Abstract
Mycobacterium leprae, an obligate intracellular pathogen, can be derived only from host tissue and thus affords the opportunity to study in vivo-expressed products responsible for the particular pathogenesis of leprosy. Despite considerable progress in the characterization of the proteins and secondary gene products of M. leprae, there is little information on the nature of the proteins associated with the cell envelope. M. leprae has been fractionated into its major subcellular components, cell wall, cytoplasmic membrane, and soluble cytosol. A number of biochemical markers, including diaminopimelic acid content, monosaccharide composition, mycolic acid, and glycolipid distribution, were applied to their characterization, and two-dimensional gel electrophoresis was used to map the component proteins. A total of 391 major proteins spots were resolved, and 8 proteins were identified based on their reactivity to a panel of monoclonal antibodies and/or relative pI size. Microsequencing of six protein spots present in the cell wall fraction allowed identification of new proteins, including the protein elongation factor EF-Tu and a homolog for the Mycobacterium tuberculosis MtrA response regulator. These results, together with previous studies, contribute to the progressive knowledge of the composition of the in vivo-expressed proteins of M. leprae.
Pathogenic mycobacteria, the causative agents of leprosy and tuberculosis, are intracellular parasites that survive inside macrophages by a poorly defined mechanism(s) (6). To better understand the molecular basis of mycobacterium-host cell interaction, we have during recent years identified several of the major proteins of armadillo-derived Mycobacterium leprae (20, 35, 36, 43). Molecular characterization of the leprosy bacillus is an attractive proposition, since it represents the only available obligate in vivo-grown mycobacterium expressing components necessary for its survival and virulence. The data derived so far indicate that the chaperonins 65-kDa GroEL-2 and 10-kDa GroES, the superoxide dismutase enzyme SodA, and an 18-kDa protein (hsp 18) related to the family of small heat shock proteins are the major proteins present in host-derived M. leprae (20). Additionally, two major membrane proteins, the 35-kDa major membrane protein I (MMP-I), with unknown function (43), and a bacterioferritin (Bfr/MMP-II), probably involved in acquisition and storage of iron, have been characterized (35). More recently, utilizing the emerging M. leprae genome sequence (18), three new expressed proteins were identified based on their N-terminal amino acid sequence: alkyl hydroperoxide (AhpC), an antioxidant enzyme; CysA, a putative sulfate sulfurtransferase; and the 50S ribosomal L7/L12 protein (36).
Despite considerable progress in the characterization of the major cellular components of M. leprae, there is little information on the nature of the proteins associated with the cell envelope. Since the cell envelope constitutes the key interface between pathogen and host, the cell wall-associated proteins are presumably crucial determinants of pathogenesis and immunogenicity. The identification of mycobacterial cell wall proteins, in general, has proved difficult, probably as a consequence of the dominant lipid environment of the cell envelope (5). Nevertheless, Anderson (2) and Ortalo-Magné et al. (32) clearly established that a major group of culture filtrate proteins had their origins in cell walls from which they were gradually released during culture. Also, the physiological roles of two major secreted proteins, the phosphate-binding 38-kDa PhoS homolog in Mycobacterium tuberculosis (7) and the mycolyltransferase antigen 85 complex (3), are in accord with their demonstrated presence in cell walls (16, 42). Additionally, a pore-forming 59-kDa protein has been identified in Mycobacterium chelonae (41). Other proteins claimed to be associated with the cell wall of mycobacteria are the 10-kDa GroES and 70-kDa DnaK homologs, SodA, alanine dehydrogenase (2), and the chaperonin 65-kDa GroEL-2 homolog (14, 20).
In this study, we developed two-dimensional (2D) maps of the major proteins of the different subcellular compartments of host-derived M. leprae, with special emphasis on proteins associated with the cell wall. Immunoblotting and amino acid sequence allowed recognition of the 65-kDa GroEL-2 protein, elongation factor Tu (EF-Tu), and a homolog of the M. tuberculosis MtrA response regulator protein as cell wall-associated proteins.
MATERIALS AND METHODS
Fractionation of M. leprae.
M. leprae was purified from irradiated and nonirradiated armadillo spleens and livers by the Draper protocol (9). The bacteria (300 mg [dry weight]) were pelleted by centrifugation at 2,000 × g for 10 min and resuspended in phosphate-buffered saline (PBS) containing 1 mM phenylmethylsulfonyl fluoride. The cells were disrupted by intermittent probe sonication (Ultrasonic Homogenizer 4710; Cole and Parmer Instruments, Chicago, Ill.) for 30 min (90-s bursts/90 s of cooling). The unbroken cells were removed by three low-speed centrifugation steps (325, 1,310, and 2,940 × g) for 5 min each time. The supernatant was digested with 10 μg of DNase and RNase per ml for 1 h at 4°C. Cell walls were obtained from the particulate material provided by centrifugation at 5,000 × g for 40 min (30). The supernatant was centrifuged at 27,000 × g for 30 min, and the supernatant from this step was recentrifuged at 100,000 × g for 2 h to provide the membrane fraction and the soluble cytosolic fraction. The crude cell wall fraction was washed three times with PBS by centrifugation at 27,000 × g for 30 min; the final pellet was resuspended in PBS and layered onto a sucrose gradient consisting of steps of 15, 30, 40, 50, and 70% (wt/vol) sucrose, which was centrifuged at 100,000 × g for 2 h, after which gradients were collected and monitored for absorption at 280 nm. Fractions corresponding to absorption peaks were pooled, diluted with water, centrifuged at 27,000 × g for 30 min, and washed repeatedly to remove sucrose. Protein concentrations of all subcellular fractions were measured by the bicinchoninic acid assay reagent (Pierce, Rockford, Ill.), and the amount of the total carbohydrate was estimated (10).
