Abstract
Genetic and functional data suggest that Pseudomonas aeruginosa exoenzyme S (ExoS), an ADP-ribosyltransferase, is translocated into eukaryotic cells by a bacterial type III secretory mechanism activated by contact between bacteria and host cells. Although purified ExoS is not toxic to eukaryotic cells, ExoS-producing bacteria cause reduced proliferation and viability, possibly mediated by bacterially translocated ExoS. To investigate the activity of translocated ExoS, we examined in vivo modification of Ras, a preferred in vitro substrate. The ExoS-producing strain P. aeruginosa 388 and an isogenic mutant strain, 388ΔexoS, which fails to produce ExoS, were cocultured with HT29 colon carcinoma cells. Ras was found to be ADP-ribosylated during coculture with 388 but not with 388ΔexoS, and Ras modification by 388 corresponded with reduction in HT29 cell DNA synthesis. Active translocation by bacteria was found to be required, since exogenous ExoS, alone or in the presence of 388ΔexoS, was unable to modify intracellular Ras. Other ExoS-producing strains caused modification of Ras, indicating that this is not a strain-specific event. ADP-ribosylation of Rap1, an additional Ras family substrate for ExoS in vitro, was not detectable in vivo under conditions sufficient for Ras modification, suggesting possible ExoS substrate preference among Ras-related proteins. These results confirm that intracellular Ras is modified by bacterially translocated ExoS and that the inhibition of target cell proliferation correlates with the efficiency of Ras modification.
Exoenzyme S (ExoS) is an ADP-ribosyltransferase produced and secreted by Pseudomonas aeruginosa (19). The production of ExoS has been associated with bacterial virulence, and the enzyme has been shown to modify functionally important mammalian proteins in vitro. However, the in vivo catalytic activity of ExoS remains uncharacterized. The purpose of our studies has been to determine whether ExoS, translocated through the P. aeruginosa type III pathway, is active in mammalian cells and can significantly alter host cell function.
ExoS was originally purified from P. aeruginosa 388 culture supernatants as an aggregate consisting of immunologically related 49- and 53-kDa forms (33). Recent analyses have revealed that although the primary structure of the two proteins is 76% identical, they are encoded by separate, coregulated genes defined as exoS, which encodes 49-kDa ExoS, and exoT, which encodes 53-kDa ExoT (alternatively referred to as Exo53) (38). ExoT was found to have a substrate specificity similar to that of ExoS in vitro but to catalyze ADP-ribosylation at 0.2% of the rate of ExoS (28, 38). The enzymatic activity of both proteins is dependent on a eukaryotic cofactor termed FAS (factor activating ExoS), which is a member of the highly conserved 14-3-3 protein family (12, 16, 28). 14-3-3 proteins have been found to modulate interactions between diverse components of cell signaling pathways (1). The requirement for a eukaryotic cofactor suggests that both ExoS and ExoT function in the eukaryotic-cell environment, although the consequences of this targeting are currently unknown.
P. aeruginosa is an opportunistic pathogen which can cause lethal infections in persons with cystic fibrosis, burn patients, immunocompromised patients, or long-term hospitalized patients (4). ExoS production by P. aeruginosa strains has been associated with increased tissue damage and bacterial dissemination in animal studies (23, 29–33) and with cell injury in tissue culture models (2, 34). However, secreted ExoS, purified from P. aeruginosa 388 culture supernatants, is not toxic to animals or to cultured cells (9, 31). Recent data indicate that the lack of effect of soluble ExoS reflects its dependence on the bacterial type III secretory pathway for translocation into target cells (15, 39). In type III systems, the synthesis and translocation of bacterial effector proteins appear to be initiated by contact between bacterial and host cells. Translocated bacterial proteins have been found to induce a variety of responses in the target cell, creating favorable conditions for bacterial survival and proliferation (18). In P. aeruginosa, three potential effector proteins have been identified as being coregulated by the type III secretory system. These include the ADP-ribosyltransferases ExoS and ExoT, as well as the cytotoxic protein ExoU, the precise activity of which has not yet been characterized. A limited survey of P. aeruginosa strains has suggested that two genetic groups exist, one possessing exoS and the other possessing exoU (encoding ExoU), with ExoU-producing strains being associated with acute cytotoxicity and epithelial injury (14). Our previous report that the coculture of eukaryotic cells with ExoS-producing strains resulted in inhibition of eukaryotic-cell proliferation and viability suggests that bacterially translocated ExoS is also able to mediate cytotoxicity (34). This has recently been confirmed by studies in which ExoS, expressed in Yersinia spp. and translocated by Yersinia type III mechanisms, was found to cause morphological alterations and reduced viability in HeLa cells (15).
Partly due to the difficulty of introducing purified ExoS into target cells, no in vivo targets of ExoS have yet been identified. In vitro studies have found that ExoS modifies multiple substrates in mammalian-cell lysates, including the cytoskeletal protein vimentin, but that it preferentially targets Ras and other GTP-binding proteins in the Ras superfamily (13). The three genetically distinct but structurally similar forms of Ras, H-, K-, and N-Ras, function as central molecular switches in cell signaling pathways. Mutant forms of Ras are associated with many human cancers, including approximately 50% of colon carcinomas (6). Subsequent studies of ExoS substrate specificity in vitro revealed that ExoS exhibits selectivity within the Ras superfamily, with recombinant proteins from the Rap, Ral, and Rab subgroups being modified, while Rho proteins and ADP-ribosylation factor (ARF) did not act as substrates (11).
We report here that Ras is ADP-ribosylated in intact HT29 colon carcinoma cells during coculture with ExoS-producing P. aeruginosa strains and that ExoS activity in these cells correlates with reduced DNA synthesis. We also note that there is no detectable modification of Rap1 proteins in HT29 cells, suggesting possible preferential targeting among Ras-related substrates in vivo.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Parental P. aeruginosa 388 (388) and the construction of the 388ΔexoS mutant (ΔS) have been described previously (3, 24). The mutant strain fails to produce 49-kDa ExoS due to allelic exchange of the majority of the structural gene with a tetracycline gene cartridge. The 388ΔexoT mutant (ΔT), which was constructed by the same strategy, produces ExoS but not ExoT (38). Other P. aeruginosa strains used were PAK (ATCC 25102) (American Type Culture Collection [ATCC], Manassas, Va.) and DG1 (8), which produce ExoS and ExoT, and PA103 (27), which produces ExoT and ExoU, but not ExoS (14). For coculture with HT29 cells, bacteria were grown in a chelated dialysate of Trypticase soy broth, as previously described (34), then washed and diluted in McCoy’s 5A medium (Gibco-BRL, Gaithersburg, Md.) containing 1.5 mM l-glutamine and 25 mM HEPES supplemented with 0.6% bovine serum albumin (McCoy’s-BSA). Bacteria were diluted to the indicated concentration, and suspensions were subsequently plated to confirm the accuracy of the dilution.
