Abstract
Dengue virus serotype 2, genotype Cosmopolitan (DENV-2-GII), is one of the most widespread DENV strains globally. In the USA, DENV-2 epidemics have been dominated by DENV-2 genotype Asian-American (DENV-2-GIII), and the first cases of DENV-2-GII were only described in 2019, in Peru, and in 2021 in Brazil. To gain new information about the circulation of DENV-2-GII in Brazil, we sequenced 237 DENV-2 confirmed cases sampled between March 2021 and March 2023 and revealed that DENV-2-GII is already present in all geographic regions of Brazil. The phylogeographic analysis inferred that DENV-2-GII was introduced at least four times in Brazil, between May 2020 and August 2022, generating multiple clades that spread throughout the country with different success. Despite multiple introductions of DENV-2-GII, analysis of the country-wide laboratory surveillance data showed that the Brazilian dengue epidemic in 2022 was dominated by DENV-1 in most states. We hypothesize that massive circulation of DENV-2-GIII in previous years in Brazil might have created a population immune barrier against symptomatic homotypic reinfections by DENV-2-GII, leading to sustained cryptic circulation in asymptomatic cases and localized outbreaks of this new genotype. In summary, our study stresses the importance of arboviral genomic surveillance to close monitoring and better understanding the potential impact of DENV-2-GII in the coming years.
Keywords: dengue virus serotype 2, genotype Cosmopolitan, Brazil, genomic surveillance, phylogeography
Research letter
Dengue virus serotype 2 (DENV-2) can be divided into five endemic/epidemic (nonsylvatic) genotypes: American, Cosmopolitan, Asian-American, Asian-II, and Asian-I (Twiddy et al. 2002), also known as genotypes I–V, respectively. In the USA, the DENV-2 American genotype (DENV-2-GI) was the first to be identified in the 1940s. However, it was later replaced by the Asian-American genotype (DENV-2-GIII) in the 1980s, and cases of DENV-2-GI were not found since then (Allicock et al. 2012). In Brazil, DENV-2-GIII was first introduced in the country in the 1990s and at least three other introductions happened between 1990 and 2014, all with origin in Caribbean or Northern South America countries (de Jesus et al. 2020; Brito et al. 2021). Besides DENV-2, in Brazil, all four DENV serotypes were already identified, presenting a complex dynamics of introductions, co-circulation, and alternation of dominance, which is associated with an increasing number of severe dengue cases and deaths in recent years (Fares et al. 2015).
In 2021, the first cases of DENV-2 Cosmopolitan genotype (DENV-2-GII) were identified in the Brazilian states of Goiás (GO) (Giovanetti et al. 2022) and Acre (AC) (Amorim et al. 2023), located in the Central-Western and Northern country regions, respectively. Phylogenetic analysis suggested that the Brazilian DENV-2-GII sequences derived from an outbreak of this genotype in Peru in 2019 (García et al. 2022) representing the first documented introduction of DENV-2-GII in the American continent. DENV-2-GII is one of the most widespread genotypes, circulating in the Asia–Pacific, the Middle East, Africa, and Oceania, substantially contributing to the global dengue burden (Yenamandra et al. 2021). Here, we present new data on the DENV-2-GII spread in Brazil, showing that shortly after multiple introductions in the country, it is already present in all geographical regions.
The genomic surveillance for DENV was performed in the Brazilian states of Amazonas (AM), Pernambuco (PE), São Paulo (SP), Paraná (PR), Santa Catarina (SC), and Rio Grande do Sul (RS) by each Central State Laboratories (LACEN) and Fiocruz laboratories. A selection of DENV-2-positive samples in molecular tests was submitted for whole-genome amplification and sequencing using Illumina’s Viral Surveillance Panel or COVIDseq Test adapted to DENV-2. The genomes were assembled in ViralFlow software (Dezordi et al. 2022) or using Geneious Prime 2022, and consensus sequences were genotyped using online tools (methods detailed in Supplementary text).
