Through integrative analyses of mouse models and clinical data, Qin et al. find that certain ALK-rearranged LUAD hold the potential toward squamous transition. Such phenotypic transition associates with TKI resistance and relies on the JAK-STAT signaling.
Abstract
Human lung adenosquamous cell carcinoma (LUAS), containing both adenomatous and squamous pathologies, exhibits strong cancer plasticity. We find that ALK rearrangement is detectable in 5.1–7.5% of human LUAS, and transgenic expression of EML4-ALK drives lung adenocarcinoma (LUAD) formation initially and squamous transition at late stage. We identify club cells as the main cell-of-origin for squamous transition. Through recapitulating lineage transition in organoid system, we identify JAK-STAT signaling, activated by EML4-ALK phase separation, significantly promotes squamous transition. Integrative study with scRNA-seq and immunostaining identify a plastic cell subpopulation in ALK-rearranged human LUAD showing squamous biomarker expression. Moreover, those relapsed ALK-rearranged LUAD show notable upregulation of squamous biomarkers. Consistently, mouse squamous tumors or LUAD with squamous signature display certain resistance to ALK inhibitor, which can be overcome by combined JAK1/2 inhibitor treatment. This study uncovers strong plasticity of ALK-rearranged tumors in orchestrating phenotypic transition and drug resistance and proposes a potentially effective therapeutic strategy.
Introduction
Transition from one pathological type to another is a phenomenon known as lineage plasticity, which has been implicated as a mechanism of resistance to targeted therapies across multiple cancer types (Hanahan, 2022; Quintanal-Villalonga et al., 2020). In non-small-cell lung cancer, lung adenosquamous cell carcinoma (LUAS) featured with mixed adenomatous and squamous pathology, and shared oncogenic mutations represent a classical example of lineage plasticity (Lin et al., 2020; Quintanal-Villalonga et al., 2021). Human LUAS accounts for about 0.7–11.4% of non-small-cell lung cancer and display the mutations of EGFR, TP53, or the dysregulation of PI3K/AKT signaling pathway (Cheng et al., 2021; Quintanal-Villalonga et al., 2021; Wang et al., 2014). Previous studies show that LUAS has a remarkable cancer plasticity and poor prognosis (Cheng et al., 2021; Quintanal-Villalonga et al., 2021; Wang et al., 2014). Given the difficulty in collecting LUAS samples and the scarcity of comprehensive genomic analyses, the key drivers and molecular alterations that drive squamous transition remain largely unknown.
The fusion between echinoderm microtubule-associated protein-like 4 (EML4) and anaplastic lymphoma receptor kinase (ALK) is one of the most common oncogenic fusions in lung cancer (Soda et al., 2007). The ALK-rearranged lung tumors tend to exhibit aggressive behaviors and associate with high malignancy (Kim et al., 2014; Paik et al., 2012). As one of the receptor tyrosine kinase family members, ALK is well established as an important therapeutic target for tyrosine kinase inhibitor (TKI) treatment (Lin et al., 2017). Most ALK-rearranged tumors are pathologically diagnosed as lung adenocarcinoma (LUAD), and studies show that these tumors display a variety of morphological features including solid, acinar, papillary, cribriform formation, mucin production, and with signet ring cell component (Kim and Chung, 2015; Kim et al., 2013; Li et al., 2013; Popat et al., 2012; Rodig et al., 2009; Yoshida et al., 2011). Previous studies also show that some ALK-rearranged LUAD expresses the squamous biomarker p63 besides the adenomatous biomarker TTF1 (gene name as NKX2-1; Li et al., 2013; Yoshida et al., 2011). Focal squamous differentiation is observed in ALK-rearranged LUAD, albeit at low frequency (Rodig et al., 2009; Yoshida et al., 2011). Several studies also show that ALK rearrangement is observed in lung tumors with mixed adenosquamous pathology (Preusser et al., 2013; Song et al., 2017; Wang et al., 2014; Zito Marino et al., 2015). These clinical observations implicate a potential link between ALK fusion and squamous transition.
Phenotypic plasticity has been observed in relapsed LUAD patients who failed ALK inhibitor therapy. Several studies have implicated a potential link between histological transition of ALK-rearranged LUAD and ALK TKI resistance (Ball et al., 2022; Caumont et al., 2016; Cha et al., 2016; Coleman et al., 2019; Fares et al., 2020; Fujita et al., 2016; Gong et al., 2019; Kaiho et al., 2020; Kobayashi et al., 2013; Levacq et al., 2016; Miyamoto et al., 2016; Ou et al., 2017; Oya et al., 2018; Park et al., 2019; Takegawa et al., 2016; Ueda et al., 2021; Wang et al., 2020; Yang et al., 2021; Zhang et al., 2021; Zhu et al., 2017). When these LUAD patients develop drug resistance, their second biopsies show the pathology of lung squamous cell carcinoma (LUSC), small-cell lung carcinoma, large cell neuroendocrine carcinoma, or even pulmonary sarcomatoid carcinoma. Importantly, most histologically transitioned tumors still retain the ALK rearrangement as detected in the initial biopsy, indicative of potential phenotypic transition. Among these reported cases, 7 out of 21 samples are proposed to undergo the adeno-to-squamous transition (AST).
Through integrative study of human lung cancer samples, EML4-ALK genetically engineered mouse models (GEMMs), and organoid system, we here uncover the dynamic progression of squamous transition in EML4-ALK lung cancer and identify the signaling importantly regulating lineage transition. Our data also provide a potential therapeutic strategy for overcoming AST-associated drug resistance.
Results
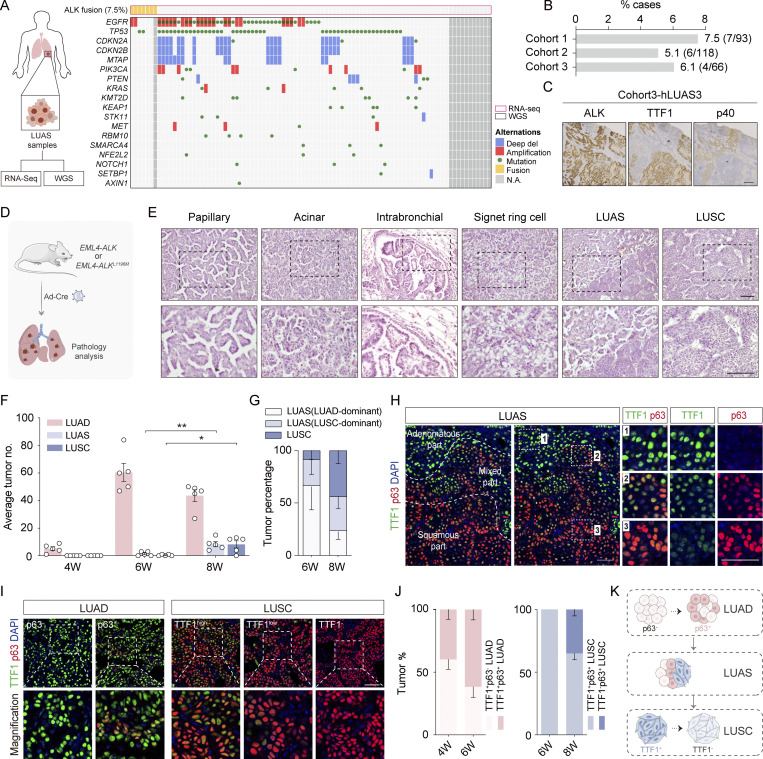
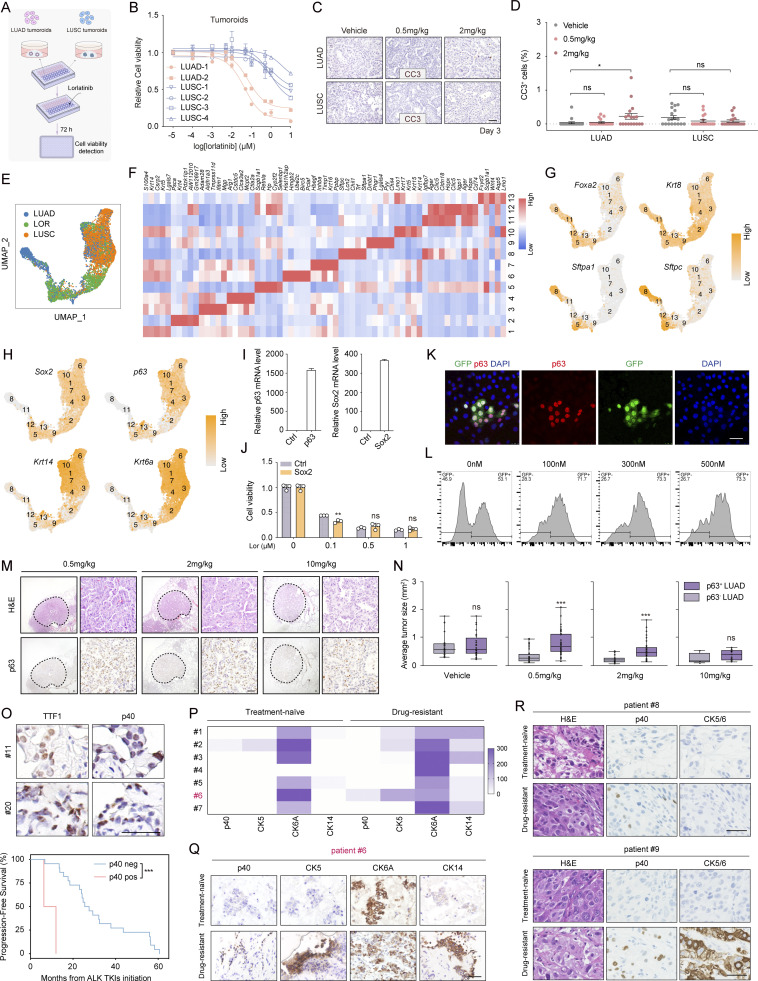
ALK rearrangements are detected in 7.5% of human LUAS specimens
Human LUAS frequently showed shared genetic alterations in adenomatous and squamous components, indicative of strong plasticity and a potential AST occurrence (Lin et al., 2020; Quintanal-Villalonga et al., 2021). Through whole genome sequencing (WGS) and RNA sequencing (RNA-seq) of Chinese LUAS specimens, we identified the mutation rates of three important oncogenic drivers EGFR mutation (39.8%, 37/93), KRAS mutation (5.4%, 5/93), and ALK fusion (7.5%, 7/93; Tang et al., 2023; Fig. 1 A). Among seven ALK fusions, six were EML4-ALK fusion, and one was KIF5B-ALK fusion. Interestingly, ALK fusion was mutually exclusive with those significantly mutated genes in human LUSC, e.g., PTEN, PIK3CA, and KMT2D, implying a potentially unique role of ALK fusion in LUAS (Fig. 1 A). To corroborate these results, we next examined another two LUAS patient cohorts (Cohort 2 containing 118 patients and Cohort 3 containing 66 patients) and found the rate of ALK rearrangements at 5.1% and 6.1%, respectively (Fig. 1 B and Fig. S1 A). Immunostaining analyses showed high ALK levels in both adenomatous and squamous components from six out of seven LUAS samples (three from Cohort 2 and four from Cohort 3; Fig. 1 C; and Fig. S1, B and C). These clinical observations collectively suggest a potential link between ALK oncogenic fusions and squamous transition.
Figure 1.
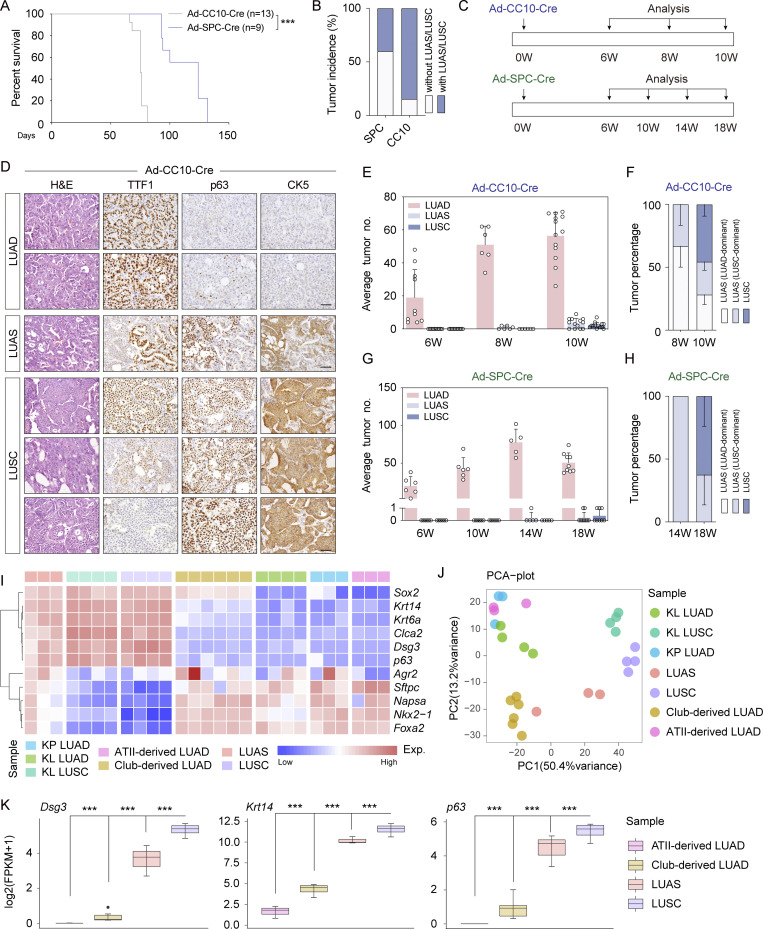
ALK fusion lung tumor heterogeneity in human LUAS and mouse models. (A) Left panel shows experimental design for WGS and RNA-seq of treatment-naïve human LUAS surgical samples (93 samples were RNA-sequenced and 81 samples were analyzed by WGS). The right panel shows the plot between somatic mutation status of significantly mutated genes from TCGA LUSC database and ALK-rearrangement events. Somatic mutations were identified through WGS analyses and gene fusion events were identified through RNA-seq data. (B) Frequency of ALK rearrangements in three different LUAS cohorts. (C) Representative ALK, p40, and TTF1 immunostaining of one human adenosquamous carcinoma tissue sample. Scale bar, 50 μm. (D) Schematic illustration of Rosa26-Loxp-Stop-Loxp-EML4-ALK and Rosa26-Loxp-Stop-Loxp-EML4-ALK L1196M mouse models. (E) Histological characteristics of ALK-rearranged tumors from EML4-ALK L1196M mice. Representative micrographs showing papillary and acinar tumors, lesions originating in intrabronchial hyperplasia, tumors with signet-ring cell pattern, adenosquamous cell carcinoma, and squamous cell carcinoma. Scale bar, 50 μm. (F) Quantification of average numbers for LUAD, LUAS, and LUSC in the EML4-ALK L1196M model. n = 5 for each time point. (G) Statistical analysis of indicated tumors in the EML4-ALK L1196M model. n = 5 for each time point. (H) Multicolor IHC staining of LUAS from the EML4-ALK L1196M model. TTF1 in green, p63 in red, nucleus in blue (DAPI staining). Scale bar, 50 μm. (I) Multicolor IHC staining of LUAD and LUSC from the EML4-ALK L1196M model. Scale bar, 50 μm. (J) Left panel shows statistical analysis of TTF1+p63−/TTF1+p63+LUAD ratio in the EML4-ALK L1196M model. Right panel shows statistical analysis of TTF1+p63+/TTF1−p63+ LUSC ratio in the EML4-ALK L1196M model. n = 5 for each time point. (K) Schematic illustration of AST process: at LUAD stage, some LUAD cells become TTF1+/p63+; at LUAS stage, LUAD cells (TTF1+/p63− or TTF1+/p63+) are mixed with LUSC cells (TTF1+/p63+); at LUSC stage, TTF1high/p63+, TTF1low/p63+, and TTF1−/p63+ LUSC cells become dominant. LUAD, LUAS, and LUSC were defined pathologically (see Materials and methods for details). Data in F, G, and J were collected from three independent experiments. *P < 0.05, **P < 0.01 by two-tailed unpaired Student’s t test (F). Data are represented as mean ± SEM. W, week.
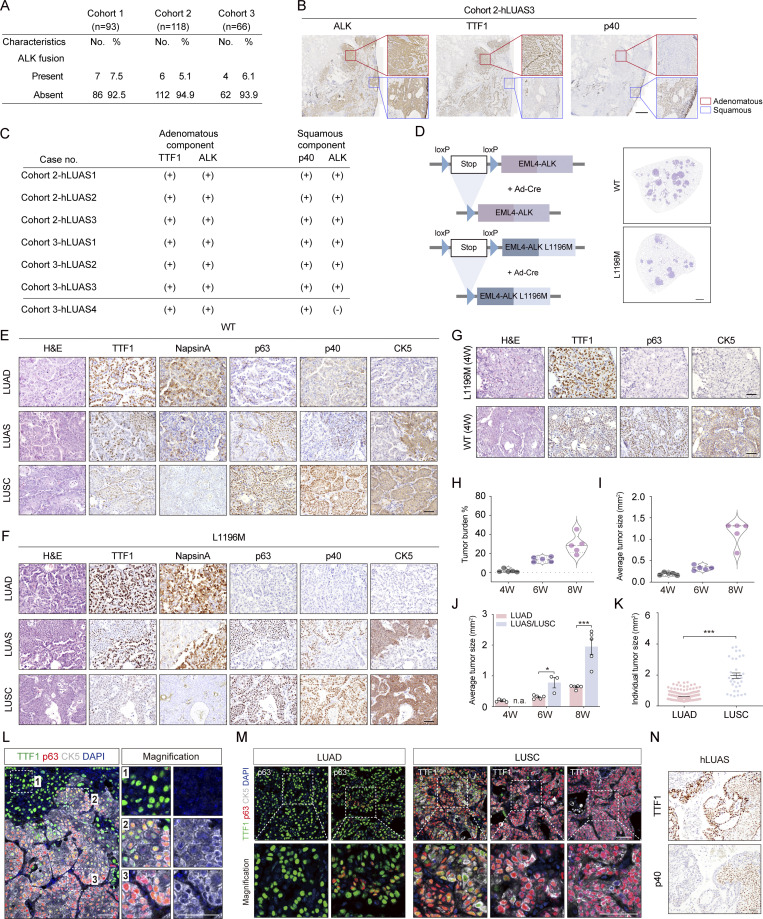
Figure S1.