HT29 cell culture.
HT29 cells (ATCC HTB 38) were cultured as specified by ATCC, at 37°C in 5% CO2–95% air in antibiotic-free McCoy’s 5A medium supplemented with 10% fetal bovine serum (McCoy’s-FBS). Cells were split 1:6 and passaged as the culture reached confluence. For use in experiments, cells were detached from the growth surface with 0.25% trypsin–1 mM EDTA (Gibco-BRL), resuspended in McCoy’s-FBS, counted, and then diluted to the appropriate density in McCoy’s-FBS and seeded in 48- or 6-well culture plates (Costar, Cambridge, Mass.).
Immunoprecipitation and detection of Ras and Rap1 proteins.
Cellular Ras was detected by immunoprecipitation from [35S]methionine-radiolabeled cells and by immunoblot analysis of immunoprecipitated Ras. Cellular Rap1 was detected by immunoprecipitation from [35S]methionine-radiolabeled cells. For [35S]methionine labeling studies, HT29 cells were seeded in 3 ml of McCoy’s-FBS at a concentration of 6 × 105 cells/ml in 6-well plates and were grown for 24 h. The monolayer was then washed with phosphate-buffered saline (PBS), and the medium was replaced with methionine-deficient Dulbecco’s modified Eagle’s medium (Gibco-BRL) containing 10% dialyzed FBS (DMEM-DFBS). After 1 h, fresh DMEM-DFBS containing 25 μCi of [35S]methionine/ml (>1,000 Ci/mmol; Amersham, Arlington Heights, Ill.) was added, and cells were radiolabeled for 18 h. Radiolabeled cells either were not exposed to bacteria or were exposed to 108 CFU of the indicated bacterial strain in 1 ml of McCoy’s-BSA. After 3 h, the medium was removed, and cells were lysed in 1 ml of PBS containing 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 0.2% sodium azide, and 1 mM sodium fluoride (PBS-TDS). Ras was immunoprecipitated from precleared lysates by mixing at 4°C overnight with 1 μg of rat monoclonal antibody Y13-259 (ATCC CRL 1742)/ml, 5 μg of rabbit anti-rat immunoglobulin G (IgG) (Sigma, St. Louis, Mo.)/ml, and ∼0.5 μg of protein A-Sepharose (Sigma)/ml. Rap1 proteins were similarly immunoprecipitated, by using 1 μg of rabbit polyclonal Rap1–Krev-1 (121) (Santa Cruz Biotechnology, Santa Cruz, Calif.)/ml mixed with protein A-Sepharose. Immunoprecipitated proteins were resolved on a 15% polyacrylamide gel by the method of Laemmli (26). Gels were treated for 30 min with Amplify (Amersham), dried, and exposed to X-ray film for 3 to 7 days.
For detection of immunoprecipitated Ras by immunoblot, HT29 cells were seeded as described above and grown for 48 h in McCoy’s-FBS. Cells were incubated with bacteria and then lysed, and Ras was immunoprecipitated as described above. Proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to membranes by the method of Towbin et al. (36). Ras proteins were detected by using mouse monoclonal anti-pan Ras Ab2 (Calbiochem, La Jolla, Calif.), followed by peroxidase-conjugated goat anti-mouse antibody (Amersham), and were visualized with the enhanced chemiluminescence (ECL) system (Amersham).
Production of recombinant ExoS and modification of proteins in HT29 cell lysates in vitro.
The gene encoding ExoS was cloned into the pET15b vector (Novagen, Madison, Wis.), and recombinant six-histidine N-terminal-tagged ExoS (r6HisExoS) was expressed and purified essentially as described elsewhere (22), except that the binding buffer contained 25 mM imidazole and the protein was eluted with 250 mM imidazole. HT29 cells were grown, and cellular proteins were labeled with [35S]methionine, as described above. Monolayers were then removed from the wells by treatment with trypsin-EDTA and centrifuged at 250 × g for 5 min. Cell pellets were washed twice in 1 ml of PBS, resuspended in 100 μl of PBS–0.1% Triton X-100, and lysed by two cycles of freeze-thawing. r6HisExoS (1 μl) was added to give a final concentration of ∼3 μg/ml in a reaction volume of 150 μl, and the ADP-ribosyltransferase reaction was allowed to proceed for 30 min at room temperature. PBS-TDS (850 μl) was then added, and proteins were immunoprecipitated, resolved by SDS-PAGE, and detected as described above.
Immunoprecipitation and detection of [3H]ADP-ribosylated Ras and Rap proteins.
The intracellular pool of NAD in HT29 cells was radiolabeled with [3H]adenosine, based on methods previously described (35). Modifications, including serum starvation and treatment with actinomycin D, were introduced into the procedure to decrease the incorporation of radiolabel into RNA. In the adapted method, HT29 cells were plated as described, then serum starved in McCoy’s 5A medium supplemented with 0.04% BSA (McCoy’s-BSA4). After a 24-h starvation period, the cells were treated for 30 min with McCoy’s-BSA4 containing 5 μg of actinomycin D (Sigma)/ml. The monolayers were then washed three times with PBS and grown for a further 16 to 18 h in McCoy’s-BSA4 containing 50 μCi of [2-3H]adenosine (21 Ci/mmol; Amersham)/ml. Bacteria were cocultured with HT29 cells as described above, after which monolayers were washed once with PBS, lysed, and processed in PBS-TDS. In some experiments, 1 mM NAD was added to PBS-TDS to preclude further addition of radiolabeled ADP-ribose to proteins during the immunoprecipitation period. Ras and Rap1 were immunoprecipitated and analyzed as described above, with dried gels exposed to X-ray film for 4 weeks.
