Abstract
Purpose:
The COXEN gene expression model with chemotherapy-specific scores (for DD-MVAC and GC) was developed to identify responders to NAC. We investigated RNA-based molecular subtypes as additional predictive biomarkers for NAC response, PFS, and OS in patients treated in S1314.
Experimental design:
237 patients were randomized between 4 cycles of ddMVAC (51%) and GC (49%). Based on Affymetrix transcriptomic data, we determined subtypes using 3 classifiers: TCGA (k=5), Consensus (k=6), and MD Anderson (MDA; k=3) and assessed subtype association with path response to NAC and determined associations with COXEN. We also tested whether each classifier contributed additional predictive power when added to a model based on pre-defined stratification factors (PS 0 vs. 1; T2 vs. T3, T4a).
Results:
155 patients had gene expression results, received at least 3 of 4 cycles of NAC and had pT-N response based on RC. TCGA 3 group classifier BS/Neuronal, Lum, Lum infiltrated and GC COXEN score yielded the largest AUCs for pT0 (0.59 p=0.28; 0.60 p=0.18, respectively). For downstaging (<pT2), the 3 category Consensus classifier (BS/NE-like, Lum, Stroma-rich) increased the AUC from 0.57 (strat factors alone) to 0.61 (p=0.10). The MDA classifier AUC was 0.63 (p=0.18) and the GC COXEN score AUC was 0.62 (p=0.23), but neither significantly improved the AUC. There was no statistically significant association of stratification factors and subtypes with PFS or OS.
Conclusion:
The Consensus classifier, based in part on the TCGA and MDA classifiers, modestly improved prediction for pathologic downstaging but subtypes were not associated with PFS or OS.
INTRODUCTION
Cisplatin-based neoadjuvant chemotherapy (NAC) is recommended for patients with MIBC before radical cystectomy (RC) and is considered the standard of care for cisplatin-eligible patients by all the major specialty societies and NCCN guidelines. SWOG 8710 established an expected pathologic complete response rate of 38% with the three cycles of MVAC (Methotrexate, Vinblastine, Adriamycin, Cisplatin)(1) and downstaging to < pT2 in 44%(2), but a significant proportion will not have a pathologic complete response and are presumed resistant to chemotherapy and at high risk recurrent and progressive disease. Coexpression extrapolation (COXEN) is a predictive biomarker approach for treatment response developed by Theodorescu and colleagues(3,4). We sought to validate the COXEN gene expression model with chemotherapy-specific scores (for DD-MVAC and GC) in a prospective rPII clinical trial (SWOG S1314) and reported that the GC score was associated with pathologic downstaging in the pooled arms(5).
mRNA-based expression molecular subtypes are associated with prognosis in non-muscle invasive and muscle invasive bladder cancer (NMIBC and MIBC). In MIBC patients, all subtyping schemes consistently stratify patients by luminal and basal(6). In the Cancer Genome Atlas (TCGA) 5 cluster solution, Luminal papillary is associated with FGFR3 alterations, low expression of miR 99a and 100, luminal markers including cytokeratin 20, have a good prognosis and are less frequently upstaged from clinical T1 or T2(7),(8). The basal squamous subtype is enriched in the female gender, primitive cytokeratins including KRT5, and associated with a worse prognosis. They are often infiltrated with lymphocytes and may prime tumors to respond to cisplatin-based chemotherapy and immunotherapy with checkpoint inhibition.
In the present study, we investigated RNA-based molecular subtypes from the TCGA(7), MD Anderson(9), and the Consensus group(10), which developed a classifier based on data from 6 classification schemes as additional predictive biomarkers for response to NAC in patients treated in S1314. We aimed to determine if one or more subtyping schemes was associated with pathologic response to NAC and overall survival and to test the interaction with the COXEN score.