Derivation of subfractions for diaminopimelic acid (DAP) analysis by GC (25).
Each cellular subfraction (100 μg [dry weight]) was hydrolyzed in 6 N HCl at 110°C for 18 h, the acid was removed by evaporation, and the samples were treated with 100 μl of a mixture of 64 μl of acetylchloride and 300 μl of 2 methyl-1-propanol at 120°C for 20 min. The 2-butyl esters were formed by heating samples at 150°C for 5 min in 100 μl of ethyl acetate and 40 μl of heptofluorobutyric anhydride. The final product was evaporated to dryness with N2 at room temperature, dissolved in ethyl acetate and analyzed by gas chromatography (GC) on a Durabond 1 (DB-1) capillary column (30 m by 0.32 mm; 0.25-mm diameter) (25). The GC temperature program was at an initial 100°C for 2 min, followed by increments of 8°C/min to 250°C. The internal standard, α-amino adipic acid (2.5 μg), was added to each sample.
GC analysis of monosaccharides.
The relative quantity of monosaccharides present in each subcellular fraction was determined by GC analysis of alditol acetates (1). Samples were methanolyzed with 0.5 M HCl in methanol at 80°C for 18 h. Fatty acid methyl esters were removed by hexane extraction and the methanol phase was neutralized with silver carbonate and re-N-acetylated with acetic anhydride for 18 h in the dark. The trimethylsilyl derivatives were obtained by reaction of samples with a mixture of bis-trimethylsilyltrifluoroacetamide and pyridine (1:1), and the products were analyzed by GC on the DB-1-fused silica column with hydrogen as the carrier gas. The column temperature was programmed from 120 to 240°C at 2°C/min. Peaks were identified through comparison with standards and quantified as described elsewhere (1).
Biphasic extraction and analysis of lipids (15, 28).
Lipids were extracted from dry whole M. leprae (50 mg) and from the different subcellular fractions by extraction with a mixture of methanol–0.3% aqueous sodium chloride (10:1) and petroleum ether (2:1) and centrifuged. The upper petroleum ether layer containing the apolar lipids was evaporated under N2, resuspended in dichloromethane, and analyzed by 2D thin-layer chromatography (TLC) as described elsewhere (28). The methanolic-saline extract (lower layer) was heated in a boiling water bath for 5 min, followed by the addition of an equal volume of chloroform–methanol–0.3% aqueous sodium chloride (9:10:3) and further mixing. Chloroform–0.3% aqueous sodium chloride (1:1) was added, the biphasic mixture was centrifuged, the upper layer was discarded, and the lower layer was evaporated to dryness, yielding the polar lipids. These were dissolved in chloroform-methanol (2:1) and analyzed by 2D TLC as described in the legend to Fig. 2. Lipids were detected by charring with α-naphthol or 5% ethanolic molybdophosphoric acid. The delipidated whole bacteria or subcellular fractions were analyzed for bound mycolic acids (15, 28). Mycolic acid methyl esters were prepared by phase transfer catalysis by heating at 100°C overnight with 5% tetrabutylammonium hydroxide. The reaction mixture was diluted with water, dichloromethane, iodomethane (2:1:0.25), stirred for 30 min, and centrifuged. The upper layer was discarded, and the lower organic phase was washed with 1 M hydrochloric acid, followed by water and dried under a stream of nitrogen. The methyl esters were dissolved in dichloromethane and analyzed by TLC (15).
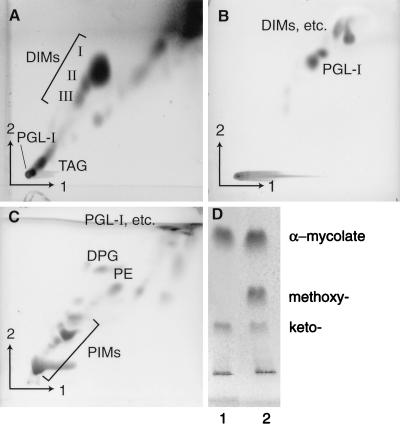
FIG. 2.
One-dimensional and 2D TLC analysis of whole M. leprae lipids. Plates were developed in various solvents. (A) First direction, petroleum ether-ethyl acetate (98:2, three times); second direction, petroleum ether-acetone (98:2); (B) first direction, chloroform-methanol-water (100:14:0.8); second direction, chloroform-methanol-acetone-water (50:60:2.5:3); (C) first direction, chloroform-methanol-water (60:30:6); second direction, chloroform-acetic acid-methanol-water (40:25:3:6); (D) petroleum ether-diethyl ether (95:5, six times). Lanes: 1, M. leprae whole-cell extract; 2, α-, methoxy-, and keto-mycolate were observed in whole-cell extracts of M. tuberculosis H37Ra, which was used as a control. The plates were charred with 5% ethanolic molybdophosphoric acid (A and D) and α-naphthol (B and C). I to III, members of DIM family; TAG, triacylglycerols.