Comparison of Ras modification in HT29 cells incubated with ExoS-producing bacteria or with secreted ExoS.
HT29 cells were incubated for 3 h with 1 ml of McCoy’s-BSA alone or McCoy’s-BSA containing 108 CFU of the indicated bacterial strains. After 3 h of coculture, the medium was removed, HT29 cells were lysed, and Ras was immunoprecipitated. Medium from cells cultured with bacteria was harvested and centrifuged, and then the supernatant was passed through a 0.2-μm-pore-size filter to remove bacteria. Aliquots were removed for ExoS activity assays, and then the ExoS-containing supernatant was applied to fresh HT29 cell monolayers for 3 h, either without bacteria or in the presence of 108 CFU of ΔS bacteria. HT29 cells were lysed, and Ras was immunoprecipitated and detected by immunoblotting. ExoS activity in 10 μl of harvested culture medium was assayed as previously described (34). The incorporation of radiolabeled ADP-ribose into substrate was reported as picomoles of ADP-ribose transferred to substrate per milliliter of culture medium.
Correlation of reduced DNA synthesis with Ras modification in HT29 cells. (i) Quantification of DNA synthesis.
[3H]thymidine uptake assays were performed as described elsewhere (34), except that additional steps were taken at the end of the coculture period to ensure the termination of bacterial effects. In these studies, bacteria were diluted and applied to monolayers in McCoy’s-BSA. After removal of bacteria, monolayers were washed three times with 0.5 ml of McCoy’s-FBS containing 200 μg of gentamicin/ml and 100 μg of ciprofloxacin/ml (McCoy’s-FBS-GC), then pulsed with 1 μCi of [methyl-3H]thymidine (Amersham)/ml in 200 μl of McCoy’s-FBS-GC for 20 h.
(ii) Measurement of modified Ras.
Immunoblots developed by ECL, as described above, were photographed, and computerized images were analyzed with NIH Image version 1.60 software. The areas and intensities of modified and unmodified Ras bands were calculated, and modified Ras was expressed as a percentage of total Ras.
RESULTS
Modification of Ras by ExoS in vitro and in vivo.
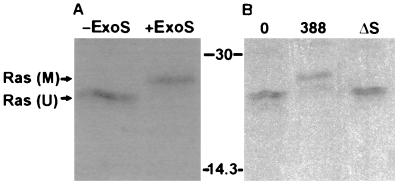
Recent studies indicate that translocation of ExoS into eukaryotic cells is mediated by a bacterial contact-dependent type III secretion system (15, 39). To confirm the contact-dependent translocation of ExoS, and to investigate whether ExoS is able to modify intracellular proteins, we examined the ADP-ribosylation of Ras in intact cells cultured with ExoS-producing P. aeruginosa bacteria. The in vitro ADP-ribosylation of Ras has previously been found to result in a large electrophoretic mobility shift when examined by SDS-PAGE (13). This mobility shift provided a potential means of detecting Ras modification in vivo. To first confirm that ADP-ribosylation by ExoS caused a shift in the electrophoretic mobility of Ras isolated from HT29 cells, purified recombinant ExoS (r6HisExoS) was added to an aliquot of [35S]methionine-labeled HT29 cell extract, and after a 30-min reaction Ras was immunoprecipitated. The apparent molecular mass of Ras immunoprecipitated from reaction mixtures containing r6HisExoS was approximately 2,500 Da greater than that of Ras from untreated aliquots of the same cell extract (Fig. 1A). To determine if a similar shift in Ras mobility, indicative of ADP-ribosylation, could be detected in intact cells, [35S]methionine-labeled HT29 cells were exposed to the ExoS-producing strain 388 or the ΔS mutant strain. A 2,500-Da increase in the apparent molecular mass was detected in Ras isolated from cells exposed to the ExoS-producing strain, whereas the electrophoretic mobility of Ras from cells exposed to the non-ExoS-producing strain was identical to that seen in Ras from untreated control cells (Fig. 1B). Unmodified Ras was consistently immunoprecipitated as two bands of slightly different mobilities. Analysis using monoclonal antibodies specific for different forms of Ras revealed that the upper band corresponded to K-Ras, while H- and N-Ras made up the lower band (data not shown). No radiolabeled bands were seen in the 19- to 30-kDa range when normal rat immunoglobulin was used as a control (data not shown).
FIG. 1.
Mobility shift of Ras in HT29 cell lysates modified by ExoS in vitro and in vivo. (A) Ras modification in vitro by purified ExoS. HT29 cell monolayers were radiolabeled with [35S]methionine for 18 h and then lysed, and extracts were incubated for 30 min in the presence of buffer (−ExoS) or purified recombinant ExoS (+ExoS). Ras was then immunoprecipitated with monoclonal antibody Y13-259 coupled to anti-rat IgG plus protein A-Sepharose. Proteins were separated by SDS-PAGE and visualized by fluorography. M, modified Ras; U, unmodified Ras. (B) Ras modification in vivo by ExoS-producing bacteria. [35S]methionine-labeled HT29 cells were incubated for 3 h with McCoy’s-BSA alone (0) or with 108 CFU of ExoS-producing (388) or non-ExoS-producing (ΔS) bacterial strains as indicated. Bacteria were removed, cells were lysed, and Ras was immunoprecipitated and detected as described for panel A. Molecular masses (in kilodaltons) are indicated.
Although it might be argued that coculture with bacteria could result in damage to the host cell membrane, enabling secreted ExoS to gain access to the host cell and modify Ras without active translocation by bacteria, this seemed unlikely because (i) dye exclusion assays, performed after 3 h of coculture with bacteria, revealed no loss of membrane integrity in cells exposed to 388 or ΔS bacteria, compared to untreated controls, and (ii) leakage of [35S]methionine-labeled proteins into the medium of bacterially treated cells was not above levels in cultures not exposed to bacteria (data not shown). To preclude the possibility that residual secreted ExoS might be modifying Ras during the in vitro immunoprecipitation period, SDS was added to the lysis-immunoprecipitation buffer at concentrations sufficient to inhibit ExoS activity (0.2 to 0.5%) (25). When these concentrations of SDS were present, no reduction in the amount of modified Ras was detected, indicating that Ras modification during the immunoprecipitation period did not contribute to the observed effects (data not shown).
