Abstract
The fate of uranium in natural systems is of great environmental importance. X-ray absorption near-edge spectroscopy (XANES) revealed that U(VI) was reduced to U(IV) in shallow freshwater sediment at an open pit in an inactive uranium mine. Geochemical characterization of the sediment showed that nitrate, Fe(III), and sulfate had also been reduced in the sediment. Observations of the sediment particles and microbial cells by scanning and transmission electron microscopy, coupled with elemental analysis by energy dispersive spectroscopy, revealed that uranium was concentrated at microbial cell surfaces. U(IV) was not associated with framboidal pyrite or nanometer-scale iron sulfides, which are presumed to be of microbial origin. Uranium concentrations were not detected in association with algal cells. Phylogenetic analyses of microbial populations in the sediment by the use of 16S rRNA and dissimilatory sulfite reductase gene sequences detected organisms belonging to the families Geobacteraceae and Desulfovibrionaceae. Cultivated members of these lineages reduce U(VI) and precipitate iron sulfides. The association of uranium with cells, but not with sulfide surfaces, suggests that U(VI) is reduced by the enzymatic activities of microorganisms. Uranium was highly enriched (760 ppm) in a subsurface black layer in unsaturated sediment sampled from a pit which was exposed to seasonal fluctuations in the pond level. XANES analysis showed that the majority of uranium in this layer was U(IV), indicating that uranium is preserved in its reduced form after burial.
The redox transformations between insoluble tetravalent uranium [U(IV)] and soluble hexavalent uranium [U(VI)] have controlled uranium mobility throughout Earth's history (6, 18, 32). The reduction of U(VI) to U(IV) occurs under reducing conditions. Uranium subsequently precipitates as minerals, often as uraninite (UO2). The oxidative dissolution of U(IV) minerals occurs rapidly when oxidizing agents such as oxygen, Fe(III), and Mn(IV) are available (22, 24, 35). The reduction of U(VI) in organic-rich sediments led to the formation of economically important uranium ore deposits (6, 18, 32), and these are the most significant modern global sinks for dissolved uranium (3, 18). Despite the importance of U(VI) reduction, the key pathways for U(VI) reduction in natural organic-rich sediments remain unclear. It has been noted that U(VI) is not reduced to the thermodynamically favored U(IV) in permanently anoxic hydrogen sulfide-bearing water columns, yet in sharp contrast, it is readily reduced in sediments. Experimental studies also suggest that U(VI) reduction is catalyzed by mineral surfaces (19, 23, 53), although U(VI) is not reduced by hydrogen sulfide (29). Alternatively, based on reports that microorganisms can directly enzymatically catalyze U(VI) reduction (29), reduction in sediments has been attributed to microbial activity.
For the development of cost-effective in situ bioremediation technologies (25, 26), the microbial reduction of U(VI) has been extensively studied in the past decade (10, 12-14, 42, 46; reference 47 and references therein). Recently, it was shown that in uranium-contaminated aquifer sediments, microbial U(VI) reduction stimulated by the injection of simple organic substrates such as acetate, formate, lactate, and glucose resulted in the removal of uranium from the groundwater (2, 10, 42). However, the stimulation of specific groups of microorganisms for the effective removal of uranium from groundwaters may differ from the natural processes of U(VI) reduction because complex, naturally occurring inorganic and organic electron donors and acceptors are competitively utilized by physiologically diverse groups of microorganisms.
We studied near-surface freshwater sediment from an open pit pond at an abandoned uranium mine. In order to reveal the occurrence of, and pathways for, U(VI) reduction in the aquatic sediment containing naturally produced organic matter, we characterized sediment samples in detail. This required direct observations of sediment particles including microbial cells by the use of electron microscopes, elemental microanalysis by energy dispersive spectroscopy (EDS), geochemical analysis of surface and pore waters and of the sediment, determinations of the in situ oxidation state of uranium by synchrotron-based spectroscopy, and molecular phylogenetic analysis of microbial populations. The relatively dry sediment adjacent to the pond was also characterized in order to determine whether reduced U(IV) persisted during seasonal fluctuations in the pit water level.
MATERIALS AND METHODS
Sampling site and sample collection.
The Midnite mine is an inactive open pit uranium mine located in Stevens County, eastern Washington. Pit 4 (P4) is open and is partially filled with water. The water in P4 comes from infiltration and precipitation. In June 2001, the surface layer of black aquatic sediment from the water-sediment interface to a sediment depth of ∼2 cm and surface water were collected. A black, shallow subsurface sediment layer 10 cm below the dry sediment-air interface and 1.5 m from the pond edge was also collected. The P4 sediments were stored on ice and transferred to the laboratory in a sterile plastic bag under aerobic conditions. A subsample of the pit sediment was stored at −80°C for the molecular biological and chemical analyses described below.
Incubation of P4 sediment under anoxic conditions.
In order to simulate the consequence of the establishment of anoxic conditions in the aquatic pit sediment, we placed the sediment (50 g) in a serum bottle (100 ml) sealed with a rubber stopper and an aluminum cover with the headspace filled with N2 and incubated it for 30 days at room temperature. Changes in elementary compositions in the pore water and the sediment without the pore water were measured as described below.
Geochemical characterizations.
The pit water sample to be used for chemical analysis was filtered through a 0.2-μm-pore-size nylon filter in polypropylene housing at the sampling site. The pHs of the pit water and pore water were measured on site. Fe(II), Fe(total), Mn, nitrate, sulfate, and sulfide were measured by use of a spectrophotometer (Hach DE/2010) on site for the surface water and in the laboratory for pore waters (water analysis handbook, Hach, Loveland, Colo.). The pore waters were extracted from the sediments anaerobically by centrifugation in the laboratory within 3 days after sample collection and immediately after the 1-month incubation. Alkalinity was measured by colorimetric titration on site (Hach). The total organic carbon (TOC) in the P4 sediments and the amount of dissolved organic carbon (DOC) in the surface and pore waters were quantified by use of a high-temperature combustion carbon analyzer (Tekmar Dohrmann DC-190 high-temperature TOC analyzer). Elemental concentrations of uranium, copper, and calcium were determined by inductively coupled plasma optical emission spectrometry (ICP-OES; Jarrell ash IRIS high-resolution spectrometer).