Electrophoresis and Western blot analysis of fractions.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of individual fractions was performed under reducing conditions (23), and the proteins were visualized by silver staining (29). A periodic acid oxidation step was included to enhance the appearance of lipoarabinomannan (LAM), lipomannan (LM), phosphatidylinositol mannosides (PIMs), and the phenolic glycolipid-I (PGL-I). Fractions were analyzed by 2D electrophoresis (31) by using the Might Small II system (Pharmacia Biotech, Uppsala, Sweden). Before running, the samples (approximately 80 μg of protein) were dialyzed against water, dried, resuspended in 20 μl of isoelectric focusing sample buffer (9 M urea, 2% Nonidet P-40, 5% β-mercaptoethanol, 5% ampholytes, pH 3 to 10; Sigma Chemical Company, St. Louis, Mo.), and incubated for 3 h at room temperature. To improve protein solubilization, the samples were sonicated in a bath sonicator for 30 s at each hour of incubation. The insoluble components were removed by centrifugation at 16,000 × g for 15 min. The supernatants were applied over the upper electrode buffer covering the top of a 6% polyacrylamide isoelectric focusing tube gel (1.5-mm inside diameter by 7.5 cm) containing 1.6% ampholytes in the pH range of 4.5 to 6.0 and 0.4% ampholytes in the pH range of 3 to 10 (Sigma). The proteins were focused for 3 h at 1 kV with 10 mM H3PO4 as lower electrode buffer (anolyte) and 20 mM NaOH as upper electrode buffer (catholyte). The tube gels were subsequently treated with sample transfer buffer (2.9% SDS, 71 mM Tris-HCl [pH 6.8], 0.003% bromophenol blue) for 30 min and placed on a preparative SDS-PAGE containing 6% stacking gel and 15 or 17.5% separating gels (1.5 mm by 7 cm by 8 cm). Quantitative analysis of 2D gel protein spots was carried out with the Phoretix 2D Gel Analysis software (Non Linear Dynamics, Newcastle Upon Tyne, England). Immunoblottings were performed as previously described (20) with monoclonal antibodies produced in this laboratory or obtained through the World Health Organization-Tropical Disease Research-Immunology of Leprosy Program (11, 22). In blottings carried out for the detection of PGL-I, Tween 20 was omitted from all solutions.
RESULTS
M. leprae crude cell wall can be fractionated into fragments of different densities in a step gradient of sucrose.
To obtain cell wall as free as possible of cytoplasmic membrane, the initial centrifugation of the sonic extract was conducted at 5,000 × g for 40 min, and further purification was accomplished by equilibrium density centrifugation in a step gradient of sucrose. Most of the material was concentrated as a white band at the interface of 30 to 40% sucrose (data not shown). Minor brownish bands were observed at the interface of 40 to 50% and 50 to 70% (data not shown), sucrose and a faint white band was detected at the 15 to 30% interface (data not shown). In some experiments, a pellet was also observed. The membrane fraction yielded a single brownish band at the 15 to 30% interface when loaded onto parallel sucrose gradients.
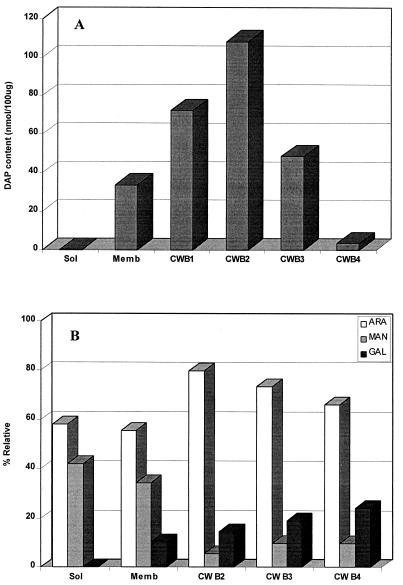
To examine the composition of the bands of different densities arising from the crude cell wall, the relative amounts of carbohydrate and protein in each were determined. Band 1 was composed of less than 20% proteins and carbohydrates, the remaining 80% obviously being lipid (5). The protein and carbohydrate contents of bands 2 and 3 were each about 40 and 60%, respectively, in agreement with their higher densities. To identify the true cell wall fraction and to examine the degree of possible cross-contamination between the various subcellular fractions, the amount of peptidoglycan, a constitutive marker of cell wall, was accessed by assaying for DAP by GC. DAP contents related to 100 μg of each of bands 1, 2, 3, and 4 were 72.3, 108.0, 48.5, and 3.45 nmol (Fig. 1A). The purified membrane fractions contained 33.7 nmol of DAP, probably due to the presence of appreciable quantities of lipid-linked peptidoglycan precursors. The soluble fraction was devoid of detectable DAP.
FIG. 1.
Analysis of M. leprae subcellular fractions for cell wall components. (A) The relative contents of DAP were measured by GC. DAP contents were related to 100 μg of each subcellular fraction. (B) The amounts of arabinose (ARA), mannose (MAN), and galactose (GAL) were estimated by GC analysis. The abundance of each monosaccharide is expressed as a percentage of combined arabinose, mannose, and galactose. Sol, soluble-cytosolic fraction; Memb, cytoplasmic membrane; CWB1, CWB2, CWB3, and CWB4, cell wall bands 1, 2, 3, and 4, respectively.
Additionally, the distribution of arabinogalactan, also exclusive to particulate cell wall, was assessed in the various particulate bands through determination of the relative molar ratios of galactose and arabinose and compared to mannose, which is reflective of PIMs, LAM, LM, GDP-mannose, polyprenyl-P-linked mannosides, and mannans, which are found primarily in membranes and cytoplasm (Fig. 1B). As expected, galactose, which as the galactofuranose of galactan is exclusive to cell walls, was predominant in the cell wall bands. However, galactose was also detected in membrane, which we attribute to the sizable quantities of lipid-linked arabinogalactan, a precursor of the insoluble form (27). Conversely, as expected, mannose was found to be preferentially located in membrane and cytosol. Similar considerations apply to arabinose; that associated with the cell wall bands arises from the insoluble arabinan bound to the galactan and peptidoglycan (5), that detected in the membrane originates mostly from LAM, and that associated with the soluble cytoplasm is probably attributable to arabinans, arabinose phosphates, and other metabolic intermediates.