Transfer of [3H]ADP-ribose to Ras in HT29 cells by ExoS-producing bacterial strains.
To confirm that ADP-ribosylation of Ras in intact cells was responsible for the observed electrophoretic-mobility shift, [3H]adenosine was used to label intracellular pools of NAD. The labeled cells were then exposed to bacterial strains, and Ras was immunoprecipitated from cell lysates. A radiolabeled band having the same electrophoretic mobility as modified Ras was detected in immunoprecipitates from cells exposed to the ExoS-producing strain but was absent in immunoprecipitates from cells exposed to the non-ExoS-producing mutant or to no bacteria (data not shown; results of a similar experiment are depicted in Fig. 6). During these studies we observed that residual levels of ExoS activity present in coculture cell lysates could catalyze the transfer of radiolabeled ADP-ribose to immunoglobulin during the immunoprecipitation reaction (data not shown). This is consistent with the report that ExoS can modify serum proteins, including immunoglobulin, in vitro (21). To ensure that residual in vitro ExoS activity did not affect the interpretation of our results, 1 mM unlabelled NAD was added to the immunoprecipitation buffer to inhibit transfer of labeled moieties to substrates present in the immunoprecipitation mixture. This treatment eliminated labeling of immunoglobulin but did not alter the amount of modified Ras detected, confirming that Ras modification was occurring in vivo and not in vitro (data not shown).
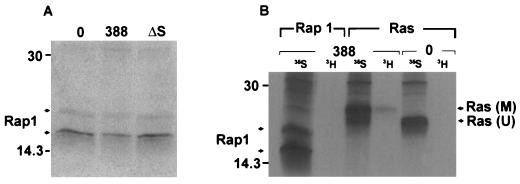
FIG. 6.
Investigation of Rap1 modification. (A) Examination of Rap1 mobility in HT29 cells following exposure to 388 or ΔS bacteria. [35S]methionine-labeled cells were prepared and cocultured with the indicated bacterial strains as described for Fig. 1B. Rap1 proteins were immunoprecipitated with rabbit polyclonal Rap1–Krev-1 (121) antibody and detected by SDS-PAGE, followed by fluorography. Arrows indicate 20- and 24-kDa Rap1 proteins. (B) Comparison of ADP-ribosylation of Ras and Rap1 in HT29 cells exposed to strain 388. Cellular proteins were labeled with [35S]methionine (35S), or intracellular NAD pools were labeled by treating HT29 monolayers with 5 μg of actinomycin D/ml to reduce RNA synthesis and then radiolabeling with [3H]adenosine (3H) for 18 h. Cells were either left untreated (0) or exposed to 108 CFU of strain 388 bacteria/ml. Ras and Rap1 were immunoprecipitated and subjected to SDS-PAGE and fluorography as described above. M, modified Ras; U, unmodified Ras. Molecular masses (in kilodaltons) are indicated.
Comparison of Ras modification by ExoS-producing bacteria and by secreted ExoS.
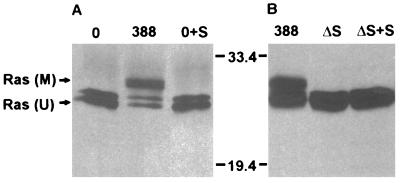
In our previous studies (34), ExoS secreted into the medium during coculture was found to have no inhibitory effect on cell proliferation. To determine whether this lack of effect correlated with the inability of secreted ExoS to modify Ras in intact cells, HT29 monolayers were exposed either to strain 388 or to ExoS-containing medium obtained following coculture with strain 388 for 3 h. As shown in Fig. 2A, mobility-shifted Ras was detected only in immunoprecipitates from cells exposed to ExoS-producing bacteria, and not in those exposed to ExoS-containing culture medium. ExoS activity in culture medium harvested from cells cultured with strain 388 was found to be 40 pmol/ml. To determine whether coculture with bacteria might indirectly cause cells to become permeable to secreted ExoS, ΔS bacteria were cultured with HT29 monolayers in the presence of ExoS-containing medium. No mobility-shifted Ras was detected in immunoprecipitates from cells exposed to the ΔS strain for 3 h, either alone or with secreted ExoS (Fig. 2B). ExoS activity in culture medium harvested from cells cultured with strain 388 or ΔS in this study was 147 or 2 pmol/ml, respectively.
FIG. 2.
Comparison of Ras modification by strain 388 and by ExoS secreted into the medium. (A) Lack of Ras modification by extracellularly secreted ExoS. Unlabeled HT29 cells were incubated for 3 h with medium alone (0) or with 108 CFU of strain 388. The 388 coculture medium was then removed, filtered, and applied to a fresh HT29 cell monolayer for 3 h (0+S). Ras was immunoprecipitated from lysates of cells cultured with strain 388 bacteria or with secreted ExoS. The electrophoretic mobility of Ras was evaluated following SDS-PAGE, immunoblotting with mouse monoclonal anti-Ras, and detection by ECL. M, modified Ras; U, unmodified Ras. (B) Lack of Ras modification by secreted ExoS in the presence of ΔS bacteria. HT29 cells were incubated for 3 h with 108 CFU of strain 388, ΔS alone, or ΔS in the presence of secreted ExoS prepared as described for panel A (ΔS + S). The electrophoretic mobility of Ras was examined as described for panel A. Molecular masses (in kilodaltons) are indicated.
Modification of Ras in vivo by other ExoS-producing P. aeruginosa strains.
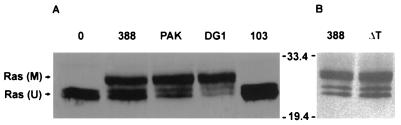
Other ExoS-producing P. aeruginosa strains, PAK and DG1, were examined for their abilities to modify Ras in coculture studies with HT29 cells. Strain PA103, which produces ExoU and ExoT, but not ExoS, was included in these studies as an ExoS-negative control. As shown in Fig. 3A, the ExoS-producing strains DG1 and PAK caused a shift in Ras mobility similar to that observed for strain 388. Densitometric analysis indicated that DG1 was able to modify 74% of intracellular Ras, while PAK and 388 modified 58 and 50%, respectively. In contrast, no modified Ras was detected following coculture with the non-ExoS-producing strain PA103. The mutant strain ΔT, which produces ExoS but not ExoT, was also examined in these studies to assess how loss of this gene affected Ras modification. The ΔT strain was able to modify Ras as effectively as the wild-type strain 388 (57% modification by ΔT versus 61% modification by strain 388), indicating that ExoT was not required for modification of Ras by ExoS in intact cells (Fig. 3B).