Methods
The SWOG S1314 clinical trial (NCT02177695) was reviewed and approved by the NCI Central Institutional Review Board (CIRB), and patients provided written, informed consent; it was conducted according to the Declaration of Helsinki guidelines. Eligible subjects 18 years or older had histologically proven urothelial carcinoma of the bladder, stage cT2-T4a N0 M0 disease, and a Zubrod performance status of 0 or 1. In addition, those with mixed histology which included a component of urothelial carcinoma, were eligible. This was a randomized Phase II trial, and 237 patients received neoadjuvant chemotherapy with either four cycles of gemcitabine/cisplatin (GC) or four cycles of dose-dense methotrexate, vinblastine, doxorubicin, and cisplatin (MVAC) at standard doses. All patients were intended to undergo radical cystectomy and bilateral pelvic lymphadenectomy within 100 days following the completion of chemotherapy.
RNA was isolated from 10-micron slides, and the samples were analyzed in 2 batches, with six patients having their samples run in both batches as a quality control measure. The whole transcriptome gene expression data used for the COXEN determination were generated using the Affymetrix direct hybridization platform and U133A GeneChip (ALMAC laboratory (Durham, North Carolina). The methodological details of data processing, the COXEN score derivations, and associations with pathologic response have been previously reported(5).
Based on Affymetrix transcriptomic data used to assign COXEN scores, we determined subtypes using three single patient classifiers: TCGA (k=5)(11), Consensus (k=6)(10), and MD Anderson (MDA; k=3)(9). Single patient classifiers were previously developed for TCGA and Consensus classification schemes by the respective research groups. MDA classification was performed based on the previously reported method (9). The primary objectives were to assess subtype association with pathologic response to NAC in the pooled arms and to determine any association with COXEN. The response to NAC was defined as complete (pT0N0) or downstaging to less than pT2N0. Now with longer follow-ups, we determined the association with the secondary endpoints of the trial for progression-free and overall survival. Across the subtyping schemes, there was alignment for luminal subtypes based on the expression of luminal markers, including KRT20, GATA3, and PPARG, with luminal papillary tumors expressing FGFR3, miRNAs associated with FGFR3 expression and papillary morphology. There was also a strong alignment of basal/squamous tumors with loss of luminal markers and expression of KRT5,6, squamous histology, and enrichment in females. Neuroendocrine or neuronal tumors are more closely aligned with Basal/Squamous but, in most subtyping schemes, consistently segregate to a unique subtype. Based on these observations and small numbers in several of the subtypes, the TCGA and Consensus classifiers were collapsed into three groups for ROC analyses. We performed a sensitivity analysis leaving out the least common subtype (NE) from TCGA and Consensus classifiers and measured changes in estimates of clinical outcomes or the association of subtype level with outcomes. We tested whether each classifier contributed additional predictive accuracy for pathologic response when added to a logistic regression model and compared it to COXEN. The AUC from the model with only the trial’s two stratification factors (Performance Status [PS] 0 vs. 1; clinical tumor stage T2 vs. T3, T4a) predicting path response was compared to the AUC from the model with the added classifier. The null hypothesis was that the change in AUC=0 was tested between the two models. For PFS and OS endpoints, Cox regression models were evaluated with the two stratification factors and each classifier. Hazard ratios, 95% confidence intervals, and p-values are reported for each classifier. Five-year estimates are derived from the Kaplan-Meier curves.
Data Availability
Processed gene expression matrix and data that support the findings of this study are available from GEO Accession number GSE87304; ID 200087304. Platform GPL22995; 305 samples; Download data: CEL.
Results
One hundred fifty-five patients had adequate tissue, and gene expression results received at least 3 of 4 cycles of NAC and had pathologic staging of primary tumor stage and lymph nodes based on the radical cystectomy. For the pre-planned stratification factors, 78% were PS=0 and 89% were clinical T2. Additional covariates were 84% male, median age 65, 51% randomized to ddMVAC, and 49% GC. Of all patients, 33% had a complete pathologic response to NAC and were pT0N,0, and 52% were downstaged (pTa,T1,TisN0). Table 1 describes the response to NAC based on the three subtyping schemes. Basal TCGA, Consensus and MDA luminal had highest pT0 rates, while Luminal across all three classifiers had the highest overall downstaging rate to < pT2N0. The sensitivity analysis removing NE tumors (3 from TCGA and 2 from Consensus) resulted in very small changes in pT0 and downstaging rates that were within the 95% CI originally reported combining these with BS (data not shown).