The original and incubated sediments were centrifuged, and supernatants (pore waters) were removed under anaerobic conditions. Using subsamples of the sediments without pore water, we measured the concentrations of HCl-extractable Fe(II), Fe(III), Mn, sulfide, and uranium by spectrophotometry and ICP-OES according to the method of Lovley and Phillips (28). The pellets of the sediments were freeze-dried. Whole dried sediments were digested with HNO3-HClO4 (6:1), followed by ICP-OES analysis.
XANES analysis of P4 sediments.
The original and incubated aquatic black sediments and the subsurface black sediment without pore waters were mounted in airtight sample holders in an anaerobic glove box (Coy, Grass Lake, Mich.) for synchrotron-based X-ray absorption near-edge structure (XANES) analysis. All X-ray absorption fine structure (XAFS) measurements were made with the MRCAT insertion device (41) at the Advanced Photon Source. The energy of the incident X-rays was selected by the use of Bragg reflection from two Si(111) crystals. Higher harmonics were rejected by use of a rhodium mirror. The incident X-ray intensity was sampled with an ion chamber filled with nitrogen gas, and the fluorescent X-ray intensity was sampled with a fluorescence detector in the Stern-Heald geometry (45) filled with Ar gas. An Sr filter of three absorption lengths was used to reduce the background signal. Linearity tests (17) indicated <0.30% nonlinearity for a 50% decrease in incident X-ray intensity. The incident X-ray intensity varied <15% throughout the energy range of the XAFS measurements. The sample was exposed to the X-ray beam for approximately 1 min for each of the measurements. Measuring several spectra at different sample locations enabled a determination of radiation-induced chemical effects on a 1-min time scale. No time-dependent change in the XAFS data was observed for any of the samples. The transmission XAFS signal of a Y-foil, as described elsewhere (5), was used as a reference to accurately align the edge-energy positions of the U(IV) (UO2) and U(VI) (UO3) standards and the P4 sediments.
Scanning and transmission electron microscopy.
The original sediment was freeze-dried and embedded in epoxy resin after removal of the pore water by anaerobic centrifugation. The polished surface of the solidified epoxy resin was coated with carbon and characterized by use of a LEO 1530 scanning electron microscope (SEM). Samples were mounted on a Ni grid coated with Formvar film and then characterized by use of a Philips CM200UT transmission electron microscope (TEM).
DNA extraction, PCR, and cloning.
Using an UltraClean Mega Prep soil DNA kit (Mo Bio Laboratories, Solana Beach, Calif.), we extracted community nucleic acids from the original pit sediment stored at −80°C. Community 16S rRNA genes were amplified by PCR in mixtures containing approximately 50 ng of DNA per μl, 1× PCR buffer (Perkin-Elmer, Norwalk, Conn.), a 200 μM concentration of each of the four deoxynucleoside triphosphates, 2.5 mM MgCl2, 350 mM (each) forward and reverse primers, and 0.025 U of AmpliTaq Gold (Perkin-Elmer) per μl. For PCRs, the reverse primer was the universal primer 1492R (5′-GGTTACCTTGTTACGACTT-3′) (21), and the forward primer was the Bacteria-specific primer 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) (21) or the Archaea-specific primer 21F (5′-TTCCGGTTGATCCYGCCGGA-3′) (8). A GenAmp 2400 thermocycler (Perkin-Elmer) was used to incubate mixtures for an initial denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, 45°C for 45 s, and 72°C for 1.5 min and completed with an extension period of 20 min at 72°C. Community dissimilatory sulfite reductase (DSR) genes were amplified by PCR in mixtures containing approximately 50 ng of DNA per μl, a 500 μM concentration of each of the four deoxynucleoside triphosphates, 2.5 mM MgCl2, 350 mM (each) forward and reverse primers, and 0.1 U of AmpliTaq Gold (Perkin-Elmer) per μl. The reverse and forward primers used for DSR gene amplification were DSR67F (5′-SCACTGGAARCACGG-3′) and DSR698R (5′-GTGTARCAGTTRCCRCA-3′), which were modified from previously designed primers (52) by the introduction of a few degeneracies. The reaction mixtures were incubated for an initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, 53°C for 45 s, and 72°C for 1.5 min and completed with an extension period of 20 min at 72°C. The products for the 16S rRNA genes were purified by the use of QIAquick PCR purification columns (QIAGEN, Valencia, Calif.). Products for the DSR genes with the expected size (1.9 kb) were excised from agarose gels and purified by use of a Gel Spin DNA purification kit (Mo Bio Laboratories). The purified products were ligated into the vector pGEM-T (insert/vector ratio of 3:1) (Promega, Madison, Wis.), and the inserted vectors were transformed into competent host cells according to the manufacturer's instructions.
RFLP screening and sequencing.
For restriction fragment length polymorphism (RFLP) screening and sequencing, the inserted 16S rRNA and DSR genes were amplified by PCR, with cloned host cells as templates. The vector-specific T7 and SP6 primers were used for PCRs. Aliquots of amplified PCR products encoding 16S rRNA and DSR genes were digested with 1 U each of the four-base-specific restriction endonucleases HinP1 and MspI in 1× NEB buffer 2 (New England Biolabs, Beverly, Mass.) and 0.01% Triton X-100 overnight at 37°C. Digested products were separated by agarose (3%) gel electrophoresis. Bands were visualized by staining with ethidium bromide and UV illumination. RFLP patterns were grouped, and representative patterns were selected for sequencing.