To complement these analyses and conclusions, the distribution of LAM, LM, and the PIMs was investigated. These lipoglycans-glycolipids are abundant end products and can readily be detected by SDS-PAGE analysis, in which they run as diffuse bands with average molecular masses of 30, 20, and 8 kDa, respectively. LAM and LM were readily observed in membranes, cytosol, and cell wall bands 1 and 2, but not in bands 3 and 4. However, the PIMs were present in membranes and all cell wall bands. Thus, the high content of mannose over galactose in the soluble and membrane fractions (Fig. 1B) can be explained by the abundance of LAM, LM, and PIMs over arabinogalactan in these fractions.
The distribution of a panel of different complex lipids, abundant in the mycobacterial cell envelope (5), was assessed by TLC. Figure 2 exemplifies the lipids identified in whole M. leprae cells by using four TLC solvent systems. The major phospholipids of mycobacteria, phosphatidylethanolamine (PE) and bis-phosphatidylglycerol (DPG), were found in all cell wall fractions and cell membranes, supporting the contention that mycobacterial cell walls have their own complement of phospholipids apart from those found in cell membranes (34, 38). PGL-I and dimycocerosylphthiocerol (DIM), considered part of a copious capsule or lipid droplets surrounding the bacilli in foci of infection (13, 19), were found within all four cell wall bands. However, the distribution of mycolic acids was special. The largest proportion of these are esterified to the distal regions of arabinan of the arabinogalactan-peptidoglycan complex. These were found in cell wall bands 1 and 2 and membranes but not in bands 3 and 4. The nature of the membrane-associated mycolic acids of Mycobacterium spp. has been described, namely, the 6-O-mycolylmannosylphosphorylheptaprenol (4). Taken together, these results demonstrate that cell wall band 2 was enriched in mycolyl-arabinogalactan-peptidoglycan, i.e., the cell wall skeleton, and was probably the innermost of the multitiered cell wall (5).
Cell wall fragments with different densities show similar 2D gel electrophoresis protein profiles.
Further characterization of cell wall fractions was performed by submitting these preparations to 2D gel electrophoresis. Dry samples were resuspended in sample buffer and incubated for 3 h at room temperature, and proteins and lipids were further solubilized by rapid sonication. The proteins were first resolved by isoelectrophoresis in the first dimension and by SDS-PAGE in the second dimension. Silver staining of the proteins of bands 1, 2, 3, and 4 showed similar protein profiles, with a typical cluster of major proteins concentrated at the range of 35 to 65 kDa (results not shown). Clearly, an increasing number of spots parallel the increasing densities of the bands. These profiles were, however, quite different from those seen in the soluble and membrane fractions (Fig. 3). A surprise in this 2D gel electrophoresis analysis was the detection of PGL-I, readily identified through comparison with the 2D gel electrophoresis profile of the purified lipid. The presence of PGL-I was confirmed by immunoblotting with monoclonal antibody CS48, which was specific to PGL-I made in the laboratory. PGL-I was also abundant in bands 1 and 2, forming a ladder at the basic side of the gel, probably as a reflection of the microheterogeneity observed in both the lipid and the carbohydrate entities (13).
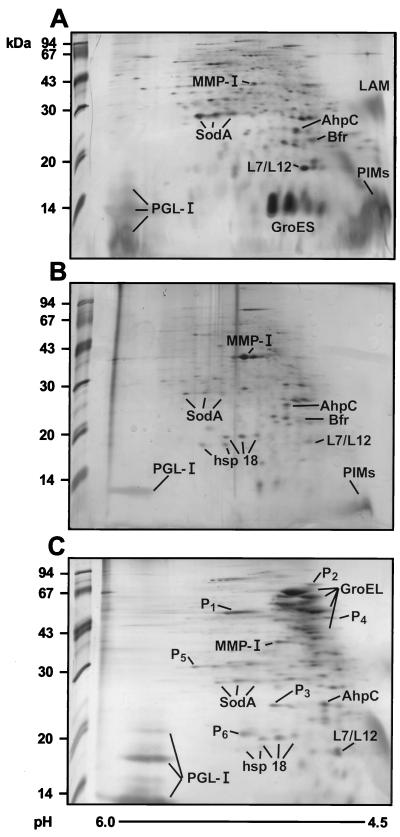
FIG. 3.
Subcellular distribution of M. leprae proteins. The cytosol-soluble (A), membrane (B), and cell wall band 2 fractions (C) were fractionated by isoelectric focusing in the first dimension (pH 4.5 to 6.0 from right to left) and by SDS-PAGE in the second dimension. The proteins were visualized by silver staining. The alkyl hydroperoxide reductase (AhpC), bacterioferritin (Bfr), and ribosomal L7/L12 proteins were identified by N-terminal sequence analysis (43). The remaining known proteins (MMP-I, SodA, the chaperonins GroES and GroEL, and the 18-kDa hsp 18) were identified by Western blotting with specific monoclonal antibodies. The spot proteins, indicated as P1 to P6, were microsequenced. The new proteins are described in Table 1.
Chaperonin 60-2 (65-kDa GroEL-2 antigen) is the major protein associated with the M. leprae cell wall.
