Abstract
BACKGROUND
The pathogenicity of Helicobacter pylori is dependent on factors including the environment and the host. Although selenium is closely related to pathogenicity as an environmental factor, the specific correlation between them remains unclear.
AIM
To investigate how selenium acts on virulence factors and reduces their toxicity.
METHODS
H. pylori strains were induced by sodium selenite. The expression of cytotoxin-associated protein A (CagA) and vacuolating cytotoxin gene A (VacA) was determined by quantitative PCR and Western blotting. Transcriptomics was used to analyze CagA, CagM, CagE, Cag1, Cag3, and CagT. C57BL/6A mice were infected with the attenuated strains subjected to sodium selenite induction, and H. pylori colonization, inflammatory reactions, and the cell adhesion ability of H. pylori were assessed.
RESULTS
CagA and VacA expression was upregulated at first and then downregulated in the H. pylori strains after sodium selenite treatment. Their expression was significantly and steadily downregulated after the 5th cycle (10 d). Transcriptome analysis revealed that sodium selenite altered the levels affect H. pylori virulence factors such as CagA, CagM, CagE, Cag1, Cag3, and CagT. Of these factors, CagM and CagE expression was continuously downregulated and further downregulated after 2 h of induction with sodium selenite. Moreover, CagT expression was upregulated before the 3rd cycle (6 d) and significantly downregulated after the 5th cycle. Cag1 and Cag3 expression was upregulated and downregulated, respectively, but no significant change was observed by the 5th cycle. C57BL/6A mice were infected with the attenuated strains subjected to sodium selenite induction. The extent of H. pylori colonization in the stomach increased; however, sodium selenite also induced a mild inflammatory reaction in the gastric mucosa of H. pylori-infected mice, and the cell adhesion ability of H. pylori was significantly weakened.
CONCLUSION
These results demonstrate that H. pylori displayed virulence attenuation after the 10th d of sodium selenite treatment. Sodium selenite is a low toxicity compound with strong stability that can reduce the cell adhesion ability of H. pylori, thus mitigating the inflammatory damage to the gastric mucosa.
Keywords: Helicobacter pylori, Sodium selenite, Virulence factors, Adherence, Inflammation
Core Tip: The situation caused by Helicobacter pylori drug resistance is critical. Selenium is one of the trace elements in the human body, and its content in the stomach is related to the degree of H. pylori infection. Initial studies have shown that sodium selenite has inhibitory effects on H. pylori, so we further investigated the mechanism of action of sodium selenite on H. pylori. This study provides an experimental basis for use of the trace element selenium in the treatment of H. pylori infection.
INTRODUCTION
Helicobacter pylori is a common gram-negative bacterium that colonizes the stomach. Half of the global population is infected with this bacterium, which causes gastritis, stomach ulcers, and other diseases. In 1994, the World Health Organization listed H. pylori as a Class I carcinogen. In 2022, the United States listed this bacterium on the list of microorganisms associated with cancer[1,2].
The control and treatment of H. pylori infections can reduce gastric cancer incidence. However, because of the extensive use of antibiotics against H. pylori, the drug resistance rate has increased, and the eradication rate has significantly decreased[3]. Therefore, new antibacterial drugs or prevention and control programs need to be developed to alleviate the public health threat posed by H. pylori. Imitating live vaccines that lower H. pylori toxicity and reduce pathogenicity serves as better control methods.
CagA and VacA, the main H. pylori virulence factors, are associated with inflammation, apoptosis, autophagy, and the epithelial–mesenchymal transformation[4]. CagA, a bacterial oncoprotein[5], and VacA, which promotes the survival of H. pylori in gastric epithelial cells, are associated with phenomena such as mitochondrial damage[4]. The cag pathogenicity island (CagPAI) contains secretory protein-encoding CagA genes and type IV secretion system (T4SS) genes including CagE, CagM, Cag3, CagT, CagX, and CagY. These genes are involved in the T4SS activity[6]. However, the mutual regulation of these genes is not well understood.
The incidence of H. pylori infection is 10%-15%. Most people infected with H. pylori do not develop disease; however, the reason for this remains unclear. The mechanism underlying the interaction between H. pylori and the environment and body needs to be further explored. Selenium (Se), a trace element in the human body[7,8], plays a major role in immunoregulation, antioxidation, and antitumor and antibacterial functions[9-12]. As an environmental factor in the stomach, Se is closely associated with H. pylori pathogenesis. Ustündağ et al[13] found that the stomach Se level is higher in the early stage of H. pylori infection but significantly decreases when H. pylori induces precancerous lesions. Burguera et al[14] found that the stomach Se content is significantly lower in patients with gastric ulcers and gastric cancer than in those with gastritis. Liu[15] found that sodium selenite can promote gastric ulcer healing[16,17], but it remains unknown why Se content in the patient’s stomach decreases and how Se promotes ulcer healing.
Therefore, in this study, we explored how the Se-rich environment acts on H. pylori virulence factors by inducing H. pylori with sodium selenite and investigated the interaction between Se and H. pylori in light of the toxicity and inflammatory injury mediated by strains in the stomach. These findings provide an experimental basis for Se application for the prevention and treatment of H. pylori infection-related diseases.
MATERIALS AND METHODS
The cells and bacteria
The human gastric mucosal epithelial cell line GES-1 (No. CC4026; Guangzhou Cellcook Biotech Co., Ltd., Guangzhou, China) and human gastric cancer cell line BGC823 (Nanjing Kaijian Biotech Co., Ltd., Nanjing) were purchased. H. pylori strains (159, 26695, G27, and NSH57) were all provided by Professor Bi Hongkai, Laboratory of Pathogen Biology, Nanjing Medical University (Nanjing, China).
Induction and culture of H. pylori cells with sodium selenite
Hp G27 and NSH57 bacterial solutions were prepared at an initial concentration of 1 × 105 colony-forming units (CFU)/mL. The sodium selenite concentration for induction was determined according to the minimal inhibitory concentration (MIC) results, and the induction time was determined according to the growth curve. Sequential induction was performed using culture medium containing the same sodium selenite concentration for continuous passages.
Quantitative PCR targeting virulence factors in H. pylori strains to determine mRNA expression after sodium selenite induction
RNA was extracted using the FastPure Cell/Tissue Total RNA Isolation Kit V2 (No. RC112-01; Vazyme Biotech Co., Ltd., Nanjing, China). cDNA was obtained through RNA reverse transcription using a dsDNase-containing reverse transcription premix (No. MR05101M; Monad Biotech Co., Ltd., Suzhou, China). The MonAmp SYBR Green qPCR Mix (No. MQ10301S; Monad Biotech) was used to configure the quantitative PCR (qPCR) system, and the Light Cycler 96 was used for amplification, with the procedures adjusted as follows: initial denaturation at 95 °C for 30 s, denaturation at 95 °C for 10 s, annealing at 60 °C for 10 s, and extension at 72 °C for 30 s; all steps were performed for 40 cycles. The corresponding calculations were made according to the following equation: 2-ΔCT (test) –ΔCT (calibrator) = relative expression. The mRNA expression of CagA, VacA, 16s RNA, interleukin 6 (IL-6), IL-8, tumor necrosis factor-alpha (TNF-α), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Cag1, Cag3, CagT, CagM, and CagE was determined. Table 1 lists the design and synthesis of the primers.
Table 1.