Among the 237 DENV-2 sequenced samples, we identified 60 DENV-2-GII genomes (Supplementary Table S1) and 177 DENV-2-GIII, representing 0.05–0.9 per cent of all DENV-2 cases detected in the studied Brazilian states between March 2021 and March 2023 (Supplementary Table S2). DENV-2-GII genomes were aligned with a representative global dataset, and the maximum likelihood phylogenetic analysis revealed that all Brazilian genomes, from this and previous studies (Giovanetti et al. 2022; Amorim et al. 2023), clustered in a monophyletic clade with sequences from Peru and Bangladesh (Supplementary Fig. S1). Our phylogeographic analysis estimated that DENV-2-GII was introduced from Bangladesh into Peru between May 2017 (December 2016—September 2017, 95 per cent high posterior density [HPD]) and April 2019 (November 2018—August 2019, 95 per cent HPD), and from there, it was introduced in Brazil at least four times (Fig. 1A). The first introduction occurred through the AC state (North) between February 2020 (June 2019—September 2020, 95 per cent HPD) and May 2020 (June 2019—November 2020, 95 per cent HPD), and from AC, DENV-2-GII disseminated to all other four Brazilian regions represented by the states of GO (Central-West), PE (Northeast), SP (Southeast), and SC (South). The oldest DENV-2-GII Brazilian samples collected in 2021 in AC (February to March), PE (July), and GO (November) clustered within this clade. A second introduction of DENV-2-GII happened in the state of SP between June 2020 (November 2019—January 2021, 95 per cent HPD) and March 2021 (September 2020—July 2021, 95 per cent HPD), and from there, DENV-2-GII spread southwards to the states of PR, SC, and RS. A third introduction of DENV-2-GII occurred between February 2020 (June 2019—September 2020, 95 per cent HPD) and January 2022 (October 2021—March 2022, 95 per cent HPD) in the state of RS, without evidence of further dissemination to other Brazilian states. Finally, the most recent introduction happened in AM state between May 2020 (July 2019—March 2021, 95 per cent HPD) and August 2022 (June 2022—October 2022, 95 per cent HPD), causing an outbreak in cities close to the border with Peru and Colombia (Naveca 2023). A previous study supports that aerial transportation of humans and/or vector mosquitoes is an important driving force for the spatial spread of DENV between Brazilian states (Nunes et al. 2014). Despite the air travel restrictions being implemented during the first year of the COVID-19 pandemic in Brazil to restrict human mobility, the introduction and dispersion of the first DENV-2-GII viruses in Brazil probably occurred in that time frame (May 2020—March 2021). This supports that new DENV strains could be introduced from abroad and establish local transmission chains in Brazil even during the periods of more restricted air flux of both international and national passengers.
Figure 1.
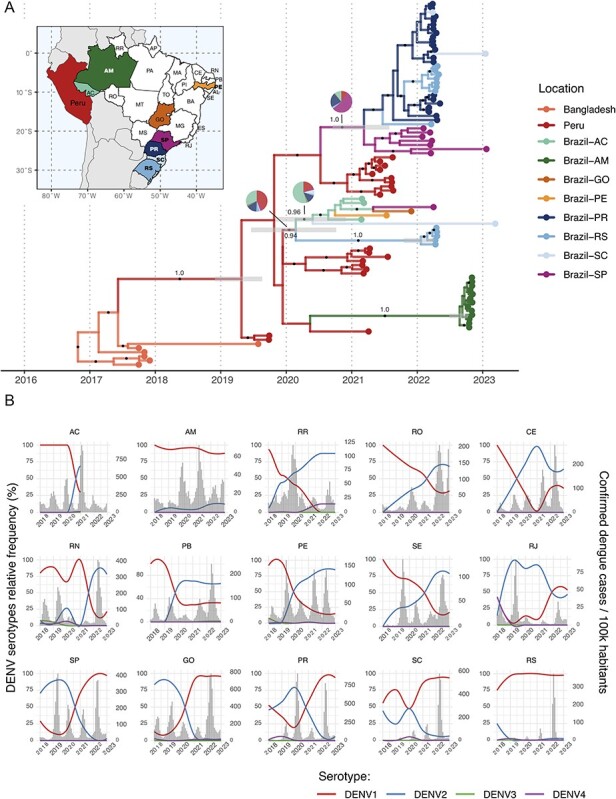
Spatiotemporal evolution of DENV-2 genotype II (Cosmopolitan) in South America and circulation dynamics of DENV serotypes in Brazil in recent years. (A) Time-scaled phylogenetic tree of eighty-eight DENV-2-GII genomes with ancestral locations inferred by the phylogeographic model. The branches are colored according to the most likely ancestral location, and pie charts show the posterior probability of each location for key nodes with high uncertainty. Error bars show the 95 per cent HPD of node heights related to introductions to Brazil. The dots on branches indicate a posterior probability higher than 0.9, and key branches are annotated with the posterior support. Inset map depicts the Brazilian states from which DENV-2 sequences were sampled. Colors on the map and phylogenetic tree correspond and the states directly sampled in this study are highlighted in bold. (B) The relative frequency of each DENV serotype (lines) and confirmed dengue cases per 100k habitants (histogram) for a selection of Brazilian states between 2018 and 2022. The acronyms for depicted states: AC—Acre, AM—Amazonas, RR—Roraima, RO—Rondônia, CE—Ceará, RN—Rio Grande do Norte, PB—Paraíba, PE—Pernambuco, SE—Sergipe, RJ—Rio de Janeiro, SP—São Paulo, GO—Goiás, PR—Paraná, SC—Santa Catarina, and RS—Rio Grande do Sul.