FIG. 3.
Comparison of Ras modification in HT29 cells by different P. aeruginosa strains. (A) HT29 cells were incubated with 108 CFU of strain 388, PAK, DG1, or PA103 (103), as indicated; then Ras was immunoprecipitated and detected by immunoblotting as described for Fig. 2A. M, modified Ras; U, unmodified Ras. (B) HT29 cells were incubated with 108 CFU of strain 388 or ΔT and analyzed as described above. Molecular masses (in kilodaltons) are indicated.
Dependence of Ras modification in vivo on duration of coculture period and bacterial concentration.
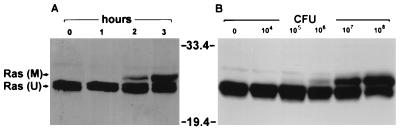
To determine optimal culture conditions for modification of Ras in HT29 cells by strain 388, the effects of both the coculture time and bacterial concentration on the efficiency of Ras modification were examined. As shown in Fig. 4A, modification of Ras was found to correlate directly with the length of exposure to ExoS-producing bacteria. Although slight further modification was detected when exposure to bacteria was allowed to proceed for 4 to 6 h, extended time of coculture with high concentrations of bacteria was also found to cause some loss of HT29 cell membrane integrity, as assessed by trypan blue exclusion (data not shown). Fig. 4B shows that modification of Ras was found to correlate with the number of bacteria in the inoculum. An initial bacterial concentration of 105 CFU/ml, corresponding to approximately 1 bacterium to 10 HT29 cells, was required for detection of minimal modification, with the degree of modification increasing proportionately as the initial ratio of bacteria to HT29 cells was increased by 10-fold increments to approximately 100:1 (108 CFU/ml).
FIG. 4.
Dependence of in vivo Ras modification on time of exposure to bacteria and bacterial concentration. (A) Time required for Ras modification. HT29 cells were incubated with no bacteria (0) or with 108 CFU of strain 388 bacteria for 1, 2, or 3 h, as indicated. Ras was immunoprecipitated from cell lysates and detected by SDS-PAGE and immunoblotting. M, modified Ras; U, unmodified Ras. (B) Bacterial concentration required for Ras modification. Cells were incubated for 3 h with 0 to 108 CFU of strain 388 bacteria, as indicated, and Ras modification was examined as for panel A. Molecular masses (in kilodaltons) are indicated.
Correlation of reduced DNA synthesis with modification of Ras.
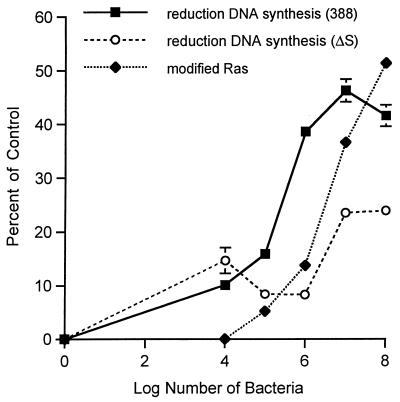
We previously observed a significant reduction in eukaryotic-cell DNA synthesis resulting from long-term exposure to strain 388 compared to ΔS (34). In the present investigation, to help assess the mechanism by which ExoS exerts inhibitory effects on cell proliferation, we compared the efficiency of Ras modification to effects on cell proliferation. For these studies, increasing concentrations of strain 388 or ΔS were applied to HT29 cells for 3 h under conditions identical to those described in the legend to Fig. 4B. Monolayers were then extensively washed, and inhibitory antibiotics were added to limit further modification of Ras by bacteria during the radiolabeled thymidine incorporation period. Figure 5 shows that the reduction in DNA synthesis associated with strain 388 becomes significantly greater than that seen with ΔS at 105 CFU/ml, the minimum bacterial concentration required for Ras modification. Thereafter, the effects of ExoS production on cell proliferation closely parallel the percentage of Ras found to be modified with increasing concentrations of strain 388 bacteria. In contrast, the lesser effect of the ΔS mutant strain on cell proliferation is apparent only when bacterial numbers exceed 106 CFU/ml and is maximal at 107 CFU/ml, without approaching the levels of inhibition caused by strain 388.
FIG. 5.
Correlation of Ras modification with reduced DNA synthesis. DNA synthesis was measured in HT29 cells which had been seeded in 48-well plates at 105 cells/well, grown for 48 h, then incubated with 0 to 108 CFU of strain 388 or ΔS/ml in McCoy’s-BSA for 3 h. At this time, monolayers were washed to remove bacteria, and McCoy’s-FBS-GC containing 1 μCi of [3H]thymidine/ml was added. DNA synthesis was determined after 18 h and is expressed as percent reduction in [3H]thymidine uptake compared to that in nonbacterially treated controls. Results are expressed as means and standard deviations of a single assay performed in quadruplicate and are representative of two independent studies. Percent reduction in DNA synthesis was compared to percent modification of Ras by incubating HT29 cells in an identical manner with increasing concentrations of strain 388 bacteria. The percentage of immunoprecipitated Ras modified was determined by densitometric analysis of a representative image.
Examination of Rap1 modification in HT29 cells.