Purified PCR products (QIAquick column; see above) were sequenced by use of a Prism Big Dye terminator sequencing kit (Applied Biosystems, Foster City, Calif.) and 50 to 100 ng of template DNA according to the manufacturer's instructions. Partial sequences were obtained by using the Bacteria-specific 27F, Archaea-specific 21F, or DSR67F primer. DNA sequences were determined by use of an Applied Biosystems 3100 genetic analyzer system (Applied Biosystems) at the University of California-Berkeley DNA Sequencing Facility.
Phylogenetic analysis.
Phylogenetic affiliations of the partial sequences were estimated with the BLAST (basic local alignment search tool) program (1) and available nucleotide databases. Sequences were aligned with the ARB FastAligner utility (http://www.arb-home.de). The similarities of partial sequences were determined by the use of ARB, and those with >98% similarity were grouped. Chimeric sequences were also checked by comparing the phylogenetic affiliations of the 5′ and 3′ halves of each sequence. The DSR gene nucleotide sequences were translated into amino acid sequences by the use of ARB. The 16S rRNA gene nucleotide sequences and inferred amino acid sequences of the DSR genes were reduced to unambiguously alignable positions by use of the Lane mask (21) and a DSR amino acid alignment mask prepared in ARB, respectively. Evolutionary analyses of alignments were performed by distance methods and parsimony by the use of PAUP for 237 amino acid positions (α-subunit) for DSR genes and 639 nucleotide positions for 16S rRNA genes (49).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences determined during this study are AY723298 to AY723313 and AY725428 to AY725438.
RESULTS
Geochemical characterization of surface and pore waters.
Naturally organic-rich sediments occur in P4 at the abandoned Midnite uranium mine in the state of Washington. By the time mining ceased in 1981, most of the uranium ore in the vicinity of P4 had been removed. The water is dilute and slightly alkaline and has maintained a uranium concentration of ∼2 to 4 ppm, only slightly higher than the average uranium content of the crust (50). In June 2001, surface water and pore waters from the original and incubated sediments were analyzed (Table 1). The surface water was slightly alkaline and contained U at 2.67 ppm and DOC at ∼3 ppm. In contrast, the pore water from the original sediment had a neutral pH and was enriched with uranium (∼8 ppm) and organic matter (DOC concentration, 787 ppm). In addition, Fe(II), sulfide, Mn, and Ca were enriched in the pore water compared to the surface water (Table 1), while nitrate and sulfate were depleted. After anoxic incubation of the sediment for 30 days, the concentration of DOC in the pore water from the incubated sediment increased significantly, accompanied with the nearly complete removal of uranium (0.22 ppm) from the pore water (Table 1). Sulfate was further removed from the pore water, with an increase in the sulfide concentration.
TABLE 1.
Geochemical characteristics of surface P4 water, in situ pore water in aquatic sediment, and pore water incubated for 30 days under anoxic conditions
| Characteristic | Value for indicated sample (mg/liter, unless indicated otherwise)a
|
||
|---|---|---|---|
| Surface pH water | In situ pore water | Incubated pore water | |
| Temperature (°C) | 19.4 | 19.6 | 23.5 |
| pH | 7.82 | 7.01 | 6.74 |
| DOC | 3.21 | 787.24 | 5,366.42 |
| Alkalinity | 42.4 | ND | ND |
| Fe(II) | 0.01 | 9.44 | 10.72 |
| Fe(total) | 0.07 | 9.48 | 10.87 |
| Mn | 0.77 | 18.22 | 5.91 |
| NO3− | 3.14 | 0.82 | 0.63 |
| SO42− | 354.20 | 246.25 | 123.12 |
| S2− | <0.001 | 0.06 | 0.16 |
| Ca | 138.01 | 198.88 | 221.96 |
| Cu | 0.135 | 0.17 | 0.16 |
| U | 2.67 | 7.30 | 0.22 |
All values are means from at least duplicate measurements and ranged within <10% error, unless indicated otherwise. ND, not determined in this study.
Characterization of P4 sediments.
The original sediment without pore water contained U at ∼500 ppm, as determined analytically after digestion with HNO3-HClO4 (Table 2). The TOC and total iron concentrations in the sediment were ∼1% and ∼34 g/kg, respectively. It was previously demonstrated that HCl-extractable Fe(III) is available for microbial Fe(III) reduction (28). In the sediment, there was 1.13 g of HCl-extractable Fe(III)/kg. The concentrations of HCl-extractable Fe(II) and sulfide were 4.15 and 0.22 g/kg, respectively. After a 1-month incubation, the concentration of uranium in the sediment increased to 750 ppm without any significant changes in the total Fe and TOC concentrations. The concentrations of HCl-extractable uranium, total iron, Fe(II), and sulfide increased after the incubation.
TABLE 2.
Chemical compositions of original and incubated aquatic sediments and of sediments in a black layer
| Element | Amt in sample (g/kg, unless indicated otherwise)a
|
|||||
|---|---|---|---|---|---|---|
| HCl extractable
|
Total
|
|||||
| Original sediment | Incubated sediment | Black-layer sediment | Original sediment | Incubated sediment | Black-layer sediment | |
| TOC (%) | ND | ND | ND | 1.03 | 1 | 1.16 |
| Fe(II) | 4.15 | 6.02 | 6.67 | ND | ND | ND |
| Fe(total) | 5.28 | 7.58 | 9.47 | 33.94 | 32.38 | 35.95 |
| Mn | 0.7 | 0.99 | 1.01 | 1.01 | 0.99 | 1.24 |
| S2− | 0.22 | 0.32 | 0.28 | ND | ND | ND |
| U | 0.38 | 0.59 | 0.58 | 0.5 | 0.75 | 0.76 |
All values are means from at least duplicate measurements and ranged within <10% error, unless indicated otherwise. ND, not determined.
