Abstract
Background
Total joint replacement for osteoarthritis is one of the most successful surgical procedures in modern medicine. However, aseptic loosening continues to be a leading cause of revision arthroplasty. The diagnosis of aseptic loosening remains a challenge as patients are often asymptomatic until the late stages. MicroRNA (miRNA) has been demonstrated to be a useful diagnostic tool and has been successfully used in the diagnosis of other diseases. We aimed to identify differentially expressed miRNA in the plasma of patients with aseptic loosening.
Methods
Adult patients undergoing revision arthroplasty for aseptic loosening and age- and gender-matched controls were recruited. Samples of bone, tissue and blood were collected, and RNA sequencing was performed in 24 patients with aseptic loosening and 26 controls. Differentially expressed miRNA in plasma was matched to differentially expressed mRNA in periprosthetic bone and tissue. Western blot was used to validate protein expression.
Results
Seven miRNA was differentially expressed in the plasma of patients with osteolysis (logFC >|2|, adj-P < 0.05). Three thousand six hundred and eighty mRNA genes in bone and 427 mRNA genes in tissue samples of osteolysis patients were differentially expressed (logFC >|2|, adj-P < 0.05). Gene enrichment analysis and pathway analysis revealed two miRNA (miR-1246 and miR-6089) had multiple gene targets in the Wnt signalling pathway in the local bone and tissues which regulate bone metabolism.
Conclusion
These results suggest that aseptic loosening may be regulated by miR-1246 and miR-6089 via the Wnt signalling pathway.
Keywords: Aseptic loosening, MicroRNA, Sequencing, Wnt pathway, Revision surgery
Introduction
Aseptic loosening around total joint replacements (TJAs) remain an ongoing challenge. Despite the success of TJA, aseptic loosening still accounts for more than 20% of all revision procedures for primary total hip and knee arthroplasty [1]. The pathophysiology of aseptic loosening is due to prosthetic wear particles eliciting an inflammatory response. This causes an imbalance in the activity of osteoclasts and osteoblasts, eventually leading to osteolysis, which may result in component loosening and pain or instability. [2]. However, this theory alone does not explain why some patients have significant wear of the prosthesis but very minimal osteolysis and vice versa. Perhaps altered gene expression in response to wear particles can predispose certain patients to osteolysis and subsequent aseptic loosening [3, 4].
Aseptic loosening can be difficult to diagnose. Early changes are often undetected due to limitations in our current diagnostic modalities. By the time patients are symptomatic, the prosthesis may have already failed, therefore often necessitating a revision procedure [5]. Currently, there is no gold-standard diagnostic test for aseptic loosening. A combination of clinical features, radiographs, cross-sectional imaging and nuclear medicine studies can be used [6]. Biochemical markers have been trialled in the past with varying results [7]. Given the multiple challenges in diagnosis and treatment, it is important to discover novel methods of diagnosis and treatment.
MicroRNA (miRNA) and small interference RNA (siRNA) are a contemporary subject of investigation, which shows promise as biomarkers and therapeutic targets [8–11]. MicroRNA is a family of small, non-coding RNA which regulates gene expression at the post-transcriptional level. They can silence genes via repressing translation or direct cleavage of messenger RNA (mRNA) [12]. There have also been reports of select miRNA upregulating target genes as an alternative method of regulation [13]. Several miRNA has been implemented in the development of diseases of the musculoskeletal system [14, 15]. MiRNA is readily detectable in bodily fluids such as blood and urine inside microvesicles known as exosomes. These exosomes protect miRNA from degradation by RNases which makes them useful as a candidate for biomarkers [16]. Furthermore, understanding the function and gene targets of miRNA can potentially lead to the development of a new class of gene therapy drug [12, 15].
Several miRNA have been shown to regulate osteogenesis and bone metabolism [17]. In vitro studies demonstrate miR-24 has an important role in osteoclast differentiation in the presence of titanium particles [18]. In vivo studies have linked miR-21 and miR-130b to osteolysis due to metal particles in mice [19, 20]. In humans, miR-106b has a role in osteoclastogenesis and osteolysis in bone tumours [21]. However, there are no studies to our knowledge exploring the role of miRNA in human patients with aseptic loosening using next-generation sequencing. A genetic test to predict the likelihood of developing aseptic loosening will allow clinicians to stratify those at risk of the disease and monitor them more closely. It can also be utilised to help diagnose patients who have the disease before any radiographic signs develop. Identifying genes which are responsible for causing increased bone resorption around the implant can be the foundations of future genetic, patient-specific therapies. Thus, we aimed to identify the miRNA expression profile in plasma of patients undergoing revision for aseptic loosening compared to controls. We hypothesise that the miRNA expression profile in patients with aseptic loosening will be different compared to controls, and that these differentially expressed miRNA have mRNA targets in the bone and tissue of aseptic loosening patients.
Materials and methods
Study cohort
Adult patients undergoing elective hip or knee arthroplasty across two centres were recruited over an 8-year period. Cases were patients undergoing revision arthroplasty for aseptic loosening (RAL), and controls were patients undergoing primary TJA for osteoarthritis (PA). The diagnosis for aseptic loosening was made by the senior orthopaedic surgeon performing the procedure using clinical findings, radiographs and CT or MRI scans demonstrating loosening or periprosthetic osteolysis. The control group was age and gender-matched patients who were undergoing primary TJA for osteoarthritis. Exclusion criteria included active infection, bloodborne viruses, inflammatory arthritis, immunomodulatory drugs, osteoporosis and metabolic bone diseases. Demographic data were collected including age, sex, BMI, ethnicity, joint affected, comorbidities, immunomodulatory medications, biochemistry studies and the Charlson Comorbidity Index.
Ethics approval was granted by the ACT Health Research Ethics and Governance Office, Human Research Ethics Committee (ETH.9.07.865) to recruit patients and collect intraoperative tissue samples and blood samples. Written informed consent was obtained from each patient.
RNA sequencing
Sample collection and RNA extraction
For RAL patients, intraoperative samples of bone and soft tissue around the prosthesis were collected by the primary surgeon. Bone samples were taken from around the areas of loosening and osteolysis. Soft tissue samples were taken from the capsule and pseudomembrane around the joint. For PA patients, intraoperative samples of bone and soft tissue around the hip or knee joint were collected by the primary surgeon. For the hip, bone was removed from the femoral head and for the knee, bone was removed from the cut surfaces of bone. Soft tissue samples were obtained from the capsule of the hip and knee joint. Samples were collected using a surgical knife, surgical curettes and/or Rongeurs and were stored in sterile containers on ice until processing. Blood samples were collected from an intravenous cannula or arterial line during the procedure and stored in EDTA tubes in ice until processing. All blood samples were processed within 1 h of collection to minimise degradation of RNA.
Bone and tissue
Bone and tissue samples were cut into small 1mm3 pieces under aseptic technique. Care was taken to remove areas of heat necrosis. Specifically for bone, we harvested bone away from the cut surface such as the centre of the femoral head. For tissue, we removed areas damaged by electrocautery and only used samples which were not visibly necrotic. These were snap frozen using liquid nitrogen and homogenised using a mortar and pestle. Total RNA was extracted from each sample using TRIzol reagent and the RNeasy Mini Kit (QIAGEN, USA) according to the manufacturer’s protocol. Samples which were not immediately processed were stored fresh in -80°C in sterile Eppendorf tubes until future use.
