh, 1994 | J. Albert |
| E. coli JM109 | F′ traD36 lacIq Δ(lacZ)M15 proA+B+/e14− (McrA−) Δ(lac-proAB)thi gyrA96 (Nalr) endA1 hsdR17 (rK− mK+) relA1 supE44 recA1 | 31 |
| Plasmids | ||
| pTrc99A | Expression plasmid, Apr | Pharmacia |
| pJM232 | pTrc99A · mshA (C6706) | This study |
| pJM249 | pTrc99A · mshA (150659, 1995) | This study |
MSHA testing.
MSHA testing was performed by slide agglutination with agar-grown vibrios and chicken erythrocytes as described previously (8).
Choleraphage techniques.
Preparation and assay of choleraphage were as described previously (19). Sensitivity to phage was determined by a plaque assay. Aliquots (0.01 ml) of serial dilutions of phage-containing samples were plated on soft Luria-Bertani (LB) agar (0.4%) overlays seeded with indicator strains. The overlays were prepared by inoculating 5 ml of 0.4% LB agar with 10 μl of a culture of the indicator strain (∼108 cells/ml) grown for ∼3 h in LB broth at 37°C with shaking. Results were observed after overnight incubation.
To examine the adsorption of the phage to indicator cells, ∼108 cells of LB broth-grown cultures of strains C6706, KHT46, and KHT52 were mixed with ∼105 PFU of the phage. LB broth mixed with the phage was used as a control. Parallel samples were incubated for 20 min at 37, 22, and 4°C, and then the bacteria were removed by centrifugation for 20 min at 1,200 × g. Supernatants were filtered through 0.22-μm-pore-size filters, and 10-fold serial dilutions were plated on C6706 and KHT46 as indicator strains. Plaque counts were determined after 16 h of incubation at 37°C.
To test the inhibition of phage infection with anti-MSHA antibody, dilutions of anti-MSHA monoclonal antibody (MAb) 2F12F1, provided by J. A. Benitez (6), in parallel with LB broth as a control, were mixed with ∼108 cells of KHT52 at a ratio of 1:2; the mixtures were incubated at 37°C for 30 min. Phage 493 then was added, and plaques were formed in 5 ml of soft agar. Plaques were counted after 16 h of incubation at 37°C.
DNA techniques.
Phage DNA was extracted according to Sambrook et al. (26). Plasmid DNA was prepared with the Wizard Plus SV Miniprep system (Promega, Madison, Wis.). DNA fragments were separated by agarose gel electrophoresis in TAE buffer (26). Labelling of virion DNA and Southern hybridizations were done with the Genius system (Boehringer Mannheim Biochemicals). Genomic DNA from three choleraphage 493-resistant strains (20049, 150659, and AJ17564) and two phage 493-sensitive strains (AJ19290 and C6706) was PCR amplified with the following primer pair, which flanks the mshA gene: MSH1, 5′-AAAAGTCGACAGCGAAAGCGAATAGTGG-3′, and MSH4, 5′-AAAAGCATGCGTGGTTACCACCGCAAAGG-3′. The resulting PCR product was Qiaquik gel purified (Qiagen), and 100 ng of this template was subjected to cycle sequencing of both DNA strands by use of the ABI Prism Dye Terminator Ready Reaction (Applied Biosystems) and 3.2 pmol of either primer MSH39 or primer MSH40. The reactions were performed in triplicate, and the reaction mixtures were column purified (Centrisep) and analyzed on an ABI model 373 stretch automated DNA sequencer (Applied Biosystems). ABI software and DNASTAR software were used for sequence analysis and alignment.
Alkaline phosphatase assay.
Alkaline phosphatase assays were performed in triplicate on mshA-phoA fusions with strains O395, JM69, C6706, and JM191 as described previously (22). Units of alkaline phosphatase activity were expressed by use of the following formula: alkaline phosphatase activity = {1,000 × [OD420 − (1.75 × OD550)]}/[time (minutes) × volume (milliliters) × OD660], where OD420 is optical density at 420 nm. Standard deviations were calculated with the EXCEL program.
RESULTS
Sensitivity of strains and mutants to choleraphage 493.
The results of plaque assays to determine the sensitivity of various strains and mutants to phage 493 are summarized in Table 2. In contrast to their wild-type MSHA-positive (MSHA+) parents, spontaneous MSHA-negative (MSHA−) mutants (selected after serial absorptions of strains 17 and 26-3 with chicken erythrocytes [13]) were resistant to phage 493, as were MSHA− Tn5 insertion mutants of MSHA+ strain JBK70 (14). These results supported the hypothesis that MSHA is the receptor for phage 493.