ALK fusion lung tumor heterogeneity in human LUAS and mouse models. (A) Statistical analyses of ALK-rearranged samples in three different LUAS cohorts. (B) Representative ALK, TTF1, and p40 immunostaining of the adenomatous components and squamous components within one human LUAS sample in Cohort 2. Red box indicates adenomatous components; blue box indicates squamous components. Scale bar, 1 mm. (C) Statistical analysis of ALK immunostaining of adenomatous components and squamous components from the available LUAS samples (n = 7) in different LUAS cohorts. (D) Left panel shows schematic illustration of Rosa26-Loxp-Stop-Loxp-EML4-ALK and Rosa26-Loxp-Stop-Loxp-EML4-ALK L1196M mouse models. Right panel shows H&E staining of lung tumors from EML4-ALK and EML4-ALK L1196M mice at 4 wk after Ad-Cre infection. WT, the wildtype EML4-ALK mice; L1196M, the EML4-ALK L1196M mice. (E) Representative H&E staining, TTF1, NapsinA, p63, p40, and CK5 immunostaining of LUAD, LUAS, and LUSC from wt EML4-ALK model. Scale bar, 50 μm. (F) Representative H&E staining, TTF1, NapsinA, p63, p40, and CK5 immunostaining of LUAD, LUAS, and LUSC from EML4-ALK L1196M model. Scale bar, 50 μm. (G) Representative H&E staining, TTF1, p63, and CK5 immunostaining from lung tumors in the wt EML4-ALK and EML4-ALK L1196M mice treated with Ad-Cre for 4 wk (W). Scale bar, 50 μm. (H–J) Statistical analyses of tumor burden (H), average tumor size (I), and tumor size for LUAD and LUAS/LUSC (J) in EML4-ALK L1196M mice at 4 wk (n = 5), 6 wk (n = 5), and 8 wk (n = 5) after Ad-Cre treatment. (K) Quantification of individual tumor size for LUAD (n = 209) and LUSC (n = 33) in the EML4-ALK L1196M mice at 8 wk after Ad-Cre treatment. (L) Multicolor IHC staining of LUAS from the same region illustrated in Fig. 1 H. TTF1 in green, p63 in red, CK5 in white, and nucleus in blue (DAPI staining). Scale bar, 50 μm. (M) Multicolor IHC staining of LUAD and LUSC from the same region illustrated in Fig. 1 I, respectively. TTF1 in green, p63 in red, CK5 in white, nucleus in blue (DAPI staining). Scale bar, 50 μm. (N) Representative TTF1 and p40 immunostaining of one human LUAS sample. Scale bar, 50 μm. Data in H–K were collected from three independent experiments. *P < 0.05, ***P < 0.001 by multiple t test (J), two-tailed unpaired Student’s t test (K). Data are represented as mean ± SEM.
EML4-ALK drives AST in GEMMs
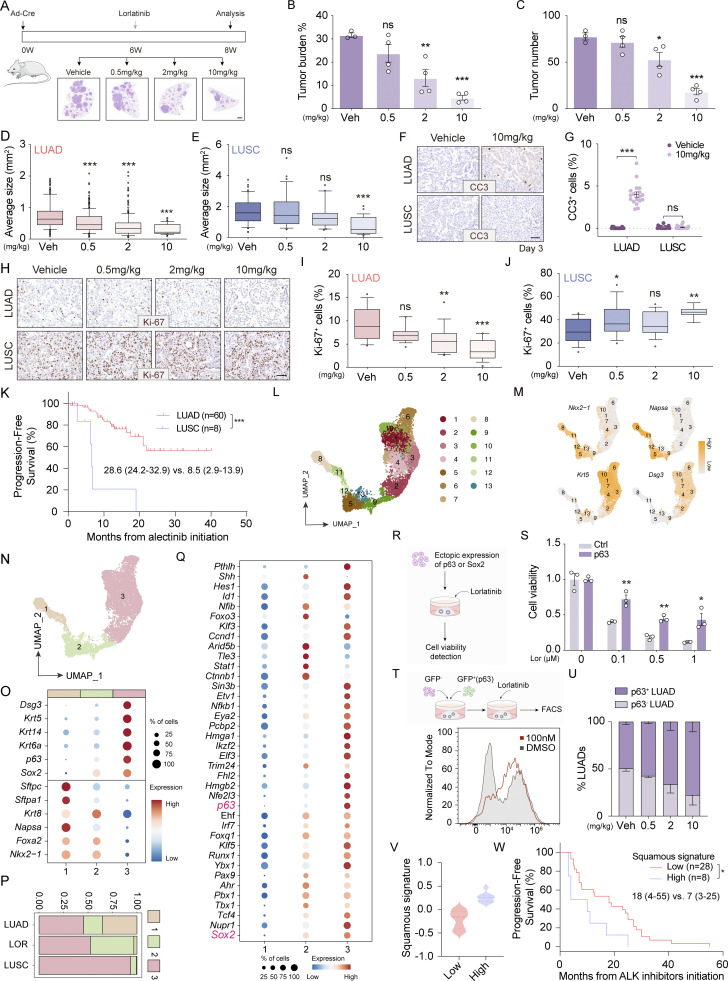
We next established two GEMMs including Rosa26-loxp-stop-loxp-wildtype (wt) EML4-ALK and Rosa26-loxp-stop-loxp-EML4-ALK L1196M with Ad-Cre nasal inhalation to test if ALK fusion can drive squamous transition (DuPage et al., 2009; Fig. 1 D and Fig. S1 D). Pathological analyses showed that most tumors displayed a classic histological pattern of LUAD, positive for adenomatous biomarkers, including TTF1 (also known as Nkx2-1) and NapsinA, and negative for squamous biomarkers cytokeratin 5 (CK5), p63, and p40 (indicative of ΔNp63 expression; Fig. S1, E and F). Consistent with previous studies (Chen et al., 2010, 2014; Maddalo et al., 2014; Pyo et al., 2017; Soda et al., 2008), these EML4-ALK LUAD displayed a variety of morphological features including papillary, acinar, intrabronchial, and with signet ring cell component (Fig. 1 E). Interestingly, we also noticed that some tumors displayed squamous pathology (Fig. 1 E; and Fig. S1, E and F). Besides, mixed pathologies in single nodules resembling human LUAS were also observed (Fig. 1 E; and Fig. S1, E and F), similar to previous findings in the well-established AST mouse model, KrasG12D; Lkb1L/L (KL; Han et al., 2014; Li et al., 2015; Zhang et al., 2017). In contrast to the wt EML4-ALK model, the EML4-ALK L1196M model developed LUSC at a relatively late time point, e.g., after 4 wk of Ad-Cre treatment (Fig. S1 G), which allows us for detailed mechanistic study. We therefore focused on the EML4-ALK L1196M mouse model for further analyses.
We next analyzed the EML4-ALK L1196M tumors at serial time points (Fig. S1, H and I). Only LUAD was detectable at 4 wk after Ad-Cre treatment whereas LUAS and a few typical LUSC began to arise at 6 wk (Fig. 1 F). At 8 wk, mixed LUAS and typical LUSC increased with a concomitant decrease of LUAD (Fig. 1 F). Consistently, a progressive trend from LUAD-dominant LUAS to LUSC-dominant LUAS to typical LUSC was observed along with cancer malignant progression (Fig. 1 G). Although LUSC arose at a late time point, their tumor sizes were evidently bigger than LUAD (Fig. S1, J and K), indicating that these nascent LUSCs were likely developed from LUAD.
Our previous studies in the KL model demonstrate that LUSC as well as the squamous components of LUAS are frequently double positive for both TTF1 and p63 (Han et al., 2014; Li et al., 2015). Using multicolor immunohistochemistry staining, we analyzed the expression of TTF1 and p63 at single-cell level in the EML4-ALK model. We observed a clear progressive pattern in LUAS tumor: the adenomatous region was positive for TTF1 only, the mixed region double positive for TTF1 and p63, and the squamous region with low TTF1 and high p63 expression (Fig. 1 H and Fig. S1 L). We also found that some LUAD were double positive for TTF1 and p63 (Fig. 1 I and Fig. S1 M). Moreover, the p63-positive LUSC exhibited various TTF1 expression patterns from negative, low to high (Fig. 1 I and Fig. S1 M). Statistical analysis revealed that the TTF1+/p63+ LUAD increased at 6 wk after Ad-Cre treatment and the TTF1−/p63+ LUSC increased at 8 wk after Ad-Cre treatment (Fig. 1 J). In agreement with these findings in mouse, we observed TTF1 and p40 double positivity in the squamous component of human ALK-rearranged LUAS (Fig. S1 N). We therefore proposed a three-stage AST evolution model: from LUAD to LUAS and eventually to LUSC. At the initial stage, some LUAD cells become TTF1+/p63+; at the intermediate stage, LUAD cells (TTF1+/p63− or TTF1+/p63+) were mixed with LUSC cells (TTF1+/p63+); at the final stage, TTF1high/p63+, TTF1low/p63+, and TTF1−/p63+ LUSC cells become dominant (Fig. 1 K).
Single-cell RNA-seq (scRNA-seq) uncovers the evolution route of AST
We further performed scRNA-seq using dissected mouse LUAD and LUSC (Fig. 2 A; and Fig. S2, A and B). We observed an increase of neutrophils with concurrent decrease of T cells in EML4-ALK LUSC (Fig. S2, C and D), and further confirmed this via flow cytometry analysis (Fig. S2, E–H), which is consistent with previous findings in KL model (Koyama et al., 2016). Uniform manifold approximation and projection (UMAP) analysis divided cancer cells (the epithelial subpopulation) into 13 clusters with distinct expression patterns (Fig. 2, B and C; and Fig. S2 I). We found that the LUAD-related genes including Sftpb, Foxa2, Nkx2-1, Napsa, and Sftpc were highly expressed in cluster 1–3, indicative of an LUAD-like state (Fig. 2, D–F; and Fig. S2 J). Interestingly, cluster 3 showed a notable decrease of Nkx2-1 expression (Fig. 2 F), and we therefore termed this cluster as the Nkx2-1low LUAD. The LUSC-related genes such as Krt5, Krt14, Dsg3, and Krt6a were enriched in clusters 7–13, indicative of a LUSC-like state (Fig. 2, D–F; and Fig. S2 J). In contrast, clusters 4–6 expressed both LUAD- and LUSC-related genes, indicative of an intermediate state (Fig. 2, D–F; and Fig. S2 J). Monocle trajectory analysis further confirmed the intermediate state (cluster 4–6) along with lineage switch from LUAD-like state to LUSC-like state (Fig. 2 G). Visualization of gene expression along the transition trajectory showed that Sftpc was the first decreased adenomatous biomarker whereas the Krt6a increased earlier than most squamous biomarkers (Fig. 2 H and Fig. S2 K). Krt8, a transitional-state biomarker from human alveolar type 2 cells (hAEC2s) to basal cell transition process (Kathiriya et al., 2022), showed slight upregulation at the intermediate state (Fig. 2 H and Fig. S2 K). These data uncover a potential transition trajectory of ALK fusion tumors at single-cell level.
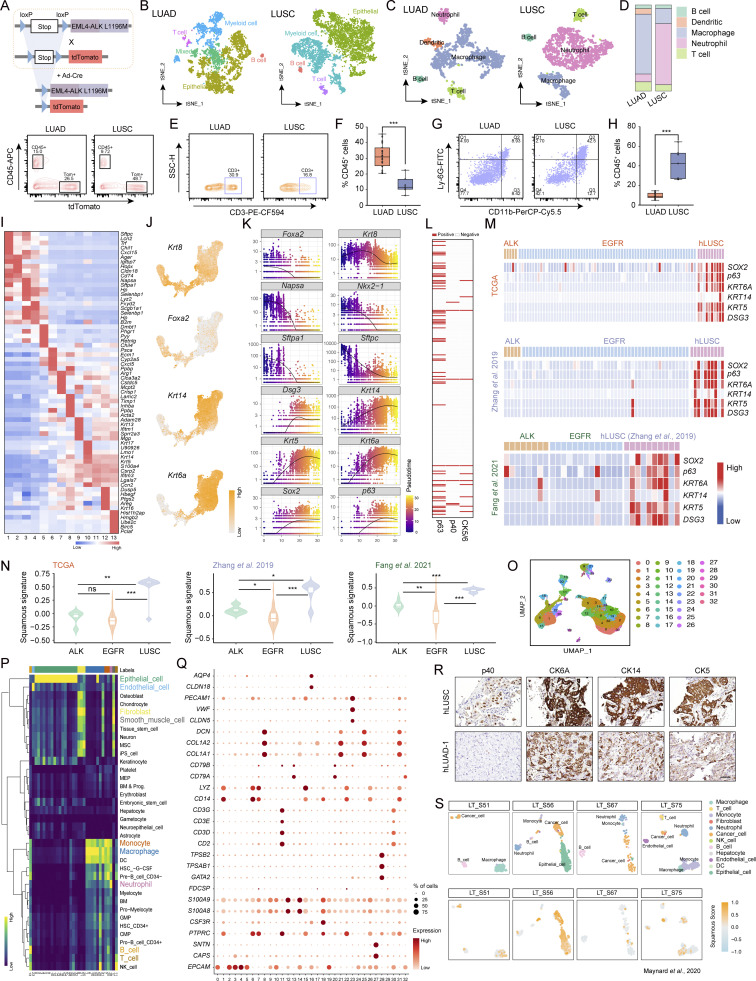
Figure 2.
Single-cell analyses of mouse and human ALK fusion tumors reveal the evolution route of AST. (A) Scheme depicting experimental design for scRNA-seq of mouse LUAD and LUSC. (B) UMAP visualization of epithelial cells (cancer cells) derived from LUAD and LUSC. (C) UMAP visualization of LUAD and LUSC cells labeled with Seurat clusters. (D) Marker gene expression across various clusters. Dot diameter indicates the proportion of cells expressing a given gene; color indicates the expression level. (E) Feature plots of LUAD-related genes (Nkx2-1, Sftpc) and LUSC-related genes (p63, Krt5). (F) Dot plot showing expression of LUAD and LUSC-related genes in each cluster. Dot diameter indicates the proportion of cells expressing a given gene; color indicates the expression level. (G) Pseudotime ordering of cancer cells using Monocle3. (H) Biomarker gene expressional dynamics over pseudotime trajectory. The curves were fitted with the Lowess method. (I) Comparison of squamous expression signature from bulk RNA-seq in 41 human (h) EML4-ALK LUAD samples and 1 human EML4-ALK LUSC sample. The bulk RNA-seq data of 10 human LUSC were downloaded from Zhang et al. (2019b). (J) Statistical analyses of p63, p40, and CK5/6 immunostaining of ALK-positive LUAD samples. (K) Scheme depicting experimental design for scRNA-seq of ALK-rearranged human LUAD biopsies. (L) UMAP visualization of cells from five patients (two LUSC samples and three ALK-rearranged LUAD samples) with cells colored based on the cell types (upper row) and squamous score using the well-established squamous biomarker genes (lower row). The minimum score is indicated by light gray and the maximum score is indicated by yellow. (M) Heat maps showing expression of LUAD-related genes (FOXA2, KRT8, SFTPC, NKX2-1, NAPSA, and SFTPA1) and LUSC-related genes (SOX2, p63, KRT5, KRT14, CLCA2, KRT6A, and DSG3) in human cancer cell clusters from scRNA-seq data.
Figure S2.
scRNA-seq of mouse and human ALK fusion tumors uncovers the evolution route of AST. (A) Top panel shows schematic illustration of tdTomato; EML4-ALK L1196M mouse model. Bottom panel shows flow cytometry sorting of CD45+ immune cells and tdTomato+ cancer cells for scRNA-seq. (B) t-Distributed Stochastic Neighbor Embedding (t-SNE) visualization of mouse LUAD and LUSC labeled by cell types annotated with LUSCancer package. (C) t-SNE visualization of immune cell types of LUAD and LUSC. Following markers were used for identification of immune subtypes: immune cell (Ptprc), B cell (Cd19, Cd79a), T cell (Cd3d, Cd3e), macrophage (Cd14, Fogr2b), dendritic (Cd22, Fscn1), neutrophil (S100a8, S100a9). (D) Bar plot showing distribution of immune subtypes in mouse LUAD and LUSC. (E) Flow cytometry analysis of T cells (CD45+CD3+) in LUAD and LUSC. (F) Quantification of T cell populations in LUAD (n = 22) and LUSC (n = 7) by flow cytometry. (G) Flow cytometry analysis of neutrophils (CD45+CD11b+Ly-6G+) in LUAD and LUSC. (H) Quantification of neutrophil populations in LUAD (n = 22) and LUSC (n = 7) by flow cytometry. (I) Heatmap showing mean expression of top differentially expressed genes in each epithelial cell cluster. (J) Feature plots of known LUAD-related genes (Krt8, Foxa2) and LUSC-related genes (Krt14, Krt6a). (K) Expression levels of LUAD- and LUSC-related genes over pseudotime trajectory. (L) Heatmap of IHC staining of p63, p40, and CK5/6 for 206 human LUAD samples with ALK rearrangements. (M) Comparison of squamous expression signatures from bulk RNA-seq in human EML4-ALK LUAD samples, human EGFR LUAD samples, and human LUSC samples. The bulk RNA-seq data were downloaded from the TCGA database and studies (Zhang et al., 2019b; Fang et al., 2021). (N) ssGSEA score of squamous signature among human EML4-ALK LUAD samples, human EGFR LUAD samples, and human LUSC samples. (O) UMAP visualization of scRNA-seq data labeled with Seurat clusters. (P) Heat map displays the scores for all cells across all reference labels annotated with SingleR. DC, dendritic cell; NK cell, natural killer cell. (Q) Expression level for markers of different cell types across various Seurat clusters. Dot diameter indicates the proportion of positive cells. Color indicates the expression level. (R) Representative p40, CK6A, CK14, and CK5 immunostaining of samples from sample hLUAD-1. One LUSC sample was used as a positive control. Scale bar, 50 μm. (S) UMAP visualization of cells of the single-cell sequencing data (Maynard et al., 2020) from four ALK-rearranged LUAD with cells colored based on the cell types and squamous score using the well-established squamous biomarker genes. The minimum score is indicated by blue; the maximum score is indicated by yellow. Data in E–H represent one experiment of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 by two-tailed unpaired Student’s t test (F and H), Wilcoxon rank sum test (N). ns: not significant. Data are represented as mean ± SEM.