Table 1:
Collapsed classifier groups and pathologic response outcomes, pooled treatment arms
| Classifier | # patients | Complete response, pT0N0 n (%; 95% CI) | Downstaging* (<pT2N0) n (%; 95% CI) |
|---|---|---|---|
| Total | 155 | 51 (33%; 26%, 41%) | 81 (52%; 44%, 60%) |
| TCGA Basal/Sq + Neuronal |
55 |
21 (38%; 25%, 52%) |
26 (47%; 34%, 61%) |
| Luminal | 77 | 25 (33%; 22%, 44%) | 43 (56%; 44%, 67%) |
| Luminal infiltrated | 23 | 5 (22%; 7%, 44%) | 12 (52%; 31%, 73%) |
| Consensus Ba, Sq, NE |
51 |
17 (33%; 21%, 48%) |
24 (47%; 33%, 62%) |
| Luminal | 80 | 27 (34%; 24%, 45%) | 46 (58%; 46%, 68%) |
| Stroma-rich | 24 | 7 (29%; 13%, 51%) | 11 (46%; 26%, 67%) |
| MDA Basal |
49 |
14 (29%; 17%, 43%) |
23 (47%; 33%, 62%) |
| Luminal | 56 | 22 (39%; 27%, 53%) | 36 (64%; 50%, 77%) |
| p53-like | 50 | 15 (30%; 18%, 45%) | 22 (44%; 30%, 59%) |
Downstaging category includes complete responders
TCGA – The Cancer Genome Atlas, Ba – Basal, Sq – Squamous, NE – neuroendocrine, MDA – MD Anderson
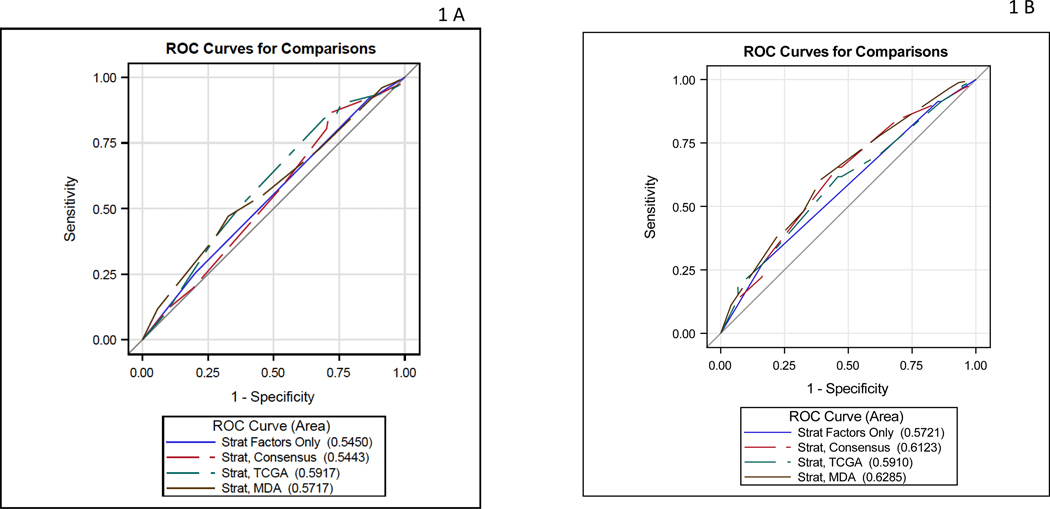
Table 2 describes the analysis of the contribution of each classifier’s association with pT0 and downstaging. Although the TCGA 3 group classifier (Basal-Squamous (BS)/Neuronal, Luminal, Luminal infiltrated) and GC COXEN score yielded modest in magnitude AUCs (0.59 and 0.60, respectively) for pT0 response, neither reached statistical significance (p=0.28, p=0.18, respectively). For downstaging (<pT2N0), the three-category Consensus classifier (BS/NE-like, Luminal, Stroma-rich) increased the AUC from 0.57 (strat factors alone) to 0.61 (p=0.10). For downstaging, the MDA classifier AUC was 0.63 (p=0.18), and the GC COXEN score AUC was 0.62 (P=0.23), but neither was statistically significant. The MVAC COXEN score did not improve the AUC beyond the stratification factors (AUC=0.56, p=0.68). This is graphically shown in the ROC curves for stratification factors alone and then combined with each classifier separately for the pT0 outcome (Figure 1A) and downstaging outcome (Figure 1B).