TABLE 2.
| Strain | Plaque assay result | Southern hybridization result |
|---|---|---|
| 17 | +++ | +++ |
| 17− | − | − |
| 26-3 | +++ | +++ |
| 26-3− | − | − |
| JBK70 | +++ | +++ |
| Tn5-1, -3, -9, and -10 | − | − |
| C6706 | +++ | +++ |
| KHT46 | − | − |
| JM137 | +++ | +++ |
| JM253 | +++ | +++ |
| AJ17564 | − | +++ |
| AJ19290 | +++ | +++ |
| 150659 | − | + |
| 20049 | − | + |
| O395 Sm | (+) | + |
| O395-mshA | (+) | + |
| NIH41 (20-A-11) | +++ | +++ |
| 20-A-11+ | +++ | +++ |
As demonstrated in plaque assays. +++, plaque formation with phage preparation diluted 10−6; (+), suppression of growth only with undiluted phage preparation; −, no plaques, or inhibition of growth.
As demonstrated by Southern hybridization following exposure to phage 493. No signal was detected prior to infection. +++, strong signal; +, very weak signal; −, no signal.
Confirmation that MSHA is the receptor for choleraphage 493.
To further examine the hypothesis that MSHA serves as the phage 493 receptor, several additional experiments were performed. Strain KHT46, which is a ΔmshA mutant of phage 493-sensitive El Tor strain C6706, was shown to be phage resistant (Table 2). Phage sensitivity was restored upon the introduction of the mshA structural gene carried on plasmid pJM232, resulting in strain JM137, thus demonstrating the specific requirement of the expression of mshA for phage 493 sensitivity.
If mshA expression is required because MSHA serves as the actual phage receptor, incubation of phage 493 with MSHA+ cells should absorb out phage and correspondingly reduce the titer of the lysate. Three consecutive absorptions of 493 lysate with strains C6706 and KHT52 (ΔtcpA10) reduced the titer from ∼105 to ∼102 PFU/ml, whereas the titer remained at ∼105 PFU/ml when the lysate was similarly absorbed with ΔmshA mutant KHT46. In addition, a monoclonal antibody specific for MSHA blocked the ability of the phage to form plaques. Incubation of strain KHT52 with a 1:1,000 final dilution of anti-MSHA MAb 2F12F1 reduced the titer 100-fold, from ∼105 to ∼102 PFU/ml, and incubation with a 1:100 dilution of the antibody resulted in a complete loss of plaque formation. Taken together, these observations indicate that MSHA is the receptor for choleraphage 493.
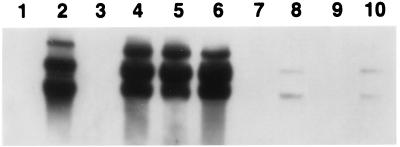
Southern hybridization with virion DNA as a probe confirmed the results of the biological assays (Table 2). The phage 493-sensitive strains described above became hybridization positive following exposure to phage 493, whereas the resistant MSHA− spontaneous, insertion, and deletion mutants were negative. The mshA-complemented deletion mutant JM137 gave a strong signal (Fig. 1).
FIG. 1.
Southern blot analysis of plasmid DNA of V. cholerae strains with total virion DNA as a probe. Lanes: 1, AJ17564; 2, AJ17564/493 (i.e., after infection with 493); 3, KHT46; 4, JM137/493; 5, JM253/493; 6, AJ27-493; 7, 150659; 8, 150659/493; 9, 20049; 10, 20049/493. Positive signals represent double-stranded phage RF.
Choleraphage 493-resistant El Tor strains have a functional mshA gene.