To assess the potential of DENV-2-GII to spread and trigger relevant outbreaks in Brazil, we analyzed the dynamics of DENV serotypes’ circulation in the country as identified through molecular tests by the laboratory surveillance and informed in the national system of diseases of compulsory notification (SINAN). Between 2018 and 2022, the Brazilian dengue epidemic was dominated by DENV-1 and DENV-2, with significant regional differences (Fig. 1B). A predominance of DENV-2 after 2020, which could indicate a genotype II outbreak, was observed in the northern states of AC (where genomic surveillance detected DENV-2-GII circulation), Rondônia and Roraima, in the southeastern state of Rio de Janeiro (RJ), and in the northeastern states of Ceará (CE), Paraíba (PB), Rio Grande do Norte (RN), Sergipe (SE), and PE. In the latter, only one sequence of DENV-2-GII was found compared to eighty sequences of DENV-2-GIII in 2021, indicating that the dominance of DENV-2 in PE at that time was not caused by genotype II, which could also be the case for the neighboring northeastern states. A high prevalence of DENV-2 was observed in several states between 2018 and 2020, but there was no evidence of circulation of DENV-2-GII in Brazil before 2021 (Supplementary Figure S2). In other states, including AM and RS, where genomic surveillance detected recent introductions of DENV-2-GII in 2022, DENV-1 was responsible for most dengue cases in the whole period supporting that DENV-2-GII only caused localized outbreaks in those states. In fact, in 2022, DENV-1 was responsible for more than 80 per cent of the dengue cases in seventeen out of twenty-five Brazilian states with data available (Supplementary Figure S3). Finally, it is important to note that our study sampled only a fraction of Brazilian states, and a more comprehensive sampling regarding DENV-2, both in time and space, will be necessary to unveil the complete picture of genotype II spread and to assess the implications in future outbreaks.
In summary, this study confirms that DENV-2-GII is circulating nationally in Brazil, expanding the findings of previous localized studies (Giovanetti et al. 2022; Amorim et al. 2023). Our phylogenetic analysis unveiled multiple introductions of this genotype into Brazil from Peru, generating multiple clades that spread in the country with different success. Despite the wide geographic dispersion of DENV-2-GII, dengue epidemics in 2021–2 in most Brazilian states were dominated by DENV-1, supporting that circulation of DENV-2-GIII in previous years probably created a higher population immunity barrier against homotypic reinfections by DENV-2-GII than against heterotypic reinfections by DENV-1. At the same time, the existence of asymptomatic homotypic reinfections (Murphy and Whitehead 2011; Tan et al. 2018; López et al. 2022) combined with the heterogeneous population immune landscape across Brazilian regions could have created an epidemic scenario for sustained cryptic circulation, rapid spread, and localized outbreaks of DENV-2-GII across the country’s territory. Indeed, DENV-2-GII was possibly responsible for most DENV-2 infections in Brazil in 2022, and this warrants close monitoring to better understand the potential impact of this genotype in the coming years.