Plasma
Whole blood was centrifuged using two cycles to separate plasma and remove cellular debris. Samples with visible haemolysis were discarded. Two hundred microlitres of plasma were used for miRNA isolation using the miRNeasy Serum/Plasma Kit (QIAGEN, USA) according to the manufacturer’s protocol.
Purified samples of RNA were stored in microfuge tubes and frozen at − 80°C until further use. The quality and concentration of RNA isolated was measured by the NanoDrop Spectrophotometer (ThermoFisher, USA), and the highest quality samples were used for quantification using the Bioanalyzer 2100 (Agilent, USA). Samples with an RNA Integrity Number > 7 were used for sequencing.
RNA sequencing
A cDNA library was synthesised from RNA samples and sequencing was conducted using a NovaSeq 6000 Sequencing System (Illumina, USA) with a 100bp run. We refer to an individual deep sequence read as a tag and the number of times it occurs as a count. The absolute counts for each miRNA were recorded onto a spreadsheet and those miRNA with very low counts (i.e. < 10) were excluded.
Statistics
Demographic data were analysed using SPSS 26 (IBM, USA). Chi-square tests were used to compare categorical variables and a student t-test for continuous variables. A P-value of < 0.05 was deemed statistically significant.
Sequencing data analysis
For tissue and bone total RNA samples, data analysis was performed using Bioconductor software in the R programming language via Rstudio. Single-end raw reads were aligned to the human genome, hg38, using hisat2. The mapped reads were assigned based on the NCBI genome annotation with FeatureCounts v2.0. Pre-processing and denoising of the data were performed using edgeR and differentially expressed genes were identified using limma with the voom method. P-values were adjusted using the Bonferroni method and an adj-P < 0.05 and a logFC >|2.0| was deemed statistically significant. Graphical representations were generated using the Glimma package. Gene-set enrichment analysis was performed with Enrichr and GSEA of C2 and C7 sets in the Molecular Signatures Database and the DisGeNET database. Gene-ontology over-representation testing was performed to identify molecular function, biological processes and cellular components.
For plasma miRNA samples, data analysis was performed using the bioinformatic tools available on Galaxy and the R programming language via RStudio. The quality of the reads was checked with FastQC. Adapters were trimmed using Cutadapt according to the instructions provided in the CATS Small RNA-seq kit (Diagenode, USA). The trimmed reads were mapped to the whole human genome, hg38 using miRDeep2, which utilises Bowtie. Reads with counts less than ten were filtered to minimise false positives. Differentially expressed genes were identified using DESeq2. P-values were adjusted using the Bonferroni method and an adj-P < 0.05 and a logFC >|2.0| was deemed statistically significant.
To determine miRNA-mRNA target expression pairs, differentially expressed miRNA genes were matched to significantly enriched C3 miRNA target gene sets in the MSigDB. TargetScan Human 7.2 was to predict gene targets of differentially expressed miRNA by matching miRNA targets and C2 canonical pathway gene sets relevant to bone metabolism: inflammation, RANKL, Wnt signalling, JAK-STAT signalling and BMP signalling. Of these, significantly up or down-expressed genes were identified according to a cutoff of adj-P < 0.05 and logFC >|2.0|. Furthermore, we applied a stricter criterion using a cutoff of adj-P < 0.01 and logFC >|3.0| to identify top target genes relevant to bone metabolism. A manual literature search was performed of the resultant target genes to confirm the genes with direct relevance to bone homeostasis, osteoblast and osteoclast function. These genes were selected for protein expression experiments.
Western Blot
Protein extraction from bone
Intraoperative samples were thawed to room temperature and homogenised using a mortar and pestle with liquid nitrogen. 1 × concentration of RIPA buffer with protease inhibitor at a volume of 10ml RIPA / gram of wet tissue. The samples were centrifuged at 10,000 × g for 15min at −4°C. The supernatant was collected and stored in −20°C until further processing. A Bradford protein assay was performed to determine concentrations of each protein sample.
Western blot
Protein samples were thawed at room temperature, and 5µg of protein were loaded into each well of the SDS-PAGE gel (12- well TruPAGE Precast Gel 4–12% 8 × 10cm, Sigma-Aldrich, USA). Gels were run for 20min at a constant 50V and at 45min at a constant 80V in Nu-Sep running buffer (Sigma-Aldrich, USA) at room temperature. Proteins were transferred to a PVDF membrane using Nu-Sep Transfer Buffer (Sigma-Aldrich, USA) at 100V for 1 h at room temperature. The PVDF membranes were blocked using blocking buffer and stained using primary and secondary antibodies. The membrane was incubated for 2 h at room temperature with the primary antibody FRAT2 Polyclonal Rabbit anti-Human IgG antibody (Bioss, USA) at a 1:500 dilution. The membrane was incubated for 1 h at room temperature with the secondary antibody Goat anti-Rabbit IgG antibody HRP (abcam, USA) at a 1:2000 dilution. Proteins were stained using the ECL Prime Western Blotting Detection Reagent (Sigma-Aldrich, USA) and detected using an Amersham 680 Imager (GE Life Sciences, USA) via the chemiluminescence method. Images were enhanced using Adobe Lightroom Classic CC (Adobe, USA).
Results
Patient demographics
A total of 50 patients were recruited to our study—24 patients in the RAL group and 26 patients in the PA group. There were no significant demographic differences between the patient groups (Table 1). Fifteen patients underwent miRNA sequencing for plasma miRNA and 35 patients underwent total RNA sequencing to identify RNA genes in bone and tissue samples.
Table 1.
Patient demographics
| Revision N = 24 | Primary N = 26 | p-value | |
|---|---|---|---|
| Age | 67 ± 2 | 72 ± 2 | 0.31 |
| Sex | 6 male 18 female | 6 male 20 female | 0.81 |
| Joint | 22 hip 2 knee | 16 hip 10 knee | 0.07 |
| Height (m) | 1.62 ± 0.03 | 1.64 ± 0.03 | 0.78 |
| Weight (kg) | 85.2 ± 6.4 | 74.5 ± 6.1 | 0.26 |
| BMI (kg/m2) | 32.3 ± 2.4 | 27.7 ± 1.7 | 0.17 |
| Charlson Comorbidity Index | 2.9 ± 0.5 | 2.7 ± 0.3 | 0.74 |
| Cardiovascular disease | 12 | 14 | 0.79 |
| Diabetes mellitus | 4 | 4 | 0.72 |
| Autoimmune disease | 0 | 0 | N/A |
| Cancer | 0 | 0 | N/A |
| Respiratory disease | 4 | 3 | 0.45 |
| Osteoporosis | 0 | 0 | N/A |
| Renal impairment | 2 | 0 | 0.18 |
| Immunomodulatory meds | 0 | 0 | 1.00 |
| Blood tests | |||
| Haemoglobin (g/L) | 130 ± 3 | 134 ± 3 | 0.36 |
| Creatinine (µmol/L) | 74 ± 10 | 62 ± 5 | 0.87 |
| eGFR | 74 ± 5 | 80 ± 3 | 0.94 |
| Calcium (mmol/L) | 2.29 ± 0.05 | 2.29 ± 0.05 | 0.87 |
| Phosphate (mmol/L) | 1.30 ± 0.30 | 1.36 ± 0.31 | 0.94 |
| Magnesium (mmol/L) | 0.81 ± 0.04 | 0.72 ± 0.03 | 0.34 |
| Potassium (mmol/L) | 4.69 ± 0.17 | 4.38 ± 0.10 | 0.20 |
The expression profile of miRNA in plasma of patients with aseptic loosening is significantly different compared to controls
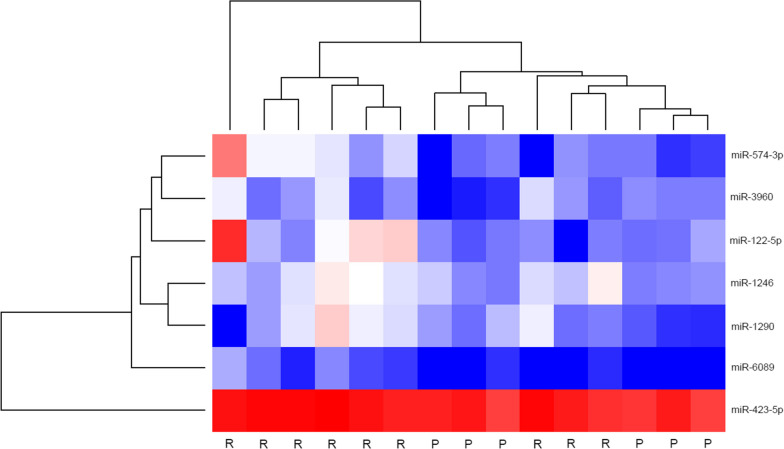
Eighty miRNA genes were identified in the plasma of our patients using miRNA sequencing. Seven miRNA were significantly up-expressed in RAL compared to PA (adj-P < 0.05) and six miRNA also had a logFC > 2.0 (Fig. 1).
