Abstract
G-quadruplexes (G4s) are reported to present on the SARS-CoV-2 RNA genome and control various viral activities. Specific ligands targeting those viral nucleic acid structures could be investigated as promising detection methods or antiviral reagents to suppress this menacing virus. Herein, we demonstrate the binding between a G4 structure in the RNA of SARS-CoV-2 and a fluorescent probe created by fusing a parallel-G4 specific RHAU53 and a cyan fluorescent protein. The specific binding of G4 in SARS-CoV-2 by RHAU peptide was easily detected under the fluorescence spectrometer. The drawbacks of this approach and potential solutions are also discussed.
Keywords: RHAU peptide, G-quadruplex (G4), SARS-CoV-2, RNA, G4-targeting
Graphical abstract
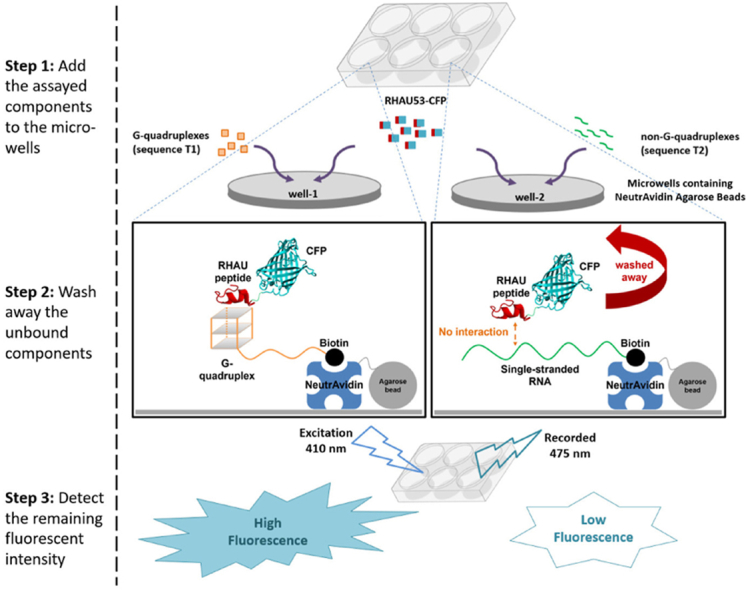
Diagram showing the presented fluorescent assay to investigate the binding between the SARS-CoV-2's RNA G4 and the RHAU53 peptide.
Highlights
-
•
The specific binding of G4 in SARS-CoV-2 by RHAU peptide was easily detected under the fluorescence spectrometer.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread globally since 2019, causing one of the deadliest pandemics in the human history, COVID-19. By September 2023, almost every countries has reported about 770 million infections and more than 6.9 million deaths (“WHO Coronavirus (COVID-19) Dashboard,” n.d.). Despite extensive international efforts for treatment and vaccination, COVID-19 morbidity and mortality remain high; and the rapid evolution of highly virulent and vaccine-resistant SARS-CoV-2 strains has sparked complicated waves of virus transmission in many countries (Telenti et al., 2022; Ciotti et al., 2022). Therefore, the development of broad-spectrum antiviral reagents with high indexes of effectiveness and safety is decisively important. Anti-SARS-CoV-2 medications generally target RNA-dependent RNA polymerases, exonucleases, proteases, viral structural proteins, and host proteins (Majumder and Minko, 2021; Zarandi et al., 2021). However, targeting a single molecule seems to be inadequate since SARS-CoV-2 mutations have been shown to occur very frequently. Combinatorial regimens that target multiple stages of the SARS-CoV-2 life cycle would increase efficacy and diminish drug-resistant variants (Akinbolade et al., 2022; Wang et al., 2022).