Supplementary Material
Acknowledgements
We acknowledge all data contributors, i.e. authors and their originating laboratories responsible for obtaining and timely sharing dengue virus genomes, including metadata, via GenBank or other public databases. We would also like to thank Dr Edson Delatorre for his help with the design of the map. We appreciate the support of FIOCRUZ Genomics Surveillance Network members.
Contributor Information
Tiago Gräf, Laboratório de Virologia Molecular, Instituto Carlos Chagas, Fundação Oswaldo Cruz, Curitiba 81350-010, Brazil.
Caroline Do Nascimento Ferreira, Laboratório de Virologia Molecular, Instituto Carlos Chagas, Fundação Oswaldo Cruz, Curitiba 81350-010, Brazil.
Gustavo Barbosa de Lima, Núcleo de Plataformas Tecnológicas (NPT), Instituto Aggeu Magalhães (IAM), FIOCRUZ-Pernambuco, Recife, Pernambuco 50740-465, Brazil.
Raul Emídio de Lima, Núcleo de Plataformas Tecnológicas (NPT), Instituto Aggeu Magalhães (IAM), FIOCRUZ-Pernambuco, Recife, Pernambuco 50740-465, Brazil.
Lais Ceschini Machado, Departamento de Entomologia, Instituto Aggeu Magalhães (IAM)-Fundação Oswaldo Cruz-FIOCRUZ, Recife, Pernambuco 50670-420, Brazil; Núcleo de Bioinformática (NBI), Instituto Aggeu Magalhães (IAM), FIOCRUZ-Pernambuco, Recife, Pernambuco 50740-465, Brazil.
Tulio de Lima Campos, Núcleo de Bioinformática (NBI), Instituto Aggeu Magalhães (IAM), FIOCRUZ-Pernambuco, Recife, Pernambuco 50740-465, Brazil.
Michelle Orane Schemberger, Laboratório de Ciências e Tecnologias Aplicadas em Saúde, Instituto Carlos Chagas, Fundação Oswaldo Cruz, Curitiba 81350-010, Brazil.
Helisson Faoro, Laboratório de Ciências e Tecnologias Aplicadas em Saúde, Instituto Carlos Chagas, Fundação Oswaldo Cruz, Curitiba 81350-010, Brazil.
Marcelo Henrique Santos Paiva, Núcleo de Ciências da Vida, Universidade Federal de Pernambuco (UFPE), Centro Acadêmico do Agreste-Rodovia BR-104, km 59-Nova Caruaru, Caruaru 55002-970, Brazil.
Matheus Filgueira Bezerra, Departamento de Microbiologia, Instituto Aggeu Magalhães (IAM), FIOCRUZ-Pernambuco, Recife, Pernambuco 50740-465, Brazil.
Valdinete Nascimento, Laboratório de Ecologia de Doenças Transmissíveis na Amazônia, Instituto Leônidas e Maria Deane, Fiocruz, Manaus 69057-070, Brazil.
Victor Souza, Laboratório de Ecologia de Doenças Transmissíveis na Amazônia, Instituto Leônidas e Maria Deane, Fiocruz, Manaus 69057-070, Brazil.
Fernanda Nascimento, Laboratório de Ecologia de Doenças Transmissíveis na Amazônia, Instituto Leônidas e Maria Deane, Fiocruz, Manaus 69057-070, Brazil.
Matilde Mejía, Laboratório de Ecologia de Doenças Transmissíveis na Amazônia, Instituto Leônidas e Maria Deane, Fiocruz, Manaus 69057-070, Brazil.
Dejanane Silva, Laboratório de Ecologia de Doenças Transmissíveis na Amazônia, Instituto Leônidas e Maria Deane, Fiocruz, Manaus 69057-070, Brazil.
Yasmin Silva de Oliveira, Laboratório de Ecologia de Doenças Transmissíveis na Amazônia, Instituto Leônidas e Maria Deane, Fiocruz, Manaus 69057-070, Brazil.