Fig. 1.
Heatmap demonstrating expression levels of differentially expressed miRNA genes in plasma of aseptic loosening patients compared to controls logFC >|2.0| ranked by adj-P value. R revision arthroplasty for aseptic loosening; P primary total joint arthroplasty (control)
The expression profile of mRNA in bone and tissue of patients with aseptic loosening is significantly different compared to controls
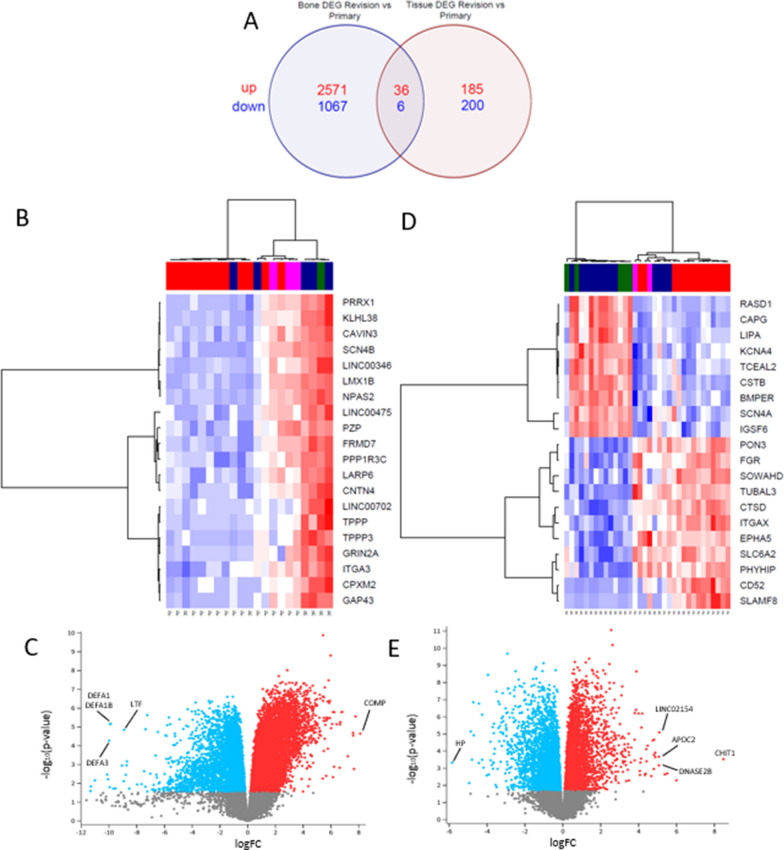
A total of 3680 differentially expressed genes were identified in the bone of RAL patients compared to PA, of these 2607 were up-expressed and 1173 were down-expressed (adj-P < 0.05 and logFC >|2.0|). A total of 427 differentially expressed genes were identified in the tissue of RAL patients compared to controls, of these 221 were up-expressed and 206 were down-expressed (adj-P < 0.05 and logFC >|2.0|). Thiry-six genes were up-expressed in both bone and tissue samples and six genes were down-expressed in both bone and tissue samples. The top genes are depicted in Fig. 2 and Table 2.
Fig. 2.
A Venn Diagram depicting number of differentially expressed genes in bone and tissue and the number of overlapping genes, up-expressed genes are in red and down-expressed genes in blue; DEG – differentially expressed genes. B Heatmap of the top 20 differentially expressed genes in bone samples with logFC >|2.0| ranked by adj-P value. C Volcano plot of differentially expressed genes in bone samples. D Heatmap of top 80 differentially expressed genes in tissue samples with logFC >|2.0| ranked by adj-P value. E Volcano plot of differentially expressed genes in tissue samples, the top three up or down-expressed mRNA genes are labelled
Table 2.
Top differentially expressed genes in bone and tissue
| Location | Gene | logFC | AveExpr | t | P Value | adj.P.Val | B |
|---|---|---|---|---|---|---|---|
| Bone | DEFA1B | − 9.87323 | 10.52257 | − 5.89125 | 6.82E-06 | 0.00011 | 3.695094 |
| DEFA1 | − 9.92204 | 10.51236 | − 5.88224 | 6.96E-06 | 0.000111 | 3.676803 | |
| LTF | − 8.9081 | 8.370686 | − 5.57556 | 1.42E-05 | 0.000161 | 3.187831 | |
| COMP | 8.113538 | 1.880959 | 5.378271 | 2.27E-05 | 0.000213 | 2.709947 | |
| DEFA3 | − 9.98611 | 8.880224 | − 5.01815 | 5.34E-05 | 0.000376 | 1.835942 | |
| Tissue | LINC02154 | 5.085439 | −2.17528 | 5.23428 | 8.12E-06 | 0.000152 | 2.337842 |
| APOC2 | 5.065549 | 0.516944 | 4.13716 | 0.000213 | 0.001447 | 0.577554 | |
| CHIT1 | 8.470192 | 3.545606 | 4.022007 | 0.000297 | 0.001844 | 0.334267 | |
| HP | − 5.8447 | 1.932185 | -3.85548 | 0.000479 | 0.00265 | − 0.08945 | |
| DNASE2B | 5.064432 | − 0.82393 | 3.731657 | 0.000681 | 0.00347 | − 0.55021 |
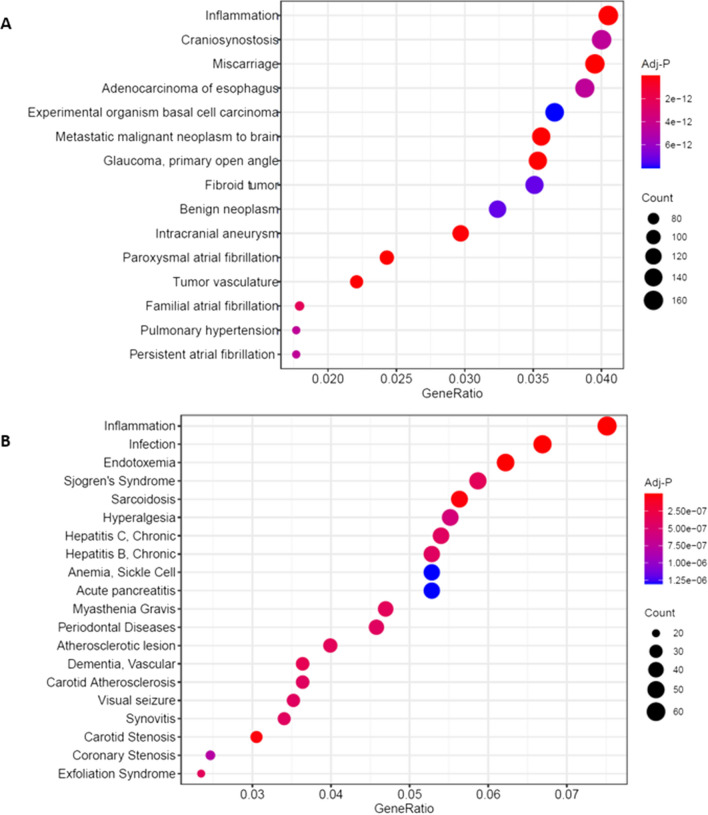
In-silico disease gene-set enrichment analysis reveals disease associations with inflammation, osteolysis and osteoporosis
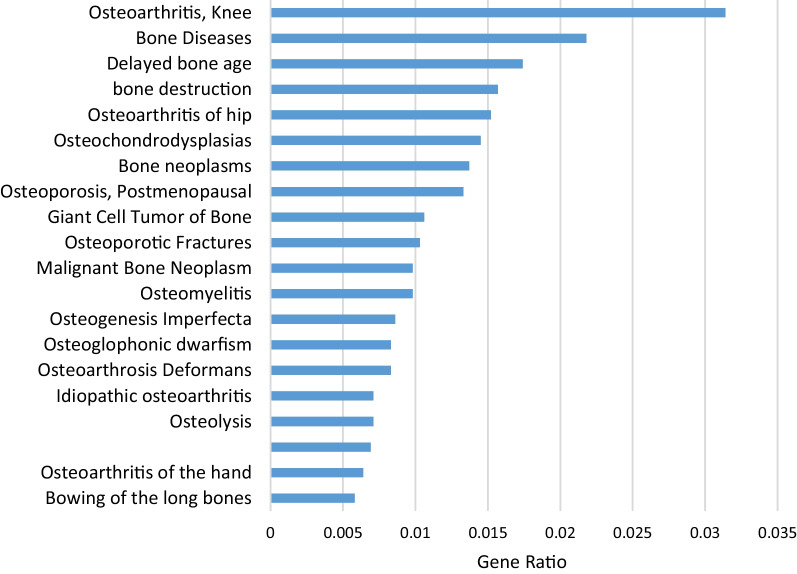
Analysis of gene sets in the DisGeNet 7.0 database demonstrate there is a significant association between patients with osteolysis and the inflammation gene set in both bone and tissue samples (Fig. 3). Of note, synovitis and infection are also in the top 20 disease associations in tissue samples but was not the case for bone samples. Interestingly, RAL patient bone samples were significantly associated with the osteolysis gene set and multiple osteoporosis gene sets. Tissue samples were also associated with multiple osteoporosis gene sets (Fig. 4).