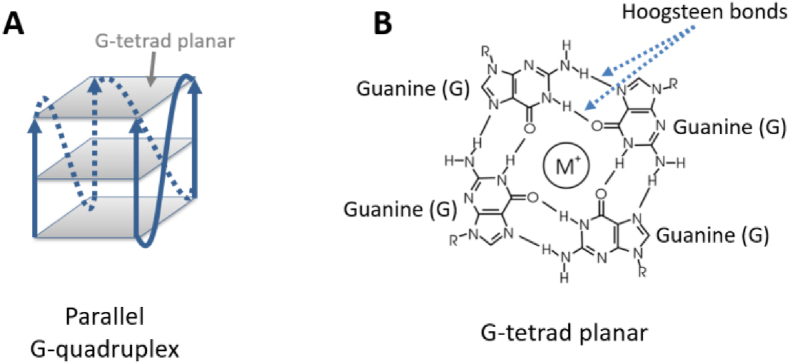
Nucleic acid in the viruses is quite conserved in terms of secondary structures, and this feature may facilitate their prospect as promising antiviral targets (Rangan et al., 2020; Vandelli et al., 2020; Lan et al., 2022). It is evident that G4s may form on guanine-rich strands of DNA or RNA, by stacking multiple layers of G-tetrad in the presence of ion Na + or K+ (Phan et al., 2006; Varshney et al., 2020). Four guanines in the G-tetrad interact with each other by Hoogsteen bonds (Fig. 1). G4s are proved to involve in the regulation of many important biological processes, not only in humans but also in yeasts, bacteria, and viruses (Rhodes and Lipps, 2015; Wu et al., 2021). As a conserved nucleic acid secondary structure, G4s are gaining recognition as potential antiviral targets for several viruses, such as HIV, Zika, Ebola, and even SARS-CoV-2 (Métifiot et al., 2014; Fleming et al., 2016; Wang et al., 2016; Qin et al., 2022). Potential quadruplex sequences (PQSs) on a significant proportion of the SARS-CoV-2 genome have been predicted (Cui and Zhang, 2020; Zhang et al., 2020; Ji et al., 2021), and their formation has also been demonstrated in vitro (Qin et al., 2022). Many existing G4 ligands have entered various preclinical trials to contain the propagation of this virus, showing promising outcomes (Qin et al., 2022; Liu et al., 2022). It is notable that the number of studies on targeting SARS-CoV-2 RNA G4s is growing quickly (Zhai et al., 2022), implying significant progresses in the future.
Fig. 1.
Representations of a G4 structure and G-tetrad.
It has been shown that there is a G4-binding motif located at the N-terminal of the RHAU protein (RNA Helicase Associated with AU-rich Element) (Meier et al., 2013; Heddi et al., 2015). Indeed, various peptides generated from that motif have been investigated as G4 ligands in a wide range of research and applications (Dang and Phan, 2016, 2019; Dang et al., 2021; Nguyen and Dang, 2022). In this study, we aim to demonstrate that the RHAU peptide can specifically bind to a G-quadruplex structure formed on the RNA genome of SARS-CoV-2, as well as suggested a simple fluorescent methodology to detect those viral G4s in certain cases. Even though the fluorescent protein probe (RHAU53-CFP) and the associating method have been already reported in a previous study (Dang and Phan, 2016), the feasibility of applying them on SARS-CoV-2 has never been proved before. This work might play as a proof-of-concept study to open a novel research direction of using RHAU peptides to handle this threatening virus.
2. Materials and methods
2.1. RNA G4 derived from the SARS-CoV-2 genome and the production of RHAU53-CFP fusion protein
An RNA fragment having the sequence 5′-rGrGrC-rUrGrGrCrArArUrGrGrCrGrG-3’ (sequence T1; Table 1), derived from the RNA region coding for the nucleocapsid (N) protein of SARS-CoV-2 (at the genome position 28,903), was reported to form a stable G4 structure in K+ solution (Qin et al., 2022). In this study, a fluorescent approach was employed to evaluate the binding between this G4 structure and the RHAU53 peptide. Particularly, biotin was incorporated into the T1 sequence at the 3′-end, flanked by an octa-rU fragment as the linker. The RHAU53 peptide was cloned and produced in E. coli as a fusion protein with CFP (cyan fluorescent protein). Specific binding between the SARS-CoV-2's RNA G4 structure (T1 sequence) and the RHAU53 would lead to the co-precipitation of fluorescent proteins with NeutrAvidin agarose beads (Thermo Fisher Scientific, USA), through the biotin-streptavidin and RHAU53-G4 interactions, resulting in the increased fluorescent signal in T1 samples as compared to the control (Fig. 2). T1 was mutated in the G4-forming motif, yielding the sequence 5′-rGrUrCrUrGrUrCrArArUrGrUrCrUrG-3’ (denoted as T2; mutations highlighted by bold case; Table 1). The sequence T2 serves as a non-G4 control of the T1. An octa-rU-biotin fragment (italic) was also incorporated into the T2 at their 3′-end. Both RNAs were purchased from Synbio Technologies (USA) and reconstituted in distilled water containing 100 mM KCl, aliquoted and kept at −80 °C for further uses.
Table 1.