Table 2:
Evaluating AUC in logistic models predicting pathologic response with stratification (strat) factors and molecular subtype classifiers in 155 evaluable patients
| (n=155) | # of Categories | pT0N0 outcome | Downstaging (< pT2N0) | ||
|---|---|---|---|---|---|
| Incremental P-value * | AUC | Incremental P-value* | AUC | ||
| 2 Strat Factors Only (PS, stage) | NA | 0.55 | NA | 0.57 | |
| Classifier plus two strat factors | |||||
| TCGA | 3 | 0.28 | 0.59 | 0.44 | 0.59 |
| Consensus | 3 | 0.98 | 0.54 | 0.10 | 0.61 |
| MD Anderson | 3 | 0.51 | 0.57 | 0.18 | 0.63 |
| Dichotomous Coxen Scores plus two strat factors | |||||
| ddMVAC Coxen | 2 | 0.93 | 0.55 | 0.68 | 0.56 |
| GC Coxen | 2 | 0.18 | 0.60 | 0.23 | 0.62 |
2-sided p-value tests if classifier increases the AUC beyond the two stratification factors: clinical stage and PS (performance status)
Figure 1:
A) ROC curve for pT0 response: stratification factors alone and then combined with each classifier B) ROC curve for downstaging response: stratification factors alone and then combined with each classifier
There was a good correlation among the subtyping classifiers (Supplementary Table 1). There was very good agreement on the diagonal between the Consensus and TCGA classifiers, and the kappa statistic that evaluates the level of agreement between the two classifiers was: 0.80 (95% CI 0.71, 0.88) (Table 3A). Even though there was strong agreement in subtype assignment, the Consensus classifier predicted downstaging better than the TCGA classifier (Table 2). Comparing the Consensus and MD Anderson classifiers, cross-classification showed a good correlation between basal/squamous/neuroendocrine-like and basal and between stroma-rich and p-53-like in Consensus and MDA, respectively. However, the Consensus luminal was divided 2:1 between MDA luminal and p-53, and the overall kappa statistic was 0.63 (95% CI 0.53, 0.73)(Table 3B). Comparing the TCGA and MD Anderson classifiers, cross-classification similarly showed a good correlation between basal/squamous/neuroendocrine-like and basal between luminal infiltrated and p-53-like in Consensus and MDA, respectively. Similar to the Consensus classifier, TCGA luminal was divided between MDA luminal and p-53-like, and the overall kappa statistic was 0.56 (95% CI 0.46, 0.66).(Table 3C)
Table 3:
Agreement between Pairs of Classifiers for 155 evaluable patients
| A | |||
|---|---|---|---|
| Consensus Classifier | TCGA Ba Sq Ne | TCGA Luminal | TCGA Luminal infiltrated |
| Ba Sq Ne | 49 | 1 | 1 |
| Luminal | 3 | 71 | 6 |
| Stroma-rich | 3 | 5 | 16 |
| Kappa statistic: 0.80 (95% CI 0.71, 0.88) | |||
| B | |||
|---|---|---|---|
| Consensus Classifier | MDA Basal | MDA Luminal | MDA p53-like |
| Ba Sq Ne | 45 | 1 | 5 |
| Luminal | 1 | 53 | 26 |
| Stroma-rich | 3 | 2 | 19 |
| Kappa statistic: 0.63 (95% CI 0.53, 0.73) | |||
| C | |||
|---|---|---|---|
| TCGA Classifier | MDA Basal | MDA Luminal | MDA p53-like |
| Ba Sq Ne | 43 | 5 | 7 |
| Luminal | 3 | 49 | 25 |