To validate these mouse model findings, we further analyzed a cohort of 687 ALK-rearranged lung cancer samples including 42 samples (41 LUAD samples and 1 LUSC sample) for RNA-seq analyses and 645 LUAD for immunostaining of squamous biomarkers. RNA-seq data analyses demonstrated that 24.4% (10/41) LUAD exhibited squamous signature to variable extents (Zhang et al., 2019b; Fig. 2 I). Consistently, 27.7% (84/303) LUAD were positive for p63 antibody (Fig. 2 J). Moreover, we found that 4% (15/379) LUAD were positive for p40 antibody and 12% (36/301) were positive for CK5/6 (Fig. 2 J). Among the 206 LUAD with three available staining data, four exhibited triple-positivity (Fig. S2 L).
Analyses of two public Chinese RNA-seq datasets also revealed an increased enrichment of squamous signature in ALK-rearranged LUAD in contrast to EGFR-mutant LUAD (Fang et al., 2021; Zhang et al., 2019b), which was not observed in The Cancer Genome Atlas (TCGA) dataset (Fig. S2, M and N). We further performed scRNA-seq of three Chinese ALK-rearranged LUAD biopsies and analyzed them together with two Chinese LUSC samples (Fig. 2, K and L). UMAP analysis classified different cell clusters into nine cell types with canonical markers (Zhang et al., 2019a; Fig. S2, O–Q). We found that hLUAD-1 was enriched with squamous signature (Fig. 2 L), e.g., its cluster 5 showed a notable increase of KRT6A, KRT5, and KRT14 (Fig. 2 M). Positive staining of CK6A, CK5, and CK14 were also observed in hLUAD-1 (Fig. S2 R). The enrichment of squamous signature was also found in another published scRNA-seq dataset (Maynard et al., 2020; Fig. S2 S). These data demonstrate that certain ALK-rearranged LUAD might undergo the process of squamous transition even pathologically resembling adenocarcinoma.
CC10+ cells serve as the major cell-of-origin for squamous transition
Previous studies in KL mouse model have identified alveolar type II (ATII) cells expressing surfactant protein C (Sftpc or SPC) or club cells expressing secretoglobin 1a1 (Scgb1a1 or CC10) as the cell-of-origin for AST (Han et al., 2014; Li et al., 2015; Zhang et al., 2017). To test this in EML4-ALK model, we generated the Sftpc-CreERT2; EML4-ALK L1196M and Scgb1a1-CreERT2; EML4-ALK L1196M mice for lineage tracing experiments (Fig. S3 A). Unexpectedly, these two mouse models spontaneously developed tumors even without tamoxifen administration (Fig. S3 B), likely due to genetic leakage. Nonetheless, we observed LUAD, LUAS as well as LUSC in the Scgb1a1-CreERT2; EML4-ALK L1196M mice but only LUAD in the Sftpc-CreERT2; EML4-ALK L1196M mice (Fig. S3, C and D). This indicates that club cells might serve as the potential cell-of-origin for AST in the EML4-ALK model.
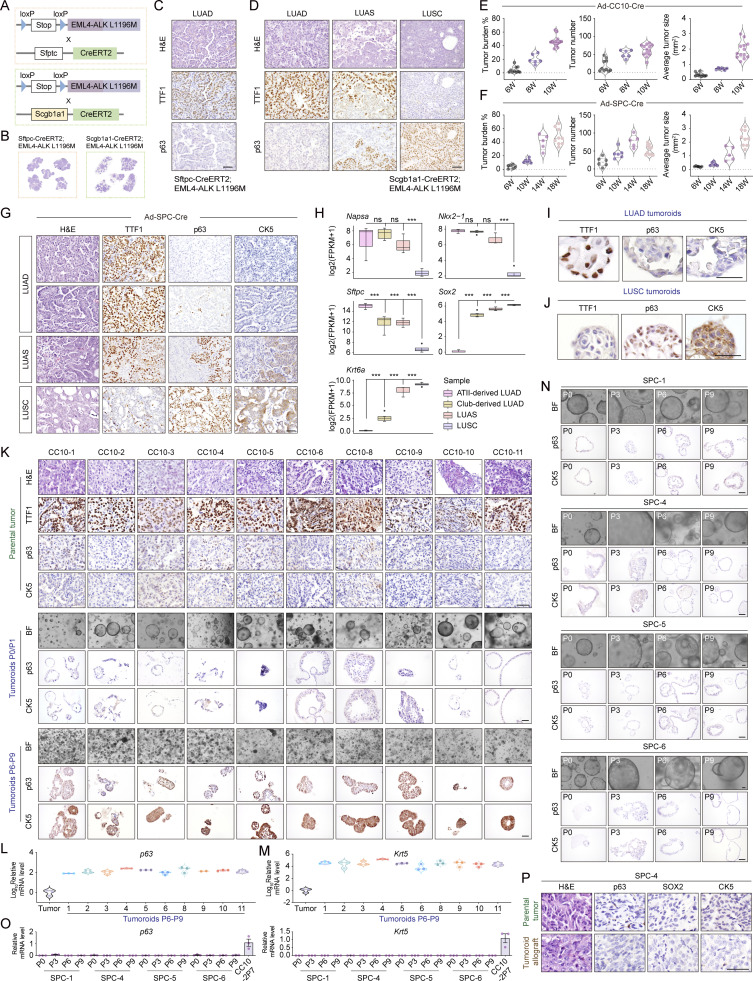
Figure S3.
Club cells serve as the major cell-of-origin of AST. (A) Schematic illustration of the Sftpc-CreERT2; EML4-ALK L1196M or Scgb1a1-CreERT2; EML4-ALK L1196M model. (B) Representative H&E staining of Sftpc-CreERT2; EML4-ALK L1196M or Scgb1a1-CreERT2; EML4-ALK L1196M mice tumors without tamoxifen induction. (C) Representative H&E staining, TTF1, and p63 immunostaining from lung tumors in the Sftpc-CreERT2; EML4-ALK L1196M mice without tamoxifen treatment. Scale bar, 50 μm. (D) Representative H&E staining, TTF1, and p63 immunostaining from lung tumors in the Scgb1a1-CreERT2; EML4-ALK L1196M mice without tamoxifen treatment. Scale bar, 50 μm. (E) Statistical analyses of tumor burden, tumor number, and average tumor size of EML4-ALK L1196M mice at 6 wk (W; n = 10), 8 wk (n = 6), and 10 wk (n = 13) after Ad-CC10-Cre treatment. (F) Statistical analyses of tumor burden, tumor number, and average tumor size of EML4-ALK L1196M mice at 6 wk (n = 6), 10 wk (n = 6), 14 wk (n = 5), and 18 wk (n = 9) after Ad-SPC-Cre treatment. (G) Representative H&E staining, TTF1, p63, and CK5 immunostaining of LUAD, LUAS, and LUSC from EML4-ALK L1196M mice after Ad-SPC-Cre treatment for 18 wk. Scale bar, 50 μm. (H) Expression (FPKM) comparison of LUAD and LUSC signature genes among ATII cell–derived LUAD, club cell–derived LUAD, LUAS, and LUSC samples. (I and J) Representative TTF1, p63, and CK5 immunostaining of tumoroids derived from LUAD (I) and LUSC (J) in the EML4-ALK L1196M model. Scale bar, 50 μm. (K) Representative H&E staining, TTF1, p63, and CK5 immunostaining from parental tumors and club cell–derived plastic LUAD tumoroids at early (P0/P1) and late (P6–P9) passages. Scale bar, 50 μm. (L and M) Real-time PCR detection of mRNA levels for p63 (L) and Krt5 (M) of club cell–derived plastic LUAD tumoroids at P6-P9. Tumor, the parental tumor (LUAD) used for organoid culture. (N) Representative photos for bright field (BF), p63, and CK5 immunostaining of four tumoroids (SPC-1, SPC-4, SPC-5, and SPC-6) derived from ATII cells at indicated passages. Scale bar, 50 μm. (O) Real-time PCR detection of p63 and Krt5 mRNA levels of four tumoroids derived from ATII cells. CC10-2P7, club cell–derived LUAD tumoroid #2 at passage 7. (P) Representative H&E staining, p63, SOX2, and CK5 immunostaining of primary tumor and allograft tumor of the ATII cell–derived non-plastic tumoroids (SPC-4). Scale bar, 50 μm. Data in E and F were collected from four independent experiments. Data in K and N were collected from three independent experiments. Data in L, M, and O represent one experiment of three independent experiments. ***P < 0.001 by DEseq2 (H). ns: not significant. Data are represented as mean ± SEM.
We further took advantage of adenovirus specifically targeting either club cells or ATII cells using Ad-CC10-Cre or Ad-SPC-Cre (Sutherland et al., 2011). In contrast to the Ad-SPC-Cre group, the Ad-CC10-Cre group showed a shorter survival (Fig. 3 A), frequent LUSC development (Fig. 3 B), and the progressive pattern from LUAD to LUAS to LUSC starting from 8 wk after viral treatment (Fig. 3, C–E; and Fig. S3 E). Moreover, we detected a progressive trend from LUAD-dominant LUAS to LUSC-dominant LUAS to typical LUSC in the Ad-CC10-Cre group (Fig. 3 F). In contrast, the Ad-SPC-Cre group showed very few LUSC even after 18 wk of viral treatment (Fig. 3, G and H; and Fig. S3, F and G).
Figure 3.
Club cells serve as the main cell-of-origin for AST in EML4-ALK model. (A) Kaplan–Meier curve shows the overall survival of the EML4-ALK L1196M mice receiving Ad-SPC-Cre (n = 9) or Ad-CC10-Cre (n = 13). (B) Statistical analysis of LUAS/LUSC incidence in the EML4-ALK L1196M mice after Ad-SPC-Cre (n = 9) or Ad-CC10-Cre (n = 13) treatment for 18 or 10 wk, respectively. (C) Scheme depicting experimental design for analysis of Ad-SPC-Cre or Ad-CC10-Cre treated mice at serial time points. W, week. (D) Representative H&E staining and immunostaining for TTF1, p63, and CK5 of LUAD, LUSC, and LUAS from the EML4-ALK L1196M mice at 10 wk after Ad-CC10-Cre treatment. Scale bar, 50 μm. (E) Quantification of average numbers of indicated tumors in the EML4-ALK L1196M mice at 6 wk (W; n = 10), 8 wk (n = 6), and 10 wk (n = 13) after Ad-CC10-Cre treatment. (F) Statistical analysis of indicated tumors from the EML4-ALK L1196M mice at 8 wk (n = 6) and 10 wk (n = 13) after Ad-CC10-Cre treatment. (G) Quantification of average numbers indicated tumors in the EML4-ALK L1196M mice at 6 wk (n = 6), 10 wk (n = 6), 14 wk (n = 5), and 18 wk (n = 9) after Ad-SPC-Cre treatment. (H) Statistical analysis of indicated tumors from the EML4-ALK L1196M mice at 14 wk (n = 5) and 18 wk (n = 9) after Ad-SPC-Cre treatment. (I) Heat maps showing gene signatures across various tumors from multiple mouse models. (J) PCA analysis of various tumors from multiple mouse models. (K) Expression (FPKM) comparison of Dsg3, Krt14, and p63 across ATII cell–derived LUAD, club cell–derived LUAD, LUAS, and LUSC from EML4-ALK L1196M mice. Data in A, B, and E–H were collected from four independent experiments. ***P < 0.001 by DEseq2 (K). Data are represented as mean ± SEM.
We next comparatively analyzed the gene expression profiling of ATII cell–derived LUAD and club cell–derived tumors including LUAD, LUAS, and LUSC together with lung tumors from KrasG12D; P53L/L (KP), and KL models (Fang et al., 2023). The EML4-ALK LUSC showed strong expression of squamous signature genes comparable with KL LUSC (Fig. 3 I). The principal component analysis (PCA) showed that club cell–derived EML4-ALK LUAD were proximal to LUAS, whereas ATII cell–derived LUAD were proximal to KP LUAD and KL LUAD (Fig. 3 J). In contrast, club cell–derived LUAD showed increased expression of squamous biomarkers including Sox2, Dsg3, p63, Krt14, and Krt6a (Fig. 3 K and Fig. S3 H). These findings collectively identify club cells as the major cell-of-origin for AST in the EML4-ALK model.
Organoid system recapitulates the AST process
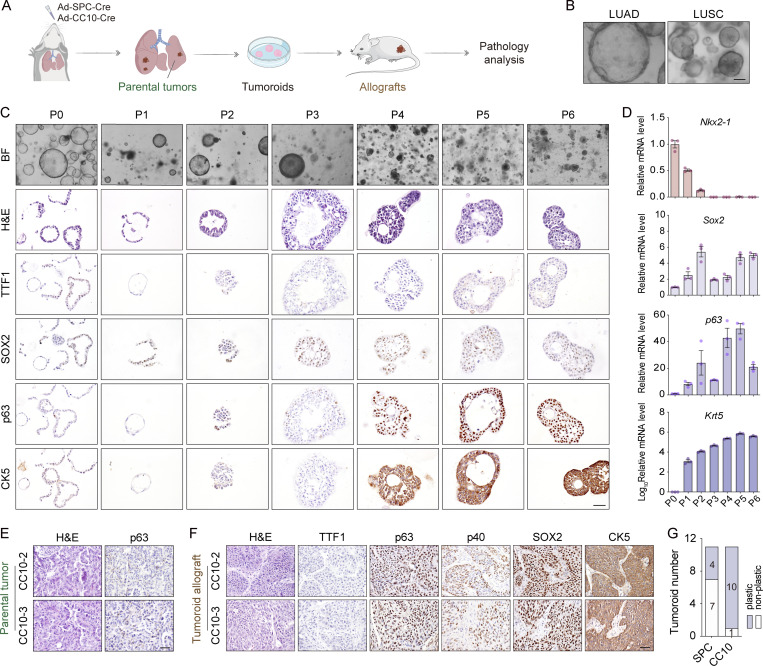
The organoid system is known to closely recapitulate tumor malignant progression, which allows for detailed dissection of molecular mechanisms and potential development of effective therapeutic strategies (Clevers, 2016). We then dissected the EML4-ALK LUAD for organoid culture to recapitulate the AST process (Fig. 4 A). Meanwhile, we also cultured primary LUSC in the organoid system. We found that the tumoroids derived from LUAD or LUSC showed different morphologies, e.g., LUAD tumoroids maintained vacuole morphology and expressed TTF1, whereas LUSC tumoroids formed solid spheres and expressed p63 and CK5 (Fig. 4 B; and Fig. S3, I and J). Despite the initial p63−/CK5− expression pattern, most club cell–derived LUAD tumoroids became p63+/CK5+ and showed solid sphere morphology at late passages (Fig. S3, K–M). We therefore referred these samples to plastic tumoroids. Immunostaining as well as real-time PCR analyses of serial passaging tumoroids also confirmed the trend of gradual increase of squamous biomarkers with concomitant decrease of adenomatous biomarkers (Fig. 4, C and D). In contrast, most ATII cell–derived tumoroids maintained the vacuole morphology without squamous biomarker expression even after long-term culture (Fig. S3, N and O). When transplanted into severe combined immunodeficiency (SCID) mice, the club cell–derived tumoroids with solid sphere morphology displayed typical LUSC characteristics, whereas the allograft tumors from ATII cell–derived tumoroids mainly displayed LUAD pathology without squamous biomarker expression (Fig. 4, E and F; and Fig. S3 P). The squamous transition penetrance for club cell–derived LUAD is ∼90% (10/11; Fig. 4 G), further supporting our lineage tracing data about the cell-of-origin for AST.
Figure 4.
Establishment of an organoid model recapitulating the AST process. (A) Schematic illustration of the organoid culture system. Primary tumors from the EML4-ALK L1196M model were dissected for organoid culture. (B) Representative photos for the LUAD or LUSC tumoroids. Scale bar, 50 μm. (C) Representative photos, H&E staining, and immunostaining for TTF1, SOX2, p63, and CK5 in club cell–derived tumoroid (CC10-3). Scale bar, 50 μm. P, passage; BF, bright field. (D) Real-time PCR detection of Nkx2-1, Sox2, p63, and Krt5 mRNA levels in club cell–derived LUAD tumoroid (CC10-3) at indicated passages. (E) Representative H&E staining and p63 immunostaining for primary tumors from Ad-CC10-Cre–treated group used for organoid culture (CC10-2 and CC10-3). Scale bar, 50 μm. (F) Representative H&E staining and immunostaining for TTF1, p63, p40, SOX2, and CK5 of allograft tumors of club cell–derived LUAD tumoroids (CC10-2 and CC10-3). Scale bar, 50 μm. (G) Statistical analysis of squamous transition incidence of LUAD tumoroids from Ad-SPC-Cre (n = 11) or Ad-CC10-Cre (n = 11) treatment. Data in G were collected from three independent experiments. Data are represented as mean ± SEM.