Luciana Gonçalves, Laboratório de Ecologia de Doenças Transmissíveis na Amazônia, Instituto Leônidas e Maria Deane, Fiocruz, Manaus 69057-070, Brazil; Fundação de Vigilância em Saúde do Amazonas—Dra Rosemary Costa Pinto, Manaus, Amazonas 69093-018, Brazil.
Tatyana Costa Amorim Ramos, Fundação de Vigilância em Saúde do Amazonas—Dra Rosemary Costa Pinto, Manaus, Amazonas 69093-018, Brazil.
Daniel Barros de Castro, Fundação de Vigilância em Saúde do Amazonas—Dra Rosemary Costa Pinto, Manaus, Amazonas 69093-018, Brazil.
Ana Ruth Arcanjo, Laboratório Central de Saúde Pública do Amazonas (LACEN-AM), Manaus, Amazonas 69020-040, Brazil.
Herton Augusto Pinheiro Dantas, Laboratório de Fronteira de Tabatinga (LAFRON-AM), Tabatinga, Amazonas 69640-000, Brazil.
Mayra Marinho Presibella, Laboratório Central de Saúde Pública do Estado do Paraná (LACEN-PR), São José dos Pinhais, Paraná 83060-500, Brazil.
Sandra Bianchini Fernandes, Laboratório Central de Saúde Pública do Estado de Santa Catarina (LACEN-SC), Florianópolis, Santa Catarina 88010-001, Brazil.
Tatiana Schaffer Gregianini, Laboratório Central de Saúde Pública do Rio Grande do Sul (LACEN-RS), Porto Alegre, Rio Grande do Sul 90610-000, Brazil.
Keilla Maria Paz E Silva, Laboratório Central de Saúde Pública de Pernambuco (LACEN-PE), Recife, Pernambuco 50050-210, Brazil.
Claudio Tavares Sacchi, Laboratorio Estrategico, Instituto Adolfo Lutz (IAL), Sao Paulo 01246-902, Brazil.
Ana Cecília Ribeiro Cruz, Department of Arbovirology and Hemorrhagic Fevers, Evandro Chagas Institute, Health and Environment Surveillance Secretariat, Ministry of Health, Ananindeua, Para 670030-000, Brazil; Institute of Biological Sciences, Federal University of Pará, Belém, Para 66075-110, Brazil.
Claudia Nunes Duarte dos Santos, Laboratório de Virologia Molecular, Instituto Carlos Chagas, Fundação Oswaldo Cruz, Curitiba 81350-010, Brazil.
Ana Maria Bispo de Filippis, Laboratório de Arbovírus e Vírus Hemorrágicos, Instituto Oswaldo Cruz, Fiocruz, Rio de Janeiro 21040-360, Brazil.
Gonzalo Bello, Laboratório de Arbovírus e Vírus Hemorrágicos, Instituto Oswaldo Cruz, Fiocruz, Rio de Janeiro 21040-360, Brazil.
Gabriel Luz Wallau, Departamento de Entomologia, Instituto Aggeu Magalhães (IAM)-Fundação Oswaldo Cruz-FIOCRUZ, Recife, Pernambuco 50670-420, Brazil; Núcleo de Bioinformática (NBI), Instituto Aggeu Magalhães (IAM), FIOCRUZ-Pernambuco, Recife, Pernambuco 50740-465, Brazil; Department of Arbovirology, Bernhard Nocht Institute for Tropical Medicine, WHO Collaborating Center for Arbovirus and Hemorrhagic Fever Reference and Research, National Reference Center for Tropical Infectious Diseases, Bernhard-Nocht-Straße 74, Hamburg 20359, Germany.
Richard Steiner Salvato, Laboratório Central de Saúde Pública do Rio Grande do Sul (LACEN-RS), Porto Alegre, Rio Grande do Sul 90610-000, Brazil.
Felipe Naveca, Laboratório de Ecologia de Doenças Transmissíveis na Amazônia, Instituto Leônidas e Maria Deane, Fiocruz, Manaus 69057-070, Brazil; Laboratório de Arbovírus e Vírus Hemorrágicos, Instituto Oswaldo Cruz, Fiocruz, Rio de Janeiro 21040-360, Brazil.
Data availability
The data for this article are available at https://github.com/akograf/Multiple_introductions_and_spread_of_DENV-2-GII_in_Brazil.git.