The membrane, soluble fraction, and cell wall band 2 proteins were separated by 2D gel electrophoresis (Fig. 3), and maps were generated with the Phoretix 2D gel analysis software (Nonlinear Dynamics). Fractions originating from nonirradiated bacilli were chosen for analysis, since additional spots, likely resulting from protein degradation, were detected in fractions from irradiated M. leprae. A total of 391 major silver-stained spots were detected in the three fractions, with each fraction showing a unique and consistent profile. Proteins were identified either by their relative size and pI and reactivity with a panel of specific antibodies or by microsequencing. Chaperonin 10 (GroES), the most abundant protein of M. leprae (20), was almost exclusively detected in the soluble fraction as three spots of close pIs representing about 30% of total protein in this fraction (Fig. 3A). The 2D map of the membrane also revealed a unique profile of proteins, with enrichment in the MMPI protein, which represented 11% of the total protein in this fraction (Fig. 3B). As mentioned before, the unique protein profile of the cell wall was dominated by the chaperonin 65-kDa GroEL-2 protein (Fig. 3C). A total of 98 spots were observed in this fraction, approximately 16 of them reacting with monoclonal antibodies specific to GroEL, and totaled 38% of the protein content of this fraction. However, most proteins could clearly be detected in more than one subcellular fraction, such as hsp 18, which was seen in the membrane and the cell wall; Bfr, which was present in the soluble and membrane fractions; and SodA, AhpC, L7/L12, and MMPI, which were detected in all fractions. The 85 complex was detected by immunoblotting mainly in cell wall band 2. However, faint traces of these proteins were also observed in the membrane and soluble fractions. None of these major M. leprae proteins were observed in the course of 2D gel analysis of uninfected armadillo tissues, with the exception of a 65-kDa protein, possibly the mitochondrial GroEL homolog, which was observed when the gels were screened with a pool of sera from lepromatous leprosy patients.
Identification of new proteins in the M. leprae cell wall.
By loading small quantities of the protein of cell wall band 2 and pooling material from several Coomassie blue-stained polyvinylidene difluoride membranes, enough selected spots were obtained for limited N-terminal sequencing (26, 40) (Fig. 3; Table 1). However, since spot 1 was apparently N-terminally blocked, this protein, instead, was digested in gel (17) and analyzed by liquid chromatography-tandem mass spectrometry to derive the sequence. No significant homologies were found in GenBank and Swissprot sequence data banks when the sequences of spots 2 (NH2-KQKRTEPLXTXN) and 4 (NH2-TLIXQRPMLXE) were used as probes. On the other hand, identification was successful for protein spots 1, 5, and 6 (Fig. 3; Table 1). Protein 1, abundantly present in cell wall band 2, proved to be EF-Tu. Due to discrepancies in the sequence of the M. leprae tuf gene as reported in the literature (39), an extensive analysis of the sequence of the EF-Tu protein was performed. The sequence of the discrepant region of the protein (Table 1) showed 100% homology to that reported previously (39). Spots 5 and 6, minor proteins, were also seen in the other bacterial compartments. Their sequences correspond, respectively, to the M. tuberculosis MtrA response regulator homolog (GenBank accession no. U01971) and to the product of M. tuberculosis gene MTCY270.28, whose function is unknown (cosmid Y270; EMBL accession no. Z95388). Spot 3 is clearly a C-terminal fragment of the native GroEL-2, with an expected molecular mass of 20.4 kDa (Table 1). Incidentally, PI, a protein sequenced in our previous work (36), is included in Table 1, since we now observe that it shows significant homology to M. tuberculosis malate synthase, recently sequenced as part of the genome project (cosmid SCY01A11; EMBL accession no. Z78020).
TABLE 1.
Amino acid sequences of cell wall-associated proteins of M. leprae
| Protein | Molecular mass (kDa) | Sequence and homology | Accession no. |
|---|---|---|---|
| 127 | |||
| M. leprae P1 | 55a | QVGVPYILVALNKSDAVDDEELLELVEMEVR | |
| ||||||||||||||||||||||||||||||| | |||
| M. leprae EF-Tu | 43.7 | QVGVPYILVALNKSDAVDDEELLELVEMEVR | GenBank L13276 |
| 127 | |||
| M. leprae P3 | 25a | NH2-VAQIRTEIENXDXDYDRE | |
| |||||||||| | ||||| | |||
| M. leprae GroEL-2 | 20.4 | VAQIRTEIENSDSDYDRE | GenBank M14341 |
| 343 | |||
| M. leprae P5 | 30a | NH2-MRQRILVVDDD | |
| ||||||||||| | |||
| M. tuberculosis MtrA | 30 | NH2-MRQRILVVDDD | GenBank U01971 |
| M. leprae P6 | 19a | NH2-KLPPDPYAALPKLPP | |
| |||||||||||| | |||
| M. tuberculosis MTCY270.28 gene product | 19 | NH2-TTSPDPYAALPKLPP | EMBL Z95388 |
| M. leprae PI | 80.4a | NH2-TDXVSAGNLGVARVLY | |
| || || ||| :||||| | |||
| M. tuberculosis malate synthase | 85 | NH2-TDRVSVGNLRIARVLY | EMBL Z78020 |
Estimated by SDS-PAGE.
DISCUSSION
M. leprae is an obligate intracellular parasite that, upon ingestion by cells of the monocyte-macrophage series, resists destruction and, in fact, proliferates within these cells. During the past years, we have investigated the major proteins present in armadillo-derived M. leprae (20, 35, 36, 43). This approach has been undertaken based on the knowledge that proteins expressed by the pathogen in vivo would reflect its life style as well as the host cell environment, thereby allowing a better understanding of the molecular basis of the mycobacterium-host cell relationship. In this study, a subcellular fractionation protocol including sonication, differential centrifugation, and fractionation in a sucrose gradient was applied allowing for the first time the definition of 2D protein profiles of the cell wall, membrane, and soluble-cytosolic fractions of M. leprae. A total of 391 major protein spots were resolved in the three fractions, and 14 proteins were identified based on their relative sizes and pIs, reactivity to a panel of monoclonal antibodies, or microsequencing. An emphasis on the cell wall compartment allowed the identification of new proteins, including the EF-Tu and a homolog for the M. tuberculosis MtrA response regulator.