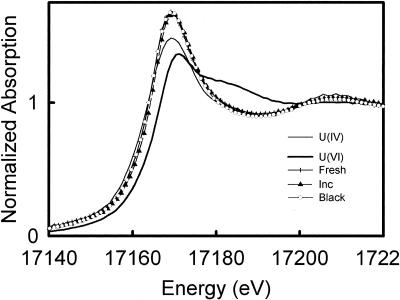
XANES analysis was performed with the original and incubated P4 sediments to determine the oxidation states of uranium in the sediments. The averaged normalized U L3-edge absorption spectra of the original and incubated sediments and the U(IV) and U(VI) standards are shown in Fig. 1. XANES analysis of the incubated sediment indicated that 70% ± 10% of the uranium was in the form of U(IV). Despite an increase in the concentration of uranium in the sediment after the incubation (Fig. 1), within the accuracy of the XANES measurement the percentages of U(IV) in both samples were equal.
FIG. 1.
Normalized U L3-edge absorption spectra for fresh and incubated aquatic black sediments and a subsurface black sediment (fresh, inc, and black, respectively) along with those for U(IV) and U(VI) standards from UO2 and UO3, respectively.
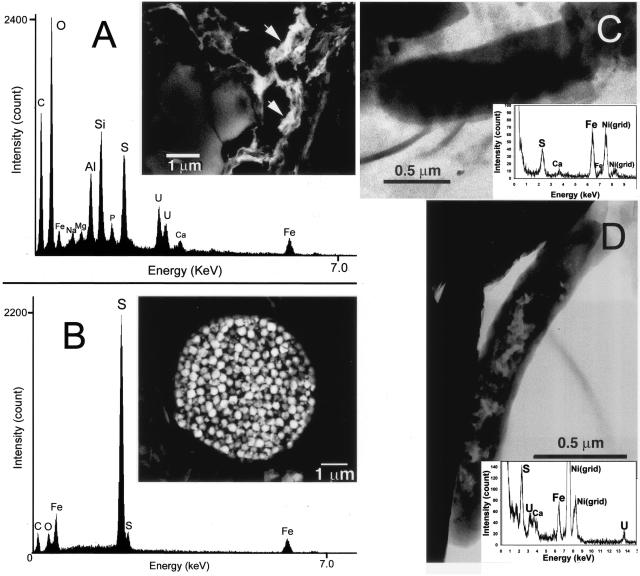
In order to clarify the pathways for U(VI) reduction in the original P4 sediment, we observed the sediment particles including microbial cells by SEM and TEM coupled to energy-dispersive X-ray spectroscopy (EDX) microanalysis. SEM imaging and a few-micrometer-scale EDS analysis of the P4 sediment embedded in epoxy showed that uranium was concentrated in loci that were also enriched with sulfur and iron (Fig. 2A). Peaks from Si and Al in the EDS spectrum were derived from the background silicate minerals (Fig. 2A). Framboidal pyrite (FeS2) (Fig. 2B) was abundant but was not associated with uranium. TEM observations coupled to submicrometer-scale EDX analysis of sediment particles mounted on a grid was conducted to clarify whether the loci enriched with iron, sulfur, and uranium were microbial cells. It was revealed that algal cells mineralized with silica and other mineral grains were not enriched with uranium. Some ovoid prokaryotic cells were coated with nanometer-sized FeS particles, but there was no colocalized uranium (Fig. 2C). Other morphologically distinct rod-shaped prokaryotic cells that precipitated FeS were enriched with uranium (Fig. 2D).
FIG. 2.
(A) Backscattered electron image of uranium-bearing phases (arrows) found in the aquatic sediment and an associated EDX spectrum. (B) Backscattered electron image of framboidal pyrite in the sediment and an associated EDX spectrum. (C) TEM image and EDX spectrum (∼30 nm spatial resolution) of a prokaryotic cell in the sediment that had accumulated iron and sulfur. (D) TEM image and EDX spectrum of a prokaryotic cell from the sediment that had accumulated uranium, iron, and sulfur.
The sampling site is hot and dry in the summer, but abundant water is supplied in the form of snow each winter. This leads to fluctuations in the pond water level and seasonal exposures of the sediments to air. We sampled a black shallow subsurface layer 10 cm below the dry sediment-air interface in June 2001. The sampling location was wet and the sediment was black in June 2000. Although the color of sediments below and above the black layer indicated that these layers were oxidized (data not shown), XANES analysis indicated that 75% ± 10% of the uranium in the subsurface black sediment remained in the U(IV) form (Fig. 1). In this sediment, organic matter, Fe(II), and sulfide were also present at concentrations comparable to those in the original and incubated aquatic sediments, as shown in Table 2. The subsurface black layer contained U at 780 ppm (Table 1), indicating that uranium was as enriched as some economically important U ore deposits (6, 32).
Phylogenetic analysis of 16S rRNA and DSR gene sequences obtained from the original P4 sediment.