Supplementary data
Supplementary data are available at Virus Evolution online.
Funding
This work was supported by funding from FAPEAM (Universal/AM call 2019; Rede Genômica de Vigilância em Saúde—REGESAM); Inova Fiocruz/Fundação Oswaldo Cruz (Inova Amazônia); Departamento de Ciência e Tecnologia (DECIT) of the Brazilian MoH; G.B. is supported by CNPq through a productivity research fellowships (304883/2020-4) and FAPERJ (Grant number E-26/202.896/2018).
Conflict of interest:
The authors report there are no competing interests to declare.
References
- Allicock O. M. et al. (2012) ‘Phylogeography and Population Dynamics of Dengue Viruses in the Americas’, Molecular Biology and Evolution, 29: 1533–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim M. T. et al. (2023) ‘Emergence of a New Strain of DENV-2 in South America: Introduction of the Cosmopolitan Genotype through the Brazilian-Peruvian Border’, Tropical Medicine and Infectious Disease, 8: 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito A. F. et al. (2021) ‘Lying in Wait: The Resurgence of Dengue Virus after the Zika Epidemic in Brazil’, Nature Communications, 12: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jesus J. G. et al. (2020) ‘Genomic Detection of a Virus Lineage Replacement Event of Dengue Virus Serotype 2 in Brazil, 2019’, Memorias Do Instituto Oswaldo Cruz, 115: e190423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezordi F. Z. et al. (2022) ‘ViralFlow: A Versatile Automated Workflow for SARS-CoV-2 Genome Assembly, Lineage Assignment, Mutations and Intrahost Variant Detection’, Viruses, 14: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares R. C. G. et al. (2015) ‘Epidemiological Scenario of Dengue in Brazil’, BioMed Research International, 2015: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García M. P. et al. (2022) ‘Emergence of the Cosmopolitan Genotype of Dengue Virus Serotype 2 (DENV2) in Madre de Dios, Peru, 2019’, Revista Peruana Medicina Experimental Salud Publica, 39: 126–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanetti M. et al. (2022) ‘Emergence of Dengue Virus Serotype 2 Cosmopolitan Genotype, Brazil’, Emerging Infectious Diseases, 28: 1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López L. et al. (2022) ‘Considering Waning Immunity to Better Explain Dengue Dynamics’, Epidemics, 41: 100630. [DOI] [PubMed] [Google Scholar]
- Murphy B. R., and Whitehead S. S. (2011) ‘Immune Response to Dengue Virus and Prospects for a Vaccine’, Annual Review of Immunology, 29: 1. [DOI] [PubMed] [Google Scholar]
- Naveca F. G. (2023), Investigação Sobre O Surto de Dengue No Segundo Semestre de 2022 Nos Municípios da Região Do Alto Solimões, Região da Tríplice-fronteira Brasil, Colômbia E Peru - Relatório Preliminar. Personal communication. <https://www.genomahcov.fiocruz.br/wp-content/uploads/2023/05/Investigacao_sobre_o_surto_de_DENV2_cosmopolita_no_Amazonas_Relatorio_preliminar_v2_assinado.pdf> accessed 18 May 2023.
- Nunes M. R. et al. (2014) ‘Air Travel is Associated with Intracontinental Spread of Dengue Virus Serotypes 1-3 in Brazil’, PLoS Negl Trop Dis, 8: e2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K. et al. (2018) ‘Autochthonous Spread of DENV-3 Genotype III in Malaysia Mitigated by Pre-existing Homotypic and Heterotypic Immunity’, Epidemiology and Infection, 146: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiddy S. S. et al. (2002) ‘Phylogenetic Relationships and Differential Selection Pressures among Genotypes of Dengue-2 Virus’, Virology, 298: 63–72. [DOI] [PubMed] [Google Scholar]
- Yenamandra S. P. et al. (2021) ‘Evolution, Heterogeneity and Global Dispersal of Cosmopolitan Genotype of Dengue Virus Type 2’, Scientific Reports, 11: 13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data for this article are available at https://github.com/akograf/Multiple_introductions_and_spread_of_DENV-2-GII_in_Brazil.git.