The level of cross-contamination between the cellular compartments seemed tolerable, as judged by the distribution of the major proteins. As previously indicated (20), the 10-kDa GroES, 35-kDa MMP-I, and 65-kDa GroEL-2 proteins were detected preferentially in the soluble, membrane, and cell wall fractions, respectively, and proved to be reliable protein-markers of these compartments. Additionally, analysis of DAP, monosaccharides, and mycolic acids proved useful as quality control markers of subcellular fractions. For instance, the failure to detect DAP, mycolic acids, and galactose within the soluble contents of the cell indicated an absence of cell wall fragments. On the other hand, the detection of DAP and mycolic acids in the membrane could imply cross-contamination with the cell wall material, although membrane-associated precursors of cell wall containing those compounds are well known (4, 27). Conventional membrane phospholipids such as PE and DPG were found in the cell wall. These may be attributable to contamination with membranes or regarded as genuine cell wall constituents as argued by others (34, 38).
Fractionation of M. leprae crude cell wall in a sucrose gradient led to the resolution of four different fractions with similar protein profiles, namely, cell walls bands 1, 2, 3, and 4, a pattern of bands consistently obtained in five independent experiments. This characteristic feature, not shared by Mycobacterium chelonae (30) or M. tuberculosis (results not shown), probably reflects differences between mycobacterial species in their cell envelope composition and organization. Band 1 was rich in lipids, and the classical lipids that constitute M. leprae capsule, PGL-I, and diacylphthiocerol, were very much in evidence. PIMs, LM, and glycans, which can be interpreted as LAM or as the arabinomannans recently described on the surface of M. tuberculosis (33), were also found in this fraction. Thus, band 1 may constitute remnants of the capsular material adhering to bacilli even after its purification. The fact that the protein profile of this band was shared by the other cell wall bands discounts the possibility of heavy contamination with membranes. Band 2 was highly enriched in DAP, mycolic acids, and galactose, indicating that it represents the cell wall skeleton, i.e., the mycolyl-arabinogalactan-peptidoglycan complex. The high-density cell wall bands 3 and 4 contain very low levels of DAP and undetectable amounts of mycolic acid and thus may represent particles originating from protein-enriched domains present in the cell wall. Alternately, the more complex protein profiles seen in these bands may be a reflection of protein denaturation-precipitation, a not unexpected occurrence in view of the low viability of armadillo-derived M. leprae.
2D gel electrophoresis of the M. leprae cell wall fraction revealed the presence of a diverse group of proteins. Most of these proteins can be extracted by SDS or Triton X-114, suggesting retention in this compartment by hydrophobic interactions. The 65-kDa GroEL-2 protein and a group of smaller products originating from this protein constitute the most abundant proteins of the cell wall. Reinforcing our present findings, immunogold electron microscopy of sections of M. leprae present in armadillo liver macrophages demonstrated the distribution of the 65-kDa GroEL-2 protein in both the envelope and cytosol (12). Besides the 65-kDa GroEL-2 protein, hsp 18, a member of a family of small heat shock proteins, was also abundant in the cell envelope and was absent from the soluble fraction. Another major spot observed was the antioxidant enzyme AhpC, which was equally distributed among the subcellular fractions. Also, a surprising observation was the preferential distribution of EF-Tu in the cell wall, as well as the detection of the ribosomal L7/L12 protein, the MtrA homolog, and malate synthase in this compartment. The presence of all of these conventional cytosolic proteins in the purified M. leprae cell wall may suggest aggregation of such proteins with cell wall components that may happen in dead or decaying bacteria or during the fractionation steps.
With respect to EF-Tu, it is a major protein in Escherichia coli which is produced in excess over the amounts seemingly required for protein synthesis (21). It plays a major role in the elongation phase of protein biosynthesis by positioning the aminoacyl tRNA onto the active site of ribosomes and belongs to the large family of G proteins. Since it is essential for growth and is susceptible to several antibiotics (24), EF-Tu may serve as a target for new drug development. Besides its cytosolic location, EF-Tu has also been found to be associated with membrane and is a dominant component of periplasmic extracts (21, 37), although its function in these locations is not clear.
Finally, the identification of the M. leprae MtrA homolog, the response regulator of a bacterial two-component signal transduction system, constitutes an exciting finding, supporting its role in mycobacterial adaptation to intracellular parasitism (8). The M. leprae P6 shows significant homology to the M. tuberculosis MTCY270.28 gene. However, we know little about the significance of this gene. Its peculiar location in the mycobacterial genome added to the similarity to a hypothetical 17.1-kDa E. coli protein, YbhB, suggests an involvement in carboxylation steps in cell wall biosynthesis.
ACKNOWLEDGMENTS
This work was supported by contract NO1 AI 55262 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and by funds from the Heiser Program for Research in Leprosy (New York Community Trust). M. A. M. Marques was a fellow of the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brasilia, Brazil.
We thank Marilyn Hein for preparation of the manuscript and Carol Marander for the graphics. We thank Delphi Chatterjee, José Oswaldo Previato, Gurdyal S. Besra, and Fauzi Silbaq for constructive scientific discussions and physical help during the course of this work.