Primer list
|
Name
|
Sequence
|
Company performing synthesis
|
| CagA | F: ACCCCTAGTCGGTAATG | Shanghai Invitrogen Biotech Co., Ltd. |
| R: GCTTTAGCTTCTGATACTGC | ||
| VacAs1a | F: GTCAGCATCACACCGCAAC | Shanghai Generay Biotech Co., Ltd. |
| R: CTGCTTGAATGCGCCAAAC | ||
| 16sRNA | F: CTGGAGAGACTAAGCCCTCC | Shanghai Invitrogen Biotech Co., Ltd. |
| R: AGGATCAAGGTTTAAGGATT | ||
| IL-6 | F: GCAGAAAAAGGCAAAGAATC | Wuhan Genecreate Biotech Co., Ltd. |
| R: CTACATTTGCCGAAGAGC | ||
| IL-8 | F: CACCGGAAGGAACCATCTCA | Wuhan Genecreate Biotech Co., Ltd. |
| R: TGGCAAAACTGCACCTTCACA | ||
| TNF-α | F: TCTTCTCGAACCCCGAGTGA | Wuhan Genecreate Biotech Co., Ltd. |
| R: CCTCTGATGGCACCACCAG | ||
| GAPDH | F: GGACCTGACCTGCCGTCTAG | Wuhan Genecreate Biotech Co., Ltd. |
| R: GTAGCCCAGGATGCCCTTGA | ||
| Cag1 | F: GCTATGGGGATTGTTGGGATAA | Shanghai Sangon Biotech Co., Ltd. |
| R: GCTTCAGTTGGTTCGTTGGTAA | ||
| Cag3 | F: GACACCTTGAATGTGAATGACAAA | Shanghai Sangon Biotech Co., Ltd. |
| R: GTTGTAATACCCATTGACTTGCTCTAA | ||
| CagT | F: TCTAAAAAGATTACGCTCATAGGCG | Shanghai Sangon Biotech Co., Ltd. |
| R: CTTTGGCTTGCATGTTCAAGTTGCC | ||
| CagE | F: GCGATTGTTATTGTGCTTGTAG | Shanghai Sangon Biotech Co., Ltd. |
| R: GAAGTGGTTAAAAAATCAATGCCCC | ||
| CagM | F: ACAAATACAAAAAAGAAAAAGAGGC | Shanghai Sangon Biotech Co., Ltd. |
| R: ATTTTTCAACAAGTTAGAAAAAGCC |
16sRNA: 16S ribosomal RNA; CagA: Cytotoxin-associated protein A; GAPDH: Glyceraldehyde 3-phosphate dehydrogenase; IL-6: Interleukin 6; TNF-a: Tumor necrosis factor-alpha; VacAs1a: Vacuolating cytotoxin gene As1a.
Western blotting of H. pylori virulence factor proteins after sodium selenite induction
In total, 3-7 mL of H. pylori bacterial solution was centrifuged at 12000 rpm for 2 min. The supernatant was removed and dosed with approximately 130 mL RIPA lysate (No. P0013B; Shanghai Beyotime Biotech Co., Ltd., Shanghai, China). Then a protease inhibitor and 0.1% EDTA were added at a 100:1 ratio. Thereafter, the solution was mixed on ice and ultrasonicated for approximately 5 min until the solution became clear. The solution was centrifuged at 13000 rpm for 5 min (4 °C) to obtain the supernatant protein solution. The protein concentration of the obtained solution was determined and the sample was divided to ensure 100 mg per tube. Then the samples were mixed with sodium dodecyl sulfate polyacrylamide gel electrophoresis (PAGE) loading buffer, denatured in boiling water at 100 °C for 5–10 min, and stored at -80 °C.
A PAGE Gel Fast Preparation Kit (10%, No. PG112; Shanghai Epizyme Biomedicine Technology Co., Ltd., Shanghai, China) was used for colloid preparation. Electrophoresis was performed using the loading protein and marker at 80 V for 30 min and 120 V for 1 h, respectively; the proteins were electrotransferred to a PVDF membrane at a constant current of 300 mA for 3 h. The protein gel was placed in a PVDF membrane for 1 h for blocking; transferred into the primary antibody dilution buffer containing CagA (A-10), VacA, and GAPDH antibodies at dilution ratios of 1:500, 1:500, and 1:6000, respectively; and incubated at 4°C overnight.
Thereafter, the PVDF membrane was washed three times with 0.1% Tween 20 Detergent (1 ×) on a shaker (10 min each time). The secondary antibody dilution buffer containing the mouse IgGκ light chain binding protein horseradish peroxidase (HRP) antibody and the goat anti-rabbit IgG (H + L) HRP antibody (both diluted at 1:6000) was added. After incubation for 2 h, the aforementioned washing steps were repeated.
Using an enhanced chemiluminescence detection kit, the image was developed in the automatic chemiluminescence image analyzer. The gray values of the bands were quantified using ImageJ software. The expression of all proteins was compared with that of GAPDH. Table 2 presents the antibody information.
Table 2.
Antibody information
|
Name
|
Art. No.
|
Company
|
| CagA (A-10) | sc-32746 | Santa Cruz |
| VacA | sc-28368 | Santa Cruz |
| m-IgGk BP-HRP | sc-516102 | Santa Cruz |
| GAPDH Ab | AF7021 | Affinity Biosciences |
| Goat anti-rabbit IgG (H + L) HPR | S0001 | Affinity Biosciences |
CagA: Cytotoxin-associated protein A; GAPDH: Glyceraldehyde 3-phosphate dehydrogenase; HPR: Horseradish peroxidase; H: Heavy chain; L: Light chain; m-IgGk BP-HRP: Mouse IgGκ light chain binding protein; VacA Vacuolating cytotoxin gene A.
Differentially expressed genes of H. pylori after sodium selenite induction were determined by transcriptome sequencing
To explore the effect of the half inhibitory concentration of sodium selenite on the Hp G27 strain, the bacterial solution was prepared (1 × 108 CFU/mL); treated with sodium selenite at concentrations of 1/8, 1/5, 1/4, 1/2, and 1 times the MIC; and cultured for 0, 2, and 8 h. Thereafter, 1 mL solution was coated over the plate and the plate was cultured for 3 d. The colonies were counted to determine the concentration of the original bacterial solution. The concentration at which the colony count remained relatively stable for 0, 2, and 8 h after sodium selenite treatment was considered the half inhibitory concentration and used as the transcriptome induction concentration.
The transcriptome analysis samples were treated with sodium selenite at a concentration of 1/5 the MIC. The intervention experiment was repeated three times to obtain three sample groups (n = 9 samples). The markers S_0, S_2, and S_8 represent the treatment groups at 0, 2, and 8 h, respectively. Sequencing and analysis were performed by Nanjing Fengzi Biomedical Technology Co., Ltd. (Nanjing, China).
Detection of the cell adhesion ability of H. pylori strains after induction with sodium selenite
Phosphate-buffered saline (PBS), H. pylori, induction, and H. pylori + 4 mol/L sodium selenite groups were used for the Alma blue experiments. GES-1 cells were cultured in a 96-well plate for 18-24 h (1 × 104 cells/well) and infected with H. pylori for 1 h (multiplicity of infection [MOI] = 300:1). Then the cells that did not adhere to the GES-1 cells were slowly removed, and the adhering cells were treated with 50 mL of 0.4% saponin solution for 5 min. Thereafter, the cells with the spent solution were removed, added to the culture medium containing 100 mL H. pylori, treated with 10 mL Alma blue, inoculated in a three-gas incubator for 4-6 h, and analyzed using a multimode reader.