EML4-ALK phase separation activates the JAK-STAT signaling to regulate AST
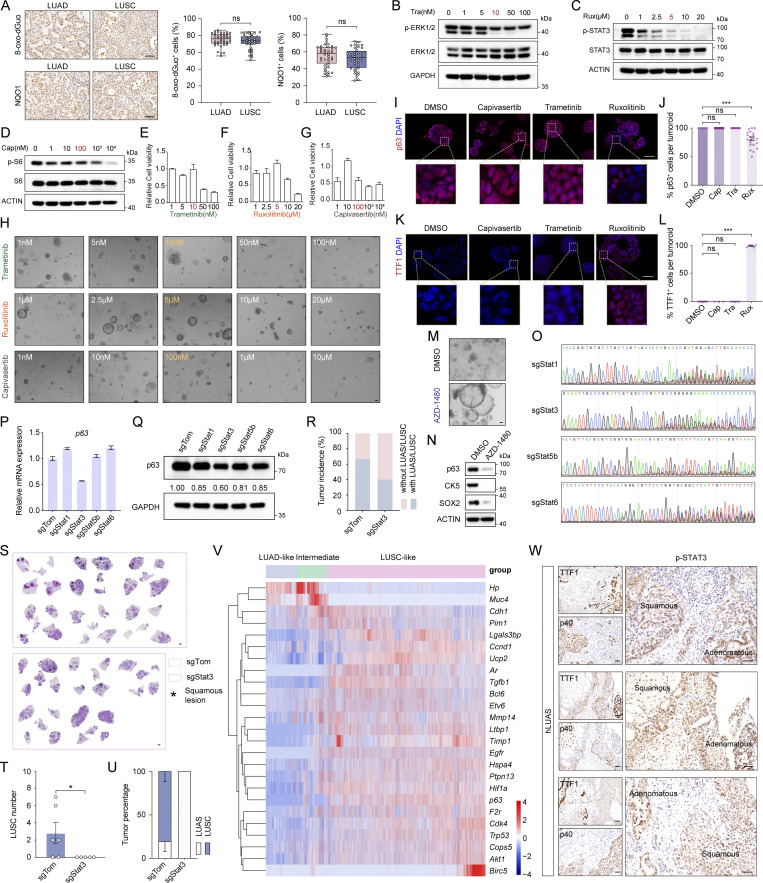
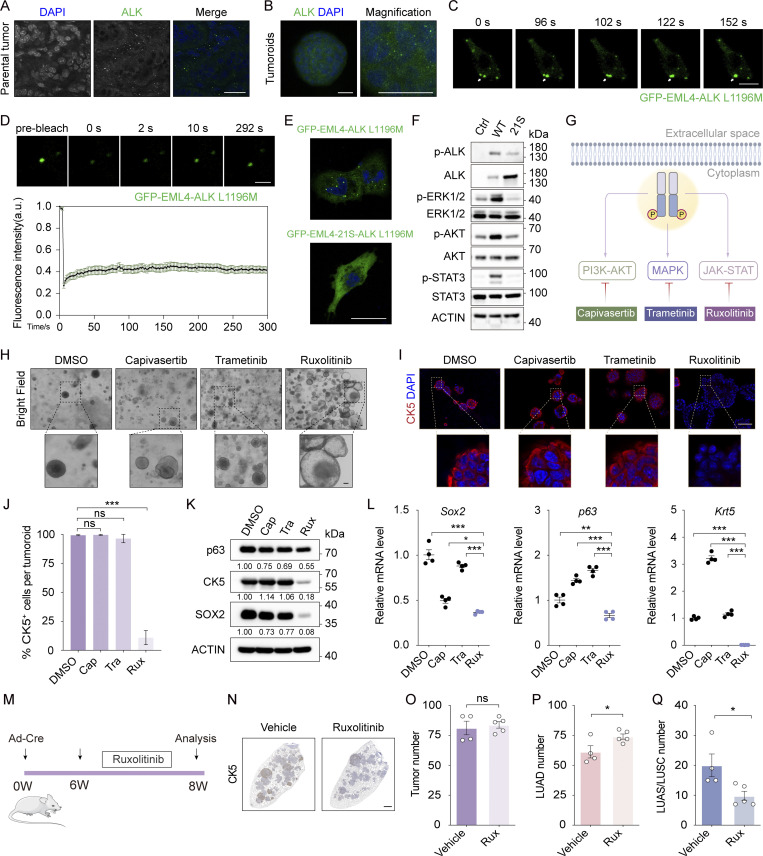
We have previously identified an important role of ROS in regulating AST in the KL model (Li et al., 2015). However, LUAD and LUSC in the EML4-ALK model showed no difference of 8-oxo-dGuo and/or NQO1 levels (Fig. S4 A), indicative of a distinct mechanism for squamous transition. We then asked how EML4-ALK regulates the AST process. We and others have previously shown that wt EML4-ALK fusion forms cytoplasmic condensates to fire downstream signaling (Qin et al., 2021; Sampson et al., 2021; Tulpule et al., 2021). Through immunofluorescence staining analyses, we found a notable protein condensate formation in both EML4-ALK L1196M tumors and tumoroids (Fig. 5, A and B). Through living cell imaging analyses and fluorescence recovery after photobleaching (FRAP) assays in human bronchial epithelial cell line BEAS-2B, we found that the GFP-EML4-ALK L1196M condensates had the liquid property and underwent dynamic exchange (Fig. 5, C and D). Similar to our previous study (Qin et al., 2021), the 21S mutant, which replaced 9 tyrosine and 12 phenylalanine with serine in EML4, significantly disrupted phase separation of EML4-ALK L1196M (Fig. 5 E). Importantly, the 21S mutant displayed reduced activation of STAT3, ERK1/2, and AKT (Fig. 5 F), indicative of the importance of phase separation in triggering downstream signaling.
Figure S4.
Inhibition of the JAK-STAT pathway blocks the AST process. (A) Representative immunostaining and statistical analyses of 8-oxo-dGuo+ and NQO1+ cells in LUAD and LUSC from EML4-ALK L1196M mice. Scale bar, 50 μm. 40 representative images for each group were counted. (B–D) Western blot analyses of p-ERK1/2 (B), p-STAT3 (C), and p-S6 (D). Club cell–derived EML4-ALK L1196M tumoroids were treated with indicated doses of trametinib (Tra), ruxolitinib (Rux), and capivasertib (Cap) for 12 h and then subjected to western blot analyses. (E–G) Cell viability of club cell–derived LUAD tumoroids treated with indicated doses of trametinib (E), ruxolitinib (F), and capivasertib (G) for 72 h. (H) Representative morphology photos of club cell–derived LUAD tumoroids at early passages (p0/1) treated with indicated doses of capivasertib, trametinib, and ruxolitinib for 4 days. Scale bar, 50 μm. (I) Representative p63 immunofluorescence staining for club cell–derived LUAD tumoroids treated with capivasertib (100 nM), trametinib (10 nM), ruxolitinib (5 μM), or DMSO for six passages. p63 in red; nucleus in blue (DAPI staining). Scale bar, 50 μm. (J) Statistical analysis of p63+ cell ratio per tumoroid. Tumoroids analyzed for DMSO, capivasertib, trametinib, and ruxolitinib groups were 8, 44, 12, and 21, respectively. (K) Representative TTF1 immunofluorescence staining for club cell–derived LUAD tumoroids treated with capivasertib (100 nM), trametinib (10 nM), ruxolitinib (5 μM), or DMSO for six passages. TTF1 in red; nucleus in blue (DAPI staining). Scale bar, 50 μm. (L) Statistical analysis of TTF1+ cell ratio per tumoroid. Tumoroids analyzed for DMSO, capivasertib, trametinib, and ruxolitinib groups were 9, 3, 6, and 6, respectively. (M) Representative morphology photos of club cell–derived LUAD tumoroids treated with 5 μM AZD-1480 for six passages. Scale bar, 50 μm. (N) Western blot analyses of p63, CK5, and SOX2. Club cell–derived EML4-ALK L1196M tumoroids were treated with AZD-1480 (5 μM) or DMSO for six passages and then subjected to western blot analyses. (O) Representative chromogram sequences showing knockout efficiency of sgStat1, sgStat3, sgStat5b, and sgStat6 in club cell–derived plastic EML4-ALK L1196M tumoroids. (P) Relative mRNA levels of p63 in club cell–derived EML4-ALK L1196M plastic tumoroids with or without Stat1, Stat3, Stat5b, or Stat6 knockout. (Q) Protein levels of p63 in club cell–derived plastic EML4-ALK L1196M tumoroids with or without Stat1, Stat3, Stat5b, or Stat6 knockout. (R) Quantification of LUAS/LUSC incidence in the EML4-ALK L1196M model with or without Stat3 knockout. n = 6 for control group (sgTomato, sgTom). n = 5 for sgStat3 group. (S) Representative whole lungs of Stat3 knockout (sgStat3) and control (sgTom) in the EML4-ALK L1196M model. Scale bar, 1 mm. (T) Quantification of LUSC number. (U) Statistical analysis of LUAS and LUSC in the EML4-ALK L1196M model with or without Stat3 knockout. (V) Heat map showing the expression dynamics of multiple STAT3 target genes from TRRUST database (https://www.grnpedia.org/trrust/) in LUAD-like, intermediate, and LUSC-like states (FDR<0.01) according to scRNA-seq data analyses. (W) Representative p-STAT3, TTF1, and p40 immunostaining of the adenomatous components and squamous components within human LUAS samples. Scale bar, 50 μm. Data in A–Q represent one experiment of two independent experiments. Data in R–U were collected from two independent experiments. *P < 0.05, ***P < 0.001 by two-tailed unpaired Student’s t test (A), one-way ANOVA (J and L), one-tailed unpaired Student’s t test (T). ns: not significant. Data are represented as mean ± SEM. Source data are available for this figure: SourceData FS4.
Figure 5.
EML4-ALK phase separation regulates AST through activating JAK-STAT signaling. (A) Immunofluorescence staining in club cell–derived EML4-ALK L1196M tumors. ALK in green, nucleus in blue (DAPI staining). Scale bar, 25 μm. (B) Immunofluorescence staining of club cell–derived EML4-ALK L1196M tumoroids. ALK in green, nucleus in blue (DAPI staining). Scale bar, 25 μm. (C) Human bronchial epithelial BEAS-2B cells were transfected with GFP-EML4-ALK L1196M for 12 h and GFP fluorescence was monitored through live imaging. Snapshots at indicated time points showed the fusion event. Scale bar, 10 μm. (D) Top panel shows representative FRAP images of GFP-EML4-ALK L1196M condensates in BEAS-2B cells. The images were taken before and after photobleaching. Scale bar, 2 μm. Bottom panel shows FRAP recovery curve of GFP-EML4-ALK L1196M condensates in BEAS-2B cells. n = 13. (E) BEAS-2B cells were transfected with GFP-EML4-ALK L1196M or GFP-EML4-21S-ALK L1196M and analyzed via confocal microscopy. Scale bar, 25 μm. (F) Western blot analyses of AKT, ERK1/2, and STAT3 phosphorylation. BEAS-2B cells stably expressing EML4-ALK L1196M or EML4-21S-ALK L1196M were deprived of serum and glucose for 2 h and then subjected to western blot analyses. Ctrl, control; WT, GFP-EML4-ALK L1196M; 21S, GFP-EML4-21S-ALK L1196M. (G) Schematic illustration of EML4-ALK downstream signaling pathways and their corresponding inhibitors. (H) Bright field photos of club cell–derived EML4-ALK L1196M tumoroids treated with DMSO, capivasertib (100 nM), trametinib (10 nM), and ruxolitinib (5 μM) for six passages. Scale bar, 50 μm. (I) Representative CK5 immunofluorescence staining in club cell–derived EML4-ALK L1196M tumoroids. CK5 in red, nucleus in blue (DAPI staining). Scale bar, 50 μm. (J) Statistical analysis of CK5+ cell ratio. The numbers of tumoroids analyzed over DMSO, capivasertib, trametinib, and ruxolitinib groups were 20, 18, 21, and 22, respectively. (K) Western blot analyses of p63, CK5, and SOX2. Club cell–derived EML4-ALK L1196M tumoroids were treated with DMSO, 100 nM capivasertib (Cap), 10 nM trametinib (Tra), and 5 μM ruxolitinib (Rux) for six passages and then subjected to western blot analyses. (L) Relative mRNA levels of LUSC signature genes. (M) Experimental design for in vivo treatment of ruxolitinib. W, week. (N) Representative CK5 immunostaining. Scale bar, 1 mm. (O–Q) Statistical analyses of tumor number (O), LUAD number (P), and LUAS/LUSC number (Q) in mice receiving ruxolitinib (n = 5) or vehicle (n = 4). Data in A–F represent one experiment of three independent experiments. Data in H–L and O–Q represent one experiment of two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA (J and L), two-tailed unpaired Student’s t test (O–Q). ns: not significant. Data are represented as mean ± SEM. Source data are available for this figure: SourceData F5.
To dissect detailed downstream pathways involved in AST, we treated plastic tumoroids with various inhibitors including trametinib (MEK1/2 inhibitor), capivasertib (pan-AKT inhibitor), or ruxolitinib (JAK1/2 inhibitor) at optimized doses effectively inhibiting individual pathways without significant cell survival inhibition (Fig. 5 G and Fig. S4, B–H). Among these inhibitors, ruxolitinib showed a dramatic impact upon AST (Fig. 5 H). Ruxolitinib-treated tumoroids maintained TTF1 expression and exhibited decreased CK5 and p63 levels (Fig. 5, I–L and Fig. S4, I–L). Using another JAK1/2 inhibitor, AZD-1480, we further validated the role of JAK-STAT signaling in AST (Fig. S4, M and N). Through ruxolitinib treatment in the EML4-ALK L1196M model, we found that the JAK-STAT inhibition preferentially decreased the number of LUSC without affecting total tumor numbers (Fig. 5, M–Q).
We then utilized CRISPR/Cas9 system to knock out multiple STATs in plastic tumoroids. We found that only Stat3 depletion significantly decreased p63 expression (Fig. S4, O–Q). Moreover, Stat3 knockout also inhibited the AST process in mouse (Fig. S4, R–U). Consistently, scRNA-seq data analyses revealed that the Stat3 target genes were upregulated at the intermediate state and LUSC-like state (Fig. S4 V). Moreover, we observed strong p-STAT3 immunostaining in human LUAS samples (Fig. S4 W). These data collectively support an important role of the JAK-STAT signaling involving Stat3 in regulating AST.
LUSC show poor TKI therapeutic responses
Clinical observation suggests a potential link between AST and drug resistance (Ball et al., 2022; Gong et al., 2019; Kaiho et al., 2020; Park et al., 2019; Ueda et al., 2021; Wang et al., 2020; Zhang et al., 2021). To prove this, we first treated non-plastic and plastic tumoroids with the third generation ALK inhibitor lorlatinib (Fig. S5 A). We found that the non-plastic tumoroids were vulnerable to lorlatinib treatment, whereas the plastic tumoroids showed significantly higher half-maximal inhibitory concentration (IC50) values (Fig. S5 B).
Figure S5.
LUSC or LUAD with squamous signature show poor TKI therapeutic responses. (A) Schematic illustration of lorlatinib treatments in tumoroids. (B) Drug response curves of the non-plastic and plastic tumoroids. Cell viability was measured after 72 h lorlatinib treatment. Non-plastic LUAD tumoroids: LUAD-1, LUAD-2. Plastic tumoroids: LUSC-1, LUSC-2, LUSC-3, and LUSC-4. (C) Representative CC3 immunostaining for LUAD and LUSC in 0.5 mg/kg, 2 mg/kg lorlatinib, and control (vehicle) groups. Scale bar, 50 μm. (D) Statistical analysis of CC3 staining. (E) UMAP visualization of epithelial cells from mouse LUAD, LUSC, and LOR (remaining tumors after 2 wk of 10 mg/kg lorlatinib treatment). Batch effect was removed by fastMNN. (F) Heat map showing mean expression of top differentially expressed genes in each epithelial cell cluster for mouse LUAD, LUSC, and LOR samples. (G) Feature plots of known LUAD-related genes including Foxa2, Krt8, Sftpa1, and Sftpc. (H) Feature plots of known LUSC-related genes including Sox2, p63, Krt14, and Krt6a. (I) Real-time PCR detection confirmed the ectopic expression of p63 and Sox2 in the non-plastic tumoroids. (J) Cell viability detection of Sox2-expressing or control tumoroids treated with lorlatinib. Cell viability was measured after 72 h lorlatinib treatment. (K) Representative immunofluorescence staining of the GFP+ (expressing p63) and GFP− (without p63 expression) mixed tumoroids. p63 in red; nucleus in blue (DAPI staining). Scale bar, 50 μm. (L) Flow cytometry analysis of the GFP+ cells. The GFP+ cells (expressing p63) and GFP− cells (without p63 expression) were mixed at 1:1 ratio and cultured with 100, 300, and 500 nM lorlatinib for 24 h before being subjected to flow cytometry analyses. (M) Representative H&E staining and p63 immunostaining of remaining LUAD in 0.5, 2, 10 mg/kg lorlatinib group. Scale bar, 50 μm. (N) Quantification of average tumor size of the p63− or p63+ LUAD in mice with vehicle, 0.5, 2, and 10 mg/kg lorlatinib treatments. (O) Top panel shows TTF1 immunostaining photos in two ALK-rearranged human LUAD biopsies with p40 positivity. Scale bar, 50 μm. Bottom panel shows the PFS of LUAD patients with or without p40 positivity. PFS was calculated from the start date of TKI treatments to the date of tumor progression. (P) Heat map of IHC score (scale 0–300) of p40, CK5, CK6A, and CK14 for seven paired treatment-naïve and drug-resistant biopsies with ALK rearrangements. (Q) The p40, CK5, CK6A, and CK14 immunostaining of treatment-naïve and drug-resistant biopsy samples from patient #6. Scale bar, 50 μm. (R) The H&E staining, p40, and CK5/6 immunostaining of treatment-naïve and drug-resistant biopsy samples from patients #8 and #9. Scale bar, 50 μm. Data in B and I–L represent one experiment of three independent experiments. Data in C, D, M, and N represent one experiment of two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA (D), multiple t test (J), two-tailed unpaired Student’s t test (N), Mantel–Cox test (O). ns: not significant. Data are represented as mean ± SEM.
We further investigated the responses of LUAD and LUSC to increased lorlatinib dosages in the EML4-ALK L1196M model (Fig. 6 A) and found that lorlatinib suppressed ALK-driven tumor growth in a dose-dependent manner (Fig. 6, B and C). Upon 0.5 and 2 mg/kg lorlatinib treatments, LUAD but not LUSC showed significant decrease in tumor sizes (Fig. 6, D and E). Treatment with 10 mg/kg lorlatinib, a relatively high dose according to previous studies (Makimoto et al., 2019; Mizuta et al., 2021; Redaelli et al., 2018), resulted in a dramatic decrease in LUAD sizes and a slight decrease in LUSC sizes (Fig. 6, D and E). Increased cleaved caspase 3 (CC3) staining and decreased Ki-67 staining were observed in LUAD but not LUSC (Fig. 6, F–J; and Fig. S5, C and D).