Fig. 3.
Dot-plot of disease associations using disease gene set enrichment analysis of genes differentially expressed in bone (A) and tissue (B) samples of patients with aseptic loosening compared to controls
Fig. 4.
Bone specific disease associations using disease gene set enrichment analysis of differentially expressed genes in patients with aseptic loosening compared to controls
Target prediction reveals plasma miRNA targets mRNA genes in bone and tissue leading to bone resorption
Analysis of miRNA targets in the C3 regulatory target gene sets in the MSigDB reveals 1116 miRNA in bone samples and 63 in tissue samples which have statistically significant gene sets (adj-P < 0.05) in RAL patients compared to controls. Five miRNA identified in our plasma had target gene sets which were significantly enriched in bone samples and two plasma miRNA had target genes which were significantly enriched in tissue samples (Table 3).
Table 3.
miRNA target prediction using C3 gene sets in MSigDB
| miRNA | GeneRatio | Adj-P | Genes | |
|---|---|---|---|---|
| Bone | miR-3960 | 15/4286 | 0.00000 | PCDHA3/PCDHA10/PCDHA6/PCDHA7/PCDHAC2/PCDHA1 2/PCDHA13/PCDHA2/PCDHA9/PCDHA8/PCDHA11/PCDHAC1/PCDHA5/PCDHA1/PCDHA4 |
| miR-574-5p | 29/4286 | 0.00014 | EPHA3/RGMA/LARP6/RERGL/EVX1/DPP4/IGDCC4/NRN1/C11orf96/TEADl/MDGAl/NEBL/THRB/FOSLl/FOXI2/ADGRB2/MYCBP/SEMA7A/NSUN5/CCDCgsc/RSC1A1/GALNT3/MKRN1/CD9 6/D G KG/ N S D 2/S 0X6/1L5 RA/S10 0 A12 | |
| miR-423-5p | 40/4286 | 0.00091 |
RIMS4/CADM3/CSMD2/COL1A1/CPEB1/LHX6/ARHGEF4/ TSPAN11/RGMA/KCNIP3/DNAL11/S100A16/NNAT/EVC/C1QTNF6/SYP/ROR2/NAV1/NRSN2/LZTS3/SLC52A3/SLC20A2/CLDN23/ADGRL1/TMEM150A/OBSL1/KLK4/DUSP8/L0C730098/0DC1/CRIP3/C20Orf27/NECTIN1/ATG4D/UBE20/SLC6A9/RAC2/PHOSPHO1/TCLIB/DMTN |
|
| miR-1290 | 65/4286 | 0.00166 | OGN/KCNK2/DPT/TMEM255A/ANTXR1/COLEC12/ANK2/EFNA5/TGFB 2/COL8A1/SYNP02/THS D7A/RGS5/EGFR/SGIP1/SORCS1/PCDH7/TEAD3/ACKR4/EMP2/PRRG1/RASAL2/CSTB/NAALADL2/VEGFD/GALNT15/PREX2/SRGAP1/ASXL3/ZFHX3/EBF1/ARHGEF26/SEMA6A/AGAP1/ERBB4/KCNAB1/MSR1/LRRTM2/CALCR/DYNC1|1/RASGRF2/TAFA5/CAPN2/HYDIN/MGME1/CAMK4/CBFA2T3/ADARB2/USP15/SMC4/FBN2/SLAIN1/AMMECR1/ITGA4/PIP5K1B/BEND 4/EPCAM/IL5RA/MN D1/PLCH1/SG 01 /TCN1/ MS4A3/SPT A1/TSP02 | |
| miR-122-5p | 28/4286 | 0.00188 | STMN2/MYOCD/CPEB1/LALMC1/RANBP3L/MECOM/RBMS2/MAP3K12/DDR2/FLNB/CTNNA3/CLIC5/MIPOL1/KCNN3/MICU3/SLC25A34/CLIC4/ADGRB2/CO40LG/TBC1D22B/S E MA4 D/G6PD/S LC7A1/RBL1/S LC05A1/ PRR11/C21orf62/KIF11 | |
| Tissue | miR-3960 | 7/881 | 0.00046 | PCDHA11/PCDHA2/PCOHA12/PCDHA1/PCDHA10/PCDHAC2/pou3p3 |
| miR-1290 | 20/881 | 0.02309 | TMEM255A/CSTB/KLHL6/MS4A3/PLCH1/ARHGEF26/ASXL3/BTG2/THRSP/SORCS1/VEGFD/WNK2/SYN2/GREM2/BEGAIN/CNTN6/ERBB4/SYT4/TRHDE/NRXN1 |
Target prediction using TargetScan 7.2 identified 432 genes targeted by plasma miRNA in bone samples, of these 88 were differentially expressed in RAL patients (adj-P < 0.05, logFC >|2|). In tissue samples, fifty-five genes were targets of plasma miRNA and seven were differentially expressed (adj-P < 0.05, logFC >|2|). The top 20 based on adj-P are shown in Table 4.