T1 and T2 RNA sequences used in this study.
| Name | Sequences (5′-3′) | Length (nt) |
|---|---|---|
| T1 | rGrGrCrUrGrGrCrArArUrGrGrCrGrGrUrUrUrUrUrUrUrU/3Bio/ | 23 |
| T2 | rGrUrCrUrGrUrCrArArUrGrUrCrUrGrUrUrUrUrUrUrUrU/3Bio/ | 23 |
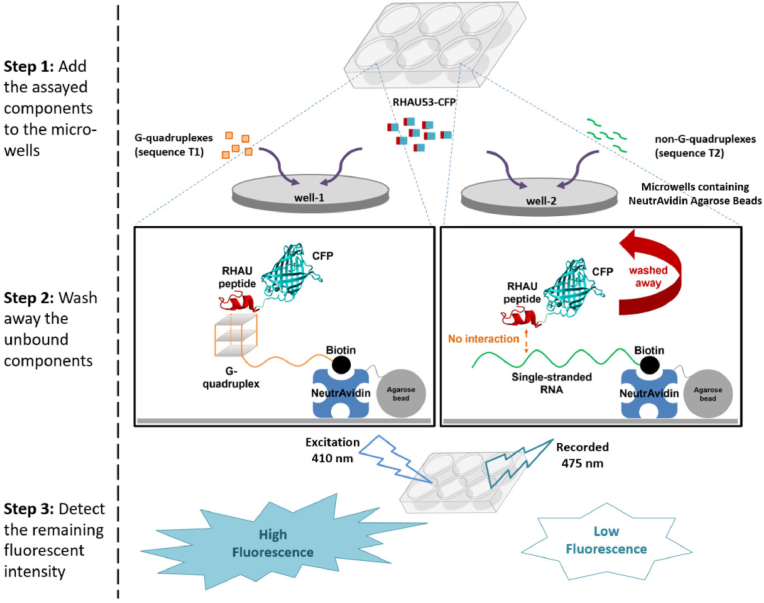
Fig. 2.
Diagram showing the presented fluorescent assay to investigate the binding between the SARS-CoV-2's RNA G4 and the RHAU53 peptide.
To produce the RHAU53-CFP fusion protein, the gene encoded for a 53-amino-acid peptide derived from the G4-binding motif of the RHAU protein (amino acid 53 to 105) was inserted into the plasmid pETDuet-1 (Addgene), in-frame, and between the 3′-end of the 6xHis-tag and the 5′-end of the CFP gene (Supporting Fig. 1). The recombinant expression was accomplished in E. coli BL21 (DE3) under the induction of 0.5 mM IPTG at 16 °C for 10 h. The fusion protein was purified by affinity chromatography on HisTrap column (GE Healthcare) in buffers containing 20 mM Na2HPO4 + NaH2PO4 (pH 7.0), 100 mM NaCl and varied concentrations of imidazole. The purified proteins were exchanged thoroughly to a storage buffer containing 20 mM Na2HPO4 + NaH2PO4 (pH 7.0), 75 mM NaCl, 5% glycerol; and were concentrated to 10–15 mg/ml to keep at −80 °C for further uses.
2.2. Investigation of the SARS-CoV-2's RNA G4 by circular dichroism (CD) spectroscopy
G-quadruplex structures could be characterized by Circular Dichroism (CD) Spectroscopy due to their chiral and polarization features. Three major topologies of G4s, namely parallel, anti-parallel and hybrid, could be readily determined according to their distinct CD spectra (Del Villar-Guerra et al., 2018), despite atomic details of a G4 structure still cannot be obtained through this technique. The molar ellipticity [θ], which represents the difference in absorbance (corrected for concentration) between the left-handed and right-handed circularly polarized lights through the chiral molecules, is measured across the range of UV wavelengths and plotted on a spectrum. It has become a standard practice to make a qualitative association between the characteristics of a CD spectrum and one of three possible G4 topologies: parallel (a positive peak at around 264 nm and a negative at around 245 nm), antiparallel (the spectrum has 2 positive peaks at around 245 nm and 295 nm, and a negative at around 264 nm), or hybrid (the spectrum has a positive peak at around 264 nm and 295 nm, and a negative at around 240 nm) (Del Villar-Guerra et al., 2018).
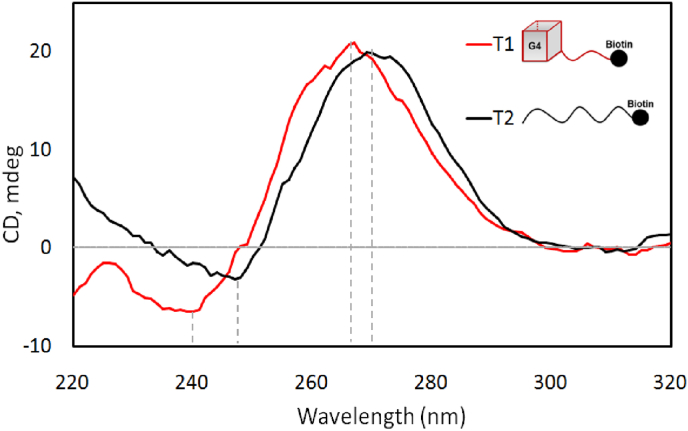
The T1 and T2 sequences were diluted to 5 μM each in a buffer containing 20 mM K2HPO4 – KH2PO4 pH 6.5 and 100 mM KCl, prior to CD spectra analyses using a JASCO-815 spectropolarimeter. The spectra were obtained in a 1-cm quartz cuvette containing 500 μl of samples at 25 °C. Bandwidth and scan rate were both set to 1 nm. The spectra of the samples were adjusted for zero at 320 nm by subtracting the spectrum of the buffer, and the subtracted spectra are shown in Fig. 3.