Figure 6.
LUSC or squamous signature–enriched LUAD show unfavorable response to ALK TKIs. (A) Top panel shows experimental design for in vivo lorlatinib treatments. Bottom panel shows representative lung H&E staining from EML4-ALK L1196M mice treated with vehicle or lorlatinib (0.5, 2, or 10 mg/kg). Scale bar, 1 mm. W, week. (B and C) Quantification of tumor burden (B) and average tumor number (C) in mice with various lorlatinib treatments. Veh, vehicle. (D and E) Quantification of average tumor size of LUAD (D) and LUSC (E) in mice with various lorlatinib treatments. (F) Representative immunostaining for CC3 of LUAD and LUSC from mice receiving vehicle or 10 mg/kg lorlatinib. Scale bar, 50 μm. (G) Statistical analysis of CC3 staining. 18 representative images for each group were counted. (H) Representative immunostaining for Ki-67 of LUAD and LUSC from mice receiving vehicle or lorlatinib. Scale bar, 50 μm. (I and J) Statistical analysis of Ki-67 staining of LUAD (I) and LUSC (J). 18 representative images for each group were counted. (K) Kaplan–Meier curves show the PFS of LUAD (n = 60) and LUSC (n = 8) patients with ALK-rearranged tumors. PFS was calculated from the start date of alectinib treatment to the date of tumor progression. (L) UMAP visualization of cancer cells labeled with Seurat clusters for mouse LUAD, LUSC, and LOR samples. LOR: after 2 wk of 10 mg/kg lorlatinib treatment. (M) Feature plots of known LUAD-related genes (Nkx2-1, Napsa) and LUSC-related genes (Krt5, Dsg3) for mouse LUAD, LUSC, and LOR samples. (N) UMAP visualization of epithelial cells labeled with LUAD-like, intermediate, and LUSC-like state. 1: LUAD-like state; 2: intermediate state; 3: LUSC-like state. (O) Dot plot showing expression of known LUAD- and LUSC-related genes in LUAD-like, intermediate, and LUSC-like state. Dot diameter indicates the proportion of cells expressing a given gene; color indicates the expression level. (P) Bar plot showing distribution of LUAD, LUSC, and LOR cells in LUAD-like, intermediate, and LUSC-like state. (Q) Dot plot showing expression of significantly upregulated transcription factors in intermediate state compared to LUAD-like state (false discovery rate [FDR] < 0.001). (R) Scheme experimental design for ectopic expression of p63 and Sox2 in non-plastic tumoroids. (S) Cell viability detection of p63-expressing or control (Ctrl) tumoroids treated with lorlatinib. Cell viability was measured after 72 h of lorlatinib treatment. (T) Top panel shows the experimental design for cell competition experiments. Bottom panel shows the flow cytometry analysis of GFP+ cells. The GFP+ cells (expressing p63) and GFP− cells (without p63 expression) were mixed at 1:1 ratio and cultured with 100 nM lorlatinib for 24 h before being subjected to flow cytometry analyses. (U) Statistical analyses of the p63−/p63+ LUAD ratio in vehicle or lorlatinib (0.5, 2, or 10 mg/kg) treatments group. (V) ssGSEA score of squamous signature of LUAD patients with ALK-rearranged tumors. (W) Kaplan–Meier curves show the PFS of LUAD patients with ALK-rearranged tumors (n = 36) according to squamous score status. According to ssGSEA score of squamous signature, patients were subjected into two groups: squamous signature high (n = 8) and squamous signature low (n = 28). PFS was calculated from the start date of TKI treatments to the date of tumor progression. Data in B–J and U represent one experiment of two independent experiments. Data in S and T represent one experiment of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA (B–E, I, and J), multiple t test (G and S), Mantel–Cox test (K and W). ns: not significant. Data are represented as mean ± SEM.
Pathology-specific responses to ALK TKI treatments were also observed in the clinic. For example, the progression-free survival (PFS) in LUSC patients with alectinib treatment is about 9.5 mo, which is significantly shorter than the median PFS, 34.8 mo in LUAD patients (Camidge et al., 2019; Shiihara et al., 2021). Consistently, we also found that the median PFS for Chinese LUSC patients receiving alectinib was 8.5 mo, in contrast to 28.6 mo in Chinese LUAD patients (Fig. 6 K; and Tables S1 and S2).
LUAD with squamous signature show poor response to ALK TKI
We further dissected the remaining tumors after 2 wk of 10 mg/kg lorlatinib treatment for scRNA-seq analysis and identified 13 clusters from the treatment-naïve and post-treatment cancer cells (Fig. 6 L; and Fig. S5, E and F). Based on adenomatous and squamous biomarker expression, we defined these cells into three different states: state 1 as LUAD-like, state 2 as intermediate, and state 3 as LUSC-like (Fig. 6, M–O; and Fig. S5, G and H). We found that the ratio of cells at LUAD-like state significantly decreased, whereas the ratio of cells at intermediate state significantly increased after lorlatinib treatment (Fig. 6 P). Comparative analyses of transcriptional factor (TF) showed that both Sox2 and p63, two major lineage TFs in LUSC, were significantly upregulated at intermediate state (Fig. 6 Q).
We next ectopically expressed p63 and Sox2 in non-plastic tumoroids to explore their potential contribution to TKI resistance (Fig. 6 R and Fig. S5 I). We found that p63 but not Sox2 increased lorlatinib resistance (Fig. 6 S and Fig. S5 J). We further labeled the p63-expressing cells with GFP and mixed them with the parental cells (GFP−) at 1:1 ratio (Fig. 6 T and Fig. S5 K) and found that lorlatinib treatment significantly increased the proportion of GFP+ cells (Fig. 6 T and Fig. S5 L). Moreover, we found that the percentage of p63-positive LUAD increased with lorlatinib dosage escalation in mouse (Fig. 6 U). The p63+ LUAD showed larger tumor sizes in contrast to those p63− LUAD (Fig. S5, M and N). These findings suggest that p63-positive LUAD with squamous signature might be resistant to ALK TKI treatment.
We next evaluated the link between squamous signature and TKI response in 36 ALK-rearranged LUAD patients. In line with mouse model findings, human LUAD patients with high squamous signature showed an unfavorable therapeutic response to ALK TKI treatments (Fig. 6, V and W; and Tables S3 and S4). In another cohort of 24 ALK-rearranged LUAD patients, we found that p40 expression was associated with shorter patient PFS (Fig. S5 O). We further performed immunostaining analyses in seven paired TKI treatment-naïve and -resistant biopsies from ALK-rearranged Chinese LUAD patients. Squamous biomarkers were detectable in most drug-resistant tumors (Fig. S5 P), whereas one sample showed positivity for all biomarkers analyzed including p40, CK5, CK14, and CK6A (Fig. S5 Q). We further collected two paired ALK TKI treatment-naïve and -resistant biopsies from Japanese patients and found that both re-biopsies were positive for p40 and one was also positive for CK5/6 (Fig. S5 R). These findings collectively indicate that human ALK-rearranged LUAD with squamous signature have unfavorable responses to TKI treatment.
Combined ruxolitinib and lorlatinib treatment results in dramatic tumor regression
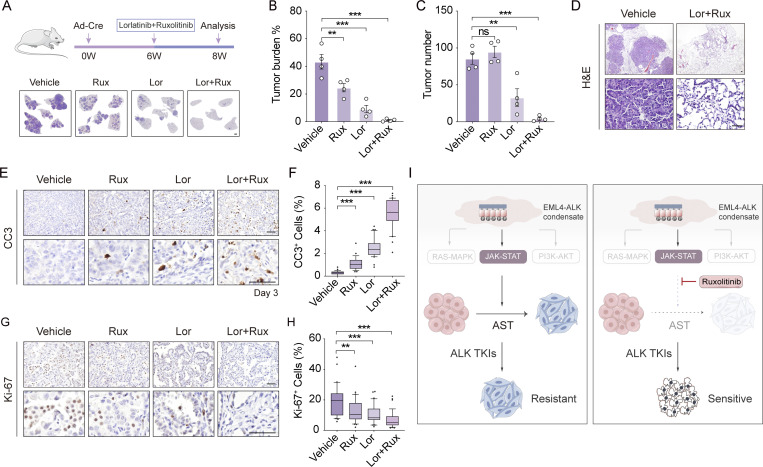
We next asked if targeting the JAK-STAT signaling is effective in treating EML4-ALK tumors. To this, we performed combined ruxolitinib and lorlatinib treatment in mouse model (Fig. 7 A). We found that combination treatment significantly inhibited tumor progression, leading to a dramatic decrease in tumor numbers and burdens, in stark contrast to single-agent treatment (Fig. 7, A–C). Extensive necrosis was detectable in those remaining tumors in the combination treatment group (Fig. 7 D). Compared with control groups, combination therapy significantly inhibited tumor cell proliferation and induced apoptosis (Fig. 7, E–H). These data suggest that combination treatment with ruxolitinib and lorlatinib overcomes drug resistance caused by squamous transition and significantly improves therapeutic efficacy.
Figure 7.
Combined lorlatinib and ruxolitinib treatment eradicates ALK-driven tumors and significantly improves therapeutic efficacy. (A) Top panel shows experimental design for in vivo combinational treatments using lorlatinib and ruxolitinib. Bottom panel shows lung H&E staining from four groups. Rux: ruxolitinib; Lor: lorlatinib; Lor+Rux: lorlatinib and ruxolitinib. Scale bar, 1 mm. W, week. (B and C) Quantification of tumor burden (B) and average tumor number (C) in mice with vehicle (n = 4), lorlatinib (n = 4), ruxolitinib (n = 4), or combined (n = 4) treatment. (D) Representative H&E staining of tumors from mice with vehicle and combined treatment. Scale bar, 50 μm. (E) Representative immunostaining for CC3 of tumors from vehicle, ruxolitinib, lorlatinib, or combined treatments group. Scale bar, 50 μm. (F) Statistical analysis of CC3 staining. 30 representative images for each group were counted. (G) Representative immunostaining for Ki-67 of tumors from vehicle, ruxolitinib, lorlatinib, or combined treatments group. Scale bar, 50 μm. (H) Statistical analysis of Ki-67 staining. 30 representative images for each group were counted. (I) Proposed model for AST and histological transition-associated TKI resistance. EML4-ALK LUAD can progressively transition into LUSC, which results in increased TKI resistance. The JAK-STAT signaling is important for driving squamous transition. Combined JAK1/2 inhibitor and TKI treatment significantly inhibits the AST process and regain the high efficacy of molecular targeted therapy. Data in B–H represent one experiment of two independent experiments. **P < 0.01, ***P < 0.001 by one-way ANOVA (B, C, F, and H). ns: not significant. Data are represented as mean ± SEM.
Taken together, we provide convincing evidence showing the cancer plasticity of EML4-ALK tumors in mouse and human. Mechanistic studies uncover that the JAK-STAT signaling activated by EML4-ALK phase separation promotes AST and further leads to ALK inhibitor resistance (Fig. 7 I). Combined ruxolitinib and lorlatinib treatment overcomes squamous transition-driven drug resistance (Fig. 7 I).
Discussion
Evidence from multiple works has highlighted the importance of tumor cell phenotypic plasticity and functional heterogeneity (Cooper et al., 2022; Hanahan, 2022). Through comprehensive analyses of three cohorts of human LUAS samples, we find that ALK rearrangement is at 5.1–7.5%. Through recapitulating human ALK-rearranged lung cancer particularly the development of LUAS, we propose the squamous transition route from LUAD (TTF1+/p63− or TTF1+/p63+) to LUAS (mixed with TTF1+/p63+ LUAD cells and p63+/TTF1+ LUSC cells) to LUSC (p63+/TTF1high, p63+/TTF1low or p63+/TTF1−). Detailed scRNA-seq analyses of mouse tumors confirm the intermediate state as well as squamous transition trajectory. Our current analyses demonstrate that certain human EML4-ALK LUAD express squamous biomarkers and/or signature, indicative of a trend toward squamous transition. This is consistent with previous reports about the observation of p63 expression in EML4-ALK LUAD (Li et al., 2013; Yoshida et al., 2011). These analyses are further supported by detailed cell subpopulation analyses using scRNA-seq data, which identifies a subpopulation enriched with squamous signature gene expression. These clinical obversions highlight the relationship between EML4-ALK and squamous transition.
In this study, we employ the EML4-ALK GEMMs as well as the organoid model to recapitulate squamous transition, which mirrors the AST process in human ALK-rearranged LUAS and enables an in-depth study of underlying mechanisms. In mouse models, we find that histological transition tends to occur at a relatively late stage during LUAD malignant progression, in line with our previous findings in the KL model (Han et al., 2014; Li et al., 2015). In clinic, most ALK-rearranged tumors are predominantly diagnosed as LUAD. It remains possible that the surgery or biopsy sampling is done at an early time point before the initiation of the AST process.
Our study identifies club cell as the major cell-of-origin for AST in the EML4-ALK mouse model. Previous study shows that club cell–derived tumors uniformly exhibit classical adenomatous pathology in KrasG12D or KP mouse models, very similar to ATII cell–derived tumors (Sutherland et al., 2014). This indicates that both the cell-of-origin and oncogenic signaling are important for AST. For example, although the ATII cell is the predominant cell-of-origin for Kras-driven LUAD (Mainardi et al., 2014; Xu et al., 2014), amplification of MAPK signaling expands the cell-of-origin to club cells (Cicchini et al., 2017). Similarly, deletion of Lkb1 in KrasG12D mice results in developing multiple types of tumors including LUAD, LUAS, LUSC, and the club cells or bronchioalveolar stem cells (BASCs) is considered to be the cell-of-origin for adenosquamous and squamous tumors (Nagaraj et al., 2017; Zhang et al., 2017). Since BASCs also express club cell marker CC10, the usage of Ad-CC10-Cre might infect BASCs in our model. Future efforts are necessary to explore the potential role of BASCs in squamous transition in context with EML4-ALK fusion.
Most previously reported EML4-ALK mouse models mainly focus on the ATII cell–derived cancer, which might not be optimal for studying the plasticity of ALK-driven tumors and the AST process (Chen et al., 2010, 2014; Pyo et al., 2017; Soda et al., 2008). The mouse model closely recapitulating human ALK-rearranged lung cancer has been generated through the CRISPR/Cas9-system by Maddalo and colleagues, and no AST phenomena have been observed (Maddalo et al., 2014). We reason the discrepancy might be due to multiple technical details including viral dosage, the CAS9 expression, and study depth.
Using an organoid system, we successfully recapitulate the AST process and find that EML4-ALK phase separation is responsible for the AST process. Phase separation of EML4-ALK is important for firing downstream signaling (Qin et al., 2021; Sampson et al., 2021; Tulpule et al., 2021). Interestingly, we find that phase separation of EML4-ALK regulates AST mainly through the JAK-STAT signaling. The JAK-STAT pathway is one of the central communication nodes and transduces multiple signals for development, homeostasis, and cancer progression (Villarino et al., 2015). Fascinatingly, recent studies demonstrate that the JAK-STAT signaling is critical in regulating lineage plasticity in prostate cancer (Chan et al., 2022; Deng et al., 2022). Deng et al. find that the JAK-STAT activation is required for prostate cancer cell lineage plasticity and mainly involves STAT1 (Deng et al., 2022). Chan et al. find that pharmacologic inhibition of JAK1/2 in combination with fibroblast growth factor receptor blockade could restore luminal differentiation and resensitize prostate tumoroids to antiandrogen therapy (Chan et al., 2022). These findings, together with our work, suggest that the JAK-STAT signaling might serve as a common mechanism in lineage switch across various genetic contexts and different tissues. Further study will be interesting to dissect how and when the JAK-STAT signaling regulates the AST process.
Accumulating evidence has linked cancer phenotypic plasticity to therapeutic resistance in lung cancer including ALK-rearranged cancer (Cooper et al., 2022). However, it remains unknown if AST is the causal or collateral factor. We find that plastic tumoroids and LUSC show notably increased resistance to lorlatinib treatment in comparison to non-plastic tumoroid and LUAD respectively. This is further supported by our clinical analyses. We find that the LUSC patients receiving ALK TKI treatment tend to have much shorter PFS in comparison with LUAD patients. We further find that both human and mouse LUAD with squamous signature show unfavorable responses to TKI treatment. Moreover, we detect the expression of squamous biomarkers in rebiopsy samples from relapsed patients, further highlighting the potential link between AST and drug resistance.
We find that combined inhibition of JAK-STAT signaling and ALK can overcome histological transition-driven ALK inhibitor resistance and significantly improve therapeutic efficacy. Ruxolitinib is reported to be well tolerated, although few cases of skin cancer development have been reported after drug treatment (Blechman et al., 2017). Further studies are required to fully understand the potential toxicity and assess the viability of combination therapy. In lung cancer, phenotypic transition also occurs in patients with acquired EGFR or KRAS inhibitor resistance. After gefitinib or erlotinib treatment, squamous transition is rarely observed in relapsed LUAD patients with EGFR-mutant tumors (Chen et al., 2019). One intriguing fact is that the rate of squamous transition dramatically increases when osimertinib, the third generation EGFR TKI, is applied in clinic, e.g., squamous transition is identified in 7% of first-line and 9% of late-line cases (Schoenfeld et al., 2020). Moreover, squamous transition is observed in two of nine LUAD patients relapsed from KRASG12C inhibitor therapy (Awad et al., 2021). This finding is further supported by the strong association between AST and resistance to KRAS inhibitors in preclinical lung cancer models (Tong et al., 2023, Preprint). Interestingly, a recent study demonstrates inhibition of KRAS promotes a quiescent ATI-like state in LUAD (Li et al., 2023). In consideration of the increasing diversity of phenotypic transition, it will be important to investigate whether the JAK-STAT signaling identified here contributes to lineage plasticity in lung cancer and beyond.