The fluorescence experiential groups were divided into the same categories as those in the Alma blue experiment, as previously described. After culturing BGC823 cells in a 24-well plate for 18-24 h (5 × 104 cells/well), the cells were fluorescently stained with SYTO9 reagent, infected with H. pylori strains (MOI = 100:1), and cultured for another 3 h. The cells adhering to H. pylori strains were observed under an inverted fluorescence microscope.
Determination of inflammatory factor levels in H. pylori-infected cells induced with sodium selenite
The GES-1, GES-1 + G27, GES-1 + G27 induced, GES-1 + G27 + Se, GES-1 + Se, and Se groups were included in this experiment, and all groups were treated with 4 mol/L sodium selenite. GES-1 cells were cultured in a 6-well plate for 18-24 h (3 × 105 cells/well). After 24 h of infection with H. pylori strains (MOI = 100:1), the levels of inflammatory factors, such as IL-6, were determined.
Determination of the pathogenicity of H. pylori after sodium selenite induction in mice
Thirty specific pathogen-free (SPF) C57BL/6J mice (age: 6-7 wk) were purchased from Changsha Tianqin Biological Co., Ltd. (No. 430726210100134118; SPF Animal Use License No. SYXK Gui 2017-0004; Animal Experiment Ethics No. 2019112501; Changsha, China). The mice were randomly divided into the following five groups: PBS, Se, NSH57, NSH57-induced, and NSH57 + Se groups. Mice in the Se group were intragastrically administered sodium selenite (4 mol/L), and those in the induction group were administered sodium selenite (4 mol/L) for six cycles. The corresponding bacterial solutions were centrifuged at 12000 rpm for 2 min, and the supernatant was removed to obtain the precipitate. Thereafter, the solutions were resuspended in fresh medium or medium containing four sodium selenite, with the concentration adjusted to 1 × 109 CFU/mL.
The mice were fasted and deprived of water for 12 h before intragastric administration. Thereafter, they were intragastrically administered the solutions (0.5 mL/mouse), once every alternate day five consecutive times, and the diets were resumed diet 4 h after intragastric administration. After all rounds of intragastric administration, the mice were kept for 3 wk and dissected following the aseptic operation procedure. The tissues collected from stomachs, livers, spleens, and kidneys were fixed in 10% formalin and stained with hematoxylin and eosin (H&E). The whole stomach was divided into two parts along its major and minor curves, with the stomach contents gently scraped away. One part was fixed in H&E staining, and a fluorescence immunoassay was conducted to measure the levels of the inflammatory factors IL-1β, IL-6, and TNF-α. The other part was placed in a sterile Eppendorf tube containing magnetic beads, and the medium was weighed and recorded before and after induction, and crushed well using a multisample tissue grinder at 50 Hz three times (3 min each time). Then 100 mL abrasive solution was diluted 10-, 100-, and 1000-fold. Thereafter, 100 mL of each of the three diluted concentrations was coated over a solid plate containing selective antibiotics and cultured for 3-4 d. Then the number of H. pylori CFU/g or the extent of H. pylori colonization in the stomach was calculated.
Statistical analysis
Statistical analysis and mapping were performed using GraphPad Prism 8.0. The continuous data are expressed as the mean ± standard deviation. Differences in means between the groups were analyzed using one-way analysis of variance. P < 0.05 was considered statistically significant.
RESULTS
Induction with sodium selenite in vitro downregulated the expression of the H. pylori virulence factors CagA and VacA
According to the MIC and cytotoxic effects of sodium selenite on H. pylori observed in Supplementary Table 1 and Figure 1, 0, 4, 8, and 16 μmol/L sodium selenite was used to induce Hp G27 (48 h as a cycle), and sequential induction was performed after the medium was changed.
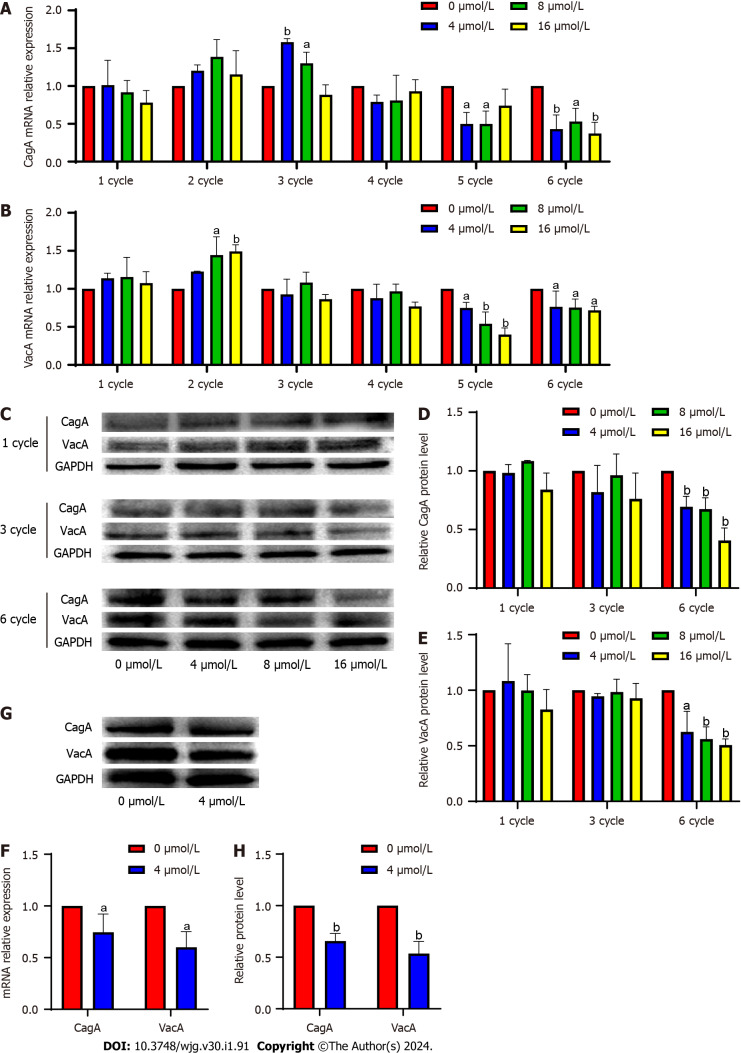
qPCR results revealed that after Hp G27 was sequentially induced with sodium selenite for 1-6 cycles, the mRNA expression of CagA was slightly downregulated in the 1st cycle, upregulated in the 2nd cycle, significantly upregulated in the 3rd cycle, downregulated again in the 4th cycle, and downregulated steadily and significantly throughout the 5th and 6th cycles (Figure 1A).
Figure 1.
Induction of Helicobacter pylori with sodium selenite downregulates cytotoxin-associated protein A and vacuolating cytotoxin gene A expression. A: Effect of induction of the Hp G27 strain with sodium selenite on cytotoxin-associated protein A (CagA) for 1-6 cycles; B: Effect of induction of the Hp G27 strain with sodium selenite on vacuolating cytotoxin gene A (VacA) for 1-6 cycles; C-E: Effect of induction of the Hp G27 strain with sodium selenite on the CagA and VacA proteins for 1, 3, and 6 cycles; F: Effect of induction of the Hp G27 strain with 4 mol/L sodium selenite on CagA and VacA of the NSH57 strain for 6 cycles; G and H: Effect of induction of the NSH57 strain with 4 μmol/L sodium selenite on CagA and VacA protein. aP < 0.05; bP < 0.01.