Fig. 3.
Circular Dichroism (CD) spectra showing the evidence of G4 in the T1 (red) and non-G4 in the T2 (black). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.3. Evaluating the binding between the SARS-CoV-2's RNA G4 and the RHAU53-CFP by a fluorescent co-precipitation assay
The specific binding between the SARS-CoV-2's RNA G4 and the RHAU53 peptide was evaluated through an adapted fashion of fluorescent assay (Fig. 2). Particularly, the T1 and T2 RNA samples (10 μM each) were separately attached to NeutrAvidin agarose beads (Thermo Fisher Scientific, USA), according to the manufacturer's protocol, as the result of interaction between biotins and avidin proteins. The RHAU53-CFP fusion protein was subsequently added into each sample to the concentration of 10 μM, incubated for 10 min at 25 °C to facilitate the interaction between RHAU peptides and G4 structures. After excessive rounds of washes by PBS 1× buffer to remove the unbound residuals, the mixtures were then dispensed into a multi-well plate (100 μl each, replicated 3 wells for each sample). All wells were then exposed to an excitation light source at the wavelength of 410 nm, and the intensity of the emission lights at the wavelength of 475 nm were recorded. The difference in fluorescent signals between the T1 and T2 samples were analyzed to indicate the presence of specific binding between the SARS-CoV-2's G4s (T1) and the RHAU53 peptides.
3. Results
3.1. Circular dichroism (CD) spectroscopy implying the presence of an RNA G4 structure in the T1 sequence
The T1 and T2 samples were subjected to CD spectroscopy to investigate the possible G4 formation. The results (Fig. 3) show that the T1's CD spectrum has a peak at around 264 nm and a trough at around 245 nm (red curve), which is evident that this RNA sequence has formed a parallel G4 structure; whereas there is a shift of about 7–10 nm to the right in the T2's CD spectrum (black curve), which indicates that the mutated sequence did not possess the G4's circular dichroism features, and might not be a G4 structure. These results were consistent with earlier investigations (Qin et al., 2022), even though there has been an octa-rU linkers and biotin incorporated at their 3′-end.
3.2. RHAU53-CFP fusion protein was effectively expressed and purified using the E. coli recombinant expression systems
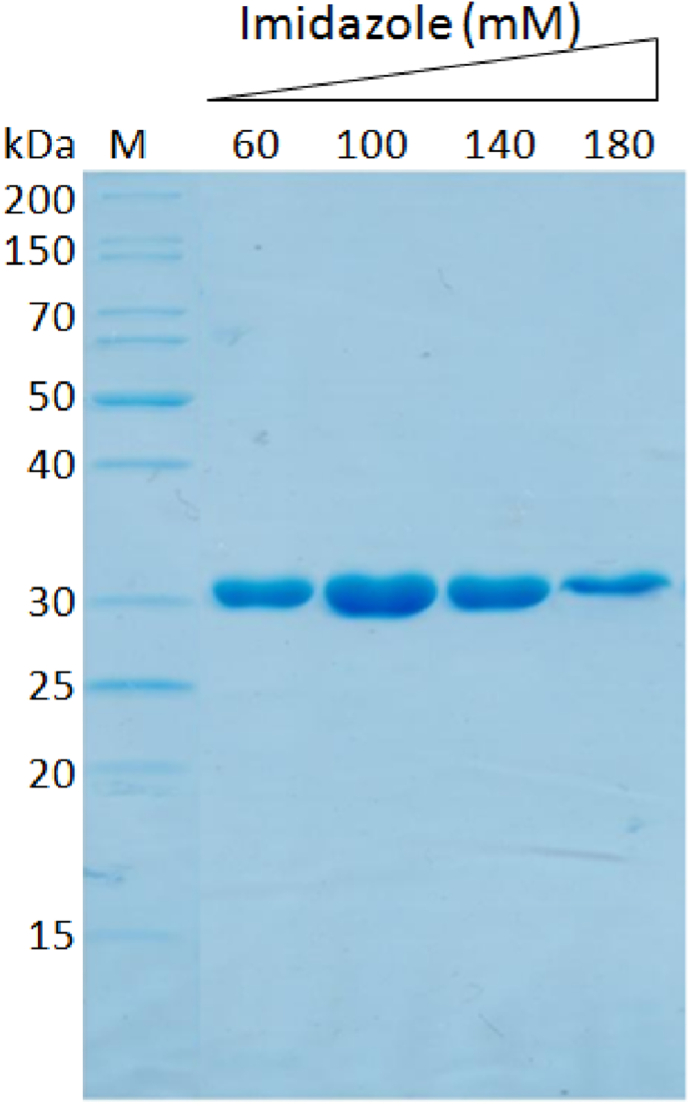
The gene encoded for RHAU53 was fused to the 5′-end of CFP gene and inserted to the plasmid pETDuet1. The recombinant fusion protein was expressed in E. coli BL21 (DE3) under the induction of IPTG on the T7 promoter; and purified by His-tag affinity chromatography (Histrap column). The purified samples were analyzed by SDS-PAGE, showing a desired molecular weight of the target protein (32 kDa). They were eluted strongly from the Histrap column in the purification buffers containing 60 mM–180 mM imidazole, yielding around 5 mg per 1 L of bacterial shaking culture with the purity of higher than 90% (Fig. 4). The pure RHAU53-CFP fusion proteins were dialyzed thoroughly (into 20 mM Na2HPO4 + NaH2PO4 (pH 7.0), 75 mM NaCl, 5% glycerol) to remove imidazole, and were subsequently used for the experiment of binding to the SARS-CoV-2's RNA G4.