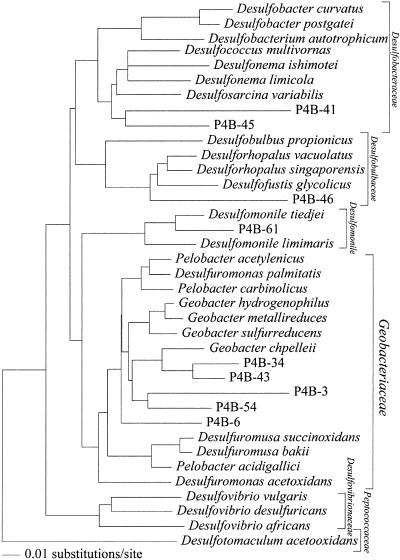
DNAs were extracted from the original P4 sediment, and 16S rRNA genes were amplified by PCRs with Archaea- and Bacteria-specific primer sets. Bacterial and archaeal rRNA gene sequences were successfully amplified from the sediment. Archaeal rRNA gene sequences related to the methanogens Methanobrevibacter arboriphilus and Methanobacterium sp. were predominant (Table 3). Among 51 bacterial clones obtained from the original P4 sediment, 24 belonged to the δ-subclass of proteobacteria (Table 3). Thirteen of the 24 δ-proteobacterial clones were clustered within the family Geobacteraceae, whose cultivated members are capable of U(VI), Fe(III), and elemental sulfur reduction (Fig. 3). The rest of the δ-Proteobacteria clones were clustered within the families Desulfobulbaceae and Desulfobacteraceae and the genus Desulfomonile, all of whose cultivated members are sulfate-reducing bacteria. Four clones were closely related to Fe(III)- and nitrate-reducing Rhodoferax ferrireducens (11) and sulfur-oxidizing Sulfuricurvum kujiense (20).
TABLE 3.
Distribution of representative clone types of archaeal and bacterial rRNAs in the original P4 sediment at the Midnite mine
| Clone name and typea | No. of rRNA clones | Database match (% identity) |
|---|---|---|
| Archaea (kingdom, division) | ||
| Euryarchaeota, Methanobacteria | ||
| P4A-2 | 10 | Methanobacterium sp. strain VeH52 (98) |
| P4A-24 | 17 | Methanobrevibacter arboriphilus (92) |
| Total archaeal rRNA clones analyzed | 33 | |
| Bacteria (division, subdivision) | ||
| Proteobacteria, Betaproteobacteria | ||
| P4B-1 | 4 | Rhodoferax ferrireducens (97) |
| Proteobacteria, Deltaproteobacteria | ||
| P4B-3 | 3 | Pelobacter venetianus (84) |
| P4B-6 | 4 | Desulfuromonas acetexigens (91) |
| P4B-34 | 2 | Geobacter chapelleii (91) |
| P4B-41 | 2 | Desulfonema magnum (85) |
| P4B-43 | 2 | Geobacter chapelleii (91) |
| P4B-45 | 5 | Desulfosarcina variabilis (88) |
| P4B-46 | 2 | Desulfocapsa thiozymogenes (90) |
| P4B-54 | 2 | Pelobacter venetianus (92) |
| P4B-61 | 2 | Desulfomonile tiedjei (89) |
| Proteobacteria, Epsillonproteobacteria | ||
| P4B-33 | 4 | Sulfuricurvum kujiense (97) |
| Firmicutes | ||
| P4B-4 | 3 | Clostridium akagii (90) |
| Bacteroidetes | ||
| P4B-25 | 4 | Cytophaga sp. strain AB015525 (91) |
| Verrucomicrobia | ||
| P4B-14 | 2 | Uncultured Verrucomicrobium DEV020 (87) |
| Total bacterial rRNA clones analyzed | 51 |
Representative rRNA clones are grouped by partial rRNA sequences having >98% similarity and being represented more than twice in the whole libraries.
FIG. 3.
Evolutionary distance dendrogram of P4 clones within the δ-Proteobacteria subdivision based on 639 nucleotides of 16S rRNA. Families and the genus Desulfomonile are bracketed to the right.
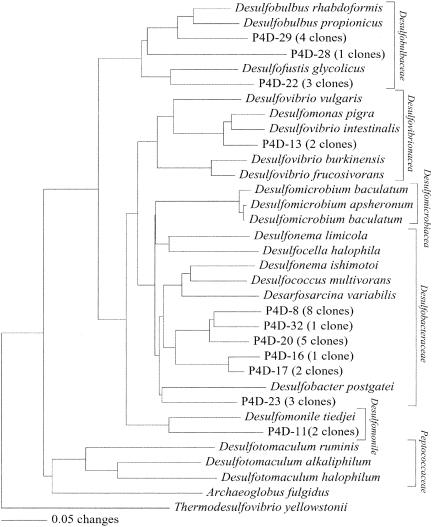
In order to better understand the community structure of sulfate-reducing bacteria, we analyzed the community DSR gene sequences obtained from the fresh aquatic sediment. Of 32 clones analyzed, 20, 8, and 2 clones were clustered within the families Desulfobacteraceae and Desulfobulbaceae and the genus Desulfomonile, respectively (Fig. 4). These results are consistent with those obtained for the 16S rRNA gene analysis. In addition, DSR gene sequences related to the family Desulfovibrionaceae were obtained, but they were minor (2 of 32 clones) (Fig. 4).
FIG. 4.
Evolutionary distance dendrogram of P4 clones based on 237 inferred amino acids of DSR genes. Families and the genus Desulfomonile are bracketed to the right. For multiple nearly identical clones (>97% identity), the number of multiples is given after the clone name of a representative clone. Accession numbers for the GenBank/EMBL/DDBJ databases are provided if the clone or organism name is not unique.
DISCUSSION
Biogeochemical processes occurring in P4 sediment.
In the shallow sediment at P4, naturally produced organic matter has stimulated a variety of biogeochemical processes. The reduction of nitrate, Fe(III), and sulfate was indicated by mineralogical data and chemical analyses (Table 1). Sulfate reduction is primarily biologically mediated in low-temperature settings (36). In the P4 sediment, several groups of sulfate-reducing bacteria were detected by molecular phylogenetic analyses, and these are inferred to have been involved in sulfate reduction. We were also able to detect bacteria closely related to the denitrifying organism R. ferrireducens (11). The reduction of Fe(III) can occur abiotically, with hydrogen sulfide as a reductant (31), or biologically, mediated by Fe(III)-reducing bacteria (25). Bacteria closely related to known Fe(III)-reducing bacteria, including R. ferrireducens (11) and the members of the Geobacteraceae family (25), probably mediate Fe(III) reduction in the P4 sediment.