Materials and methods
Mouse models
The Rosa26-loxp-stop-loxp-wt EML4-ALK, Rosa26-loxp-stop-loxp-EML4-ALK L1196M, and Rosa26-loxp-stop-loxp-Cas9 knock-in mouse lines were generated by knocking CAG-loxp-stop-loxp-wt EML4-ALK variant1, CAG-loxp-stop-loxp-EML4-ALK variant1 with L1196M mutation, or CAG-loxp-stop-loxp-Cas9 into the Rosa26 locus. The Sftpc-CreERT2, Scgb1a1-CreERT2 mice, and Rosa26-loxp-stop-loxp-tdTomato reporter mice were reported previously (Madisen et al., 2010; Rawlins et al., 2009; Rock et al., 2011). The KrasG12D; P53L/L and KrasG12D; Lkb1L/L mice were originally provided by Dr. Tyler Jacks (Cambridge, MA, USA) and Dr. Ranold DePinho (Houston, TX, USA), respectively. All mice were kept in a specific pathogen–free environment of the Shanghai Institute of Biochemistry and Cell Biology, treated in strict accordance with protocols (SIBCB-2101008) approved by the Institutional Animal Care and Use Committee of the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. Mice were treated with Ad-Cre virus (5 × 104 plaque forming unit, p.f.u.) or Ad-CC10-Cre virus (1 × 107 p.f.u.) or Ad-SPC-Cre (1 × 107 p.f.u.) via nasal inhalation at 6–8 wk of age. The Ad-CC10-Cre and Ad-SPC-Cre virus were reported previously (Ferone et al., 2016) and provided by the University of Iowa Gene Transfer Vector Core. For knockout of Stat3, EML4-ALK L1196M; Cas9 mice were treated with the lentivirus of sgTomato (sgTom) or sgStat3 (5 × 105 p.f.u.) and analyzed at 12 wk after treatment.
Lorlatinib (HY-12215; MCE) or ruxolitinib (HY-50856; MCE) were formulated in 10% DMSO, 40% PEG300, and 50% saline. For compound treatments, the EML4-ALK L1196M mice at 6 wk after Ad-Cre infection were given either lorlatinib (0.5, 2, 10 mg/kg/day) or ruxolitinib (45 mg/kg/day) or both for 2 wk via intraperitoneal injection. The control mice were given vehicle (10% DMSO: 40% PEG300: 50% saline). All mice were sacrificed for gross inspection and histopathological examination and tumors were dissected for bulk RNA-seq and scRNA-seq. Tumor number, tumor burden, and tumor size were analyzed using ImageJ software.
Human lung cancer specimen collection
Surgical or biopsy sample collection was approved by the Medical Ethics Committee of Fudan University Shanghai Cancer Center, Shanghai Pulmonary Hospital, Hunan Cancer Hospital, and National Cancer Center Hospital East. All patients in this study gave written informed consents. All cases were re-reviewed by pathologists for confirmation of tumor pathology. PFS was calculated from the start date of ALK inhibitor treatment to the date of tumor progression.
A total of 105 LUAS surgical samples (from Fudan University Shanghai Cancer Center) were used for genomic and transcriptomic sequencing: 93 tumors with paired adjacent normal tissues were sequenced with WGS (tumor 60×; normal tissue 30×) and another set of 93 tumors with four normal lungs were RNA sequenced. Among 105 samples, 81 samples were analyzed with both WGS and RNA-seq. A total of 645 ALK-rearranged treatment-naïve LUAD surgical samples (from Fudan University Shanghai Cancer Center) were collected for immunostaining analyses. A total of 24 treatment-naïve LUAD biopsies and seven paired treatment-naïve and ALK inhibitor-resistant LUAD biopsies with ALK rearrangements (patient #1 to patient #7) were collected (from Shanghai Pulmonary Hospital) for immunostaining analyses. Three LUAD biopsies with ALK rearrangements (from Shanghai Pulmonary Hospital) were used for scRNA-seq. The PFS information of 68 patients with ALK-rearranged tumors (60 LUAD and 8 LUSC) with alectinib treatment was collected (from Shanghai Pulmonary Hospital). Detailed information of these 68 patients is included in Tables S1 and S2. A total of 41 LUAD and 1 LUSC samples with ALK rearrangements were collected (from Hunan Cancer Hospital) for RNA-seq. A total of 36 LUAD of these patients were subjected to evaluate the relationship between squamous signature score and PFS. The detailed information of 36 LUAD patients were included in Tables S3 and S4. Two paired treatment-naïve and ALK inhibitor-resistant LUAD biopsies with ALK rearrangements (patient #8 and patient #9) were collected (from National Cancer Center Hospital East, Japan) for immunostaining analyses.
Histological examination
Mouse lungs were inflated with formalin, fixed overnight, and dehydrated in ethanol, embedded in paraffin, sectioned (5 µm) followed by staining with hematoxylin and eosin (H&E). The LUAD and LUSC were pathologically defined according to H&E staining. The LUAS wase defined as lung tumors with >5% of either adenomatous or squamous components. The LUAS (LUAD-dominant) or LUAS (LUSC-dominant) was defined as tumors containing over 50% adenomatous or squamous components, respectively.
IHC (immunohistochemistry) staining was performed as previously described (Ji et al., 2007). Paraffin-embedded tissues were incubated with following antibodies: CK5 (BS1208, 1:1,000; Bioworld), p63 (ab124762, 1:5,000; Abcam), SOX2 (ab92494, 1:500; Abcam), TTF1 (ab133638, 1:500; Abcam), Ki-67 (NB500-170, 1:1,000; Novus), CC3 (9664, 1:1,000; CST), p-STAT3 (9145, 1:400; CST), p40 (RMA-1006, 1:250; Maxim), CK6A (D220238, 1:4,000; Sangon Biotech), CK14 (PRB-155P, 1:5,000; BioLegend), and ALK (3633S, 1:250; CST). Multiplex IHC staining was conducted using the PANO 4-plex IHC Kit (Panovue). Paraffin-embedded tissues were incubated with following antibodies: CK5 (BS1208, 1:1,000; Bioworld), p63 (ab124762, 1:5,000; Abcam), and TTF1 (ab133638, 1:500; Abcam). Confocal images were captured using a Leica TCS SP8 WLL confocal microscope.
The cutoff of immunofluorescence positivity for p63 was 5%: “negative” means the staining was only seen in <5% of cells. The TTF1 immunofluorescence scores were determined according to the staining intensity of individual cells using a scale of 0–10 (with 0 indicating a lack of positive immune reactivity and 10 reflecting intensive staining). Slides were evaluated and the mean was calculated, and the results were converted as follows: 0–1 score was defined as “TTF1−,” 2–5 was defined as “TTF1low,” and 5–10 was defined as “TTF1high.”
For IHC staining analyses of human samples, p40, CK6A, CK5, and CK14 IHC staining intensity was scored in four categories: “0” for no staining, “1” for weak staining, “2” for intermediate staining, and “3” for strong staining. The IHC score (range 0–300) was calculated with the following formula: percentage of positive cells × staining intensity.
Organoid culture and tumoroid allograft transplantation
Mouse lung tumors were collected and rinsed with advanced DMEM/F12 containing 1×Penicillin/Streptomycin Solution (P/S) twice, and then digested with dissociation medium (Advanced DMEM/F12 containing 5 mg/ml Collagenase Type II, 1×HEPES, 1×GlutaMax-I, 1×P/S, 1×primocin, and 10 μM Y27632) and incubated at 37 °C for 60 min. Cell suspensions were washed twice and resuspended in “mouse lung tumor culture medium” and mixed with Matrigel at 1:1 and plated in 24-well plate (5,000–10,000 cells/per well). The recipe for mouse lung tumor culture medium was as follows: advanced DMEM/F12 supplemented with 1×P/S (15140-122; Invitrogen), 1×HEPES (15630-056; Invitrogen), 1×primocin (ant-pm-1; Invitrogen), 1×B27 supplement (17504-044; Invitrogen), 1.56 mM N-Acetylcysteine (A9165-5G; Sigma-Aldrich), 500 nM A-83-01 (2939; Tocris), 10 ng/ml EGF (PMG8043; Invitrogen), 10 µM Y-27632 (S1049; Selleckchem), 1×GlutaMax-I (35050-079; Invitrogen), R-spondin, and Noggin. The tumoroids were passaged at a 1:3 dilution every 4 days. For allograft transplantation, the tumoroids were digested into single-cell suspensions and mixed with Matrigel (1:1) for subcutaneous transplantation in immune-deficient SCID mice.
Immunofluorescence and fluorescent microscopy
Immunofluorescence staining was performed as previously described (Qin et al., 2021). The cells or tumoroids were incubated with the following antibodies: ALK (3633S, 1:250; CST), p63 (ab124762, 1:5,000; Abcam), TTF1 (ab133638, 1:500; Abcam), CK5 (BS1208, 1:1,000; Bioworld). Confocal images were captured using a Leica TCS SP8 system with a HC PL APO CS2 63×/1.40 OIL objective.
Live-cell imaging
Cells were seeded in 35-mm glass-bottom dishes (D35-20-1.5-N; Cellvis). For imaging the droplet fusion, the BEAS-2B cells were transfected with plasmids for 12 h. Images were captured at 2-s intervals with a Zeiss LSM880 Airyscan microscope equipped with a 63× OIL immersion objective.
FRAP
FRAP experiments in cells were carried out with following settings: region of interest was bleached using a 405-nm diode, pre-bleach and post-bleach images were acquired with a 488-nm laser. Fluorescence recovery of GFP-EML4-ALK L1196M was monitored for 10 min with a time resolution of 2 s. Images were captured at 2-s intervals with a Zeiss LSM880 Airyscan microscope equipped with a 63× OIL immersion objective.
Plasmid construction
Full-length EML4-ALK L1196M was amplified and inserted into GFP-3×linker or pCDH-EF1-Puro vectors. EML4-21S-ALK L1196M mutant plasmid was synthesized by gene synthesis technology in Generay Biotech Co., Ltd. All the constructs were verified by sequencing. The sgRNA of Tomato was chosen as previously reported (Wu et al., 2018). The sgRNA of Stat1, Stat3, Stat5b, and Stat6 was designed using optimized CRISPR design (https://chopchop.cbu.uib.no/). The sequences of all sgRNAs and the primers for knockout efficiency detection were included in Table S5.
Lentivirus production and infection
The cell lines BEAS-2B (ATCC) were maintained in RPMI-1640 supplemented with 8% FBS. For stable overexpression of EML4-ALK L1196M or EML4 21S-ALK L1196M, the BEAS-2B cells were virally infected and maintained in medium containing puromycin (2 µg/ml; Sigma-Aldrich). For p63 and Sox2 stable expression, the non-plastic LUAD tumoroids were virally infected and maintained in medium containing puromycin (2 µg/ml). For sgRNA knockout, the club cell–derived plastic tumoroids were virally infected and maintained in medium containing puromycin (2 µg/ml).
Drug treatment and cell viability assay
Cell viability was determined using a CellCounting-Lite (Vazyme) Luminescent Cell Viability Assay. 2,000 cells from tumoroids were mixed with 5 μl Matrigel and seeded in a 96-well plate. For ruxolitinib, trametinib, and capivasertib concentration optimization, the tumoroids from club cell–derived LUAD were treated with different concentrations of ruxolitinib, trametinib, and capivasertib for 72 h. For treatments with optimum doses of various inhibitors, 5 μM ruxolitinib (HY-50856; MCE), 10 nM trametinib (HY-10999; MCE), 100 nM capivasertib (HY-15431; MCE), or 5 μM AZD-1480 (HY-10193; MCE) was given to the club cell–derived LUAD tumoroids for six passages before immunofluorescence staining, western blot analyses, and real-time PCR analyses. For ALK inhibitor treatments, plastic, non-plastic tumoroids or tumoroids with ectopic p63 or Sox2 expression were treated with different concentrations of lorlatinib for 72 h. The luminescence was measured via a Synergy Neo multimode microplate reader (BioTek). Luminescence values were normalized to control and the relative values were plotted in Prism GraphPad. All cell viability assays were conducted in triplicates. For cell competition experiments, the GFP+ cells (expressing p63) and GFP− cells (without p63 expression) were mixed at 1:1 ratio and cultured with 100, 300, and 500 nM lorlatinib for 24 h before subjected to flow cytometry analyses.
Western blot
Protein samples were probed with specific antibodies against ALK (3633S, 1:2,000; CST), p-ALK (3341S, 1:1,000; CST), ERK (9102, 1:1,000; CST), p-ERK (4370, 1:1,000; CST), AKT (9272, 1:1,000; CST), p-AKT (4070, 1:1,000; CST), STAT3 (9139, 1:1,000; CST), p-STAT3 (9145, 1:1,000; CST), ACTIN (AC026, 1:50,000; Abclonal), p63 (ab124762, 1:1,000; Abcam), CK5 (BS1208, 1:1,000; Bioworld), SOX2 (ab92494, 1:1,000; Abcam), and TTF1 (ab133638, 1:1,000; Abcam). Protein expression was assessed by Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) and detected on SAGECREATION (Sage Creation Science Co.).
Real-time PCR analyses
Total RNA prepared was retro-transcribed into first-strand cDNA using PrimeScript RT Reagent Kit (TaKaRa) and then used for real-time PCR on a LightCycler 96 System (Roche) using SYBR Green I Master (Roche). β-Actin served as internal control. The primers for PCR were used as previously described (Han et al., 2014).
Sample preparation for bulk cell RNA-seq and scRNA-seq
The ATII cell–derived LUAD, club cell–derived LUAD samples, LUAS samples and LUSC samples, KP LUAD samples, and KL LUAD and KL LUSC samples were freshly dissected and prepared for bulk RNA-seq. Histological type of LUAD and LUSC were confirmed by H&E and IHC staining before sequencing. The library preparation and sequencing were performed according to the standard Illumina RNA-Seq protocol (NovaSeq 6000; Berry Genomics, Inc.).
Lung tumors including three LUAD and three LUSC from EML4-ALK L1196M; tdTomato mice at 8 wk after Ad-Cre infection were collected for scRNA-seq. Lung tumors from EML4-ALK L1196M; tdTomato mice treated with lorlatinib (10 mg/kg) for 2 wk were also collected for scRNA-seq analyses. Briefly, tumors were minced and dissociated with 0.5 ml dissociation medium (Advanced DMEM/F12 containing 5 mg/ml Collagenase Type II, 1×HEPES, 1×GlutaMax-I, 10 μM Y27632), incubated at 37°C for 60 min, and pipetted up and down every 20 min. The dissociated cell suspension was filtered with a 70-μm mesh filter and spun down at 300 g for 5 min at 4°C and then the supernatant was removed. After washing with FACS buffer (1×PBS containing 2% FBS, 10 μM Y27632), the pellet was resuspended in 50 μl FACS buffer and stained with CD45 (103112, 1:100; BioLegend) for 30 min at 4°C. DAPI was added before sorting. CD45−; tdTomato+ tumor cells or CD45+ immune cells were sorted via FACSAria Fusion (BD). Tumor cells and immune cells were artificially mixed at a 1:1 ratio before cell loading.
For tissue dissociation of human samples, three ALK-rearranged LUAD samples were collected. Briefly, tumor samples were minced into small pieces and digested in 2 ml GEXSCOPE Tissue Dissociation Solution (Singleron Biotechnologies) at 37°C for 15 min with continuous agitation. The dissociated cell suspension was filtered with a 40-μm mesh filter and spun down at 300 g for 5 min at 4°C, and then the supernatant was removed. Cell suspensions were counted with TC20 automated cell counter (Bio-Rad) to determine cell concentration and viability.
Bulk RNA-seq data and WGS data analyses
Raw FASTQ data were aligned to mm10 reference genome with STAR (2.5.2b; Dobin et al., 2013). Genes expressed as zero in >80% of samples were filtered out. Differentially expressed genes were calculated with DESeq2 package. Log2 transformed fragments per kilobase million (FPKM) data was used for further visualization and Multiple Dimensional Scaling analysis.
WGS data were analyzed to characterize somatic mutations and Cancer Genome Interpreter (Tamborero et al., 2018) was used to annotate whether the mutation was loss of function or GOF (gain of function). Fusioncatcher (Nicorici et al., 2014, Preprint), Fusionmap (Ge et al., 2011), and Prada (Torres-García et al., 2014) were performed simultaneously to identify gene fusion events through RNA-seq data. Fusion events detected in at least two algorithms were chosen for further analyses.
Heatmap showing the expression level of squamous signature genes were drawn with R package ComplexHeatmap, and potential batch effects among different datasets were removed with Combat function in an SVA package. Single-sample Gene Set Enrichment Analysis (ssGSEA) was used to calculate enrichment scores of squamous signature genes of each sample and differences among various conditions were calculated by Wilcoxon rank-sum test. The cutoff of squamous signature was 0.1: “low” means the ssGSEA score was <0.1; “high” means the ssGSEA score was >0.1.
scRNA-seq data analysis
For three human ALK-rearranged LUAD samples and two LUSC samples, hLUSC-1, hLUSC-2, and hLUAD-1 were previously described (GSE148071; Wu et al., 2021). Generation of single-cell gene expression matrices was conducted as previously described (Wu et al., 2021), and low-quality cells were filtered (expressing <200 genes or >5,000 genes, <1,000 gene counts, and >20% mitochondrial reads). As a result, 20,089 genes in a total of 2,895 cells were detected in hLUAD-1; 21,553 genes in a total of 2,683 cells were detected in hLUAD-2; and 20,482 genes in a total of 2,664 cells were detected in hLUAD-3.
For analyses of mouse LUAD and LUSC, epithelial cells from LUAD and LUSC samples were merged and normalized with Seurat package (v4.0.4; Stuart et al., 2019). For analysis of the LOR (remaining tumors after 2 wk of 10 mg/kg lorlatinib treatment) sample, we combined the epithelial cells of LOR together with treatment-naïve LUAD and LUSC and cells were normalized with Seurat package. For analyses of human ALK-rearranged samples, epithelial cells were merged and normalized with Seurat package.
FastMNN function (Haghverdi et al., 2018) of batchelor package (v1.8.1) was run to remove potential batch effect across different samples, and UMAP was produced based on reduction of “mnn.” Reclustering of epithelial cells was generated by Findcluster function of Seurat with a resolution of 0.8, and FindMarkers function was used to calculate differentially expressed genes between each cluster. Clusters that were mixed with immune cells after reclassification were removed in further analyses. Monocle trajectory analysis was performed using Monocle3 (v1.0.0; Cao et al., 2019) package by importing the counts from Seurat object.