Fig. 4.
SDS-PAGE result indicating the production and purity of recombinant RHAU53-CFP fusion proteins.
3.3. RHAU53 can effectively bind to the SARS-CoV-2's RNA G4 structure
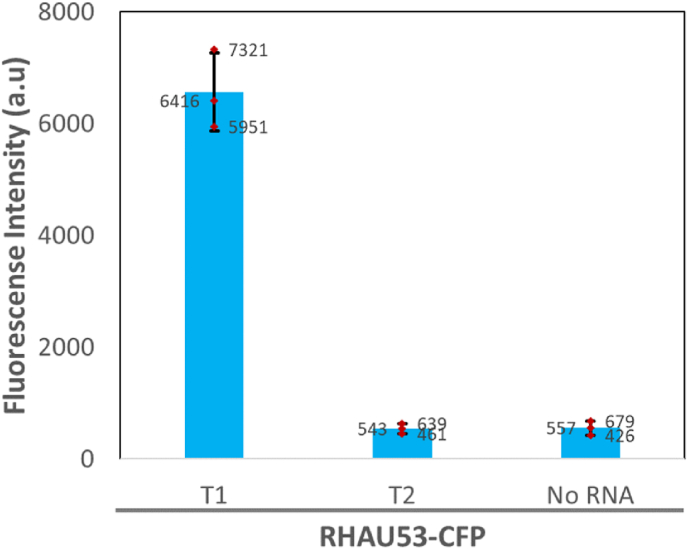
The T1 and T2 samples, with and without G4 structures, respectively (CD results), were subjected to the co-precipitation experiments with RHAU-CFP proteins through NeutrAvidin agarose beads (Fig. 2). When there are specific bindings between RHAU53 and the viral RNA G4, the fluorescent proteins will be co-precipitated with the agarose beads as the results of specific biotin-avidin and RHAU53-G4 interactions, leading to the increased fluorescent signals in the corresponding samples. On the other hand, an insignificant fluorescence might indicate a low extent of interactions between those components, particularly between the fusion protein and the RNA. The results showed that, there was a high intensity of emission lights recorded only in the micro-wells containing the T1 samples, whereas the T2 samples and the blanks (without any RNA) exhibited insignificant signals (Fig. 5). These findings indicate that the RHAU53-CFP fusion proteins had been captured only by the T1 samples but not by the T2 and the blank, suggesting a strong interaction between RHAU peptides and one of the putative G4 structures presented on the SARS-CoV-2 RNA genome (T1 sequence). It was also found that the G4 structure of T1 is possibly in a parallel form (CD results from previous report (Qin et al., 2022) and this study), which might explain why RHAU53, a peptide with parallel-G4 selectivity (Heddi et al., 2015), could effectively and specifically bind to them.
Fig. 5.
Bar chart suggesting the specific binding between RHAU53-CFP and the RNA G4 structure in the T1 sequence, indicated by a high co-precipitated fluorescent signal. Error bars: standard deviations of three replicates.