VacA mRNA expression was slightly upregulated in the 1st cycle, significantly upregulated in the 2nd cycle, downregulated again throughout the 3rd and 4th cycles, and downregulated steadily and significantly throughout the 5th and 6th cycles (Figure 1B). Western blotting results revealed that CagA protein expression changed in a manner consistent with that of mRNA in the 1st cycle. In the 3rd cycle, the amount of mRNA at 4 and 8 mol/L was significantly increased, whereas the protein expression basically remained unchanged. In the 6th cycle, the protein expression was downregulated in a manner consistent with the decreased mRNA expression.
VacA mRNA expression increased in the 1st and 3rd cycles, whereas its protein expression remained unchanged. In the 6th cycle, the protein expression was downregulated in a manner consistent with the decreased mRNA expression (Figure 1C-E). The mRNA expression of CagA and VacA was upregulated, whereas their protein expression remained unchanged. These results indicated that sodium selenite was possibly acting on biological processes posttranscriptionally or posttranslationally. The same results were obtained after the sequential induction of NSH57 strains for six cycles, and the protein expression was downregulated in a manner consistent with the decreased mRNA expression (Figure 1F-H). However, the aforementioned results suggested that the doses of 4, 8, and 16 μmol/L could induce virus attenuation, with 16 μmol/L being the best concentration.
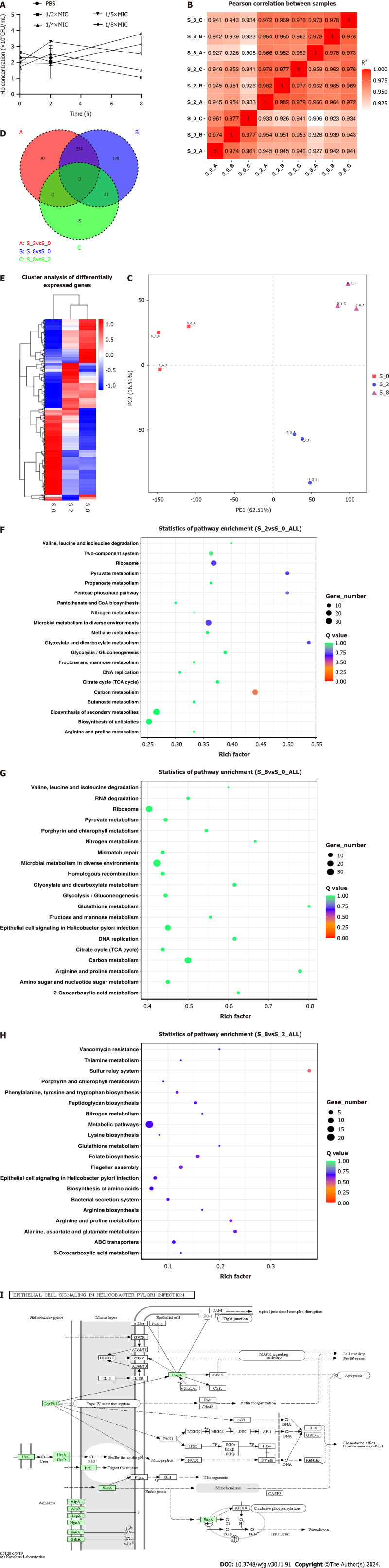
Changes in the H. pylori transcriptome after sodium selenite induction
H. pylori induced with sodium selenite at a concentration of 1/5 the MIC exhibited only slight growth. These results suggested that this concentration could be used to achieve a half-maximal inhibitory effect (Figure 2A). Therefore, this concentration was selected for induction. The RNA sequencing (RNA-seq) correlation test of the transcriptional group revealed that R2 values in the repeated intervention group were all > 0.9, thereby exhibiting good repeatability among the samples. The differences in R2 values were noted among the different intervention groups (Figure 2B). Principal component analysis revealed significant differences between the repeated and non-repeated samples (Figure 2C). The transcriptome analysis results are as follows: (1) 329 differentially expressed genes (DEGs) were detected between S_2 and S_0; (2) 466 DEGs were detected between S_8 and S_0; and (3) 85 DEGs were detected between S_8 and S_2. Overall, 247, 25, and 54 identical genes were detected between (1) and (2), (1) and (2), and (2) and (3), respectively, and 13 identical DEGs were detected between (1), (2), and (3) (Figure 2D and E). Among the top 20 pathways where the DEGs of (1), (2), and (3) were enriched, signal transduction pathways of epithelial cells associated with H. pylori infection were found to be associated with virulence factors (as shown in Figure 2F-H). The “CagPAI-CagA” pathway related to CagA virulence was further identified (Figure 2I). Thereafter, follow-up verification experiments were conducted on the genes with upregulated expression, Cag1, Cag3 and CagT, and the genes with downregulated expression, CagM and CagE, that had been identified based on significant differences, repeatability of differential expression between (1) and (2), and whether they encoded unknown proteins, as shown in Tables 3 and 4. Therefore, the transcriptional analysis suggested that virulence factors of H. pylori after sodium selenite induction were mainly enriched in H. pylori infection-associated epithelial cell signal transduction pathways. The main virulence factors of CagPAI included CagA, CagM, CagE, Cag1, Cag3, and CagT, with the urease gene and VacA also being key genes.
Figure 2.
Effect of sodium selenite on the Helicobacter pylori transcriptome. A: Half inhibitory concentration of sodium selenite given to Helicobacter pylori; B: RNA sequencing correlation examination; C: Principal component analysis of the transcriptome; D: The Venn diagram of differentially expressed genes (DEGs); E: Cluster analysis of DEGs; F-H: Gene set enrichment analysis of DEGs between treatment groups; I: Epithelial cell signal transduction pathways associated with H. pylori infection.
Table 3.
Identified differentially expressed genes (S_2 vs S_0)
|
Gene_id
|
Readcount_S_2
|
Readcount_S_0
|
Log2 fold change
|
P value
|
Padj
|
Gene name
|
Description
|
| Gene509 | 376.5506144 | 256.0954025 | 0.55616 | 0.0066152 | 0.035684 | HPG27_RS02500 | Cag pathogenicity island protein Cagl |
| Gene521 | 1368.459246 | 801.0552768 | 0.77258 | 5.49E-05 | 0.00089411 | HPG27_RS02560 | Type IV secretion system apparatus protein CagT |
| Gene511 | 1617.425068 | 1154.535615 | 0.48639 | 0.005033 | 0.28476 | HPG27_RS02510 | Type IV secretion system apparatus protein Cag3 |
| Gene540 | 181710.1299 | 131892.6179 | 0.46228 | 0.0039181 | 0.023777 | HPG27_RS02655 | Type IV secretion system oncogenic effector CagA |
| Gene536 | 1945.873607 | 3339.614831 | -0.77926 | 0.00032024 | 0.0036791 | HPG27_RS02635 | VirB4 family type IV secretion/conjugal transfer ATPase |
Table 4.