Since MSHA is the receptor for phage 493, how can the 1994 to 1996 isolates of El Tor biotype strains—which are MSHA+—be resistant to phage 493? One possibility is that there was a mutation in mshA which affected phage binding but not hemagglutination function. To examine this idea, we performed DNA sequence analysis of the mshA genes from three phage 493-resistant (AJ17564, 150659, and 20049) and two phage 493-sensitive (C6706 and AJ19290) El Tor biotype strains and found that the sequences of all five strains were identical. These results demonstrated that phage 493 resistance in these three particular El Tor strains is not due to alterations in the mshA pilin subunit gene sequence. To investigate further the structural integrity and function of the MSHA pilin subunit from a phage 493-resistant strain of V. cholerae, the mshA genes from the phage 493-resistant strain 150659 and the phage 493-sensitive strain C6706 were cloned and expressed in KHT46 (ΔmshA). The hemagglutination defect as well as the phage 493 sensitivity (Table 2) of the resulting strain was complemented by either plasmid pJM232 harboring mshA from the phage-sensitive strain C6706 or plasmid pJM249 harboring mshA from the phage-resistant strain 150659 (Table 2), demonstrating that the MSHA pilin subunit from the phage 493-resistant strain can be assembled into a functional pilus structure. The combined sequence and functional analyses indicated that phage 493 resistance is not due to any defect within MSHA itself. Another mechanism, yet to be defined, must be involved.
“Resistant” strains can be infected with choleraphage 493.
In our earlier work (19), with Southern hybridization with virion DNA as a probe, we demonstrated that homologous phage DNA was present in phage 493-sensitive El Tor strains after but not before infection. Applying the same technique, we found that the double-stranded phage replicative form (RF) was present in a 493-resistant 1994 isolate, AJ17564, after exposure to 493 (Table 2). These results indicated that resistance to 493 as measured in plaque assays is not necessarily equivalent to resistance to infection. Thus, even though this strain was negative in plaque assays, it had the capability of being infected and reproducing phage DNA. For infected 1995 to 1996 El Tor strains 150659 and 20049, however, only a weak signal was detected by Southern hybridization (Fig. 1 and Table 2). The phage 493-resistant 1995 to 1996 strains clearly differed from the 493-resistant 1994 strain AJ17564 in efficiency of phage DNA replication (Fig. 1).
Innate resistance of classical biotype strains.
We were curious about whether the introduction of the MSHA structural gene would alter the “resistance” of classical biotype strains to phage 493. When plasmid pJM232 was transferred into classical biotype strain O395, both O395-mshA and the parent strain were still resistant to phage 493 in the plaque assay, and neither hemagglutinated chicken erythrocytes. However, Southern hybridization revealed weak RF signals in both O395 and O395-mshA after exposure to 493 (Table 2). The signal levels were similar to those observed with the 493-resistant 1995 to 1996 El Tor strains (Fig. 1). The introduction of mshA into the classical biotype strain did not change its relevant phenotypic properties. Rare strains of classical V. cholerae O1, such as NIH41 (20-A-11), can produce MSHA (4, 8). Both MSHA+ 20-A-11 and its parent NIH41 (which is variably MSHA+) were found to be highly sensitive to 493 in the plaque assay (Table 2). The phage RF was easily detectable in lysogenized cells by agarose gel electrophoresis. The results cumulatively indicated that MSHA may be exposed variably by classical biotype vibrios.
The presence of the MSHA structural gene in classical cholera vibrios was confirmed by PCR (Fig. 2). The size of the amplified fragments of the mshA region from El Tor strain C6706 was identical to that of classical strain O395, 857 bp (Fig. 2). To estimate the level of expression of the mshA gene in classical strain O395, we constructed an mshA-phoA gene fusion strain, JM69, which contains the E. coli alkaline phosphatase gene fused in frame to the mshA gene of O395. Amplification of the mshA-phoA gene fusions from strains JM69 and JM191 resulted in identical 3.4-kb fragments (Fig. 2). The level of expression of the mshA gene of strain O395 in strain JM69, as measured by an alkaline phosphatase assay, was 30% that of the mshA gene of the El Tor strain in strain JM191 (Table 3). These data demonstrated that classical strain O395 can express mshA, albeit at low levels.
FIG. 2.
PCR analysis of El Tor and classical mshA and mshA-phoA gene fusions. Chromosomal DNA was PCR amplified with the MSH1-MSH4 primer pair. Lanes: 1, O395 Sm; 2, JM69; 3, JM191; 4, C6706 str2; 5, distilled H2O control; 6, lambda HindIII ladder.
TABLE 3.