4. Discussions
SARS-CoV-2 infection could be treated by targeting viral proteases, nucleases, structural proteins or host receptors such as ACE2 or TMPRSS222 (Majumder and Minko, 2021; Zarandi et al., 2021; Akinbolade et al., 2022; Wang et al., 2022). However, the treatments are still restrained in terms of effectivity, due to the frequent and rapid evolutionary mutation of the virus genome, which could essentially lead to the generation of alternative targets that might adequately evade current immunization and medication strategies (Telenti et al., 2022; Ciotti et al., 2022). Nevertheless, secondary structures of viral RNA were reported to be highly conserved, and they themselves could be the more promising targets for novel broad-spectrum antiviral agents rather than their translated products (Rangan et al., 2020; Vandelli et al., 2020; Lan et al., 2022). G4s are proven to take part in the regulation of various biological processes in most organisms including viruses, such as the RNA transcription in HIV (Métifiot et al., 2014) or the DNA replication in Zaire ebolavirus (EBOV) (Wang et al., 2016). There is an increasing number of interests in the discovery of antiviral agents that target the viral G4s to suppress the virus transmission (Métifiot et al., 2014). It has also been reported that stabilizing G4s in the SARS-CoV-2's RNA can inhibit the nucleocapsid phosphoproteins expression, one of the critical stages in the life cycles of this virus (Qin et al., 2022; Zhao et al., 2021). More importantly, SARS-CoV-2's RNA also possesses a great number of undefined G4s, which were predicted to be abundant and crucial for virus activities (Cui and Zhang, 2020; Zhang et al., 2020), making G4-targeted compounds absolutely rational for development to effectively contain the current ‘metamorphosis’ coronavirus pandemic as well as possible future outbreaks.
In this study, we have demonstrated a specific binding of the RHAU53 peptide, one of the renowned peptide-based ligands for parallel G4s (Heddi et al., 2015; Dang and Phan, 2016) to an RNA G4 structure derived from the RNA genome of SARS-CoV-2 (Qin et al., 2022) by using a fluorescent co-precipitation method. The biotinylated version of that RNA sequence was still favored for in vitro G4 formation, confirmed by CD spectroscopy; and maintained the specific recognition by RHAU53. Such specific binding should be further verified using the entire RNA genome of SARS-CoV-2 rather than just a synthesized short fragment (e.g., the T1 sequence in this study). A possible approach is using RT-qPCR assays with the SARS-CoV-2's RNA extracted from clinical specimens as the templates, and the primer pairs spanning the G4 regions (Fig. 6). When there is a possible instance that the RHAU peptides stabilize the indicated G4 structure on the viral RNA genome (Fig. 6), there should be a reduced quantitative PCR signal if using the R2 primer for cDNA synthesis as compared to using the R1 primer, since the RHAU peptide and the G4 structure probably hinder the progression of the reverse transcription enzymes.
Fig. 6.
Diagram showing the procedure of SARS-CoV-2 G4 detection by RT-qPCR. Note: using the primer SARS-CoV-2-R1 and R2 for cDNA synthesis right before real-time PCR.
There is a major drawback in the uses of G4 ligands to inhibit the virus propagation due to off-target effects. G4s also ubiquitously present in the human genome and transcriptome, and many of them exhibit vital functions for the human cells. G4s regulate DNA replication, gene expression, and telomere preservation (Rhodes and Lipps, 2015). Therefore, ligands for SARS-CoV-2 G4s may also negatively influence a large portion of biological processes in the host. However, there are still remedies for minimizing those side effects. A rising number of studies has been shown that the virus significantly replicates its RNA to generate a large number of viral G4s that transcend the cellular G4s by many logs per cell (Bar-On et al., 2020; Abiri et al., 2021; Mautner et al., 2022). This feature suggests that the use of only moderately specific ligands or low concentration of ligands could still potentially display a strong suppression of SARS-CoV-2 infection as there are higher chances of ligand binding to abundant viral G4s rather than the host G4s, limiting off-target effects. Besides, antiviral RHAU peptides would also be more effective and safer if being incorporated to a specific oligonucleotide targeting a G4-juxtaposed region on the virus genome. The complementary interaction of the designed oligonucleotides and the virus RNA may guide the RHAU peptides exactly to the virus genome rather than the human genome and transcriptome, therefore potentially minimizing the side effects. Finally, structural studies on the molecular details of specific interaction between RHAU peptides and SARS-CoV-2 G4s may also benefit the rational drug-design approaches to invent novel ligand derivatives with further enhanced viral specificity and reduced off-target effects when targeting this virus in human bodies.
5. Conclusions
In conclusion, G4s are crucial to SARS-CoV-2 life cycles and may represent a potential COVID-19 therapeutic target since they are relatively conserved in terms of molecular structure. This study demonstrated the specific binding between a G4 structure in the SARS-CoV-2's RNA genome and one of the potential peptide-based G4 ligands, RHAU53. This study might play as a proof-of-concept piece of work that potentially opens a novel research direction of using RHAU peptides to rationally identify and synthesize pharmacological candidates with great selectivity for SARS-CoV-2 genomic secondary structures as an alternative approach for COVID-19 treatment.