Identified differentially expressed genes (S_8 vs S_0)
|
Gene_id
|
Readcount_S_8
|
Readcount_S_0
|
Log2 fold change
|
P value
|
Padj
|
Gene name
|
Description
|
| Gene509 | 494.7491661 | 231.643609 | 1.0948 | 1.57E-06 | 3.04E-05 | HPG27_RS02500 | Cag pathogenicity island protein Cagl |
| Gene521 | 1519.085968 | 724.4396343 | 1.0683 | 2.78E-07 | 7.47E-06 | HPG27_RS02560 | Type IV secretion system apparatus protein CagT |
| Gene511 | 1562.119955 | 1044.031209 | 0.58134 | 0.0021884 | 0.010463 | HPG27_RS02510 | Type IV secretion system apparatus protein Cag3 |
| Gene529 | 311.1908412 | 460.4755565 | -0.56532 | 0.0049482 | 0.020333 | HPG27_RS02595 | Type IV secretion system apparatus protein CagM |
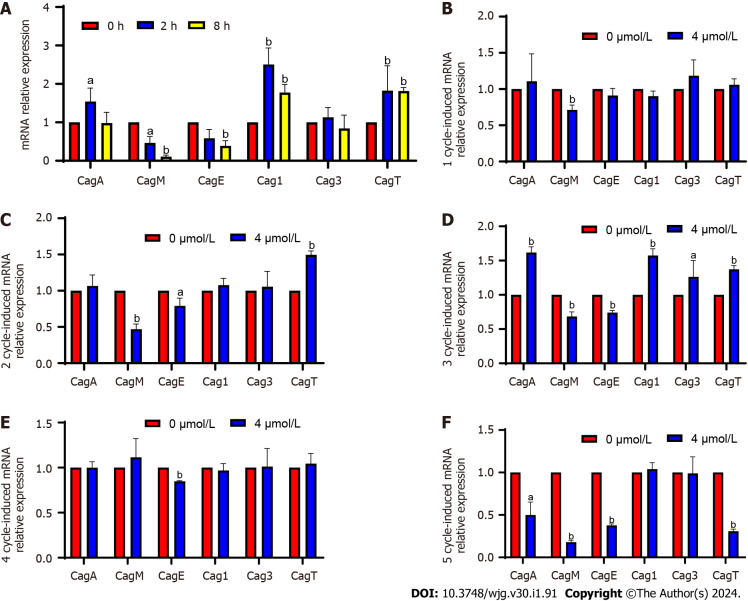
Verification of and changes in DEGs following induction
The main virulence genes of CagPAI were verified through qPCR (Figure 3). After the induction, CagM expression was significantly downregulated at 2 and 8 h and from the 1st to the 3rd cycles, upregulated again in the 4th cycle, and significantly downregulated in the 5th cycle (4-10 d). After the induction, CagE expression was significantly downregulated at 2 and 8 h with no significant change noted in the 1st cycle (2 d) and significantly downregulation from the 2nd to 5th cycles (4-10 d). The expression of these two genes was upregulated at some stages, but was significantly and continuously downregulated overall. CagM expression was upregulated at some stages but significantly and continuously downregulated overall; CagM and CagE expression was continuously and significantly downregulated at the fastest rate. Cag1 expression was significantly upregulated at 2 and 8 h and in the 3rd cycle after induction, but no significant change was observed at the other time points. Although Cag3 expression was significantly upregulated in the 3rd cycle after induction, no significant change was observed at other time points. Because no uniform trend of change was observed in the two genes, they might not be considered key genes in attenuation. CagT expression was significantly upregulated at 2 and 8 h and in the 2nd and 3rd cycles after induction, downregulated in the 4th cycle, and significantly downregulated in the 5th cycle. However, the expression at most of the detection sites was consistent with the result of the transcriptome analysis; the expression was upregulated, but whether the expression would continue to be downregulated after the 5th wk remains to be determined.
Figure 3.
Verification of differentially expressed genes and changes in screened differentially expressed genes following induction. A: Quantitative PCR verification of differentially expressed genes (DEGs); B-F: Changes in DEGs after induction with 4 μmol/L sodium selenite for 1-5 cycles. aP < 0.05; bP < 0.01.
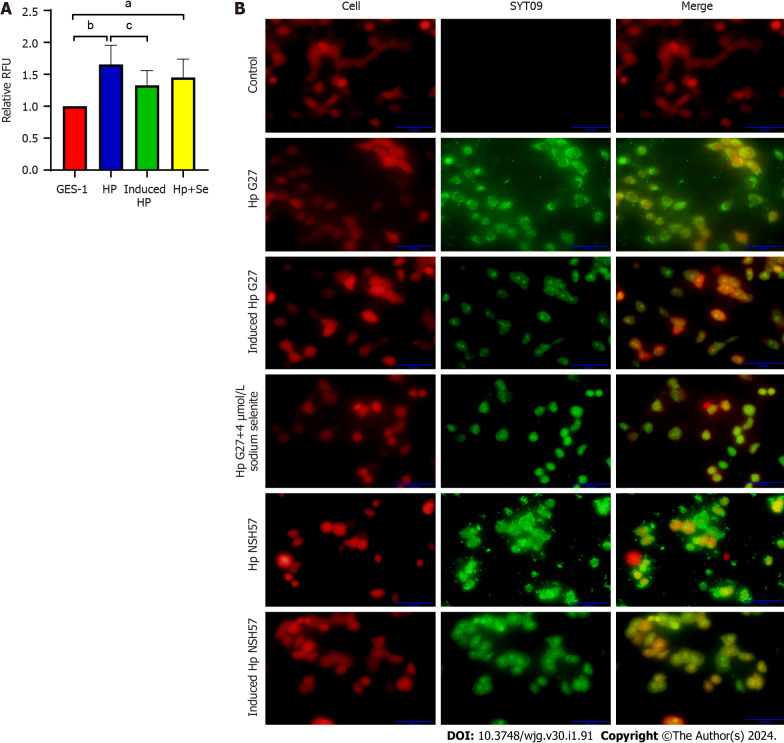
Induction with sodium selenite in vitro reduced the cell adhesion ability of H. pylori
The cell adhesion ability of H. pylori, an important prerequisite for bacterial colonization and pathogenicity, was evaluated by inducing H. pylori with 4 μmol/L sodium selenite for 6 cycles. The Alamar Blue assay revealed that the number of Hp G27 cells adhering to GES-1 cells after induction was significantly decreased (P < 0.05) and that of Hp G27 cells adhering to cells after induction with 4 mol/L sodium selenite exhibited a decreasing trend (P > 0.05) (Figure 4A). The fluorescence experiment demonstrated that the number of Hp G27 cells that adhered to cells in the induction group was significantly reduced (the red fluorescence represents BGC823 cells, and the green fluorescence represents H. pylori strains). The cell adhesion ability of NSH57 strains induced under the same conditions was the same as that of Hp G27 strains (Figure 4B). In conclusion, six induction cycles of different H. pylori strains with sodium selenite significantly reduced the cell adhesion ability of these strains.
Figure 4.
Induction with sodium selenite decreased the adhesion ability of Helicobacter pylori strains. A: Number of cells adhering to Helicobacter pylori strains was determined using a fluorescence microscope; B: Number of cells adhering to H. pylori strains was determined with the SYTO 9 stain. aP < 0.05; bP < 0.01; cP < 0.05.