To extend our knowledge of ExoS activity in vivo, we investigated modification of Rap1, another in vitro substrate of ExoS, following coculture with strain 388 bacteria. Among the Ras superfamily, Rap proteins are the most closely related to Ras structurally, and like Ras, they are found in most cell types. Rap proteins consist of two families, Rap1 and Rap2, with each family having both A and B subtypes (5). The immunoprecipitating antibody used in these studies recognizes both Rap1A and Rap1B, which are 95% identical at the amino acid level. Taking advantage of the previous report that modified recombinant Rap1A, like Ras, exhibited a gel mobility shift when modified by ExoS in vitro (11), we first examined whether Rap proteins were modified in vivo by ExoS by analyzing the electrophoretic mobility of Rap1 immunoprecipitated from [35S]methionine-labeled HT29 cells exposed to ExoS-producing bacteria. As shown in Fig. 6A, proteins of approximately 20 and 24 kDa were immunoprecipitated from control cells, and no alterations in the mobilities of these proteins were detected following exposure to the ExoS-producing strain 388. When Rap1 from [35S]methionine-labeled HT29 cell lysates was subsequently examined for altered mobility following in vitro modification with purified recombinant ExoS, no alteration in mobility could be detected. Additional in vitro analyses, however, confirmed that (i) ExoS was able to ADP-ribosylate recombinant Rap1A fused to glutathione S-transferase (GST), although no gel mobility shift accompanied modification, and (ii) the Rap1–Krev-1 antibody was able to bind normal and ADP-ribosylated GST-Rap1A in immunoblot reactions (data not shown). Since these studies suggest that a shift in mobility might not be an accurate means of detecting Rap1 modification in HT29 cells, we next examined Rap1 modification in vivo, using [3H]adenosine to radiolabel intracellular NAD pools. In these studies, the relative modification of Rap1 and Ras by ExoS was directly compared by immunoprecipitating Rap1 or Ras from identically treated cells. Also, to confirm the efficiency of the immunoprecipitation reaction, studies were run in parallel immunoprecipitating Ras or Rap1 from [35S]methionine-labeled HT29 cells. As shown in Fig. 6B, ADP-ribosylated Ras, corresponding to mobility-shifted [35S]methionine-labeled Ras, was detected in cells labeled with [3H]adenosine and exposed to strain 388. In comparison, under identical conditions, no [3H]adenosine-labeled Rap1 was detected. However, Rap1 proteins of 20 and 24 kDa were immunoprecipitated from cells labeled with [35S]methionine, indicating that the absence of ADP-ribosylated species was not due to inefficient immunoprecipitation. No proteins in the 14- to 30-kDa range were seen when normal rabbit IgG was used as a control (data not shown). The undetectable modification of Rap1, under conditions where Ras modification is detected, suggests an in vivo substrate preference of ExoS among proteins within the Ras superfamily.
DISCUSSION
We have previously reported the development of a bacterial-eukaryotic cell coculture system which allowed recognition of the inhibitory effect of ExoS-producing bacteria on host cell viability and proliferation (34). This system has now been adapted to help identify mechanisms by which ExoS affects cell function and is used here to identify the cell signaling protein, Ras, as an in vivo substrate of ExoS. Considerable care was taken in these studies to ensure that Ras modification by ExoS occurred following its translocation by bacterial contact. Of primary concern was the possibility that coculture with bacteria might cause eukaryotic-cell membrane damage, allowing passive transfer and modification of Ras by secreted ExoS. Membrane permeability analyses, however, revealed that HT29 cellular membranes remained intact during the modification of Ras. Furthermore, Ras was not modified by secreted ExoS in the presence of non-ExoS-producing bacteria, confirming that direct contact with bacteria expressing ExoS was required for Ras modification. The results support the hypothesis that modification of Ras by ExoS is an intracellular event, dependent on direct contact with ExoS-producing bacteria.
Analysis of the ADP-ribosylation of Ras by different bacterial strains found Ras modification to be dependent on ExoS production but independent of ExoT or ExoU production. P. aeruginosa strains express different combinations of three type III effector proteins. Strains 388 and PAK produce ExoS and ExoT but lack the ExoU gene. DG1 also produces ExoS and ExoT, while ExoU production has not yet been characterized. Consistent with their expression of ExoS, these strains modify Ras. In contrast, no modification of Ras was detected with strain PA103, which produces the ExoS homolog ExoT and the cytotoxic factor ExoU. Although the coordinate roles of ExoS, ExoT, and ExoU are not currently understood, the apparent selection for expression of ExoS or ExoU (14) could reflect a detrimental effect on the bacterium of production of both proteins or the redundancy of producing two coregulated cytotoxic factors. While the activity of ExoT in the host cell remains unclear, the inability to detect modified Ras in HT29 cells cultured with the ΔS mutant strain, which produces ExoT, differentiates the functions of ExoS and ExoT in vivo. Although the inefficient modification of Ras by ExoT may reflect its low ADP-ribosyltransferase activity in vitro, it remains possible that ExoT might be targeting alternative intracellular substrates. Our detection of slightly increased inhibition of HT29 cell DNA synthesis following coculture with the ΔT mutant, compared to that with the ExoS- and ExoT-producing, wild-type strain 388 (28a), also suggests that ExoT might be modulating ExoS activity, possibly via competition for substrate, cofactor, or translocation intermediates.
In examining the modification of Ras by P. aeruginosa strains, different levels of efficiency in different strains were noted. In the cases of 388, ΔT, and PAK, these differences were slight, but DG1 was able to modify a significantly higher proportion of immunoprecipitable Ras. We have consistently detected higher levels of ExoS activity in DG1 culture supernatants than in 388 and PAK culture supernatants (28a); therefore, more efficient Ras modification may reflect increased production of ExoS by this strain. It was also notable that modification of HT29 cellular Ras, under optimal conditions, never exceeded 75%. This may reflect modification of only a portion of Ras in each individual cell, reduced modification among a subpopulation of cells, or a combination of both factors. Questions to be addressed in this regard include whether the activation state or intracellular localization of Ras affects its ability to be modified by ExoS, and whether some cells possess properties conferring resistance to the effects of ExoS. In assessing the contribution of Ras modification to the effects of ExoS on cell function, we found conditions sufficient for modification of Ras in HT29 cells to correspond closely to those causing reduction of DNA synthesis. This indicates that modification of Ras is an accurate index of the effects of ExoS on cell proliferation. Since Ras is involved in relaying growth factor-induced proliferative signals to the nucleus (6), our findings raise the intriguing possibility that some of the inhibitory effects of ExoS might be due to modification of Ras function. Coburn and Gill (9, 11, 13) reported that the modification of Ras by ExoS in vitro had no effect on the ability of Ras to exchange and hydrolyze guanine nucleotides or to interact with GTPase activating and guanine nucleotide exchange proteins. These findings imply that if ExoS alters the cellular activity of Ras, it likely occurs at the level of its interaction with effector proteins. Although an increasing number of Ras-effector interactions are being identified (recently reviewed in reference 20), the best characterized of these is the Ras–Raf-1 kinase interaction, which results in the activation of Raf-1 in association with its localization to the plasma membrane. Activated Raf-1 then initiates a proliferative signal to the nucleus, through the cascade of serine-threonine kinases known as the mitogen-activated protein (MAP) kinase pathway (7). Consistent with the possibility that this pathway may be affected by ExoS is the shared interaction of Raf-1 kinase and ExoS with 14-3-3 proteins (17, 40). Interpretations of the effects of ExoS on Raf-1 activation and MAP kinase activity in HT29 cells, however, have been complicated by non-ExoS-associated effects of the bacteria on HT29 cell signaling processes. The precise characterization of the effects of ExoS on the MAP kinase pathway is therefore being deferred to future studies with an alternative epithelial-cell line which has been found to be minimally affected by exposure to bacteria while showing the same effects of ExoS on cell function.