REFERENCES
- 1.Albersheim P, Nevins D J, English P D, Karr A. A method for the analysis of sugars in plant cell-wall polysaccharides by gas-chromatography. Carbohydr Res. 1967;5:340–345. [Google Scholar]
- 2.Anderson P. The T cell response to secreted antigens of Mycobacterium tuberculosis. Immunobiology. 1994;191:537–547. doi: 10.1016/S0171-2985(11)80460-2. [DOI] [PubMed] [Google Scholar]
- 3.Belisle J T, Vissa V D, Sievert T, Takayama K, Brennan P J, Besra G S. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science. 1997;276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 4.Besra G S, Sievert T, Lee R E, Slayden R A, Brennan P J, Takayama K. Identification of the apparent transmembrane carrier in mycolic acid synthesis. Proc Natl Acad Sci USA. 1994;91:12735–12739. doi: 10.1073/pnas.91.26.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan P J, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 6.Britton W J, Roche P W, Winter N. Mechanisms of persistence of mycobacteria. Trends Microbiol. 1994;2:284–288. doi: 10.1016/0966-842x(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 7.Chang Z, Choudhary A, Lathigra R, Quiocho F A. The immunodominant 38-kDa lipoprotein antigen of Mycobacterium tuberculosis is a phosphate-binding protein. J Biol Chem. 1994;269:1956–1958. [PubMed] [Google Scholar]
- 8.Curcic L, Via E, Mudd M H, Bhandayuthapani S, Ulmer R J, Deretic V. Elements of signal transduction in Mycobacterium tuberculosis: in vitro phosphorylation and in vivo expression of the response regulator MtrA. J Bacteriol. 1996;178:3314–3321. doi: 10.1128/jb.178.11.3314-3321.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Draper P. Purification of M. leprae. Annex 4 of Report of the Fifth Meeting of the Scientific Working Group on the Immunology of Leprosy (IMMLEP), Geneva, 24 to 26 June 1980. World Health Organization document TDR/IMMLEP-SWG (5)/80.3. Geneva, Switzerland: World Health Organization; 1980. [Google Scholar]
- 10.Dubois M, Gilles K A, Hamilton J K, Rebers P A, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 11.Engers H D, Houba V, Bennedsen J, Buchanan T M, Chaparas S D, Kadival G, Closs O, David J R, Van Embden J D A, Godal T, Mustafa S A, Ivanyi J, Young D B, Kaufmann S H E, Khomenko A G, Kolk A H J, Kubin M, Louis J A, Minden P, Shinnick T M, Trnka L, Young R A. Results of a World Health Organization-sponsored workshop to characterize antigens recognized by Mycobacterium-specific monoclonal antibodies. Infect Immun. 1986;51:718–720. doi: 10.1128/iai.51.2.718-720.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esaguy N, Aguas A P. Subcellular localization of the 65-kDa heat shock protein in mycobacteria by immunoblotting and immunogold ultracytochemistry. Submicrosc Cytol Pathol. 1997;29:85–90. [PubMed] [Google Scholar]
- 13.Gaylord H, Brennan P J. Leprosy and the leprosy bacillus: recent developments in characterization of antigens and immunology of the disease. Annu Rev Microbiol. 1987;41:645–675. doi: 10.1146/annurev.mi.41.100187.003241. [DOI] [PubMed] [Google Scholar]
- 14.Gillis T P, Miller R A, Young D B, Khanolkar S R, Buchanan T M. Immunochemical characterization of protein associated with Mycobacterium leprae cell wall. Infect Immun. 1985;49:371–377. doi: 10.1128/iai.49.2.371-377.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamid M E, Minnikin D E, Goodfellow M. A simple test to distinguish mycobacteria from other mycolic-acid-containing actinomycetes. J Gen Microbiol. 1993;139:2203–2213. doi: 10.1099/00221287-139-9-2203. [DOI] [PubMed] [Google Scholar]
- 16.Harboe M, Wiker H G. The 38-kDa protein of Mycobacterium tuberculosis: A review. J Infect Dis. 1992;166:874–884. doi: 10.1093/infdis/166.4.874. [DOI] [PubMed] [Google Scholar]
- 17.Hellman U, Wernstedt C, Gonez J, Heldin C-H. Improvement of an in-gel digestion procedure for micropreparation of internal protein fragments for amino acid sequencing. Anal Biochem. 1995;224:451–455. doi: 10.1006/abio.1995.1070. [DOI] [PubMed] [Google Scholar]
- 18.Honoré N, Bergh S, Chanteau S, Doucet-Populaire F, Eiglmeier K, Garnier T, Georges C, Launois P, Limpaiboon T, Newton S, Niang K, del Portillo K P, Ramesh G R, Reddi P, Ridel P R, Sittisombut N, Hunter S W, Cole S T. Nucleotide sequence of the first cosmid from the Mycobacterium leprae genome project: structure and function of the Rif-Str regions. Mol Microbiol. 1990;7:207–214. doi: 10.1111/j.1365-2958.1993.tb01112.x. [DOI] [PubMed] [Google Scholar]
- 19.Hunter S W, Brennan P J. A novel phenolic glycolipid from Mycobacterium leprae possibly involved in immunogenicity and pathogenicity. J Bacteriol. 1981;147:728–735. doi: 10.1128/jb.147.3.728-735.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter S W, Rivoire B, Mehra V, Bloom B R, Brennan P J. The major native proteins of the leprosy bacillus. J Biol Chem. 1990;257:14065–14068. [PubMed] [Google Scholar]
- 21.Jacobson G R, Rosenbusch J P. Abundance and membrane association of elongation factor Tu in E. coli. Nature. 1976;261:23–26. doi: 10.1038/261023a0. [DOI] [PubMed] [Google Scholar]