Induction with sodium selenite reduced the inflammatory response triggered by H. pylori
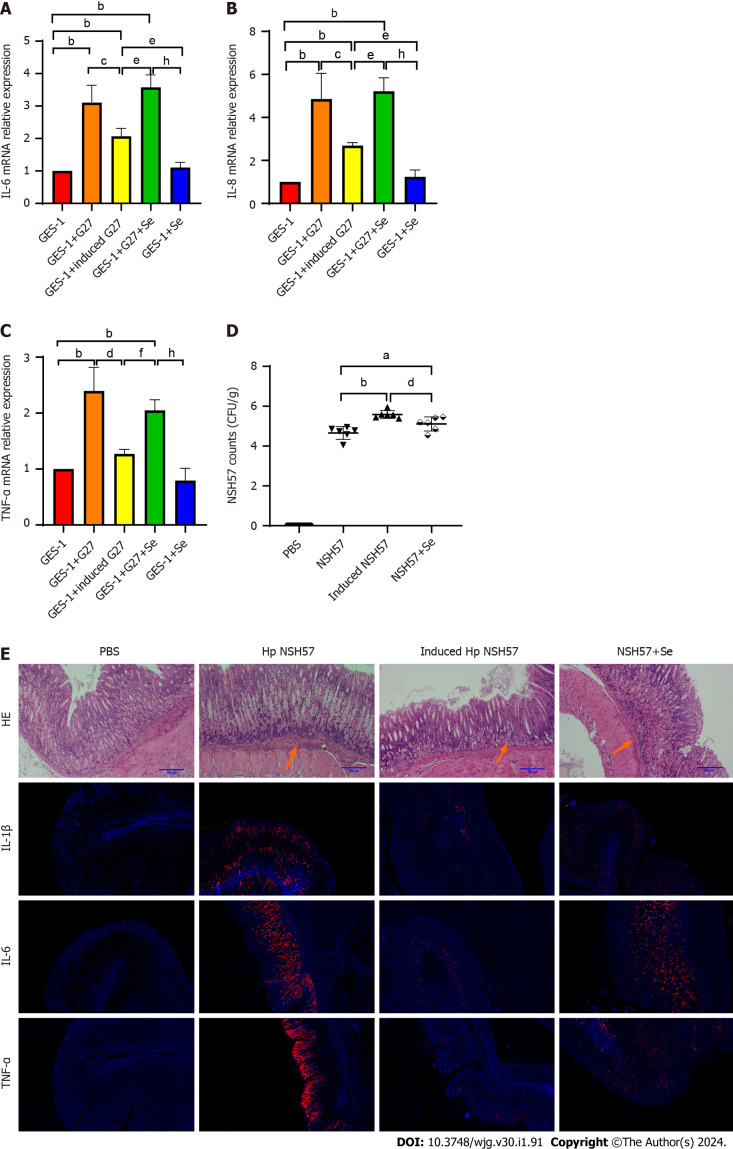
After sodium selenite induction, GES-1 cells and mice were infected with H. pylori strains. The results of the cell infection experiment suggested that the relative mRNA expression of the inflammatory factors IL-6, IL-8, and TNF-α was significantly downregulated (Figure 5A-C). In the experiment including infected mice, the extent of H. pylori colonization in the H. pylori-induced groups was significantly increased (P < 0.0001), and inflammation was significantly reduced. However, inflammation was reduced in the mice in the noninduced H. pylori groups that had received sodium selenite intragastrically compared with that of those that had not in the same group. Furthermore, the degree of inflammation reduction in the noninduced group was not as good as that in the induced group. The gastric mucosal expression of IL-1β, IL-6, and TNF-α was reduced, similar to the degree of inflammation (Figure 5D and E).
Figure 5.
Induction with sodium selenite decreased Helicobacter pylori-induced inflammation. A-C: Expression of IL-1β (A), IL-6 (B), and TNF-α (C) in Helicobacter pylori-infected mice after induction; D: Gastric mucosal inflammation in mice with acute gastritis; E: H. pylori colonization in the stomach of mice with acute gastritis. aP < 0.05; bP < 0.01; cP < 0.05; dP < 0.01; eP < 0.05; fP < 0.01; hP < 0.01.
The aforementioned results indicated that the inflammatory effect and virulence of H. pylori on cells and mice were significantly weakened after sodium selenite induction. However, the increased colonization might be a result of the weakened immune reaction in mice due to the reduced virulence of H. pylori.
DISCUSSION
Whether H. pylori infection causes disease depends on the bacteria, the environment, and the infected organism. Sodium selenite, a common inorganic Se, is also a common form of Se used for supplementation. Se has antibacterial effects[18,19]. The MIC of Se against H. pylori is 185 μmol/L, which is not strong enough to inhibit growth and makes it difficult to achieve a therapeutic effect, and high doses of Se can damage gastric mucosal cells. In addition, some infected people with high Se content in the body do not show any symptoms, although they carried H. pylori for a long time. This also reveals that the antibacterial effect of Se in the body is not the main factor and there are other factors affecting the development of H. pylori infection. In this study, after creating a Se-enriched environment and inducing H. pylori cells with sodium selenite, the expression of the H. pylori virulence factors CagA and VacA was significantly downregulated, and the downregulation of VacA expression was basically in line with the reports of Duan et al[20] and Shao et al[21]. However, as toxicity was not rapidly reduced after induction with sodium selenite, the reason and potential mechanism for the reduction in H. pylori virulence by sodium selenite is not clear.
In H. pylori, the CagPAI-encoded T4SS has 15-16 components. These components are mainly involved in CagA protein transport and play a certain role in immunogenicity[22]. T4SS can be divided into the core complex, inner membrane component protein, and outer membrane component protein. The core complex includes CagT, CagX, and CagY, the inner membrane component proteins include CagE, CagW, and CagV, and the outer membrane component proteins encompass CagM and Cag3[23]. Regarding the T4SS of H. pylori, studies have been conducted to determine changes in CagPAI expression to understand the relationship between this gene and diseases and the integrity of virulence genes to study the size of virulence factors[24,25]. CagPAI deletion caused changes in the T4SS, thereby weakening H. pylori toxicity; however, the bacterium was not completely detoxified[26,27]. Kumari et al[28] found for the first time that CagW could interact with CagA and play a crucial role in secretion while affecting the expression of flagellar components such as CagL. Similarly, Cag1 deletion could lead to a decrease in CagA heterotopia function[29].
The DEGs of epithelial cell pathways were all screened from CagPAI. Meanwhile, VacA, which is outside CagPAI, was analyzed. CagA and VacA expression first increased and then decreased after sodium selenite induction and significantly decreased after 10 d. After sodium selenite induction for 2 h, CagM and CagE expression was continuously downregulated, whereas CagT expression was upregulated before the 6th d and downregulated significantly after the 10th d; while Cag1 and Cag3 expression was upregulated and downregulated, with no significant change noted on the 10th d. The above results suggest that these five genes may be key genes in the reduction of H. pylori virulence by sodium selenite in a Se-enriched environment and that the time needed to induce a reduction in H. pylori virulence is at least 10 d.
After induction of H. pylori by sodium selenite, its adhesion ability was tested in vitro, and its colonization and inflammatory damage to the gastric mucosa of mice was tested in vivo, both results demonstrated that sodium selenite could significantly reduce the adhesion ability and toxicity of inflammatory damage, and H. pylori stability was better in a Se-enriched environment. The application of sodium selenite to reduce the toxicity of H. pylori is an ideal method for the development of attenuated vaccines[30].
CONCLUSION
In this study, we investigated how a sodium-rich environment acts on H. pylori virulence factors, verified that sodium selenite could reduce H. pylori toxicity, and confirmed that a Se-rich environment could significantly reduce H. pylori pathogenicity. We also indicated that further studies are needed to elucidate the underlying molecular mechanisms. This study offers an experimental basis for the use of Se, a trace element, in H. pylori infection treatment in the future and a reference for ensuring that humans coexist with bacteria without developing diseases. Sodium selenite treatment is a potential method that will allow attenuated vaccines to alleviate severe drug resistance in H. pylori.