Expression of an mshA-phoA gene fusion in classical and El Tor biotype strains
| Strain | Biotype | Genotype | Alkaline phosphatase activity (mean U ± SD) |
|---|---|---|---|
| O395 Sm | Classical | mshA+ | 16 ± 7 |
| JM69 | Classical | mshA-phoA | 360 ± 30 |
| C6706 | El Tor | mshA+ | 12 ± 2 |
| JM191 | El Tor | mshA-phoA | 1,240 ± 90 |
DISCUSSION
In the present work, we have established that the MSHA type IV pilus is the receptor for filamentous choleraphage 493, isolated from a 1994 strain of V. cholerae O139. We had previously hypothesized that phage could play an important role in cholera vibrio population dynamics and territoriality (19), such as the rapid rise and subsequent decline of O139 in Bangladesh and India. This hypothesis was supported by the observation that recent isolates of biotype El Tor cholera vibrios in Bangladesh, where the El Tor biotype had reemerged, were resistant to 493 in plaque assays. Like older isolates, the new El Tor isolates produced MSHA. We considered that the MSHA in the resistant strains might have mutated to a form which was still active as a hemagglutinin but was unable to serve as a receptor for 493. However, the DNA sequence of the mshA gene from a 493-resistant 1994 El Tor isolate, AJ17564, was identical to that of the mshA gene from 493-sensitive strain C6706. Further, the mshA gene from a 493-resistant El Tor isolate, 150659, restored 493 sensitivity to a 493-resistant ΔmshA mutant, KHT46. It thus became clear that a change in MSHA is not the reason for the resistance of the new El Tor isolates. It is possible that the new El Tor isolates have a resident phage which inhibits the replication of 493 or that the new El Tor isolates lack an element which is essential for 493 replication.
Our study also demonstrated that although the new El Tor isolates are apparently resistant to phage 493, as evaluated in plaque assays, they are still susceptible to infection with 493. Southern hybridization revealed double-stranded phage RFs—albeit at lower levels than in earlier sensitive strains—in plaque-resistant isolates following exposure to 493. Resistance revealed in plaque assays is not necessarily an indication of resistance to infection, which is better demonstrated by Southern hybridization. The new isolates are susceptible to infection but deficient in phage reproduction. Further, there is a difference in this regard between plaque-resistant 1994 isolates and 1995 to 1996 isolates.
The distinction between phage sensitivity and susceptibility to infection again became evident when we reexamined the resistance of classical biotype strains to phage 493 in plaque assays. Transfer of the MSHA structural gene to a classical biotype strain resulted in neither the appearance of sensitivity to 493 in the plaque assay nor the expression of MSHA, as detected in the slide hemagglutination test. However, as reported previously (4, 8), rare strains of the classical biotype do express MSHA. Jonson et al., using a MAb against MSHA from an El Tor biotype strain, detected antigen by immunoblotting, but not on the surface, of classical biotype vibrios (17). Unlike O395, MSHA+ classical biotype strain NIH41 was found to be highly sensitive to 493 in the plaque assay, and phage RF was easily detected in lysogenized cells. The presence of the MSHA structural gene in classical biotype strain O395 was confirmed by PCR. mshA-phoA fusion studies demonstrated that O395 expressed the MSHA pilin subunit gene at lower levels than El Tor vibrios: the amount of MSHA available was undetectable in the hemagglutination test but was sufficient to serve as a phage receptor on the cell surface. Like the recent El Tor strains, strain O395 is also limited in its ability to support phage reproduction. Our observations thus suggest that phage reproduction is a property of individual recipient strains and is independent of the quantity of the receptor.
The finding that mshA is present in classical biotype cholera vibrios as well as in El Tor vibrios but has limited expression is somewhat reminiscent of the history of the El Tor hemolysin or cytotoxin (8, 16). Like MSHA, the El Tor hemolysin was traditionally regarded as an exclusive characteristic—the sine qua non—of El Tor vibrios as opposed to classical biotype vibrios. During the course of the current pandemic, the El Tor vibrios exhibited markedly reduced expression of the hemolysin. However, the structural gene, hlyA, encoding the hemolysin is present in nonhemolytic classical biotype, El Tor, and some non-O1 vibrio strains (5).