Since ExoS has been found to modify multiple proteins in cell lysates, an understanding of its role in vivo will require characterization of its total intracellular substrate specificity. Our finding that HT29 cell Rap1 proteins do not appear to be modified efficiently (if at all) indicates that, among Ras-related proteins, ExoS may exhibit a more limited substrate preference in vivo than in vitro. Preliminary studies, using [3H]adenosine to radiolabel HT29 intracellular NAD pools, indicate that substrates in addition to Ras are likely to be modified by ExoS in vivo. In this regard, recent investigations by Frithz-Lindsten et al. (15), using the type III secretory system of Yersinia pseudotuberculosis to introduce ExoS into the cytosol of HeLa cells, found that ExoS induces a cytotoxic effect characterized by a rounding up of cells and disruption of actin microfilaments. We previously observed rounding up and detachment of Detroit fibroblasts following coculture with strain 388 (34), and in this study we found rounding up of HT29 cells to be associated with ExoS modification of Ras and its effects on cell proliferation. It is presently unknown whether the effects of ExoS on the cytoskeleton reflect additional substrate specificities of ExoS in vivo or relate directly or indirectly to effects of ExoS on Ras effector pathways. However, modification of the cytoskeletal protein vimentin, an in vitro substrate for ExoS (10), may not play a crucial role in this process, since HT29 cells, which exhibit rounding up and reduced proliferation, do not appear to express vimentin (28a, 37).
In summary, the contact-dependent delivery of ExoS by P. aeruginosa into eukaryotic cells has led to the identification of Ras as an in vivo substrate of ExoS. The efficiency of Ras modification by ExoS has also been found to correlate with the effects of ExoS on cell proliferation, suggesting a relationship between the ADP-ribosylation of Ras and effects of ExoS on cell function. The finding that ExoS has the potential to modulate eukaryotic-cell signaling pathways in vivo provides an impetus for further investigation of the role of ExoS, both in the communication between bacterium and host during the infectious process and as a tool for probing signaling pathways in the eukaryotic cell.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI30558 from the National Institute of Allergy and Infectious Diseases and by the Medical University of South Carolina Institutional Research Funds of 1997-98.
We thank Lawrence A. Quilliam for kindly providing the pGex vector expressing GST-Rap1A and Mark Willingham, Barry Ledford, Donna Jacobs, and Katherine Dolan for helpful discussions.
REFERENCES
- 1.Aitken A. 14-3-3 and its possible role in co-ordinating multiple signaling pathways. Trends Cell Biol. 1996;6:341–347. doi: 10.1016/0962-8924(96)10029-5. [DOI] [PubMed] [Google Scholar]
- 2.Apodaca G, Bomsel M, Lindstedt R, Engel J, Frank D, Mostov K E, Wiener-Kronish J. Characterization of Pseudomonas aeruginosa-induced MDCK cell injury: glycosylation-defective host cells are resistant to bacterial killing. Infect Immun. 1995;63:1541–1551. doi: 10.1128/iai.63.4.1541-1551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjorn M J, Pavlovskis O R, Thompson M R, Iglewski B H. Production of exoenzyme S during Pseudomonas aeruginosa infections of burned mice. Infect Immun. 1979;24:837–842. doi: 10.1128/iai.24.3.837-842.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodey G P, Bolivar R, Fainstein V, Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1993;5:270–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- 5.Bokoch G M. Biology of the Rap proteins, members of the ras superfamily of GTP-binding proteins. Biochem J. 1993;289:17–24. doi: 10.1042/bj2890017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos J L. p21ras: an oncoprotein functioning in growth factor-induced signal transduction. Eur J Cancer. 1995;31A:1051–1054. doi: 10.1016/0959-8049(95)00168-i. [DOI] [PubMed] [Google Scholar]
- 7.Burgering B M T, Bos J L. Regulation of Ras-mediated signaling: more than one way to skin a cat. Trends Biochem Sci. 1995;20:18–22. doi: 10.1016/s0968-0004(00)88944-6. [DOI] [PubMed] [Google Scholar]
- 8.Cash H A, Woods D E, McCullough B, Johanson W G, Jr, Bass D A. A rat model of chronic respiratory infection with Pseudomonas aeruginosa. Am Rev Resp Dis. 1979;119:453–459. doi: 10.1164/arrd.1979.119.3.453. [DOI] [PubMed] [Google Scholar]
- 9.Coburn J. Pseudomonas aeruginosa exoenzyme S. Curr Top Microbiol Immunol. 1992;175:133–143. doi: 10.1007/978-3-642-76966-5_7. [DOI] [PubMed] [Google Scholar]
- 10.Coburn J, Dillon S T, Iglewski B H, Gill D M. Exoenzyme S of Pseudomonas aeruginosa ADP-ribosylates the intermediate filament protein vimentin. Infect Immun. 1989;57:996–998. doi: 10.1128/iai.57.3.996-998.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coburn J, Wyatt R T, Iglewski B H, Gill D M. Several GTP-binding proteins, including p21c-H-ras, are preferred substrates of Pseudomonas aeruginosa exoenzyme S. J Biol Chem. 1989;264:9004–9008. [PubMed] [Google Scholar]
- 12.Coburn J, Kane A V, Feig L, Gill D M. Pseudomonas aeruginosa exoenzyme S requires a eukaryotic protein for ADP-ribosyltransferase activity. J Biol Chem. 1991;266:6438–6446. [PubMed] [Google Scholar]
- 13.Coburn J, Gill D M. ADP-ribosylation of p21ras and related proteins by Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1991;59:4259–4262. doi: 10.1128/iai.59.11.4259-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finck-Barbançon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish J P, Fleiszig S M J, Wu C, Mende-Mueller L, Frank D W. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 15.Frithz-Lindsten E, Du Y, Rosqvist R, Forsberg A. Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol Microbiol. 1997;25:1125–1139. doi: 10.1046/j.1365-2958.1997.5411905.x. [DOI] [PubMed] [Google Scholar]