| Luminal infiltrated | 3 | 2 | 18 |
| Kappa statistic: 0.56 (95% CI 0.46, 0.66) | |||
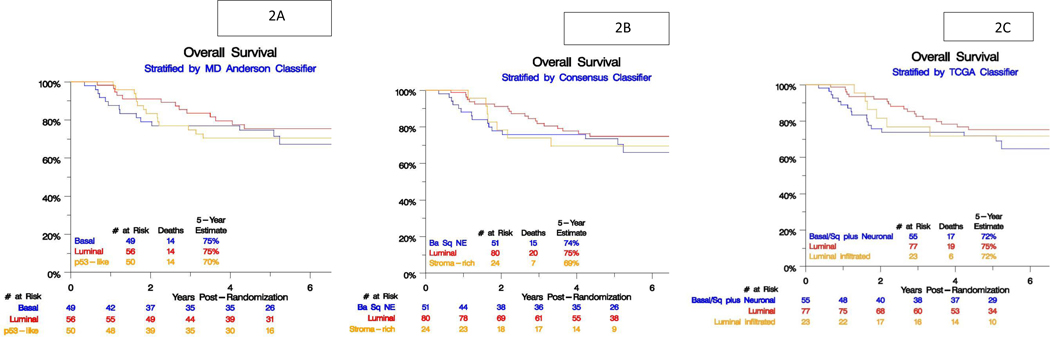
We did not find any statistically significant association between each subtype classification and PFS and OS (Table 4; Figure 2 A,B,C)
Table 4:
Association of Classifiers with Progression-Free and Overall Survival (n=155)
| Classifier | N/# deaths/# PFS events | 5-yr PFS estimate | PFS HR (95% CI), p-value* | 5-yr OS estimate | OS HR (95% CI), p-value* |
|---|---|---|---|---|---|
| TCGA | |||||
| Basal/Sq/Neuronal | 55/17/24 | 57% | 1.0 (reference) | 72% | 1.0 (reference) |
| Luminal | 77/19/28 | 63% | 0.68(0.40, 1.18), P=0.17 | 75% | 0.67 (0.35, 1.29), p=0.23 |
| Luminal inf. | 23/6/7 | 67% | 0.58 (0.25, 1.36), P=0.21 | 72% | 0.84 (0.33, 2.13) p=0.71 |
| Consensus | |||||
| Ba/Sq/NE | 51/15/21 | 60% | 1.0 (reference) | 74% | 1.0(reference) |
| Luminal | 80/20/29 | 63% | 0.75 (0.43, 1.31), P=0.31 | 75% | 0.73 (0.37, 1.43) p=0.36 |
| Stroma-rich | 24/7/9 | 61% | 0.84 (0.38, 1.85), P=0.67 | 69% | 1.03 (0.42, 2.55) p=0.95 |
| MDA | |||||
| Basal | 49/14/19 | 62% | 1.0 (reference) | 75% | 1.0 (reference) |
| Luminal | 56/14/20 | 65% | 0.81 (0.43, 1.52), P=0.51 | 75% | 0.73 (0.35, 1.55) p=0.41 |
| p53-like | 50/14/20 | 57% | 0.99 (0.53, 1.86), P=0.98 | 70% | 0.97 (0.46, 2.03) p=0.93 |
adjusted for two stratification factors, 2-sided p value
Figure 2 :
Subtype association with overall survival (OS) for A) Consensus classifier, B) MD Anderson classifier, C) TCGA classifier
Discussion
Current guidelines recommendations for patients with non-metastatic curable MIBC are for cisplatin-based neoadjuvant chemotherapy when considering radical cystectomy for definitive locoregional control. Prospective randomized phase III trials established a benchmark for expected pathologic tumor response and suggested that at least one-half of patients may not benefit from NAC. The COXEN trial was designed to validate an RNA-based predictive biomarker for NAC response. Locked-down COXEN scores for GC and ddMVAC were tested for their association with pCR and downstaging and did not predict response to their respective chemotherapy regimen, but the GC score was statistically significantly associated with pathologic downstaging in the pooled arms (GC and ddMVAC) with an OR of 2.33 (P = 0.02; 95% CI, 1.11–4.89). RNA-based expression subtypes reflect the biologic heterogeneity of MIBC and are associated with prognosis in muscle invasive bladder cancer, risk of upstaging of T stage, and response to cisplatin-based chemotherapy in multiple retrospective analyses(9,12,13). Therefore, we sought to determine subtype association with pT0 response, downstaging, and overall survival in the SWOG S1314 prospective randomized phase II trial designed to validate unique COXEN locked-down signatures for response to ddMVAC or GC.