CRediT authorship contribution statement
Le Tuan Anh Nguyen: performed experiments under the supervision of Dung Thanh Dang, All authors wrote the manuscript. Thao Thu Thi Nguyen: conceived the idea, performed experiments under the supervision of Dung Thanh Dang. Dung Thanh Dang: conceived the idea.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank Prof. Anh Tuân Phan (Nanyang Technological University, Singapore) for providing plasmid and scientific discussions.
Handling Editor: Dr A Wlodawer
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2024.100126.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- Abiri A., Lavigne M., Rezaei M., Nikzad S., Zare P., Mergny J.-L., Rahimi H.-R. Unlocking G-quadruplexes as antiviral targets. Pharmacol. Rev. 2021;73:897–923. doi: 10.1124/pharmrev.120.000230. [DOI] [PubMed] [Google Scholar]
- Akinbolade S., Coughlan D., Fairbairn R., McConkey G., Powell H., Ogunbayo D., Craig D. Combination therapies for COVID-19: an overview of the clinical trials landscape. Br. J. Clin. Pharmacol. 2022;88:1590–1597. doi: 10.1111/bcp.15089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-On Y.M., Flamholz A., Phillips R., Milo R. SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciotti M., Ciccozzi M., Pieri M., Bernardini S. The COVID-19 pandemic: viral variants and vaccine efficacy. Crit. Rev. Clin. Lab Sci. 2022;59:66–75. doi: 10.1080/10408363.2021.1979462. [DOI] [PubMed] [Google Scholar]
- Cui H., Zhang L. G-quadruplexes are present in human Coronaviruses including SARS-CoV-2. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.567317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang D.T., Nguyen L.T.A., Truong T.T.T., Nguyen H.D., Phan A.T. Construction of a G-quadruplex-specific DNA endonuclease. Chem Commun. 2021;57:4568–4571. doi: 10.1039/d0cc05890d. [DOI] [PubMed] [Google Scholar]
- Dang D.T., Phan A.T. Development of a ribonuclease containing a G4-specific binding motif for programmable RNA cleavage. Sci. Rep. 2019;9:7432. doi: 10.1038/s41598-019-42143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang D.T., Phan A.T. Development of fluorescent protein probes specific for parallel DNA and RNA G-quadruplexes. Chembiochem. 2016;17:42–45. doi: 10.1002/cbic.201500503. [DOI] [PubMed] [Google Scholar]
- Del Villar-Guerra R., Trent J.O., Chaires J.B. G-quadruplex secondary structure obtained from circular dichroism spectroscopy. Angew Chem. Int. Ed. Engl. 2018;57:7171–7175. doi: 10.1002/anie.201709184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A.M., Ding Y., Alenko A., Burrows C.J. Zika virus genomic RNA possesses conserved G-quadruplexes characteristic of the flaviviridae family. ACS Infect. Dis. 2016;2:674–681. doi: 10.1021/acsinfecdis.6b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddi B., Cheong V.V., Martadinata H., Phan A.T. Insights into G-quadruplex specific recognition by the DEAH-box helicase RHAU: solution structure of a peptide–quadruplex complex. Proc. Natl. Acad. Sci. U. S. A. 2015;112:9608–9613. doi: 10.1073/pnas.1422605112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji D., Juhas M., Tsang C.M., Kwok C.K., Li Y., Zhang Y. Discovery of G-quadruplex-forming sequences in SARS-CoV-2. Brief Bioinform. 2021;22:1150–1160. doi: 10.1093/bib/bbaa114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan T.C.T., Allan M.F., Malsick L.E., Woo J.Z., Zhu C., Zhang F., Khandwala S., Nyeo S.S.Y., Sun Y., Guo J.U., Bathe M., Näär A., Griffiths A., Rouskin S. Secondary structural ensembles of the SARS-CoV-2 RNA genome in infected cells. Nat. Commun. 2022;13:1128. doi: 10.1038/s41467-022-28603-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Du W., Sang X., Tong Q., Wang Y., Chen G., Yuan Y., Jiang L., Cheng W., Liu D., Tian Y., Fu X. RNA G-quadruplex in TMPRSS2 reduces SARS-CoV-2 infection. Nat. Commun. 2022;13:1444. doi: 10.1038/s41467-022-29135-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder J., Minko T. Recent developments on therapeutic and diagnostic approaches for COVID-19. AAPS J. 2021;23:14. doi: 10.1208/s12248-020-00532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mautner L., Hoyos M., Dangel A., Berger C., Ehrhardt A., Baiker A. Replication kinetics and infectivity of SARS-CoV-2 variants of concern in common cell culture models. Virol. J. 2022;19:76. doi: 10.1186/s12985-022-01802-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier M., Patel T.R., Booy E.P., Marushchak O., Okun N., Deo S., Howard R., McEleney K., Harding S.E., Stetefeld J., McKenna S.A. Binding of G-quadruplexes to the N-terminal recognition domain of the RNA helicase associated with AU-rich element (RHAU) J. Biol. Chem. 2013;288:35014–35027. doi: 10.1074/jbc.M113.512970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métifiot M., Amrane S., Litvak S., Andreola M.