Since the sediment was in contact with the surface oxygenated water, there was a steep redox transition in the sediment. Abiotic Fe(II) oxidation to Fe(III) by oxygen is kinetically fast (33, 34), as is abiotic oxidation of hydrogen sulfide to elemental sulfur, polysulfides, thiosulfate, and sulfate by oxygen and Fe(III) (31). Hydrogen sulfide may have been oxidized abiotically or by bacteria closely related to S. kujiense (20).
Pathway for U(VI) reduction and subsequent sequestration of uranium.
In contrast to previous studies showing organic-rich shallow sulfidogenic sediments containing only 25% U(IV) (9), the majority of uranium was in the reduced form in the P4 sediment. Algal cells and pyrite, which have previously been suggested to sequester uranium through adsorption and subsequent reduction (7, 53), were not associated with uranium accumulation in the P4 sediment. The microbial cells coated by nanometer-scale iron sulfide particles did not concentrate uranium. Since only a subset of microbial cells that precipitated iron sulfides was involved in uranium accumulation, we infer that U(VI) reduction is mediated by some groups of microorganisms rather than by iron sulfide particles in the P4 sediment.
Uranium was enriched in the pore water compared to the surface water. This was attributed to a partial reoxidation of U(IV)-bearing solid phases enriched in the sediment by O2 and/or Fe(III). Since it was recently reported that the cultivated members of the Geobacteraceae family are capable of U(IV) and Fe(II) oxidation coupled to nitrate reduction (11), bacteria belonging to the Geobacteraceae family might contribute to the oxidation of U(IV) and Fe(II) and to nitrate reduction. Although laboratory incubation of the P4 sediment under strictly anoxic conditions led to the removal of uranium from solution, a substantial portion of uranium was partitioned into solids as U(VI). Previous studies have demonstrated that the microbial reduction of U(VI) sorbed to solids in naturally occurring sediments is slower and less extensive than the microbial reduction of U(VI) from solution (13, 15). In the P4 sediment, U(VI) sorbed to solids might have persisted even during anoxic incubation.
Microbial populations responsible for U(VI) reduction in P4 sediment.
The members of the family Geobacteraceae that are available in pure culture are known to enzymatically reduce Fe(III), elemental sulfur, and U(VI), coupled to the oxidation of short-chain fatty acids such as acetate and aromatic compounds (25). In many subsurface settings, microorganisms that fall into the Geobacteraceae family are predominant and play important roles in Fe(III) reduction (39, 40, 43, 44). In a recent study, U(VI) reduction occurred when microorganisms in the Geobacteraceae family were specifically stimulated by amendment with acetate in field-collected aquifer sediments contaminated with uranium (14). Since bacteria represented by 16S rRNA gene sequences clustered within the Geobacteraceae family were also dominant in the P4 sediment, we postulate that they were mainly involved in U(VI) reduction.
Lovley et al. tested 11 strains of sulfate-reducing bacteria, including five strains of the Desulfovibrionaceae, three strains of the Desulfobacteraceae, and one strain each of the Desulfobulbaceae, Desulfoarculaceae, and Peptococcaceae, for U(VI) reduction capability (27). While all strains of the Desulfovibrionaceae tested were capable of reduction of U(VI), the rest of the strains in the other families were incapable of U(VI) reduction activity (27). These results indicate that among sulfate-reducing bacteria, only the members of the Desulfovibrionaceae are able to reduce U(VI) via an enzymatic pathway involving cytochrome c3 (30). To date, there are only two exceptions. Two strains of Peptococcaceae have been reported to reduce U(VI) (46-48, 51). Thus, it is suggested that in fresh sediment, sulfate-reducing bacteria belonging to the Desulfobacteraceae and Desulfobulbaceae families and the Desulfomonile genus are involved in FeS precipitation, but not in U(VI) reduction.
Preservation of microbially reduced U(IV) during subsequent burial.
The fate of microbially reduced U(IV) must be understood before in situ bioremediation technology is directly applied to contaminated settings. However, there are only a few studies addressing the fate of microbially reduced U(IV) in complex natural settings. In aquatic sediments, microbially reduced U(IV) is subsequently buried or reoxidized. The amounts of U(IV) preserved in sediments are determined by the intricate interplay between abiological and biological processes that influence the supply of oxidants. It is clear that oxygen is completely consumed several millimeters below the water-sediment interface, where abundant organic matter fuels the activities of aerobic heterotrophs and hydrogen sulfide consumes oxygen in abiotic or biologically mediated oxidation reactions (16, 37). During the transition from the aquatic sediment to the buried subsurface sediment at P4, it is likely that the surface sediment was exposed to air during drying, leading to a loss of U(IV) via oxidative dissolution, and that in the sediment layer underneath the dried surface, the water content and microbial activities had been maintained relatively well and prevented U(IV) from oxidation.
Relevance of uranium and the formation of sedimentary uranium ore deposits to global biogeochemical cycling.
Our results indicate that in the P4 sediment, U(VI) reduction was catalyzed by microbial enzymes rather than mineral surfaces. Uranium removal from the pore waters in near-shore marine sediments that receive terrestrial organic matters is the largest sink in the global budget of uranium (3, 18). Although the salinity of the P4 water is obviously different from that of ocean water, the terminal electron-accepting processes and TOC content in P4 are similar to those in typical marine sediments (4). Thus, the pathway for U(VI) reduction in the P4 sediment might be analogous to that occurring in near-shore marine sediments. In sedimentary uranium deposits, U(IV)-bearing minerals are frequently associated with iron sulfide minerals within the range of sulfur isotopic compositions (δ-34S), indicating that sulfide is produced by microbial sulfate reduction (38). Based on this finding, it has been suggested that U(VI) reduction mediated by aqueous hydrogen sulfide and/or iron sulfides produced by sulfate-reducing bacteria lead to the formation of uranium ore deposits. The results from our study indicate that U(IV) production may be mediated via direct reduction by enzymatic activities of microorganisms rather than indirect reduction by bacterially produced metabolites.