The possible role of MSHA in the pathogenesis of cholera is a disputed issue. Whereas earlier studies (25) regarded MSHA as an important biotype-specific adherence or colonization factor, inactivation of mshA did not significantly affect colonization in the infant mouse model (2, 18, 29) and, in contrast to earlier studies, antibodies to MSHA were not protective in that model (2). The evidence is quite solid that the toxin-coregulated pilus is essential for colonization in people (15) and that it serves the dual function of being the essential and unique receptor for CTXφ carrying the cholera toxin virulence cassette (30). Whether or not MSHA is a component of virulence in V. cholerae or contributes to the enhanced endemicity of El Tor biotype cholera vibrios (9), it has another important function of being a widely distributed receptor for filamentous phages, such as 493. Filamentous phages have the potential of introducing large amounts of DNA, which could encode pathogenicity islands (12), such as the O139-specific antigen and polysaccharide capsule (11), or other factors, which could result in quantum leaps in both identity and pathogenicity simultaneously. MSHA not only is present in O1 classical and El Tor biotypes and in V. cholerae O139 but also is found in other non-O1 vibrios (4). As we observed previously (19), a 493-related phage was found in a pre-O139 encapsulated V. cholerae O31 strain, NRT36S. Although O31 and O139 are not immunologically related and therefore NRT36S could not be the progenitor of O139 strains, Mooi and Bik (24) noted that O139 DNA-related sequences are present among other non-O1 strains. Thus, the possibility exists for DNA transfer from non-O1 vibrios to El Tor vibrios via 493 or a 493-like phage. It is widely accepted that O139 is the result of horizontal transmission of genetic information from another vibrio to an El Tor biotype recipient strain. What is unique about O139 is that it retained the epidemic capability of O1 vibrios. Our present observations allow us to suggest that 493 and the MSHA type IV pilus receptor are potentially valuable tools for further examination of mechanisms of horizontal transfer of genetic information, such as that which led to the creation of O139 and could lead to the formation of another epidemic cholera vibrio, such as “Bengal.” All the essential components are available.
ACKNOWLEDGMENTS
We appreciate the technical assistance of Don Carpenter as well as the generosity of M. J. Albert in providing strains.
This work was supported by U.S. Public Health Service grants AI17312 to R.A.F. and AI25096 to R.K.T. from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Albert M J. Vibrio cholerae O139 Bengal. J Clin Microbiol. 1994;32:2345–2349. doi: 10.1128/jcm.32.10.2345-2349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attridge S R, Manning P A, Holmgren J, Jonson G. Relative significance of mannose-sensitive hemagglutinin and toxin-coregulated pili in colonization of infant mice by Vibrio cholerae El Tor. Infect Immun. 1996;64:3369–3373. doi: 10.1128/iai.64.8.3369-3373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bik E M, Bunschoten A E, Gouw R D, Mooi F R. Genesis of the novel epidemic Vibrio cholerae strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 1995;14:209–216. doi: 10.1002/j.1460-2075.1995.tb06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booth B A, Finkelstein R A. Presence of hemagglutinin/protease and other potential virulence factors in O1 and non-O1 Vibrio cholerae. J Infect Dis. 1986;154:183–186. doi: 10.1093/infdis/154.1.183. [DOI] [PubMed] [Google Scholar]
- 5.Brown M H, Manning P A. Haemolysin genes of Vibrio cholerae: presence of homologous DNA in non-haemolytic O1 and haemolytic non-O1 strains. FEMS Microbiol Lett. 1985;30:197–201. [Google Scholar]
- 6.Falero G, Rodriguez B L, Valmaseda T, Perez M E, Perez J L, Fando R, Robert A, Campos J, Silva A, Sierra G, Benitez J A. Production and characterization of a monoclonal antibody against mannose-sensitive hemagglutinin of Vibrio cholerae. Hybridoma. 1998;17:63–67. doi: 10.1089/hyb.1998.17.63. [DOI] [PubMed] [Google Scholar]
- 7.Faruque A S G, Fuchs G J, Albert M J. Changing epidemiology of cholera due to Vibrio cholerae O1 and O139 Bengal in Dhaka, Bangladesh. Epidemiol Infect. 1996;116:275–278. doi: 10.1017/s0950268800052572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finkelstein R A, Mukerjee S. Hemagglutination: a rapid method for differentiating Vibrio cholerae and El Tor vibrios. Proc Soc Exp Biol Med. 1963;112:355–359. [Google Scholar]