Statistical analysis
For comparing means of two groups, differences were analyzed by Student’s t test (two-tailed) and performed by Prism GraphPad software. For comparing means of three or more than three groups, differences among groups were analyzed by one-way ANOVA performed by Prism GraphPad software. P value <0.05 was considered statistically significant. Error bars were represented with SEM. PFS analysis was performed using the Kaplan–Meier method.
Online supplemental material
Fig. S1 shows the heterogeneity of ALK fusion lung tumors in human LUAS and mouse models (related to Fig. 1). Fig. S2 shows the evolution route of AST derived from scRNA-seq data of mouse and human ALK fusion tumors (related to Fig. 2). Fig. S3 demonstrates that club cells serve as the major cell-of-origin of AST (related to Figs. 3 and 4). Fig. S4 shows that inhibition of the JAK-STAT pathway effectively halts the progression of AST (related to Fig. 5). Fig. S5 demonstrates that LUSC or LUAD with squamous signature exhibit limited therapeutic responses to TKIs (related to Fig. 6). Tables S1 and S2 contain the detailed information of 68 patients (60 LUAD and 8 LUSC) with alectinib treatment. Tables S3 and S4 contain detailed information of 36 LUAD patients who underwent assessment to determine the correlation between squamous signature scores and PFS. Table S5 contains a list of the sequences of all sgRNAs and the primers for knockout efficiency detection.
Supplementary Material
shows clinical characteristics of ALK-rearranged LUAD (n = 60) and LUSC (n = 8) patients.
shows PFS of ALK-rearranged LUAD (n = 60) and LUSC (n = 8) patients after alectinib treatments.
shows PFS of ALK-rearranged LUAD (n = 36) patients after ALK TKI treatments.
shows sample information and squamous signature status of ALK-rearranged LUAD (n = 36) patients.
shows sgRNA sequences and the primers for knockout efficiency detection.
is the source file for Fig. 5.
is the source file for Fig. S4.
Acknowledgments
We thank Dr. Tyler Jacks (Massachusetts Institute of Technology, Cambridge, MA, USA) for the KP mouse model and Dr. Ranold DePinho (The University of Texas MD Anderson Cancer Center, Houston, TX, USA) for the KL mouse model. We thank Dr. Anton Berns (The Netherlands Cancer Institute, Amsterdam, Netherlands) for Ad-CC10-Cre and Ad-SPC-Cre virus. We are grateful to Drs. Dong Gao (Center for Excellence in Molecular Cell Science, Shanghai, China) and Hua Zhang (New York University Langone Health, New York, NY, USA) for helpful discussion and technical support.
This work was supported by the National Key Research and Development Program of China (2022YFA1103900 to H. Ji, 2020YFA0803300 to H. Ji, 2022YFA1004800 to L.-N. Chen); the National Natural Science Foundation of China (82303039 to Z. Qin, 82341002 to H. Ji, 32293192 to H. Ji, 82030083 to H. Ji, 82303575 to S. Tang, 31930022 to L.-N. Chen, 12131020 to L.-N. Chen, T2341007 to L.-N. Chen, T2350003 to L.-N. Chen); the Science and Technology Commission of Shanghai Municipality (23YF1452900 to Z. Qin); the Basic Frontier Scientific Research Program of Chinese Academy of Science (ZDBS-LY-SM006 to H. Ji); and the Innovative Research Team of High-level Local Universities in Shanghai (SSMU-ZLCX20180500 to H. Ji).
Author contributions: H. Ji conceived the idea and designed the experiments. Z. Qin and M. Yue performed all experiments and analyzed the data. H. Sun performed the experiments related to phase separation. S. Tang and Z. Fang performed the bioinformatics analyses. S. Ren, F. Wu, and W. Wang kindly provided human biopsy specimens, scRNA-seq data, and related information. Y. Zhang and Y. Fang kindly provided human LUAD RNA-seq data and related information. H. Chen and Y. Li provided surgical clinical samples and related information. H. Izumi, S. Mori, T. Taki, K. Goto, and S.S. Kobayashi kindly provided paired human biopsy specimens. H. Huang, Y. Xue, X. Tong, Y. Jin, F. Li, F.-M. Li, Y. Gao, L. Hu, X. Yan, G. Xu, K.-K. Wong, A. Ventura, X. Zhu, L.-N. Chen, and L. Chen provided technical assistance and helpful comments. H. Ji and Z. Qin wrote the manuscript. All authors approved the final version.
Data availability
RNA-seq data and scRNA-seq data of mouse tumors, RNA-seq data, and WGS data of human LUAS samples have been deposited in the National Omics Data Encyclopedia (accession no. OEP003086, OEP001032). RNA-seq data of human ALK-rearranged tumors and scRNA-seq data of human LUAD have been deposited in the OMIX, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (accession no. OMIX002552, OMIX002554).
References
- Awad, M.M., Liu S., Rybkin I.I., Arbour K.C., Dilly J., Zhu V.W., Johnson M.L., Heist R.S., Patil T., Riely G.J., et al. 2021. Acquired resistance to KRASG12C inhibition in cancer. N. Engl. J. Med. 384:2382–2393. 10.1056/NEJMoa2105281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball, M., Christopoulos P., Kirchner M., Allgauer M., Brandt R., Winter H., Heussel C.P., Herth F., Frohling S., Savai R., et al. 2022. Histological and molecular plasticity of ALK-positive non-small-cell lung cancer under targeted therapy: A case report. Cold Spring Harb Mol Case Stud. 8:a006156. 10.1101/mcs.a006156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechman, A.B., Cabell C.E., Weinberger C.H., Duckworth A., Leitenberger J.J., Zwald F.O., and Russell M.A.. 2017. Aggressive skin cancers occurring in patients treated with the janus kinase inhibitor ruxolitinib. J. Drugs Dermatol. 16:508–511. [PubMed] [Google Scholar]
- Camidge, D.R., Dziadziuszko R., Peters S., Mok T., Noe J., Nowicka M., Gadgeel S.M., Cheema P., Pavlakis N., de Marinis F., et al. 2019. Updated efficacy and safety data and impact of the EML4-ALK fusion variant on the efficacy of Alectinib in Untreated ALK-positive advanced non-small Cell Lung cancer in the global phase III ALEX study. J. Thorac. Oncol. 14:1233–1243. 10.1016/j.jtho.2019.03.007 [DOI] [PubMed] [Google Scholar]
- Cao, J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., et al. 2019. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 566:496–502. 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caumont, C., Veillon R., Gros A., Laharanne E., Bégueret H., and Merlio J.P.. 2016. Neuroendocrine phenotype as an acquired resistance mechanism in ALK-rearranged lung adenocarcinoma. Lung Cancer. 92:15–18. 10.1016/j.lungcan.2015.12.001 [DOI] [PubMed] [Google Scholar]
- Cha, Y.J., Cho B.C., Kim H.R., Lee H.J., and Shim H.S.. 2016. A case of ALK-rearranged adenocarcinoma with small cell carcinoma-like transformation and resistance to crizotinib. J. Thorac. Oncol. 11:e55–e58. 10.1016/j.jtho.2015.12.097 [DOI] [PubMed] [Google Scholar]
- Chan, J.M., Zaidi S., Love J.R., Zhao J.L., Setty M., Wadosky K.M., Gopalan A., Choo Z.N., Persad S., Choi J., et al. 2022. Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science. 377:1180–1191. 10.1126/science.abn0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Tang W.Y., Tong X., and Ji H.. 2019. Pathological transition as the arising mechanism for drug resistance in lung cancer. Cancer Commun.. 39:53. 10.1186/s40880-019-0402-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Akbay E., Mikse O., Tupper T., Cheng K., Wang Y., Tan X., Altabef A., Woo S.A., Chen L., et al. 2014. Co-clinical trials demonstrate superiority of crizotinib to chemotherapy in ALK-rearranged non-small cell lung cancer and predict strategies to overcome resistance. Clin. Cancer Res. 20:1204–1211. 10.1158/1078-0432.CCR-13-1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Sasaki T., Tan X., Carretero J., Shimamura T., Li D., Xu C., Wang Y., Adelmant G.O., Capelletti M., et al. 2010. Inhibition of ALK, PI3K/MEK, and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer Res. 70:9827–9836. 10.1158/0008-5472.CAN-10-1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y., Zhang Y., Yuan Y., Wang J., Liu K., Yu B., Xie L., Ou-Yang C., Wu L., and Ye X.. 2021. The comprehensive analyses of genomic variations and assessment of TMB and PD-L1 expression in Chinese lung adenosquamous carcinoma. Front. Genet. 11:609405. 10.3389/fgene.2020.609405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchini, M., Buza E.L., Sagal K.M., Gudiel A.A., Durham A.C., and Feldser D.M.. 2017. Context-dependent effects of amplified MAPK signaling during lung adenocarcinoma initiation and progression. Cell Rep. 18:1958–1969. 10.1016/j.celrep.2017.01.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers, H. 2016. Modeling development and disease with organoids. Cell. 165:1586–1597. 10.1016/j.cell.2016.05.082 [DOI] [PubMed] [Google Scholar]
- Coleman, N., Wotherspoon A., Yousaf N., and Popat S.. 2019. Transformation to neuroendocrine carcinoma as a resistance mechanism to lorlatinib. Lung Cancer. 134:117–120. 10.1016/j.lungcan.2019.05.025 [DOI] [PubMed] [Google Scholar]
- Cooper, A.J., Sequist L.V., and Lin J.J.. 2022. Third-generation EGFR and ALK inhibitors: Mechanisms of resistance and management. Nat. Rev. Clin. Oncol. 19:499–514. 10.1038/s41571-022-00639-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, S., Wang C., Wang Y., Xu Y., Li X., Johnson N.A., Mukherji A., Lo U.G., Xu L., Gonzalez J., et al. 2022. Ectopic JAK-STAT activation enables the transition to a stem-like and multilineage state conferring AR-targeted therapy resistance. Nat. Cancer. 3:1071–1087. 10.1038/s43018-022-00431-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage, M., Dooley A.L., and Jacks T.. 2009. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 4:1064–1072. 10.1038/nprot.2009.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Y., Wang Y., Zeng D., Zhi S., Shu T., Huang N., Zheng S., Wu J., Liu Y., Huang G., et al. 2021. Comprehensive analyses reveal TKI-induced remodeling of the tumor immune microenvironment in EGFR/ALK-positive non-small-cell lung cancer. OncoImmunology. 10:1951019. 10.1080/2162402X.2021.1951019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Z., Han X., Chen Y., Tong X., Xue Y., Yao S., Tang S., Pan Y., Sun Y., Wang X., et al. 2023. Oxidative stress-triggered Wnt signaling perturbation characterizes the tipping point of lung adeno-to-squamous transdifferentiation. Signal Transduct. Target. Ther. 8:16. 10.1038/s41392-022-01227-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares, A.F., Lok B.H., Zhang T., Cabanero M., Lau S.C.M., Stockley T., Patel D., Bradbury P.A., Sacher A., Yasufuku K., et al. 2020. ALK-rearranged lung adenocarcinoma transformation into high-grade large cell neuroendocrine carcinoma: Clinical and molecular description of two cases. Lung Cancer. 146:350–354. 10.1016/j.lungcan.2020.06.005 [DOI] [PubMed] [Google Scholar]
- Ferone, G., Song J.Y., Sutherland K.D., Bhaskaran R., Monkhorst K., Lambooij J.P., Proost N., Gargiulo G., and Berns A.. 2016. SOX2 is the determining oncogenic switch in promoting lung squamous cell carcinoma from different cells of origin. Cancer Cell. 30:519–532. 10.1016/j.ccell.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, S., Masago K., Katakami N., and Yatabe Y.. 2016. Transformation to SCLC after treatment with the ALK inhibitor alectinib. J. Thorac. Oncol. 11:e67–e72. 10.1016/j.jtho.2015.12.105 [DOI] [PubMed] [Google Scholar]
- Ge, H., Liu K., Juan T., Fang F., Newman M., and Hoeck W.. 2011. FusionMap: Detecting fusion genes from next-generation sequencing data at base-pair resolution. Bioinformatics. 27:1922–1928. 10.1093/bioinformatics/btr310 [DOI] [PubMed] [Google Scholar]
- Gong, J., Gregg J.P., Ma W., Yoneda K., Moore E.H., Daly M.E., Zhang Y., Williams M.J., and Li T.. 2019. Squamous cell transformation of primary lung adenocarcinoma in a patient with EML4-ALK fusion variant 5 refractory to ALK inhibitors. J. Natl. Compr. Canc. Netw. 17:297–301. 10.6004/jnccn.2019.7291 [DOI] [PubMed] [Google Scholar]
- Haghverdi, L., Lun A.T.L., Morgan M.D., and Marioni J.C.. 2018. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36:421–427. 10.1038/nbt.4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X., Li F., Fang Z., Gao Y., Li F., Fang R., Yao S., Sun Y., Li L., Zhang W., et al. 2014. Transdifferentiation of lung adenocarcinoma in mice with Lkb1 deficiency to squamous cell carcinoma. Nat. Commun. 5:3261. 10.1038/ncomms4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. 2022. Hallmarks of cancer: New dimensions. Cancer Discov. 12:31–46. 10.1158/2159-8290.CD-21-1059 [DOI] [PubMed] [Google Scholar]
- Ji, H., Ramsey M.R., Hayes D.N., Fan C., McNamara K., Kozlowski P., Torrice C., Wu M.C., Shimamura T., Perera S.A., et al. 2007. LKB1 modulates lung cancer differentiation and metastasis. Nature. 448:807–810. 10.1038/nature06030 [DOI] [PubMed] [Google Scholar]
- Kaiho, T., Nakajima T., Iwasawa S., Yonemori Y., and Yoshino I.. 2020. ALK rearrangement adenocarcinoma with histological transformation to squamous cell carcinoma resistant to alectinib and ceritinib. OncoTargets Ther. 13:1557–1560. 10.2147/OTT.S236706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiriya, J.J., Wang C., Zhou M., Brumwell A., Cassandras M., Le Saux C.J., Cohen M., Alysandratos K.D., Wang B., Wolters P., et al. 2022. Human alveolar type 2 epithelium transdifferentiates into metaplastic KRT5+ basal cells. Nat. Cell Biol. 24:10–23. 10.1038/s41556-021-00809-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H., and Chung J.H.. 2015. Overview of clinicopathologic features of ALK-rearranged lung adenocarcinoma and current diagnostic testing for ALK rearrangement. Transl. Lung Cancer Res. 4:149–155. 10.3978/j.issn.2218-6751.2014.12.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H., Jang S.J., Chung D.H., Yoo S.B., Sun P., Jin Y., Nam K.H., Paik J.H., and Chung J.H.. 2013. A comprehensive comparative analysis of the histomorphological features of ALK-rearranged lung adenocarcinoma based on driver oncogene mutations: Frequent expression of epithelial-mesenchymal transition markers than other genotype. PLoS One. 8:e76999. 10.1371/journal.pone.0076999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.J., Park C.K., Yeo C.D., Park K., Rhee C.K., Kim J., Kim S.J., Lee S.H., Lee K.Y., and Yoon H.K.. 2014. Simultaneous diagnostic platform of genotyping EGFR, KRAS, and ALK in 510 Korean patients with non-small-cell lung cancer highlights significantly higher ALK rearrangement rate in advanced stage. J. Surg. Oncol. 110:245–251. 10.1002/jso.23646 [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Sakao Y., Ito S., Park J., Kuroda H., Sakakura N., Usami N., Mitsudomi T., and Yatabe Y.. 2013. Transformation to sarcomatoid carcinoma in ALK-rearranged adenocarcinoma, which developed acquired resistance to crizotinib and received subsequent chemotherapies. J. Thorac. Oncol. 8:e75–e78. 10.1097/JTO.0b013e318293d96f [DOI] [PubMed] [Google Scholar]
- Koyama, S., Akbay E.A., Li Y.Y., Aref A.R., Skoulidis F., Herter-Sprie G.S., Buczkowski K.A., Liu Y., Awad M.M., Denning W.L., et al. 2016. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T-cell activity in the lung tumor microenvironment. Cancer Res. 76:999–1008. 10.1158/0008-5472.CAN-15-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levacq, D., D’Haene N., de Wind R., Remmelink M., and Berghmans T.. 2016. Histological transformation of ALK rearranged adenocarcinoma into small cell lung cancer: A new mechanism of resistance to ALK inhibitors. Lung Cancer. 102:38–41. 10.1016/j.lungcan.2016.10.012 [DOI] [PubMed] [Google Scholar]
- Li, F., Han X., Li F., Wang R., Wang H., Gao Y., Wang X., Fang Z., Zhang W., Yao S., et al. 2015. LKB1 inactivation elicits a redox imbalance to modulate non-small cell lung cancer plasticity and therapeutic response. Cancer Cell. 27:698–711. 10.1016/j.ccell.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Pan Y., Wang R., Sun Y., Hu H., Shen X., Lu Y., Shen L., Zhu X., and Chen H.. 2013. ALK-Rearranged lung cancer in Chinese: A comprehensive assessment of clinicopathology, IHC, FISH and RT-PCR. PLoS One. 8:e69016. 10.1371/journal.pone.0069016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Zhuang X., Pan C.H., Yan Y., Thummalapalli R., Hallin J., Torborg S.R., Singhal A., Chang J.C., Manchado E., et al. 2023. Alveolar differentiation drives resistance to KRAS inhibition in lung adenocarcinoma. Cancer Discov. 10.1158/2159-8290.CD-23-0289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, G., Li C., Li P.S., Fang W.Z., Xu H.P., Gong Y.H., Zhu Z.F., Hu Y., Liang W.H., Chu Q., et al. 2020. Genomic origin and EGFR-TKI treatments of pulmonary adenosquamous carcinoma. Ann. Oncol. 31:517–524. 10.1016/j.annonc.2020.01.014 [DOI] [PubMed] [Google Scholar]