We found only modest improvement in predicting pathologic complete response or downstaging following neoadjuvant chemotherapy when accounting for clinical tumor stage and performance status. In prior studies, the MDACC and Decipher (derived from TCGA) basal subtype was associated with longer disease-specific and overall survival in patients treated with NAC(13). Sjodahl and colleagues, though, recently reported differential responses to NAC based on the Lund Taxonomy such that genomically unstable (GU) and urothelial-like (Uro) cancers were associated with the better pathologic response rates compared to basal/squamous cancers(14). A total of 58% of patients were clinical stage T3 or worse. In a multivariable analysis, this had an odds ratio of 0.19 (p=0.00011) for pCR following NAC. Similarly, a majority of the patients included in the original MD Anderson study had high-risk tumors (9). In the COXEN trial, a minority of patients (11%) were cT3–4. The challenges in accuracy of clinical staging of MIBC suggest that we are not ready to parse out cT2–4a disease for enriching a target population for further study. We should continue to stratify patients by clinical stage and test MRI as this may provide additional information to improve precision of clinical staging. In an editorial, Jong and Gibb suggest that the lack of response attributed to Lund basal/squamous cancers may be due to a preponderance of claudin-low tumors, which have a higher proportion of immune infiltration(15).
Taber and colleagues reported an integrated multi-omics analysis of patients with MIBC treated with either NAC or chemotherapy for locally advanced or metastatic disease(16). They reported that the basal squamous subtype using the consensus classifier was associated with a lower response to NAC. However, the response rate reported was just under 50%, and overall, there were no significant differences in response rates among subtypes. The overall response rate is higher than previously reported in prospective trials, and they used fresh frozen tumors for this analysis on 120 patients.
The Vesper trial randomized patients to ddMVAC and GC perioperative chemotherapy and did not meet the pre-specified hazard ratio for 3-year progression-free survival though there did appear to be improved PFS with ddMVAC in patients treated in the neoadjuvant setting (17). Groeneveld and colleagues assigned subtypes in 296 patients from the Vesper trial using the Consensus classifier and found that while mixed subtypes were associated with lower pathologic complete response rates, there was no significant association between subtypes and pCR (18). They found that patients with basal/squamous or mixed subtypes had worse PFS than other subtypes. In the COXEN trial, the individual GC or MVAC scores were not predictive for PFS or OS, but the GC score was prognostic for OS in the combined treatment arms. Subtypes determined with the three classification schemes in the present study were also not associated with PFS or OS. The COXEN trial is likely limited in power due to smaller numbers in a phase II trial design and fewer deaths.