ARTICLE HIGHLIGHTS
Research background
Helicobacter pylori is a common gram-negative bacterium that colonizes the stomach and is currently recognized as a class I carcinogen. The H. pylori infection rate is as high as approximately 60%, and the H. pylori eradication rate was markedly decreased with increasing rate of drug resistance. Finding new antimicrobial drugs or control regimens to mitigate the threat of H. pylori to human health and mimicking live vaccines is crucial. The preparation of live vaccines with reduced virulence and reduced pathogenicity is a promising control method. Selenium (Se) is one of the essential trace elements in the human body. Se has been proven to have an attenuating effect, but whether it has the same effect on H. pylori is unknown. The interaction among H. pylori, Se and its mechanism in the stomach has rarely been studied H. pylori has coexisted with humans for 100000 years, and eliminating this coexisting relationship is extremely difficult, this makes studying the effect of Se on H. pylori difficult. The influence of Se on the interaction between H.pylori and the organism may provide new ideas and an experimental basis for the prevention and treatment of H.pylori.
Research motivation
Currently, H. pylori is a persistent threat to humans, and the increase in drug resistance makes it increasingly difficult to eliminate this threat. Studying the mechanism of the attenuating effect of the trace element Se on H. pylori and its interaction with the organism after attenuation will help to better apply Se to the development of H. pylori attenuated vaccines and alleviate the problem of H. pylori drug resistance.
Research objectives
The aim of this study was to investigate the effect and mechanism of action of the trace element Se on the virulence factors of H. pylori and to provide an experimental basis for the use of the trace element Se in the prevention and treatment of H. pylori.
Research methods
A Se-enriched environment was created with sodium selenite to induce H. pylori, and the effect of the Se-enriched environment on the virulence of H. pylori and its potential mechanisms were evaluated by real-time fluorescence quantitative polymerase chain reaction, protein immunoblotting, transcriptome gene sequencing, Alma blue assay, and cell adhesion assay. A mouse gastritis model was established to understand the attenuation of H. pylori in terms of the changes in virulence in vitro and in vivo.
Research results
Se-enriched environments may lead to a reduction in the virulence factors CagA and VacA and significantly attenuate the pathogenicity of H. pylori by affecting the CagPAI-encoded type IV secretion systems of H. pylori.
Research conclusions
The mechanism of action of sodium selenite leading to the reduction of H. pylori virulence was shown to be through the downregulation of virulence factor expression, which led to a significant reduction in adhesion capacity, as well as inflammatory damage, thus inhibiting the growth of H. pylori and presenting better stability under a Se-rich environment.
Research perspectives
This study demonstrated the antibacterial mechanism of sodium selenite against H. pylori in a Se-enriched environment through in vitro and in vivo experiments. These results provide theoretical support for further research and development of sodium selenite in the preparation of attenuated vaccines and contributes to the alleviation of drug resistance in H. pylori.
Footnotes
Institutional animal care and use committee statement: All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee of the Youjiang Medical University for Nationalities Institutional Review Board.
Conflict-of-interest statement: The authors have no conflicts of interest to declare.
ARRIVE guidelines statement: The authors have read the ARRIVE Guidelines, and the manuscript was prepared and revised according to the ARRIVE Guidelines.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Peer-review started: October 26, 2023
First decision: November 13, 2023
Article in press: December 13, 2023
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Marescaux J, France; Mazher SA, United States S-Editor: Lin C L-Editor: Filipodia P-Editor: Yuan YY
Contributor Information
Chun Qin, Guangxi Technology Innovation Cooperation Base of Prevention and Control Pathogenic Microbes with Drug Resistance, Youjiang Medical University for Nationalities, Baise 533000, Guangxi Zhuang Autonomous Region, China; Department of Digestive Diseases, Affiliated Hospital of Youjiang Medical University for Nationalities, Baise 533000, Guangxi Zhuang Autonomous Region, China.
Gan-Rong Huang, Guangxi Technology Innovation Cooperation Base of Prevention and Control Pathogenic Microbes with Drug Resistance, Youjiang Medical University for Nationalities, Baise 533000, Guangxi Zhuang Autonomous Region, China; Key Laboratory of the Prevention and Treatment of Drug Resistant Microbial Infecting, Youjiang Medical University for Nationalities, Baise 533000, Guangxi Zhuang Autonomous Region, China.
Ai-Xing Guan, Guangxi Technology Innovation Cooperation Base of Prevention and Control Pathogenic Microbes with Drug Resistance, Youjiang Medical University for Nationalities, Baise 533000, Guangxi Zhuang Autonomous Region, China; Department of Digestive Diseases, Affiliated Hospital of Youjiang Medical University for Nationalities, Baise 533000, Guangxi Zhuang Autonomous Region, China.
Wen-Ting Zhou, Guangxi Technology Innovation Cooperation Base of Prevention and Control Pathogenic Microbes with Drug Resistance, Youjiang Medical University for Nationalities, Baise 533000, Guangxi Zhuang Autonomous Region, China; Key Laboratory of the Prevention and Treatment of Drug Resistant Microbial Infecting, Youjiang Medical University for Nationalities, Baise 533000, Guangxi Zhuang Autonomous Region, China.
Hao Chen, Department of Pathology, Wannan Medical College, Wuhu 241002, Anhui Province, China.
Pei-Pei Luo, Department of Gastroenterology, Wujin People’s Hospital affiliated to Jiangsu University, Changzhou 213004, Jiangsu Province, China.
Xian-Ke Luo, Department of Gastroenterology, Guangzhou Liwan District People's Hospital, Guangzhou 510370, Guangdong Province, China.
Yan-Qiang Huang, Guangxi Technology Innovation Cooperation Base of Prevention and Control Pathogenic Microbes with Drug Resistance, Youjiang Medical University for Nationalities, Baise 533000, Guangxi Zhuang Autonomous Region, China; Key Laboratory of the Prevention and Treatment of Drug Resistant Microbial Infecting, Youjiang Medical University for Nationalities, Baise 533000, Guangxi Zhuang Autonomous Region, China.
Zan-Song Huang, Guangxi Technology Innovation Cooperation Base of Prevention and Control Pathogenic Microbes with Drug Resistance, Youjiang Medical University for Nationalities, Baise 533000, Guangxi Zhuang Autonomous Region, China; Department of Digestive Diseases, Affiliated Hospital of Youjiang Medical University for Nationalities, Baise 533000, Guangxi Zhuang Autonomous Region, China. 1019846481@qq.com.
Data sharing statement
No additional data are available.