Acknowledgments
We thank D. Bruce and E. Hale for assistance with sample collections, E. J. O'Loughlin and J. H. Terry for assistance with XAFS measurements, and G. Tyson and B. Baker for laboratory assistance with constructing clone libraries. P. Hugenholtz and Y. Sekiguchi are acknowledged for modifying DSR primers and helping with phylogenetic analysis using ARB, respectively.
This research and the Advanced Photon Source are supported by grants from the U.S. Department of Energy Basic Energy Sciences Geoscience Program, the Department of Energy Biological and Environmental Research NABIR program, and the National Science Foundation. Y.S. thanks Yoshida Scholarship for additional financial support.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Basic local alignment search tool. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, A. Peacock, D. C. White, M. Lowe, and D. R. Lovley. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes, C. E., and J. K. Cochran. 1993. Uranium geochemistry in estuarine sediments: controls on removal and release processes. Geochim. Cosmochim. Acta 57:555-569. [Google Scholar]
- 4.Berner, R. A. 1982. Burial of organic carbon and pyrite sulfur in the modern ocean: its geochemical and environmental significance. Am. J. Sci. 282:451-473.
- 5.Cross, J. L., and A. I. Frenkel. 1998. Use of scattered radiation for absolute energy calibration. Rev. Sci. Instrum. 70:38-40. [Google Scholar]
- 6.Dahlkamp, F. J. 1993. Uranium ore deposits. Springer-Verlag, Berlin, Germany.
- 7.Degens, E. T., F. Khoo, and W. Michaelis. 1977. Uranium anomaly in black sea sediments. Nature 269:566-569. [Google Scholar]
- 8.Delong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duff, M. C., C. Amrhein, P. M. Bertsch, and D. B. Hunter. 1997. The chemistry of uranium in evaporation pond sediment in the San Joaquin Valley, California, USA, using X-ray fluorescence and XANES techniques. Geochim. Cosmochim. Acta 61:73-81. [Google Scholar]
- 10.Elias, D. A., L. R. Krumholz, D. Wong, P. E. Long, and J. M. Suflita. 2003. Characterization of microbial activities and U reduction in a shallow aquifer contaminated with uranium mill tailings. Microb. Ecol. 46:83-91. [DOI] [PubMed] [Google Scholar]
- 11.Finneran, K. T., C. V. Johnsen, and D. R. Lovley. 2003. Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe(III). Int. J. Syst. Evol. Microbiol. 53:669-673. [DOI] [PubMed] [Google Scholar]
- 12.Finneran, K. T., M. E. Housewright, and D. R. Lovley. 2002. Multiple influences of nitrate on uranium solubility during bioremediation of uranium contaminated subsurface sediments. Environ. Microbiol. 4:510-516. [DOI] [PubMed] [Google Scholar]
- 13.Finneran, K. T., R. T. Anderson, K. P. Nevin, and D. R. Lovley. 2002. Potential for uranium bioremediation by microbial U(VI) reduction. Soil. Sed. Contam. 11:339-357. [Google Scholar]
- 14.Holmes, D. E., K. T. Finneran, R. A. O'Neil, and D. R. Lovley. 2002. Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 68:2300-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeon, O. H., S. D. Kelly, K. M. Kemner, M. O. Barnett, W. D. Burgos, B. A. Dempsey, and E. E. Roden. 2004. Microbial reduction of U(VI) at the solid-water interface. Environ. Sci. Technol. 38:5649-5655. [DOI] [PubMed] [Google Scholar]
- 16.Jorgensen, B. B. 1982. Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature 296:643-645. [Google Scholar]
- 17.Kemner, K. M., A. J. Kropf, and B. A. Bunker. 1994. A low-temperature total electron yield detector for X-ray absorption fine structure spectra. Rev. Sci. Instrum. 65:3667-3669. [Google Scholar]
- 18.Klinkhammer, G. P., and M. R. Palmer. 1991. Uranium in the ocean: where it goes and why. Geochim. Cosmochim. Acta 55:1799-1806. [Google Scholar]
- 19.Kochenov, A. V., K. G. Korolev, V. T. Dubinchuk, and T. L. Medvedev. 1978. Experimental data on the conditions of precipitation of uranium from aqueous solutions. Geochem. Int. 14:82-87. [Google Scholar]
- 20.Kodama, Y., and K. Watanabe. 2003. Isolation and characterization of a sulfur-oxidizing chemolithotroph growing on crude oil under anaerobic conditions. Appl. Environ. Microbiol. 69:107-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, N.Y.
- 22.Langmuir, D. 1978. Uranium solution-mineral equilibria at low temperatures with applications to sedimentary ore deposits. Geochim. Cosmochim. Acta 42:547-569. [Google Scholar]
- 23.Liger, E., L. Charlet, and P. V. Cappellen. 1999. Surface catalysis of uranium(VI) reduction by iron(II). Geochim. Cosmochim. Acta 63:2939-2955. [Google Scholar]
- 24.Liu, C., J. M. Zachara, J. K. Fredrickson, D. Q. Kennedy, and A. Dohnalkova. 2002. Modeling the inhibition of the bacterial reduction of U(VI) by β-MnO2(s). Environ. Sci. Technol. 36:1452-1459. [DOI] [PubMed] [Google Scholar]
- 25.Lovley, D. R. 2000. Fe(III)- and Mn(IV)-reducing prokaryotes. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.