- 9.Finkelstein R A. Cholera. Crit Rev Microbiol. 1973;2:553–623. [Google Scholar]
- 10.Finkelstein R A, Boesman-Finkelstein M, Sengupta D K, Page W J, Stanley C M, Phillips T E. Colonial opacity variations among the choleragenic vibrios. Microbiology. 1997;143:23–34. doi: 10.1099/00221287-143-1-23. [DOI] [PubMed] [Google Scholar]
- 11.Finlay R B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Microreview: pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 13.Hanne L F, Finkelstein R A. Characterization and distribution of the hemagglutinins produced by Vibrio cholerae. Infect Immun. 1982;36:209–214. doi: 10.1128/iai.36.1.209-214.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Häse C C, Bauer M E, Finkelstein R A. Genetic characterization of mannose-sensitive hemagglutinin (MSHA)-negative mutants of Vibrio cholerae by Tn5 mutagenesis. Gene. 1994;150:17–25. doi: 10.1016/0378-1119(94)90852-4. [DOI] [PubMed] [Google Scholar]
- 15.Herrington D A, Hall R H, Losonsky G, Mekalanos J J, Taylor R K, Levine M M. Toxin, the toxin co-coregulated pili and the toxB regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honda T, Finkelstein R A. Purification and characterization of a hemolysin produced by Vibrio cholerae biotype El Tor: another toxic substance produced by cholera vibrios. Infect Immun. 1979;26:1020–1027. doi: 10.1128/iai.26.3.1020-1027.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonson G, Holmgren J, Svennerholm A-M. Identification of mannose-binding pilus on Vibrio cholerae El Tor. Microb Pathog. 1991;11:433–441. doi: 10.1016/0882-4010(91)90039-d. [DOI] [PubMed] [Google Scholar]
- 18.Jonson G, Lebens M, Holmgren J. Cloning and sequencing of Vibrio cholerae mannose-sensitive hemagglutinin pili gene: localization of mshA within a cluster of type IV pilin genes. Mol Microbiol. 1994;13:109–118. doi: 10.1111/j.1365-2958.1994.tb00406.x. [DOI] [PubMed] [Google Scholar]
- 19.Jouravleva E A, McDonald G A, Garon C F, Boesman-Finkelstein M, Finkelstein R A. Characterization and possible functions of a new filamentous bacteriophage from Vibrio cholerae O139. Microbiology. 1998;144:315–324. doi: 10.1099/00221287-144-2-315. [DOI] [PubMed] [Google Scholar]
- 20.Kaper J B, Morris J G, Jr, Levine M M. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine M M, Kaper J K, Herrington D, Losinsky G L, Morris J G, Clements M L, Black R E, Tall B, Hall M. Volunteer studies of deletion mutants of Vibrio cholerae O1 prepared by recombinant techniques. Infect Immun. 1988;56:161–167. doi: 10.1128/iai.56.1.161-167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsh J W, Sun D, Taylor R K. Physical linkage of the Vibrio cholerae mannose-sensitive hemagglutinin secretory and structural subunit gene loci: identification of the mshG coding sequence. Infect Immun. 1996;64:460–465. doi: 10.1128/iai.64.2.460-465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mel S F, Mekalanos J J. Modulation of horizontal gene transfer in pathogenic bacteria by in vivo signals. Cell. 1996;87:795–798. doi: 10.1016/s0092-8674(00)81986-8. [DOI] [PubMed] [Google Scholar]
- 24.Mooi F R, Bik E M. The evolution of epidemic Vibrio cholerae strains. Trends Microbiol. 1997;5:161–165. doi: 10.1016/S0966-842X(96)10086-X. [DOI] [PubMed] [Google Scholar]
- 25.Osek J, Jonson G, Svennerholm A-M, Holmgren J. Role of antibodies against biotype-specific Vibrio cholerae pili in protection against experimental classical and El Tor cholera. Infect Immun. 1994;62:2901–2907. doi: 10.1128/iai.62.7.2901-2907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. p. 2.80. [Google Scholar]
- 27.Sharma C, Nair G B, Mukhopadhyay A K, Bhattacharya S K, Ghosh R K, Ghosh A. Molecular characterization of Vibrio cholerae O1 biotype El Tor strains isolated between 1992 and 1995 in Calcutta, India: evidence for the emergence of a new clone of the El Tor biotype. J Infect Dis. 1997;175:1134–1141. doi: 10.1086/516453. [DOI] [PubMed] [Google Scholar]
- 28.Stroeher U H, Jedani K E, Dredge B K, Morona R, Brown M H, Karageorgos L E, Albert M J, Manning P A. Genetic rearrangements in the rfb regions of Vibrio cholerae O1 and O139. Proc Natl Acad Sci USA. 1995;92:10374–10378. doi: 10.1073/pnas.92.22.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thelin K H, Taylor R K. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waldor M K, Mekalanos J J. Lysogenic conversion by filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 31.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]