- 16.Fu H, Coburn J, Collier R J. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc Natl Acad Sci USA. 1993;90:2320–2324. doi: 10.1073/pnas.90.6.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu H, Xia K, Pallas D C, Cui C, Conroy K, Narsimhan R P, Mamon H, Collier R J, Roberts T M. Interaction of the protein kinase Raf-1 with 14-3-3 proteins. Science. 1994;266:126–129. doi: 10.1126/science.7939632. [DOI] [PubMed] [Google Scholar]
- 18.Galan J E, Bliska J B. Cross talk between bacterial pathogens and their host cells. Annu Rev Cell Dev Biol. 1996;12:221–255. doi: 10.1146/annurev.cellbio.12.1.221. [DOI] [PubMed] [Google Scholar]
- 19.Iglewski B H, Sadoff J, Bjorn M J, Maxwell E S. Pseudomonas aeruginosa exoenzyme S: an adenosine diphosphate ribosyltransferase distinct from toxin A. Proc Natl Acad Sci USA. 1978;75:3211–3215. doi: 10.1073/pnas.75.7.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katz M E, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 21.Knight D A, Barbieri J T. Ecto-ADP-ribosyltransferase activity of Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1997;65:3304–3309. doi: 10.1128/iai.65.8.3304-3309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knight D A, Finck-Barbançon V, Kulich S M, Barbieri J T. Functional domains of Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1995;63:3182–3186. doi: 10.1128/iai.63.8.3182-3186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kudoh I, Wiener-Kronish J P, Hashimoto S, Pittet J-F, Frank D. Exoproduct secretions of Pseudomonas aeruginosa strains influence severity of alveolar epithelial injury. Am J Physiol Lung Cell Mol Physiol. 1994;267:L551–L556. doi: 10.1152/ajplung.1994.267.5.L551. [DOI] [PubMed] [Google Scholar]
- 24.Kulich S M, Frank D W, Barbieri J T. Expression of recombinant exoenzyme S of Pseudomonas aeruginosa. Infect Immun. 1995;63:1–8. doi: 10.1128/iai.63.1.1-8.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulich S M, Yahr T L, Mende-Mueller L M, Barbieri J T, Frank D W. Cloning the structural gene for the 49-kDa form of exoenzyme S (exoS) from Pseudomonas aeruginosa strain 388. J Biol Chem. 1994;269:10431–10437. [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Liu P V. The roles of various fractions of Pseudomonas aeruginosa in its pathogenesis. III. Identity of the lethal toxins produced in vitro and in vivo. J Infect Dis. 1966;116:481–489. doi: 10.1093/infdis/116.4.481. [DOI] [PubMed] [Google Scholar]
- 28.Liu S, Yahr T L, Frank D W, Barbieri J T. Biochemical relationships between the 53-kilodalton (Exo53) and the 49-kilodalton (ExoS) forms of exoenzyme S of Pseudomonas aeruginosa. J Bacteriol. 1997;179:1609–1613. doi: 10.1128/jb.179.5.1609-1613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.McGuffie, E. M., and J. C. Olson. Unpublished data.
- 29.Nicas T I, Bradley J, Lochner J E, Iglewski B H. The role of exoenzyme S in infections with Pseudomonas aeruginosa. J Infect Dis. 1985;152:716–721. doi: 10.1093/infdis/152.4.716. [DOI] [PubMed] [Google Scholar]
- 30.Nicas T I, Iglewski B H. Isolation and characterization of transposon-induced mutants of Pseudomonas aeruginosa deficient in production of exoenzyme S. Infect Immun. 1984;45:470–474. doi: 10.1128/iai.45.2.470-474.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicas T I, Iglewski B H. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can J Microbiol. 1985;31:387–392. doi: 10.1139/m85-074. [DOI] [PubMed] [Google Scholar]
- 32.Nicas T I, Iglewski B H. Contribution of exoenzyme S to the virulence of Pseudomonas aeruginosa. Antibiot Chemother. 1985;36:40–48. doi: 10.1159/000410470. [DOI] [PubMed] [Google Scholar]
- 33.Nicas T I, Frank D W, Stenzel P, Lile J D, Iglewski B H. Role of exoenzyme S in chronic Pseudomonas aeruginosa lung infections. Eur J Clin Microbiol. 1985;4:175–179. doi: 10.1007/BF02013593. [DOI] [PubMed] [Google Scholar]
- 34.Olson J C, McGuffie E M, Frank D W. Effects of differential expression of the 49-kilodalton exoenzyme S by Pseudomonas aeruginosa on cultured eukaryotic cells. Infect Immun. 1997;65:248–256. doi: 10.1128/iai.65.1.248-256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staddon J M, Bouzyk M M, Rozengurt E. A novel approach to detect toxin catalyzed ADP-ribosylation in intact cells: its use to study the action of Pasteurella multocida toxin. J Cell Biol. 1991;115:949–958. doi: 10.1083/jcb.115.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trainer D L, Kline T, McCabe F L, Faucette L F, Feild J, Chaikin M, Anzano M, Rieman D, Hoffstein S, Li D-J, Gennaro D, Buscarino C, Lynch M, Poste G, Greig R. Biological characterization and oncogene expression in human colorectal carcinoma cell lines. Int J Cancer. 1988;41:287–296. doi: 10.1002/ijc.2910410221. [DOI] [PubMed] [Google Scholar]
- 38.Yahr T L, Barbieri J T, Frank D W. Genetic relationship between the 53- and 49-kilodalton forms of exoenzyme S from Pseudomonas aeruginosa. J Bacteriol. 1996;178:1412–1419. doi: 10.1128/jb.178.5.1412-1419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yahr T L, Goranson J, Frank D W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Wang H, Liu D, Liddington R, Fu H. Raf-1 kinase and exoenzyme S interact with 14-3-3ζ through a common site involving lysine 49. J Biol Chem. 1997;272:13717–13724. doi: 10.1074/jbc.272.21.13717. [DOI] [PubMed] [Google Scholar]