- 26.Lovley, D. R. 2001. Anaerobes to the rescue. Science 294:1444-1446. [DOI] [PubMed] [Google Scholar]
- 27.Lovley, D. R., E. E. Roden, E. J. P. Phillips, and J. C. Woodward. 1993. Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Mar. Geol. 113:41-53. [Google Scholar]
- 28.Lovley, D. R., and E. J. P. Phillips. 1987. Rapid assay for microbially reducible ferric iron in aquatic sediments. Appl. Environ. Microbiol. 53:1536-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovley, D. R., E. J. P. Phillips, Y. A. Gorby, and E. R. Landa. 1991. Microbial reduction of uranium. Nature 350:413-416. [Google Scholar]
- 30.Lovely, D. R., P. K. Widman, J. C. Woodward, and E. J. P. Phillips. 1993. Reduction of uranium by cytochrome c3 of Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 59:3572-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luther, G. W., III, B. T. Glazer, J. I. Popp, M. Taillefert, T. F. Rozan, P. J. Brendel, S. M. Therberge, and D. B. Nuzzio. 2001. Sulfur speciation monitored in situ with solid gold amalgam voltammetric microelectrodes: polysulfides as a special case in sediments, microbial mats and hydrothermal vent waters. J. Environ. Monit. 2:61-66. [DOI] [PubMed] [Google Scholar]
- 32.Maynard, J. B. 1983. Geochemistry of sedimentary ore deposits. Springer-Verlag, New York, N.Y. [DOI] [PubMed]
- 33.Millero, F. J., S. Hubinger, and M. Fernandez. 1987. Oxidation of H2S in seawater as a function of temperature, pH, and ionic strength. Environ. Sci. Technol. 21:439-443. [DOI] [PubMed] [Google Scholar]
- 34.Millero, F. J., S. Sotolongo, and M. Izaguirre. 1987. The oxidation kinetics of Fe(II) in seawater. Geochim. Cosmochim. Acta 51:793-801. [Google Scholar]
- 35.Nevin, K. P., and D. R. Lovley. 2000. Potential for nonenzymatic reduction of Fe(III) during microbial oxidation of organic matter coupled to Fe(III) reduction. Environ. Sci. Technol. 34:2472-2478. [Google Scholar]
- 36.Ohmoto, H., and A. C. Lasaga. 1982. Kinetics of reactions between aqueous sulfates and sulfides in hydrothermal systems. Geochim. Cosmochim. Acta 46:1727-1745. [Google Scholar]
- 37.Revsbech, N. P., B. B. Jorgensen, and T. H. Blackburn. 1980. Oxygen in the sea bottom measured with a microelectrode. Science 207:1355-1356. [Google Scholar]
- 38.Reynolds, R. L., M. B. Goldhaber, and D. J. Carpenter. 1982. Biogenic and nonbiogenic ore-forming processes in the South Texas uranium district: evidence from the Panna Maria deposit. Econ. Geol. 77:541-556. [Google Scholar]
- 39.Roling, W. F., B. M. van Breukelen, M. Braster, B. Lin, and H. W. van Verseveld. 2001. Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl. Environ. Microbiol. 67:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 2001. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segre, C. U., N. E. Leyarovska, L. D. Chapman, W. M. Lanender, P. W. Plag, A. S. King, A. J. Kropf, B. A. Bunker, K. M. Kemner, P. Dutta, R. S. Druan, and J. Kaduk. 2001. The MRCAT insertion device beamline at the Advanced Photon Source. Synch. Rad. Instrum. 521:419-422. [Google Scholar]
- 42.Senko, J. M., J. D. Istok, J. M. Suflita, and L. R. Krumholz. 2002. In-situ evidence for uranium immobilization and remobilization. Environ. Sci. Technol. 36:1491-1496. [DOI] [PubMed] [Google Scholar]
- 43.Snoeyenbos-West, O. L., K. P. Nevin, R. T. Anderson, and D. R. Lovley. 2000. Enrichment of Geobacter species in response to stimulation of Fe(III) reduction in sandy aquifer sediments. Microb. Ecol. 39:153-167. [DOI] [PubMed] [Google Scholar]
- 44.Stein, L. Y., M. T. La Duc, T. J. Grundl, and K. H. Nealson. 2001. Bacterial and archaeal populations associated with freshwater ferromanganous micronodules and sediments. Environ. Microbiol. 3:10-18. [DOI] [PubMed] [Google Scholar]
- 45.Stern, E. A., M. Newville, B. Ravel, Y. Yacoby, and D. Haskel. 1995. The UWXAFS analysis package: philosophy and details. Physica B 208-209:117-120. [Google Scholar]
- 46.Suzuki, Y., S. D. Kelly, K. M. Kemner, and J. F. Banfield. 2002. Nanometer-size products of uranium bioreduction. Nature 419:134. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki, Y., S. D. Kelly, K. M. Kemner, and J. F. Banfield. 2003. Microbial populations stimulated for hexavalent uranium reduction in uranium mine sediment. Appl. Environ. Microbiol. 69:1337-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki, Y., S. D. Kelly, K. M. Kemner, and J. F. Banfield. 2004. Enzymatic reduction of U(VI) by Desulfosporosinus spp. Radiochim. Acta 92:11-16. [Google Scholar]
- 49.Swofford, D. L. 1999. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.02. Sinauer Associates, Sunderland, Mass.
- 50.Taylor, S. R. 1964. Abundance of chemical elements in the continental crust: a new table. Geochim. Cosmochim. Acta 28:1273-1284. [Google Scholar]
- 51.Tebo, B., and A. Y. Obraztsova. 1998. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol. Lett. 162:193-198. [Google Scholar]
- 52.Wagner, M., A. D. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wersin, P., M. F. Hochella, Jr., P. Persson, R. Redden, J. O. Leckie, and D. Harris. 1994. Interaction between aqueous uranium(VI) and sulfide minerals: spectroscopic evidence for sorption and reduction. Geochim. Cosmochim. Acta 58:2829-2843. [Google Scholar]