- Lin, J.J., Riely G.J., and Shaw A.T.. 2017. Targeting ALK: Precision medicine takes on drug resistance. Cancer Discov. 7:137–155. 10.1158/2159-8290.CD-16-1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddalo, D., Manchado E., Concepcion C.P., Bonetti C., Vidigal J.A., Han Y.C., Ogrodowski P., Crippa A., Rekhtman N., de Stanchina E., et al. 2014. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 516:423–427. 10.1038/nature13902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen, L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainardi, S., Mijimolle N., Francoz S., Vicente-Dueñas C., Sánchez-García I., and Barbacid M.. 2014. Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. Proc. Natl. Acad. Sci. USA. 111:255–260. 10.1073/pnas.1320383110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makimoto, G., Ohashi K., Tomida S., Nishii K., Matsubara T., Kayatani H., Higo H., Ninomiya K., Sato A., Watanabe H., et al. 2019. Rapid acquisition of alectinib resistance in ALK-positive lung cancer with high tumor mutation burden. J. Thorac. Oncol. 14:2009–2018. 10.1016/j.jtho.2019.07.017 [DOI] [PubMed] [Google Scholar]
- Maynard, A., McCoach C.E., Rotow J.K., Harris L., Haderk F., Kerr D.L., Yu E.A., Schenk E.L., Tan W., Zee A., et al. 2020. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell. 182:1232–1251.e22. 10.1016/j.cell.2020.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, S., Ikushima S., Ono R., Awano N., Kondo K., Furuhata Y., Fukumoto K., and Kumasaka T.. 2016. Transformation to small-cell lung cancer as a mechanism of acquired resistance to crizotinib and alectinib. Jpn. J. Clin. Oncol. 46:170–173. 10.1093/jjco/hyv173 [DOI] [PubMed] [Google Scholar]
- Mizuta, H., Okada K., Araki M., Adachi J., Takemoto A., Kutkowska J., Maruyama K., Yanagitani N., Oh-Hara T., Watanabe K., et al. 2021. Gilteritinib overcomes lorlatinib resistance in ALK-rearranged cancer. Nat. Commun. 12:1261. 10.1038/s41467-021-21396-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj, A.S., Lahtela J., Hemmes A., Pellinen T., Blom S., Devlin J.R., Salmenkivi K., Kallioniemi O., Mäyränpää M.I., Närhi K., and Verschuren E.W.. 2017. Cell of origin links histotype Spectrum to immune microenvironment diversity in non-small-cell lung cancer driven by mutant Kras and loss of Lkb1. Cell Rep. 18:673–684. 10.1016/j.celrep.2016.12.059 [DOI] [PubMed] [Google Scholar]
- Nicorici, D., Şatalan M., Edgren H., Kangaspeska S., Murumägi A., Kallioniemi O., Virtanen S., and Kilkku O.. 2014. FusionCatcher – a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 10.1101/011650 (Preprint posted November 19, 2014). [DOI] [Google Scholar]
- Ou, S.I., Lee T.K., Young L., Fernandez-Rocha M.Y., Pavlick D., Schrock A.B., Zhu V.W., Milliken J., Ali S.M., and Gitlitz B.J.. 2017. Dual occurrence of ALK G1202R solvent front mutation and small cell lung cancer transformation as resistance mechanisms to second generation ALK inhibitors without prior exposure to crizotinib. Pitfall of solely relying on liquid re-biopsy? Lung Cancer. 106:110–114. 10.1016/j.lungcan.2017.02.005 [DOI] [PubMed] [Google Scholar]
- Oya, Y., Yoshida T., Uemura T., Murakami Y., Inaba Y., and Hida T.. 2018. Serum ProGRP and NSE levels predicting small cell lung cancer transformation in a patient with ALK rearrangement-positive non-small cell lung cancer: A case report. Oncol. Lett. 16:4219–4222. 10.3892/ol.2018.9158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik, J.H., Choi C.M., Kim H., Jang S.J., Choe G., Kim D.K., Kim H.J., Yoon H., Lee C.T., Jheon S., et al. 2012. Clinicopathologic implication of ALK rearrangement in surgically resected lung cancer: A proposal of diagnostic algorithm for ALK-rearranged adenocarcinoma. Lung Cancer. 76:403–409. 10.1016/j.lungcan.2011.11.008 [DOI] [PubMed] [Google Scholar]
- Park, S., Han J., and Sun J.M.. 2019. Histologic transformation of ALK-rearranged adenocarcinoma to squamous cell carcinoma after treatment with ALK inhibitor. Lung Cancer. 127:66–68. 10.1016/j.lungcan.2018.11.027 [DOI] [PubMed] [Google Scholar]
- Popat, S., Gonzalez D., Min T., Swansbury J., Dainton M., Croud J.G., Rice A.J., and Nicholson A.G.. 2012. ALK translocation is associated with ALK immunoreactivity and extensive signet-ring morphology in primary lung adenocarcinoma. Lung Cancer. 75:300–305. 10.1016/j.lungcan.2011.07.017 [DOI] [PubMed] [Google Scholar]
- Preusser, M., Berghoff A.S., Ilhan-Mutlu A., Magerle M., Dinhof C., Widhalm G., Dieckmann K., Marosi C., Wöhrer A., Hackl M., et al. 2013. ALK gene translocations and amplifications in brain metastases of non-small cell lung cancer. Lung Cancer. 80:278–283. 10.1016/j.lungcan.2013.01.019 [DOI] [PubMed] [Google Scholar]
- Pyo, K.H., Lim S.M., Kim H.R., Sung Y.H., Yun M.R., Kim S.M., Kim H., Kang H.N., Lee J.M., Kim S.G., et al. 2017. Establishment of a conditional transgenic mouse model recapitulating EML4-ALK-positive human non-small cell lung cancer. J. Thorac. Oncol. 12:491–500. 10.1016/j.jtho.2016.10.022 [DOI] [PubMed] [Google Scholar]
- Qin, Z., Sun H., Yue M., Pan X., Chen L., Feng X., Yan X., Zhu X., and Ji H.. 2021. Phase separation of EML4-ALK in firing downstream signaling and promoting lung tumorigenesis. Cell Discov. 7:33. 10.1038/s41421-021-00270-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanal-Villalonga, Á., Chan J.M., Yu H.A., Pe’er D., Sawyers C.L., Sen T., and Rudin C.M.. 2020. Lineage plasticity in cancer: A shared pathway of therapeutic resistance. Nat. Rev. Clin. Oncol. 17:360–371. 10.1038/s41571-020-0340-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanal-Villalonga, A., Taniguchi H., Zhan Y.A., Hasan M.M., Chavan S.S., Meng F., Uddin F., Allaj V., Manoj P., Shah N.S., et al. 2021. Comprehensive molecular characterization of lung tumors implicates AKT and MYC signaling in adenocarcinoma to squamous cell transdifferentiation. J. Hematol. Oncol. 14:170. 10.1186/s13045-021-01186-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins, E.L., Okubo T., Xue Y., Brass D.M., Auten R.L., Hasegawa H., Wang F., and Hogan B.L.. 2009. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 4:525–534. 10.1016/j.stem.2009.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redaelli, S., Ceccon M., Zappa M., Sharma G.G., Mastini C., Mauri M., Nigoghossian M., Massimino L., Cordani N., Farina F., et al. 2018. Lorlatinib treatment elicits multiple on- and off-target mechanisms of resistance in ALK-driven cancer. Cancer Res. 78:6866–6880. 10.1158/0008-5472.CAN-18-1867 [DOI] [PubMed] [Google Scholar]
- Rock, J.R., Barkauskas C.E., Cronce M.J., Xue Y., Harris J.R., Liang J., Noble P.W., and Hogan B.L.. 2011. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc. Natl. Acad. Sci. USA. 108:E1475–E1483. 10.1073/pnas.1117988108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodig, S.J., Mino-Kenudson M., Dacic S., Yeap B.Y., Shaw A., Barletta J.A., Stubbs H., Law K., Lindeman N., Mark E., et al. 2009. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin. Cancer Res. 15:5216–5223. 10.1158/1078-0432.CCR-09-0802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, J., Richards M.W., Choi J., Fry A.M., and Bayliss R.. 2021. Phase-separated foci of EML4-ALK facilitate signalling and depend upon an active kinase conformation. EMBO Rep. 22:e53693. 10.15252/embr.202153693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld, A.J., Chan J.M., Kubota D., Sato H., Rizvi H., Daneshbod Y., Chang J.C., Paik P.K., Offin M., Arcila M.E., et al. 2020. Tumor analyses reveal squamous transformation and off-target alterations as early resistance mechanisms to first-line osimertinib in EGFR-mutant lung cancer. Clin. Cancer Res. 26:2654–2663. 10.1158/1078-0432.CCR-19-3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiihara, J., Ohyanagi F., Amari H., Toda M., Tahara H., Yuzawa M., Maeda Y., Nomura M., Mizushina Y., Nagai Y., et al. 2021. Dramatic response to alectinib in a patient with ALK-rearranged squamous cell lung cancer. Thorac. Cancer. 12:2420–2423. 10.1111/1759-7714.14092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda, M., Choi Y.L., Enomoto M., Takada S., Yamashita Y., Ishikawa S., Fujiwara S., Watanabe H., Kurashina K., Hatanaka H., et al. 2007. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 448:561–566. 10.1038/nature05945 [DOI] [PubMed] [Google Scholar]
- Soda, M., Takada S., Takeuchi K., Choi Y.L., Enomoto M., Ueno T., Haruta H., Hamada T., Yamashita Y., Ishikawa Y., et al. 2008. A mouse model for EML4-ALK-positive lung cancer. Proc. Natl. Acad. Sci. USA. 105:19893–19897. 10.1073/pnas.0805381105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Z., Yu X., and Zhang Y.. 2017. Clinicopathological characteristics and survival of ALK, ROS1 and RET rearrangements in non-adenocarcinoma non-small cell lung cancer patients. Cancer Biol. Ther. 18:883–887. 10.1080/15384047.2016.1235660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., W.M. Mauck, 3rd, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive integration of single-cell data. Cell. 177:1888–1902 e1821. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland, K.D., Proost N., Brouns I., Adriaensen D., Song J.Y., and Berns A.. 2011. Cell of origin of small cell lung cancer: Inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 19:754–764. 10.1016/j.ccr.2011.04.019 [DOI] [PubMed] [Google Scholar]
- Sutherland, K.D., Song J.Y., Kwon M.C., Proost N., Zevenhoven J., and Berns A.. 2014. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc. Natl. Acad. Sci. USA. 111:4952–4957. 10.1073/pnas.1319963111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takegawa, N., Hayashi H., Iizuka N., Takahama T., Ueda H., Tanaka K., Takeda M., and Nakagawa K.. 2016. Transformation of ALK rearrangement-positive adenocarcinoma to small-cell lung cancer in association with acquired resistance to alectinib. Ann. Oncol. 27:953–955. 10.1093/annonc/mdw032 [DOI] [PubMed] [Google Scholar]
- Tamborero, D., Rubio-Perez C., Deu-Pons J., Schroeder M.P., Vivancos A., Rovira A., Tusquets I., Albanell J., Rodon J., Tabernero J., et al. 2018. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med. 10:25. 10.1186/s13073-018-0531-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, S., Xue Y., Qin Z., Fang Z., Sun Y., Yuan C., Pan Y., Zhao Y., Tong X., Zhang J., et al. 2023. Counteracting lineage-specific transcription factor network finely tunes lung adeno-to-squamous transdifferentiation through remodeling tumor immune microenvironment. Natl. Sci. Rev. 10:nwad028. 10.1093/nsr/nwad028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, X., Patel A.S., Kim E., Li H., Chen Y., Li S., Liu S., Dilly J., Kapner K.S., Xue Y., et al. 2023. Adeno-to-squamous transition drives resistance to KRAS inhibition in LKB1 mutant lung cancer. bioRxiv. 10.1101/2023.09.07.556567 (Preprint posted September 10, 2023) [DOI] [PubMed] [Google Scholar]
- Torres-García, W., Zheng S., Sivachenko A., Vegesna R., Wang Q., Yao R., Berger M.F., Weinstein J.N., Getz G., and Verhaak R.G.. 2014. Prada: Pipeline for RNA sequencing data analysis. Bioinformatics. 30:2224–2226. 10.1093/bioinformatics/btu169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulpule, A., Guan J., Neel D.S., Allegakoen H.R., Lin Y.P., Brown D., Chou Y.T., Heslin A., Chatterjee N., Perati S., et al. 2021. Kinase-mediated RAS signaling via membraneless cytoplasmic protein granules. Cell. 184:2649–2664.e18. 10.1016/j.cell.2021.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, S., Shukuya T., Hayashi T., Suzuki M., Kondo A., Arai Y., Takeshige T., Ninomiya H., and Takahashi K.. 2021. Transformation from adenocarcinoma to squamous cell lung carcinoma with MET amplification after lorlatinib resistance: A case report. Thorac. Cancer. 12:715–719. 10.1111/1759-7714.13829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino, A.V., Kanno Y., Ferdinand J.R., and O’Shea J.J.. 2015. Mechanisms of Jak/STAT signaling in immunity and disease. J. Immunol. 194:21–27. 10.4049/jimmunol.1401867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F., Qin J., Xie F., Wu Q., and Lu H.. 2020. Transformation of EML4-ALK fusion-positive adenocarcinoma into squamous cell carcinoma in association with acquired resistance to crizotinib. Lung Cancer. 140:118–120. 10.1016/j.lungcan.2020.01.001 [DOI] [PubMed] [Google Scholar]
- Wang, R., Pan Y., Li C., Zhang H., Garfield D., Li Y., Ye T., Hu H., Luo X., Li H., et al. 2014. Analysis of major known driver mutations and prognosis in resected adenosquamous lung carcinomas. J. Thorac. Oncol. 9:760–768. 10.1097/JTO.0b013e3182a406d1 [DOI] [PubMed] [Google Scholar]
- Wu, F., Fan J., He Y., Xiong A., Yu J., Li Y., Zhang Y., Zhao W., Zhou F., Li W., et al. 2021. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat. Commun. 12:2540. 10.1038/s41467-021-22801-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Q., Tian Y., Zhang J., Tong X., Huang H., Li S., Zhao H., Tang Y., Yuan C., Wang K., et al. 2018. In vivo CRISPR screening unveils histone demethylase UTX as an important epigenetic regulator in lung tumorigenesis. Proc. Natl. Acad. Sci. USA. 115:E3978–E3986. 10.1073/pnas.1716589115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Huang L., Futtner C., Schwab B., Rampersad R.R., Lu Y., Sporn T.A., Hogan B.L., and Onaitis M.W.. 2014. The cell of origin and subtype of K-Ras-induced lung tumors are modified by Notch and Sox2. Genes Dev. 28:1929–1939. 10.1101/gad.243717.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., Shen Y., Jiang J., Liu Z., and Mu C.. 2021. Transformation of two cases of lung adenocarcinoma into pulmonary sarcomatoid carcinoma following treatment. Can. Respir. J. 2021:6661772. 10.1155/2021/6661772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, A., Tsuta K., Nakamura H., Kohno T., Takahashi F., Asamura H., Sekine I., Fukayama M., Shibata T., Furuta K., and Tsuda H.. 2011. Comprehensive histologic analysis of ALK-rearranged lung carcinomas. Am. J. Surg. Pathol. 35:1226–1234. 10.1097/PAS.0b013e3182233e06 [DOI] [PubMed] [Google Scholar]
- Zhang, H., Fillmore Brainson C., Koyama S., Redig A.J., Chen T., Li S., Gupta M., Garcia-de-Alba C., Paschini M., Herter-Sprie G.S., et al. 2017. Lkb1 inactivation drives lung cancer lineage switching governed by Polycomb Repressive Complex 2. Nat. Commun. 8:14922. 10.1038/ncomms14922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Lan Y., Xu J., Quan F., Zhao E., Deng C., Luo T., Xu L., Liao G., Yan M., et al. 2019a. CellMarker: A manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 47:D721–D728. 10.1093/nar/gky900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.C., Wang J., Shao G.G., Wang Q., Qu X., Wang B., Moy C., Fan Y., Albertyn Z., Huang X., et al. 2019b. Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat. Commun. 10:1772. 10.1038/s41467-019-09762-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Qin Y., Xu H., Yao Q., Gao Y., Feng Y., and Ren J.. 2021. Case report: A case report of a histological transformation of ALK-rearranged adenocarcinoma with high expression of PD-L1 to squamous cell carcinoma after treatment with alectinib. Pathol. Oncol. Res. 27:637745. 10.3389/pore.2021.637745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y.C., Liao X.H., Wang W.X., Xu C.W., Zhuang W., Zhong L.H., Du K.Q., Chen Y.P., Chen G., and Fang M.Y.. 2017. Patients harboring ALK rearrangement adenocarcinoma after acquired resistance to crizotinib and transformation to small-cell lung cancer: A case report. OncoTargets Ther. 10:3187–3192. 10.2147/OTT.S139718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito Marino, F., Liguori G., Aquino G., La Mantia E., Bosari S., Ferrero S., Rosso L., Gaudioso G., De Rosa N., Scrima M., et al. 2015. Intratumor heterogeneity of ALK-Rearrangements and homogeneity of EGFR-mutations in mixed lung adenocarcinoma. PLoS One. 10:e0139264. 10.1371/journal.pone.0139264 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows clinical characteristics of ALK-rearranged LUAD (n = 60) and LUSC (n = 8) patients.
shows PFS of ALK-rearranged LUAD (n = 60) and LUSC (n = 8) patients after alectinib treatments.
shows PFS of ALK-rearranged LUAD (n = 36) patients after ALK TKI treatments.
shows sample information and squamous signature status of ALK-rearranged LUAD (n = 36) patients.
shows sgRNA sequences and the primers for knockout efficiency detection.
is the source file for Fig. 5.
is the source file for Fig. S4.
Data Availability Statement
RNA-seq data and scRNA-seq data of mouse tumors, RNA-seq data, and WGS data of human LUAS samples have been deposited in the National Omics Data Encyclopedia (accession no. OEP003086, OEP001032). RNA-seq data of human ALK-rearranged tumors and scRNA-seq data of human LUAD have been deposited in the OMIX, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (accession no. OMIX002552, OMIX002554).