Table 4.
Top 20 miRNA targets based on adj-P value and logFC
| miRNA | Bone/tissue | Gene | Direction | Adj-P | logFC |
|---|---|---|---|---|---|
| miR-1246 | Tissue | C7 | Down | 0.0003 | −3.6 |
| Bone | LINC00346 | Up | 0.0008 | 3.3 | |
| Bone | PPP1R3C | Up | 0.0008 | 3.6 | |
| Bone | HOXC10 | Up | 0.0008 | 4.0 | |
| Bone | KIAA0930 | Up | 0.0008 | 3.6 | |
| Bone | PCDHB2 | Up | 0.0009 | 3.3 | |
| Bone | THBS2 | Up | 0.0009 | 3.6 | |
| Bone | SIX4 | Up | 0.0009 | 3.5 | |
| Bone | LINC00346 | Up | 0.0008 | 3.3 | |
| Bone | DIXDC1 | Up | 0.0008 | 4.0 | |
| Bone | KLHL30 | Up | 0.0008 | 3.2 | |
| Bone | ANTXR1 | Up | 0.0008 | 3.3 | |
| Bone | APLN | Up | 0.0008 | 3.3 | |
| Bone | KIAA0930 | Up | 0.0008 | 3.6 | |
| Bone | CLDN1 | Up | 0.0009 | 3.1 | |
| Bone | PCDHB2 | Up | 0.0009 | 3.3 | |
| miR-3960_8072 | Bone | NTN1 | Up | 0.0008 | 3.7 |
| miR-423-5p | Bone | NTN1 | Up | 0.0008 | 3.7 |
| miR-574-3p | Tissue | MMP3 | Down | 0.0005 | -3.5 |
| miR-6089 | Bone | PPP1R3C | Up | 0.0008 | 3.6 |
| Bone | ACE | Up | 0.0008 | 3.2 | |
| Bone | DIXDC1 | Up | 0.0008 | 4.0 | |
| Bone | KLHL30 | Up | 0.0008 | 3.2 | |
| Bone | PPIC | Up | 0.0008 | 3.1 | |
| Bone | NTN1 | Up | 0.0008 | 3.7 | |
| Bone | APLN | Up | 0.0008 | 3.3 | |
| Bone | KIAA0930 | Up | 0.0008 | 3.6 | |
| Bone | CCDC3 | Up | 0.0008 | 3.9 |
Several gene targets relevant to the C2 pathways relating to inflammation, RANKL, Wnt signalling, JAK-STAT signalling and TGFB signalling were also identified. Seventy-eight genes in bone and 88 genes in tissue were identified which were direct targets of plasma miRNA. Of these, 10 genes in bone and zero genes in tissue were differentially expressed in RAL patients (adj-P < 0.05, logFC >|2|) (Table 5 and Fig. 5).
Table 5.
Differentially expressed miRNA and their gene targets based on C2 pathways
| miRNA | Target tissue | Pathway | Gene | Direction | Adj-P | logFC |
|---|---|---|---|---|---|---|
| miR-1246 | Bone | Wnt | FZD7 | Up | 0.0038 | 2.1 |
| Bone | Wnt | PRKCB | Down | 0.0032 | -2.5 | |
| Bone | Wnt | DIXDC1 | Up | 0.0007 | 4.0 | |
| Bone | Wnt | SFRP4 | Up | 0.0014 | 3.6 | |
| Bone | Inflammation | TGFB3 | Up | 0.0014 | 2.4 | |
| Bone | TGFB | TGFB3 | Up | 0.0014 | 2.4 | |
| Bone | Wnt | FSTL1 | Up | 0.0037 | 2.4 | |
| miR-574-3p | Bone | JAK-STAT | IL6 | Up | 0.0080 | 2.4 |
| miR-6089 | Bone | Wnt | RAC2 | Down | 0.0089 | − 2.3 |
| Bone | Wnt | FRAT2 | Down | 0.0059 | − 2.2 | |
| Bone | Wnt | DIXDC1 | Up | 0.0008 | 4.0 | |
| Bone | Wnt | WNT9A | Up | 0.0021 | 2.4 |
Fig. 5.
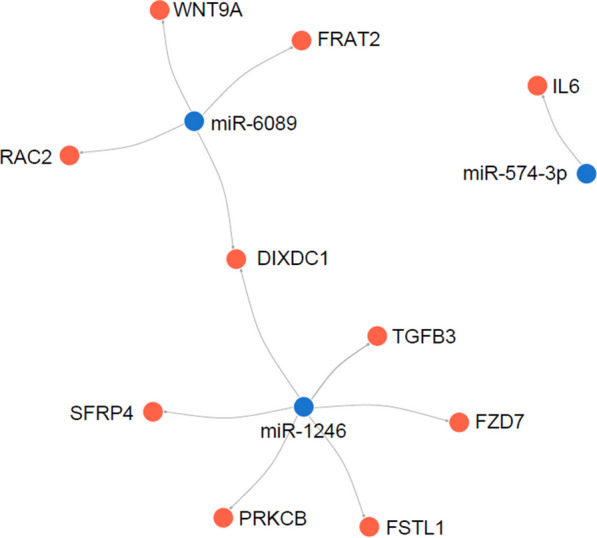
Network visualisation plot demonstrating differentially expressed miRNA in plasma with corresponding targets of differentially expressed genes in the bone of aseptic loosening patients compared to controls
FRAT2 is down-expressed in patients with aseptic loosening
Western blot analysis demonstrates FRAT2 is significantly down-expressed in the bone of patients in the RAL group compared to PA group (Fig. 6).
Fig. 6.
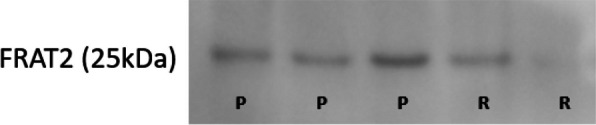
Western blot analysis of FRAT2 protein in bone samples. R revision arthroplasty for aseptic loosening; P primary total joint arthroplasty (control)
Discussion
Our results demonstrate that RAL patients have a significantly different gene expression profile in the periprosthetic bone and soft tissue. The expression of miRNA in the plasma of patients with RAL is also significantly different compared to normal controls. Furthermore, these miRNA have targets genes in bone and tissue which are linked to inflammation, bone homeostasis and osteoblast and osteoclast function.
Upregulation of genes by miRNA could explain aseptic loosening
Interestingly, several miRNA gene targets were upregulated in RAL compared to PA patients (Table 4 and 5). The most widely accepted mechanism of action for miRNA is the silencing of genes by translational repression or by direct cleavage of mRNA [22]. However, there is emerging evidence to suggest other mechanisms of action of miRNA, including the upregulation of genes [23]. In proliferating cells miRNA tend to repress translation whereas in the G1/G0 arrest phase, they activate translation [24]. There are numerous ways by which miRNA can enhance gene expression, including directly by interactions of miRNA polyribosomes with target genes and indirectly by disengaging inhibitory miRNA from their targets, thereby increasing gene expression [13].