We collapsed subtypes into three categories for each of the Consensus and TCGA classifiers based on small numbers in some of the respective subtypes. Cross-comparison of the three subtyping schemes used in this analysis suggested a good correlation between TCGA and Consensus classifiers. However, the luminal subtype for each was divided 2:1 between luminal and p-53-like MDA subtypes. This is consistent with what Kamoun et al. showed, that Consensus luminal subtypes were comprised in part of MDA p53-like subtypes with good correlation for basal/squamous between all three subtyping schemes(10).
Conclusion
The Consensus classifier, based partly on the TCGA and MDA classifiers, modestly improved prediction for pathologic downstaging when adjusted for clinical stage and performance status but was not associated with PFS or survival. A low number of events likely impacted the power to detect associations with progression-free and overall survival. These observations are not practice changing and prospective clinical trials are warranted to test the association of subtypes with outcomes, and in particular, whether subtype directed therapy can improve response to neoadjuvant therapy.
Supplementary Material
TRANSLATIONAL RELEVANCE.
The COXEN trial was designed as a validation study of predictive biomarkers for response to the two most commonly used standard of care cisplatin-based neoadjuvant chemotherapy regimens prior to radical cystectomy. The GC score was associated with pathologic downstaging but not PFS or OS. RNA-based molecular subtypes parse out the complex heterogeneity of urothelial bladder cancer and may offer a precision medicine approach to selecting appropriate therapy for patients in the neoadjuvant space. We describe the association of three published classifiers with pathologic downstaging, PFS and OS in order to determine the added value to COXEN and known co-variates associated with outcomes including clinical tumor stage and performance status. These data set the stage for testing a subtype directed approach to neoadjuvant therapy selecting patients for chemotherapy, immune- or targeted therapy.
Acknowledgments:
Support from NIH/NCI grants U10CA180888, U10CA180819, UG1CA233196, UG1CA233328, UG1CA233320, UG1CA233324, UG1CA180830, UG1CA233160. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
CONFLICT OF INTEREST STATEMENT
Drs. Lerner, McConkey and Meeks are co-holders of a patent pending on the TCGA classifier.
Drs. McConkey and Choi are inventors of the MDA classifier.
Drs. Theodorescu and Gustafson are co-inventors of the COXEN classifier.
The remaining co-authors have no relevant conflicts of interest.
References
- 1.Grossman HB, Natale RB, Tangen CM, Speights VO, Vogelzang NJ, Trump DL, et al. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. N Engl J Med. 2003. Aug 28;349(9):859–66. [DOI] [PubMed] [Google Scholar]
- 2.Sonpavde G, Goldman BH, Speights VO, Lerner SP, Wood DP, Vogelzang NJ, et al. Quality of pathologic response and surgery correlate with survival for patients with completely resected bladder cancer after neoadjuvant chemotherapy. Cancer. 2009. Sep 15;115(18):4104–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JK, Havaleshko DM, Cho H, Weinstein JN, Kaldjian EP, Karpovich J, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci U S A. 2007. Aug 7;104(32):13086–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith SC, Baras AS, Lee JK, Theodorescu D. The COXEN principle: translating signatures of in vitro chemosensitivity into tools for clinical outcome prediction and drug discovery in cancer. Cancer Res. 2010. Mar 1;70(5):1753–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flaig TW, Tangen CM, Daneshmand S, Alva A, Lerner SP, Lucia MS, et al. A Randomized Phase II Study of Coexpression Extrapolation (COXEN) with Neoadjuvant Chemotherapy for Bladder Cancer (SWOG S1314; NCT02177695). Clin Cancer Res Off J Am Assoc Cancer Res. 2021. May 1;27(9):2435–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lerner SP, McConkey DJ, Hoadley KA, Chan KS, Kim WY, Radvanyi F, et al. Bladder Cancer Molecular Taxonomy: Summary from a Consensus Meeting. Bladder Cancer Amst Neth. 2016. Jan 7;2(1):37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell. 2017. Oct 19;171(3):540–556.