-L. G-quadruplexes in viruses: function and potential therapeutic applications. Nucleic Acids Res. 2014;42:12352–12366. doi: 10.1093/nar/gku999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.T.A., Dang D.T. RHAU peptides specific for parallel G-quadruplexes: potential applications in chemical biology. Mol. Biotechnol. 2022 doi: 10.1007/s12033-022-00552-7. [DOI] [PubMed] [Google Scholar]
- Phan A.T., Kuryavyi V., Patel D.J. DNA architecture: from G to Z. Curr. Opin. Struct. Biol., Nucleic. Acids/Sequences Topol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Zhao C., Liu Y., Zhang C., Yang G., Yang J., Wang Z., Wang C., Tu C., Guo Z., Ren J., Qu X. RNA G-quadruplex formed in SARS-CoV-2 used for COVID-19 treatment in animal models. Cell Discov. 2022;8:86. doi: 10.1038/s41421-022-00450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan R., Zheludev I.N., Hagey R.J., Pham E.A., Wayment-Steele H.K., Glenn J.S., Das R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: a first look. RNA. 2020;26:937–959. doi: 10.1261/rna.076141.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Lipps H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenti A., Hodcroft E.B., Robertson D.L. The evolution and biology of SARS-CoV-2 variants. Cold Spring Harb. Perspect Med. 2022;12 doi: 10.1101/cshperspect.a041390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandelli A., Monti M., Milanetti E., Armaos A., Rupert J., Zacco E., Bechara E., Delli Ponti R., Tartaglia G.G. Structural analysis of SARS-CoV-2 genome and predictions of the human interactome. Nucleic Acids Res. 2020;48:11270–11283. doi: 10.1093/nar/gkaa864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney D., Spiegel J., Zyner K., Tannahill D., Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020;21:459–474. doi: 10.1038/s41580-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-R., Zhang Q.-Y., Wang J.-Q., Ge X.-Y., Song Y.-Y., Wang Y.-F., Li X.-D., Fu B.-S., Xu G.-H., Shu B., Gong P., Zhang B., Tian T., Zhou X. Chemical targeting of a G-quadruplex RNA in the Ebola virus L gene. Cell Chem. Biol. 2016;23:1113–1122. doi: 10.1016/j.chembiol.2016.07.019. [DOI] [PubMed] [Google Scholar]
- Wang X., Sacramento C.Q., Jockusch S., Chaves O.A., Tao C., Fintelman-Rodrigues N., Chien M., Temerozo J.R., Li X., Kumar S., Xie W., Patel D.J., Meyer C., Garzia A., Tuschl T., Bozza P.T., Russo J.J., Souza T.M.L., Ju J. Combination of antiviral drugs inhibits SARS-CoV-2 polymerase and exonuclease and demonstrates COVID-19 therapeutic potential in viral cell culture. Commun. Biol. 2022;5:154. doi: 10.1038/s42003-022-03101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Coronavirus (COVID-19) Dashboard https://covid19.who.int [WWW Document], n.d. URL. 11.30.22.
- Wu F., Niu K., Cui Y., Li C., Lyu M., Ren Y., Chen Y., Deng H., Huang L., Zheng S., Liu L., Wang J., Song Q., Xiang H., Feng Q. Genome-wide analysis of DNA G-quadruplex motifs across 37 species provides insights into G4 evolution. Commun. Biol. 2021;4:98. doi: 10.1038/s42003-020-01643-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarandi P.K., Zinatizadeh M.R., Zinatizadeh M., Yousefi M.H., Rezaei N. SARS-CoV-2: from the pathogenesis to potential anti-viral treatments. Biomed. Pharmacother. 2021;137 doi: 10.1016/j.biopha.2021.111352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L.-Y., Su A.-M., Liu J.-F., Zhao J.-J., Xi X.-G., Hou X.-M. Recent advances in applying G-quadruplex for SARS-CoV-2 targeting and diagnosis: a review. Int. J. Biol. Macromol. 2022;221:1476–1490. doi: 10.1016/j.ijbiomac.2022.09.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Xiao K., Gu Y., Liu H., Sun X. Whole genome identification of potential G-quadruplexes and analysis of the G-quadruplex binding domain for SARS-CoV-2. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.587829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Qin G., Niu J., Wang Z., Wang C., Ren J., Qu X. Targeting RNA G-quadruplex in SARS-CoV-2: a promising therapeutic target for COVID-19? Angew Chem. Int. Ed. Engl. 2021;60:432–438. doi: 10.1002/anie.202011419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.