Aseptic loosening is regulated by miR-1246 and miR-6089 via the Wnt signalling pathway
Merging gene targets in TargetScan to C2 gene sets in MSigDB reveal that the Wnt signalling pathway had multiple genes that were differentially expressed in RAL patients. Two miRNA, namely miR-1246 and miR-6089 have gene targets in the Wnt pathway (Table 4). Specifically, the Wnt genes targeted by miR-1246 include DIXDC1, SFRP4, PRKCB, FSTL1 and FZD7. The Wnt genes targeted by miR-6089 include DIXDC1, WNT9A, RAC2 and FRAT2.
The Wnt signalling pathway is one of the main pathways regulating bone formation and resorption. The primary role of the canonical Wnt pathway is to stimulate bone formation by activation of osteoblastogenesis [25]. The non-canonical pathway activates osteoclastogenesis and also has implications in inflammatory diseases [26]. Wnt proteins bind to Frizzled (FZD) receptors activating either a β-catenin-dependent canonical pathway or a β-catenin-independent non-canonical pathway, which in-turn regulate downstream genes [27].
Secreted frizzled-receptor proteins (sFRPs) are antagonists of the Wnt pathway by binding directly with FZD receptors, thereby inhibiting the interaction between Wnt and FZD [28]. SFRP4 overexpression has been demonstrated to inhibit osteoblasts, resulting in a 30% decrease in trabecular bone mass in mouse models [29]. Although sFRP4 is implicated in Pyle’s disease, a rare genetic condition characterised by thinning of cortical bone and increased fractures, sFRP4 knockout results in significantly greater trabecular bone mass in mouse models [30]. In patients with osteolysis, there is also a loss of trabecular bone adjacent to the prosthesis, ultimately leading to loosening of the prosthesis [31]. Cortical bone is usually preserved until the later stages. This loss of trabecular bone and preservation of cortical bone is apparent on radiographs as endosteal scalloping [32].
Follistatin-related protein 1 (FSTL1) is encoded by the FSTL gene with a variety of functions in the development of human diseases [33]. It has a pro-inflammatory effect and has been shown to be elevated in chronic inflammatory diseases such as rheumatoid arthritis [34]. Furthermore, overexpression of FSTL1 in bone-marrow derived macrophages positively regulates osteoclast differentiation via RANKL and FSTL1 knockout results in a decrease in osteoclast precursor proliferation [35]. In our RAL patient group, FSTL1 was significantly up-expressed, which suggest FSTL1 has a role in the development of osteolysis and aseptic loosening via an inflammatory response.
FZD7 has been extensively studies in carcinogenesis and has been shown to activate both the canonical and non-canonical Wnt signalling pathways [36]. Non-canonical pathways can lead to osteoclast activation thereby increasing bone resorption [37]. FZD7 was significantly upregulated in AL patients, suggesting a potential role for its role in the pathogenesis of osteolysis and loosening.
FRAT proteins and protein kinase C (PRKCB) are important regulators of the Wnt signal transduction pathway as they bind and deactivate glycogen synthase kinase-3β (GSK-3β) [38, 39]. In the absence of Wnt, GSK-3β phosphorylates β-catenin and stops the activation of the canonical Wnt pathway. Binding of Wnt to FZD receptors results in the phosphorylation and deactivation of GSK-3β, therefore increasing β-catenin, leading to downstream gene regulation [40]. In our AL group, FRAT2 was significantly down-expressed compared to controls, suggesting that FRAT2 downregulation could be implicated in inactivation of the canonical Wnt signalling pathway, leading to decreased osteoblastogenesis.
The RAC proteins are required for normal bone development and homeostasis. Osteoblastic differentiation from mesenchymal stem cells and knockout mice demonstrated marked reductions in bone size and trabecular bone formation [41]. Other groups have demonstrated that RAC deletions lead to increased bone formation due to defects in osteoclastic function [42, 43].
Two genes in the Wnt signalling pathway which did not support our findings were DIXDC1 and Wnt9A. DIXDC1 activates the canonical Wnt pathway as demonstrated in various cancer cell lines [44, 45]. Wnt9A was significantly up-expressed in the periprosthetic bone of RAL patients compared to controls. Wnt9A is crucial for the regulation of endochondral ossification in utero [46]. Wnt9A knockout mice have various bone abnormalities including reduction in bone length and decreased area of mineralised regions. These changes were more pronounced in the proximal bones including the femur and ilium [47]. However, there are no studies to our knowledge which have examined the role of Wnt9A in mature osteoblasts and osteoclasts.
Taken together, these results suggest that RAL is potentially regulated by miR-1246 and miR-6089 via the Wnt pathway. The genes SFRP4, PRKCB, FSTL1, RAC2, FRAT2 and WNT9A have been demonstrated to affect this process based on previous studies (Fig. 7).
Fig. 7.
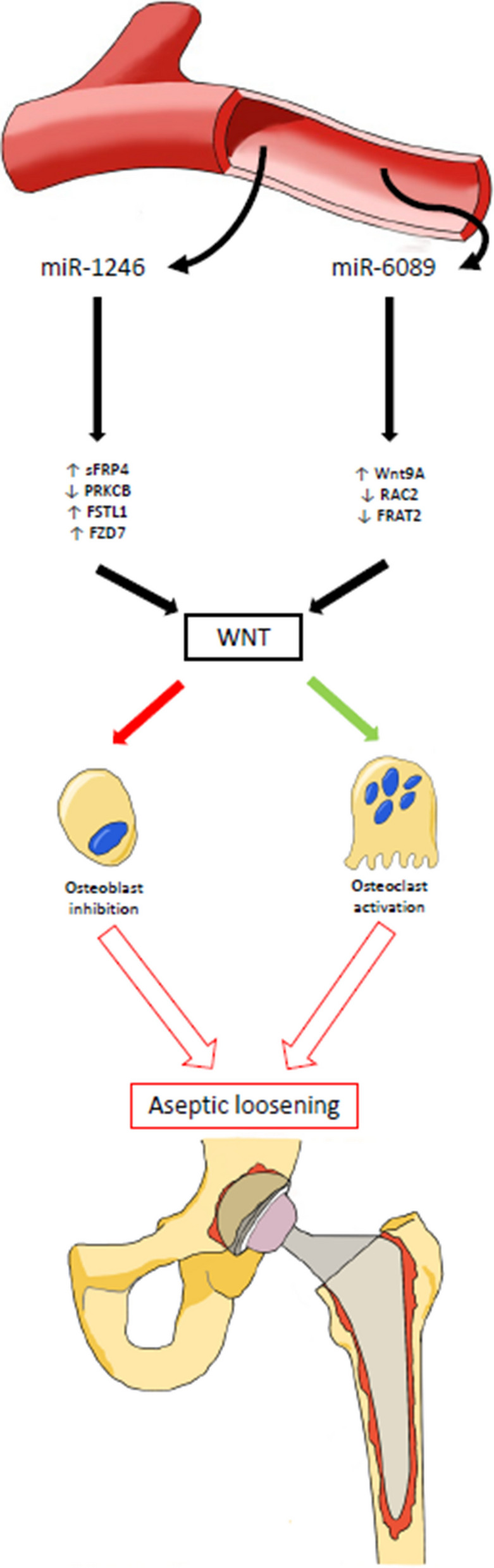
Schematic diagram demonstrating circulating miRNA targeting genes in the periprosthetic bone leading to osteoblast inhibition and osteoclast activation and subsequent aseptic loosening. Circulating miRNA in plasma (miR-1246 and miR-6089) of aseptic loosening patients with target genes in the local periprosthetic bone and soft tissues. These genes, some upregulated, some downregulated, act on the Wnt signalling pathway, which in-turn results in the inhibition of osteoblasts and activation of osteoclasts. Over time, this process results in bone resorption around the prosthesis, osteolysis and aseptic loosening
There are several limitations which must be discussed. Firstly, the control group we used to collect bone samples were patients with osteoarthritis undergoing primary TJR. Ideally, the control group would be patients with well-functioning joint replacements not requiring revision surgery or normal healthy patients free of arthritic disease. However, the significant risks associated with taking samples of bone and tissue around a normal joint replacement or from a normal patient preclude us from using this cohort as a control. Secondly, our study cohort had an uneven number of patients having hip arthroplasty and knee arthroplasty. Although there are subtle differences in the disease process of hip and knee arthritis, the underlying biological processes of osteoarthritis and the development of aseptic loosening should remain the same. As hip and knee arthroplasty are the most common forms of arthroplasty performed, we believe combining the two groups make the results more applicable to a wider population. Finally, despite our best efforts, it can be difficult to completely separate bone and tissue samples, especially in revision cases where anatomical layers are distorted, and sample volume is small. This may result in minute amounts of tissue within our bone samples and vice versa.