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lotan Y, Boorjian SA, Zhang J, Bivalacqua TJ, Porten SP, Wheeler T, et al. Molecular Subtyping of Clinically Localized Urothelial Carcinoma Reveals Lower Rates of Pathological Upstaging at Radical Cystectomy Among Luminal Tumors. Eur Urol. 2019. Aug;76(2):200–6. [DOI] [PubMed] [Google Scholar]
- 9.Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014. Feb 10;25(2):152–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamoun A, de Reyniès A, Allory Y, Sjödahl G, Robertson AG, Seiler R, et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur Urol. 2020. Apr;77(4):420–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Kwiatkowski D, McConkey DJ, Meeks JJ, Freeman SS, Bellmunt J, et al. The Cancer Genome Atlas Expression Subtypes Stratify Response to Checkpoint Inhibition in Advanced Urothelial Cancer and Identify a Subset of Patients with High Survival Probability. Eur Urol. 2019. Jun;75(6):961–4. [DOI] [PubMed] [Google Scholar]
- 12.Sjödahl G, Lauss M, Lövgren K, Chebil G, Gudjonsson S, Veerla S, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res Off J Am Assoc Cancer Res. 2012. Jun 15;18(12):3377–86. [DOI] [PubMed] [Google Scholar]
- 13.Seiler R, Ashab HAD, Erho N, van Rhijn BWG, Winters B, Douglas J, et al. Impact of Molecular Subtypes in Muscle-invasive Bladder Cancer on Predicting Response and Survival after Neoadjuvant Chemotherapy. Eur Urol. 2017. Oct;72(4):544–54. [DOI] [PubMed] [Google Scholar]
- 14.Sjödahl G, Abrahamsson J, Holmsten K, Bernardo C, Chebil G, Eriksson P, et al. Different Responses to Neoadjuvant Chemotherapy in Urothelial Carcinoma Molecular Subtypes. Eur Urol. 2022. May;81(5):523–32. [DOI] [PubMed] [Google Scholar]
- 15.de Jong JJ, Gibb EA. Sjödahl Re: Gottfrid, Abrahamsson Johan, Holmsten Karin, et al. Different Responses to Neoadjuvant Chemotherapy in Urothelial Carcinoma Molecular Subtypes. Eur Urol. 2022;81:316–7.: Neoadjuvant Chemotherapy Response in Muscle-invasive Bladder Cancer: Differences in Intrinsic Biology or Subtyping Nomenclature? Eur Urol. 2022 Apr;81(4):e90–1. [DOI] [PubMed] [Google Scholar]
- 16.Taber A, Christensen E, Lamy P, Nordentoft I, Prip F, Lindskrog SV, et al. Molecular correlates of cisplatin-based chemotherapy response in muscle invasive bladder cancer by integrated multi-omics analysis. Nat Commun. 2020. Sep 25;11(1):4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfister C, Gravis G, Fléchon A, Soulié M, Guy L, Laguerre B, et al. Randomized Phase III Trial of Dose-dense Methotrexate, Vinblastine, Doxorubicin, and Cisplatin, or Gemcitabine and Cisplatin as Perioperative Chemotherapy for Patients with Muscle-invasive Bladder Cancer. Analysis of the GETUG/AFU V05 VESPER Trial Secondary Endpoints: Chemotherapy Toxicity and Pathological Responses. Eur Urol. 2021. Feb;79(2):214–21. [DOI] [PubMed] [Google Scholar]
- 18.Groeneveld CDS, Harter V, Culine S, Krucker C, Dixon V, de Reynies A, et al.1736MO Pure or mixed basal/squamous tumours present decreased outcomes after neoadjuvant chemotherapy in the GETUG-AFU V05 VESPER trial. Annals of Oncology 33(supp7):S1330, 2022 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Processed gene expression matrix and data that support the findings of this study are available from GEO Accession number GSE87304; ID 200087304. Platform GPL22995; 305 samples; Download data: CEL.