References
- 1.Frost F, Kacprowski T, Rühlemann M, Bang C, Franke A, Zimmermann K, Nauck M, Völker U, Völzke H, Biffar R, Schulz C, Mayerle J, Weiss FU, Homuth G, Lerch MM. Helicobacter pylori infection associates with fecal microbiota composition and diversity. Sci Rep. 2019;9:20100. doi: 10.1038/s41598-019-56631-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishaq S, Nunn L. Helicobacter pylori and gastric cancer: a state of the art review. Gastroenterol Hepatol Bed Bench. 2015;8:S6–S14. [PMC free article] [PubMed] [Google Scholar]
- 3.Lee YC, Dore MP, Graham DY. Diagnosis and Treatment of Helicobacter pylori Infection. Annu Rev Med. 2022;73:183–195. doi: 10.1146/annurev-med-042220-020814. [DOI] [PubMed] [Google Scholar]
- 4.Baj J, Forma A, Sitarz M, Portincasa P, Garruti G, Krasowska D, Maciejewski R. Helicobacter pylori Virulence Factors-Mechanisms of Bacterial Pathogenicity in the Gastric Microenvironment. Cells. 2020;10 doi: 10.3390/cells10010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chmiela M, Karwowska Z, Gonciarz W, Allushi B, Stączek P. Host pathogen interactions in Helicobacter pylori related gastric cancer. World J Gastroenterol. 2017;23:1521–1540. doi: 10.3748/wjg.v23.i9.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cover TL, Lacy DB, Ohi MD. The Helicobacter pylori Cag Type IV Secretion System. Trends Microbiol. 2020;28:682–695. doi: 10.1016/j.tim.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang WF. Research progress of selenium and selenoprotein on animal immunity. Siliao Yanjiu. 2020;43:103–105. [Google Scholar]
- 8.Schweizer U, Fradejas-Villar N. Why 21? The significance of selenoproteins for human health revealed by inborn errors of metabolism. FASEB J. 2016;30:3669–3681. doi: 10.1096/fj.201600424. [DOI] [PubMed] [Google Scholar]
- 9.Labunskyy VM, Hatfield DL, Gladyshev VN. Selenoproteins: molecular pathways and physiological roles. Physiol Rev. 2014;94:739–777. doi: 10.1152/physrev.00039.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He T, Dong YB, Wang CP, Wei CB. Meta-Analysis of Organic Selenium and Inorganic Selenium on Performance of Laying Hens and Selenium Content in Eggs. Dongwu Yingyang Xuebao. 2022;34:2654–2666. [Google Scholar]
- 11.Vundela SR, Kalagatur NK, Nagaraj A, Kadirvelu K, Chandranayaka S, Kondapalli K, Hashem A, Abd Allah EF, Poda S. Multi-Biofunctional Properties of Phytofabricated Selenium Nanoparticles From Carica papaya Fruit Extract: Antioxidant, Antimicrobial, Antimycotoxin, Anticancer, and Biocompatibility. Front Microbiol. 2021;12:769891. doi: 10.3389/fmicb.2021.769891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hashem AH, Salem SS. Green and ecofriendly biosynthesis of selenium nanoparticles using Urtica dioica (stinging nettle) leaf extract: Antimicrobial and anticancer activity. Biotechnol J. 2022;17:e2100432. doi: 10.1002/biot.202100432. [DOI] [PubMed] [Google Scholar]
- 13.Ustündağ Y, Boyacioğlu S, Haberal A, Demirhan B, Bilezikçi B. Plasma and gastric tissue selenium levels in patients with Helicobacter pylori infection. J Clin Gastroenterol. 2001;32:405–408. doi: 10.1097/00004836-200105000-00009. [DOI] [PubMed] [Google Scholar]
- 14.Burguera JL, Villasmil LM, Burguera M, Carrero P, Rondon C, de Abel de la Cruz AM, Brunetto MR, Gallignani M. Gastric tissue selenium levels in healthy persons, cancer and non-cancer patients with different kinds of mucosal damage. J Trace Elem Med Biol. 1995;9:160–164. doi: 10.1016/S0946-672X(11)80041-6. [DOI] [PubMed] [Google Scholar]
- 15.Liu XG. Serum and tissue copper, zinc and selenium levels in patients with gastric carcinoma. Zhonghua Zhong Liu Za Zhi. 1991;13:93–96. [PubMed] [Google Scholar]
- 16.Kumar BS, Tiwari SK, Manoj G, Kunwar A, Amrita N, Sivaram G, Abid Z, Ahmad A, Khan AA, Priyadarsini KI. Anti-unlcer and antimicrobial activities of sodium selenite against Helicobacter pylori: in vitro and in vivo evaluation. Scand J Infect Dis. 2010;42:266–274. doi: 10.3109/00365540903493707. [DOI] [PubMed] [Google Scholar]
- 17.Tian YX, Wang DM, Chen HB. The hepreventive effect of selenium on chronic gastritis in rats. Yingyang Xuebao. 2004:348–350+353. [Google Scholar]
- 18.Wang YL, Yang ZH, Zhu MX. Progress in researches on antiviral effect of selenium: a review. Zhongguo Gonggong Weisheng. 2021;37:1580–1584. [Google Scholar]
- 19.Guillin OM, Vindry C, Ohlmann T, Chavatte L. Selenium, Selenoproteins and Viral Infection. Nutrients. 2019;11 doi: 10.3390/nu11092101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan XJ, Wang WK, Shao SH, Liu JB. Effects of selenium on VacA and recombinanted VacA of Helicobacter pylori. Zhongguo Gonggong Weisheng. 2008:833–835. [Google Scholar]
- 21.Shao SH, Liu JB, Duan XJ, Wang H. The immunologic protection of selenium on gastric mucosal lesions of mice infected by helicobacter pylori. Yingyang Xuebao. 2005;27:394–396. [Google Scholar]
- 22.Sgouras DN, Trang TT, Yamaoka Y. Pathogenesis of Helicobacter pylori Infection. Helicobacter. 2015;20 Suppl 1:8–16. doi: 10.1111/hel.12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu W, Wise MJ, Tay CY, Windsor HM, Marshall BJ, Peacock C, Perkins T. Comparative analysis of the full genome of Helicobacter pylori isolate Sahul64 identifies genes of high divergence. J Bacteriol. 2014;196:1073–1083. doi: 10.1128/JB.01021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawamura O, Murakami M, Araki O, Yamada T, Tomizawa S, Shimoyama Y, Minashi K, Maeda M, Kusano M, Mori M. Relationship between gastric disease and deletion of cag pathogenicity island genes of Helicobacter pylori in gastric juice. Dig Dis Sci. 2003;48:47–53. doi: 10.1023/a:1021726213159. [DOI] [PubMed] [Google Scholar]
- 25.Ahmadzadeh A, Ghalehnoei H, Farzi N, Yadegar A, Alebouyeh M, Aghdaei HA, Molaei M, Zali MR, Pour Hossein Gholi MA. Association of CagPAI integrity with severeness of Helicobacter pylori infection in patients with gastritis. Pathol Biol (Paris) 2015;63:252–257. doi: 10.1016/j.patbio.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Baghaei K, Shokrzadeh L, Jafari F, Dabiri H, Yamaoka Y, Bolfion M, Zojaji H, Aslani MM, Zali MR. Determination of Helicobacter pylori virulence by analysis of the cag pathogenicity island isolated from Iranian patients. Dig Liver Dis. 2009;41:634–638. doi: 10.1016/j.dld.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yakoob J, Jafri W, Abbas Z, Abid S, Khan R, Jafri N, Ahmad Z. Low prevalence of the intact cag pathogenicity island in clinical isolates of Helicobacter pylori in Karachi, Pakistan. Br J Biomed Sci. 2009;66:137–142. doi: 10.1080/09674845.2009.11730260. [DOI] [PubMed] [Google Scholar]
- 28.Kumari R, Shariq M, Sharma S, Kumar A, Mukhopadhyay G. CagW, a VirB6 homologue interacts with Cag-type IV secretion system substrate CagA in Helicobacter pylori. Biochem Biophys Res Commun. 2019;515:712–718. doi: 10.1016/j.bbrc.2019.06.013. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Ling F, Wang H, Yu M, Zhu H, Chen C, Qian J, Liu C, Zhang Y, Shao S. The Helicobacter pylori Cag Pathogenicity Island Protein Cag1 is Associated with the Function of T4SS. Curr Microbiol. 2016;73:22–30. doi: 10.1007/s00284-016-1016-x. [DOI] [PubMed] [Google Scholar]
- 30.Huang GL, Huang ZS, Zhou XH. Research Development of Vaccine for Helicobacter pylori. Yixue Zongshu. 2016;22:866–870. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.