In conclusion, RAL patients have a significantly different transcriptome compared to controls. Furthermore, there are several miRNA which are detectable in the blood of RAL patients which have gene targets in the bone and soft tissue around the prosthesis. Our findings allow us to understand the role of miRNA in the regulation of genes associated with aseptic loosening.
Author contributions
YD made substantial contributions to conception and design of the work, the acquisition, data analysis and interpretation of data and drafted the work to be published. KP made contributions to the acquisition, analysis and interpretation of data. ZF made substantial contributions to the analysis of data. PS and RL contributed substantially to the conception and design of the study, the acquisition, analysis and interpretation of data and substantively revised the final manuscript for publication. All authors have approved the submitted version (and any substantially modified versions of the paper) and agree to be personally accountable for the authors’ own contributions and ensure that questions related to the accuracy and integrity of any part of the work are appropriately investigated, resolved and documented.
Funding
No external funding was sought for this project.
Availability of data and materials
Datasets and code used for RNA-sequencing data analysis available from the corresponding author by request.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Ethics approval was granted by the ACT Health Research Ethics and Governance Office, Human Research Ethics Committee (ETH.9.07.865) to recruit patients, collect intraoperative tissue samples and blood samples. Written informed consent was obtained from each patient.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Australian Orthopaedic Association National Joint Replacement Registry (AOANJRR): Hip, Knee & Shoulder Arthroplasty: 2021 Annual Report. Adelaide, South Australia, Australia: AOA, 2021.
- 2.O'Neill SC, Queally JM, Devitt BM, Doran PP, O'Byrne JM. The role of osteoblasts in peri-prosthetic osteolysis. Bone Joint J. 2013;95-B(8):1022–1026. doi: 10.1302/0301-620X.95B8.31229. [DOI] [PubMed] [Google Scholar]
- 3.Veronesi F, Tschon M, Fini M. Gene expression in osteolysis: review on the identification of altered molecular pathways in preclinical and clinical studies. Int J Mol Sci. 2017 doi: 10.3390/ijms18030499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Del Buono A, Denaro V, Maffulli N. Genetic susceptibility to aseptic loosening following total hip arthroplasty: a systematic review. Br Med Bull. 2012;101:39–55. doi: 10.1093/bmb/ldr011. [DOI] [PubMed] [Google Scholar]
- 5.Kandahari AM, Yang X, Laroche KA, Dighe AS, Pan D, Cui Q. A review of UHMWPE wear-induced osteolysis: the role for early detection of the immune response. Bone Res. 2016;4:16014. doi: 10.1038/boneres.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stulberg BN, Della Valle AG. Implant Wear Symposium Clinical Work G What are the guidelines for the surgical and nonsurgical treatment of periprosthetic osteolysis. J Am Acad Orthop Surg. 2008;16:20–25. doi: 10.5435/00124635-200800001-00006. [DOI] [PubMed] [Google Scholar]
- 7.Talmo CT, Shanbhag AS, Rubash HE. Nonsurgical management of osteolysis: challenges and opportunities. Clin Orthop Relat Res. 2006;453:254–264. doi: 10.1097/01.blo.0000246531.59876.a8. [DOI] [PubMed] [Google Scholar]
- 8.Lam JK, Chow MY, Zhang Y, Leung SW. siRNA Versus miRNA as therapeutics for gene silencing. Mol Ther Nucl Acids. 2015;4(9):e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gargano G, Oliviero A, Oliva F, Maffulli N. Small interfering RNAs in tendon homeostasis. Br Med Bull. 2021;138(1):58–67. doi: 10.1093/bmb/ldaa040. [DOI] [PubMed] [Google Scholar]
- 10.Gargano G, Oliva F, Oliviero A, Maffulli N. Small interfering RNAs in the management of human rheumatoid arthritis. Br Med Bull. 2022;142(1):34–43. doi: 10.1093/bmb/ldac012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gargano G, Asparago G, Spiezia F, Oliva F, Maffulli N. Small interfering RNAs in the management of human osteoporosis. Br Med Bull. 2023;148(1):58–69. doi: 10.1093/bmb/ldad023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 13.Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliviero A, Della Porta G, Peretti GM, Maffulli N. MicroRNA in osteoarthritis: physiopathology, diagnosis and therapeutic challenge. Br Med Bull. 2019;130(1):137–147. doi: 10.1093/bmb/ldz015. [DOI] [PubMed] [Google Scholar]
- 15.Giordano L, Porta GD, Peretti GM, Maffulli N. Therapeutic potential of microRNA in tendon injuries. Br Med Bull. 2020;133(1):79–94. doi: 10.1093/bmb/ldaa002. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Zhang L. Circulating Exosomal miRNA as diagnostic biomarkers of neurodegenerative diseases. Front Mol Neurosci. 2020;13:53. doi: 10.3389/fnmol.2020.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang X, Xiong X, Liu J, Zhao Z, Cen X. MicroRNAs-containing extracellular vesicles in bone remodeling: an emerging frontier. Life Sci. 2020;254:117809. doi: 10.1016/j.lfs.2020.117809. [DOI] [PubMed] [Google Scholar]
- 18.Hu W, Yu Y, Sun Y, Yuan F, Zhao F. MiR-25 overexpression inhibits titanium particle-induced osteoclast differentiation via down-regulation of mitochondrial calcium uniporter in vitro. J Orthop Surg Res. 2022;17(1):133. doi: 10.1186/s13018-022-03030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y, Liu Y, Cheng L. miR-21 expression is related to particle-induced osteolysis pathogenesis. J Orthopaed Res Official Publication Orthopaed Res Soc. 2012;30(11):1837–1842. doi: 10.1002/jor.22128. [DOI] [PubMed] [Google Scholar]
- 20.Zheng DZ, Bu YM, Wang L, Liu J. MicroRNA-130b promotes wear particle-induced osteolysis via downregulating frizzled-related protein (FRZB) Curr Neurovasc Res. 2017;14(1):32–38. doi: 10.2174/1567202614666161123112409. [DOI] [PubMed] [Google Scholar]