- 22.Khanolkar-Young S, Kolk A H J, Andersen A B, Bennedsen J, Brennan P J, Rivoire B, Kuijper S, McAdam K P W J, Abe C, Batra H V, Chaparas S D, Damiani G, Singh M, Engers H D. Results of the third immunology of leprosy/immunology of tuberculosis antimycobacterial monoclonal antibody workshop. Infect Immun. 1992;60:3925–3927. doi: 10.1128/iai.60.9.3925-3927.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli V K. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Landini P, Bandera M, Goldstein B P, Ripamonti F, Saffientini A, Islam K, Denaro M. Inhibition of bacterial protein synthesis by elongation-factor-Tu-binding antibiotics MDL 62,879 and efrotomycin. Biochem J. 1992;283:649–652. doi: 10.1042/bj2830649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacKenzie S L, Tenaschuk D. Rapid formation of amino acid isobutyl esters for gas chromatography. J Chromatogr. 1975;111:413–415. doi: 10.1016/s0021-9673(00)99292-6. [DOI] [PubMed] [Google Scholar]
- 26.Matsudaira P. A practical guide to protein and peptide purification for microsequencing. San Diego, Calif: Academic Press, Inc.; 1989. [Google Scholar]
- 27.Mikusová K, Mikus M, Besra G S, Hancock I, Brennan P J. Biosynthesis of the linkage region of the mycobacterial cell wall. J Biol Chem. 1996;271:7820–7828. doi: 10.1074/jbc.271.13.7820. [DOI] [PubMed] [Google Scholar]
- 28.Minnikin D E, O’Donnell A G, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett J H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods. 1984;2:233–241. [Google Scholar]
- 29.Morrissey J H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- 30.Nikaido H, Kim S-H, Rosenberg E Y. Physical organization of lipids in the cell wall of Mycobacterium chelonae. Mol Microbiol. 1993;8:1025–1030. doi: 10.1111/j.1365-2958.1993.tb01647.x. [DOI] [PubMed] [Google Scholar]
- 31.O’Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 32.Ortalo-Magné A, Dupont M A, Lemassu A, Andersen A B, Gounon P, Daffé M. Molecular definition of the outermost capsular material of the tubercle bacillus. Microbiology. 1995;141:1609–1620. doi: 10.1099/13500872-141-7-1609. [DOI] [PubMed] [Google Scholar]
- 33.Ortalo-Magné A, Andersen A B, Daffé M. The outermost capsular arabinomannans and other mannoconjugates of virulent and avirulent tubercle bacillus. Microbiology. 1996;142:927–935. doi: 10.1099/00221287-142-4-927. [DOI] [PubMed] [Google Scholar]
- 34.Ortalo-Magné A, Lemassu A, Laneelle M A, Bardou F, Silve G, Gounon P, Marchal G, Daffé M. Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J Bacteriol. 1996;178:456–461. doi: 10.1128/jb.178.2.456-461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pessolani M C V, Smith D R, Rivoire B, McCormick J, Hefta S A, Cole S T, Brennan P J. Purification, characterization, gene sequence, and significance of bacterioferritin from Mycobacterium leprae. J Exp Med. 1994;180:319–327. doi: 10.1084/jem.180.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pessolani M C V, Brennan P J. Further molecular definition of the leprosy bacillus: identification of new proteins. Infect Immun. 1996;64:5425–5427. doi: 10.1128/iai.64.12.5425-5427.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porcella S F, Belland R J, Judd R C. Identification of an EF-Tu protein that is periplasm-associated and processed in Neisseria gonorrhoeae. Microbiology. 1987;142:2481–2489. doi: 10.1099/00221287-142-9-2481. [DOI] [PubMed] [Google Scholar]
- 38.Rastogi N. Recent observations concerning structure and function relationships in the mycobacterial cell envelope: elaboration of a model in terms of mycobacterial pathogenicity, virulence and drug-resistance. Res Microbiol. 1991;142:464–476. doi: 10.1016/0923-2508(91)90121-p. [DOI] [PubMed] [Google Scholar]
- 39.Silbaq F, Bercovier H. Nucleotide sequence of Mycobacterium leprae elongation factor (EF-Tu) gene. Nucleic Acids Res. 1993;21:3327. doi: 10.1093/nar/21.14.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stahl D C, Swiderek K M, Davis M T, Lee T D. Data-controlled automation of liquid chromatography/tandem mass spectrometry analysis of peptide mixtures. J Am Soc Mass Spectrom. 1996;7:532–540. doi: 10.1016/1044-0305(96)00057-8. [DOI] [PubMed] [Google Scholar]
- 41.Trias J, Benz R. Characterization of channel formed by the mycobacterial porin in lipid bilayer membranes. J Biol Chem. 1993;268:6234–6240. [PubMed] [Google Scholar]
- 42.Wiker H G, Harboe M. The antigen 85 complex: a major secretion product of Mycobacterium tuberculosis. Microbiol Rev. 1992;56:648–661. doi: 10.1128/mr.56.4.648-661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winter N, Triccas J A, Rivoire B, Pessolani M C V, Eiglmeier K, Hunter S W, Brennan P J, Britton W J. Isolation and characterization of the gene encoding the major membrane protein I (MMPI) of Mycobacterium leprae. Mol Microbiol. 1995;16:865–876. doi: 10.1111/j.1365-2958.1995.tb02314.x. [DOI] [PubMed] [Google Scholar]