- 21.Wang T, Yin H, Wang J, Li Z, Wei H, Liu Z, et al. MicroRNA-106b inhibits osteoclastogenesis and osteolysis by targeting RANKL in giant cell tumor of bone. Oncotarget. 2015;6(22):18980–18996. doi: 10.18632/oncotarget.4223.PubMedPMID:26053181;PubMedCentralPMCID:PMCPMC4662469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895.PubMedPMID:21532838;PubMedCentralPMCID:PMCPMC3048316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaschetto LM. miRNA activation is an endogenous gene expression pathway. RNA Biol. 2018;15(6):826–828. doi: 10.1080/15476286.2018.1451722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 25.Moorer MC, Riddle RC. Regulation of Osteoblast Metabolism by Wnt Signaling. Endocrinol Metab (Seoul). 2018;33(3):318–330. doi: 10.3803/EnM.2018.33.3.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sen M, Lauterbach K, El-Gabalawy H, Firestein GS, Corr M, Carson DA. Expression and function of wingless and frizzled homologs in rheumatoid arthritis. Proc Natl Acad Sci U S A. 2000;97(6):2791–2796. doi: 10.1073/pnas.050574297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi Y, Maeda K, Takahashi N. Roles of Wnt signaling in bone formation and resorption. Japan Dental Sci Rev. 2008;44(1):76–82. doi: 10.1016/j.jdsr.2007.11.002. [DOI] [Google Scholar]
- 28.Shi Y, He B, You L, Jablons DM. Roles of secreted frizzled-related proteins in cancer. Acta Pharmacol Sinica. 2007;28(9):1499–1504. doi: 10.1111/j.1745-7254.2007.00692.x. [DOI] [PubMed] [Google Scholar]
- 29.Nakanishi R, Akiyama H, Kimura H, Otsuki B, Shimizu M, Tsuboyama T, et al. Osteoblast-targeted expression of Sfrp4 in mice results in low bone mass. J Bone Miner Res. 2008;23(2):271–277. doi: 10.1359/jbmr.071007. [DOI] [PubMed] [Google Scholar]
- 30.Kiper POS, Saito H, Gori F, Unger S, Hesse E, Yamana K, et al. Cortical-bone fragility-insights from sFRP4 deficiency in Pyle's disease. N Engl J Med. 2016;374(26):2553–2562. doi: 10.1056/NEJMoa1509342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulvihill BM, McNamara LM, Prendergast PJ. Loss of trabeculae by mechano-biological means may explain rapid bone loss in osteoporosis. J R Soc Interface. 2008;5(27):1243–1253. doi: 10.1098/rsif.2007.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabbri N, Rustemi E, Masetti C, Kreshak J, Gambarotti M, Vanel D, et al. Severe osteolysis and soft tissue mass around total hip arthroplasty: description of four cases and review of the literature with respect to clinico-radiographic and pathologic differential diagnosis. Eur J Radiol. 2011;77(1):43–50. doi: 10.1016/j.ejrad.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 33.Mattiotti A, Prakash S, Barnett P, van den Hoff MJB. Follistatin-like 1 in development and human diseases. Cell Mol Life Sci. 2018;75(13):2339–2354. doi: 10.1007/s00018-018-2805-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi DL, Shi GR, Xie J, Du XZ, Yang H. MicroRNA-27a inhibits cell migration and invasion of fibroblast-like synoviocytes by targeting follistatin-like protein 1 in rheumatoid arthritis. Mol Cell. 2016;39(8):611–618. doi: 10.14348/molcells.2016.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim HJ, Kang WY, Seong SJ, Kim SY, Lim MS, Yoon YR. Follistatin-like 1 promotes osteoclast formation via RANKL-mediated NF-kappaB activation and M-CSF-induced precursor proliferation. Cell Signal. 2016;28(9):1137–1144. doi: 10.1016/j.cellsig.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 36.Phesse T, Flanagan D, Vincan E. Frizzled7: a promising Achilles' heel for targeting the Wnt receptor complex to treat cancer. Cancers (Basel). 2016 doi: 10.3390/cancers8050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabokbar A, Mahoney DJ, Hemingway F, Athanasou NA. Non-Canonical (RANKL-Independent) pathways of osteoclast differentiation and their role in musculoskeletal diseases. Clin Rev allerg Immunol. 2016;51(1):16–26. doi: 10.1007/s12016-015-8523-6. [DOI] [PubMed] [Google Scholar]
- 38.van Amerongen R, Nawijn MC, Lambooij JP, Proost N, Jonkers J, Berns A. Frat oncoproteins act at the crossroad of canonical and noncanonical Wnt-signaling pathways. Oncogene. 2010;29(1):93–104. doi: 10.1038/onc.2009.310. [DOI] [PubMed] [Google Scholar]
- 39.Murray NR, Davidson LA, Chapkin RS, Clay Gustafson W, Schattenberg DG, Fields AP. Overexpression of protein kinase C betaII induces colonic hyperproliferation and increased sensitivity to colon carcinogenesis. J Cell Biol. 1999;145(4):699–711. doi: 10.1083/jcb.145.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitoh T, Moriwaki J, Koike J, Takagi A, Miwa T, Shiokawa K, et al. Molecular cloning and characterization of FRAT2, encoding a positive regulator of the WNT signaling pathway. Biochem Biophys Res Commun. 2001;281(3):815–820. doi: 10.1006/bbrc.2001.4421. [DOI] [PubMed] [Google Scholar]
- 41.Lane SW, De Vita S, Alexander KA, Karaman R, Milsom MD, Dorrance AM, et al. Rac signaling in osteoblastic cells is required for normal bone development but is dispensable for hematopoietic development. Blood. 2012;119(3):736–744. doi: 10.1182/blood-2011-07-368753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Itokowa T, Zhu ML, Troiano N, Bian J, Kawano T, Insogna K. Osteoclasts lacking Rac2 have defective chemotaxis and resorptive activity. Calcif Tissue Int. 2011;88(1):75–86. doi: 10.1007/s00223-010-9435-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu M, Sun BH, Saar K, Simpson C, Troiano N, Dallas SL, et al. Deletion of rac in mature osteoclasts causes osteopetrosis, an age-dependent change in osteoclast number, and a reduced number of osteoblasts in vivo. J Bone Miner Res. 2016;31(4):864–873. doi: 10.1002/jbmr.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan C, Qiao F, Wei P, Chi Y, Wang W, Ni S, et al. DIXDC1 activates the Wnt signaling pathway and promotes gastric cancer cell invasion and metastasis. Mol Carcinog. 2016;55(4):397–408. doi: 10.1002/mc.22290. [DOI] [PubMed] [Google Scholar]
- 45.Xin H, Li C, Wang M. DIXDC1 promotes the growth of acute myeloid leukemia cells by upregulating the Wnt/beta-catenin signaling pathway. Biomed Pharm Biomed Pharm. 2018;107:1548–1555. doi: 10.1016/j.biopha.2018.08.144. [DOI] [PubMed] [Google Scholar]
- 46.Ling IT, Rochard L, Liao EC. Distinct requirements of wls, wnt9a, wnt5b and gpc4 in regulating chondrocyte maturation and timing of endochondral ossification. Dev Biol. 2017;421(2):219–232. doi: 10.1016/j.ydbio.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spater D, Hill TP, O'Sullivan RJ, Gruber M, Conner DA, Hartmann C. Wnt9a signaling is required for joint integrity and regulation of Ihh during chondrogenesis. Development (Cambridge, England). 2006;133(15):3039–3049. doi: 10.1242/dev.02471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets and code used for RNA-sequencing data analysis available from the corresponding author by request.