Abstract
Small lipases of Bacillus species, such as LipA from Bacillus subtilis, have a high potential for industrial applications. Recent studies showed that deletion of six AT-rich islands from the B. subtilis genome results in reduced amounts of extracellular LipA. Here we demonstrate that the reduced LipA levels are due to the absence of four genes, skfABCD, located in the prophage 1 region. Intact skfABCD genes are required not only for LipA production at wild-type levels by B. subtilis 168 but also under conditions of LipA overproduction. Notably, SkfA has bactericidal activity and, probably, requires the SkfB to SkfD proteins for its production. The present results show that LipA is more prone to proteolytic degradation in the absence of SkfA and that high-level LipA production can be improved significantly by employing multiple protease-deficient B. subtilis strains. In conclusion, our findings imply that SkfA protects LipA, directly or indirectly, against proteolytic degradation. Conceivably, SkfA could act as a modulator in LipA folding or as a protease inhibitor.
Lipases are produced by a wide variety of gram-positive and gram-negative bacterial species (26, 28). Application areas include the organic synthesis of chiral drugs, the production of washing detergent additives and baking ingredients, and the enhancement of flavor in the dairy industry (27, 40, 41, 45). Generally, bacterial lipases range in size from about 30 to 75 kDa (31). It is thought that many of these lipases act on their substrates at the lipid-water interface. This process, called interfacial activation, is most likely enhanced by a lid-like polypeptide, which covers the active site of the lipase in the absence of lipid-water interfaces. These enzymes are referred to as “true lipases” (3, 28). Notably, not all lipases show this interfacial activation. There are lipase family members that lack the lid that covers their active site and thus do not show activation at oil-water interfaces. This applies to the lipases of Bacillus subtilis and Bacillus pumilus, which are actually the smallest lipases known. These small, lidless lipases seem to be well-suited for biotechnological applications, the synthesis of chiral drugs in particular (7, 26, 36, 42).
The mature extracellular form of B. subtilis lipase A, which is encoded by the lip gene (lipA is referred to as lip in the SubtiList database [http://genolist.pasteur.fr/SubtiList/], has a molecular mass of 19 kDa (7). It is synthesized with a signal peptide of 34 amino acids, which is cleaved off during or shortly after protein translocation via the Sec pathway (30). Lipase A is regarded as an alkaliphilic lipase, because it is stable under highly alkaline conditions and is optimally active at pH 10. It was classified as a lipase rather than an esterase, because the enzyme displays more activity towards long-chain triacylglycerides (tricaprylyl glycerol) than towards triacetyl glycerol (36). In addition, B. subtilis contains a second gene, lipB, which encodes an extracellular lipolytic protein of 182 amino acid residues sharing 74% amino acid sequence identity with LipA (53). LipB lacks activity towards long-chain triacylglyceride substrates but can hydrolyze triacylglycerol esters and p-nitrophenyl esters of fatty acids with short-chain lengths of ≤10 carbon atoms (13). A common feature of LipA and LipB is that the active site consists of a Ser-His-Asp catalytic triad, which is similar to that of serine proteases, indicating that the reaction mechanisms are comparable (26, 53). Notably, LipA and LipB are differentially expressed, depending on the composition of the growth medium. LipA is produced in rich and minimal media, whereas LipB is produced only in rich media. Furthermore, lipB transcription is upregulated after the addition of hydrophobic components to the growth medium, whereas lipA transcription is not (14). These Bacillus lipases share an α-β hydrolase fold for the protein core, but they have different substrate specificities because their protein surfaces differ. This suggests that different physiological functions exist for the two enzymes (14, 53).
Whereas the secretion and folding of certain lipases from gram-negative bacteria, especially Pseudomonas species, have been studied in great detail (5, 11, 16-18, 24), relatively little is known about lipase secretion and folding in gram-positive bacteria. The folding of lipases from gram-negative bacteria requires specific chaperones, the so-called lipase-specific foldases. In contrast, no evidence for the presence of lipase-specific folding catalysts in gram-positive bacteria, such as Bacillus species, has been documented. This might relate to the different secretion mechanisms of these lipases. Whereas in gram-negative bacteria two membranes have to be crossed, a secreted lipase of a gram-positive bacterium crosses only one membrane (48).
B. subtilis and related Bacillus species are well known for their high secretion potential. Therefore, they are used for the commercial production of a great variety of secretory proteins (15, 19, 29, 52). Nevertheless, further improvement of B. subtilis as a cell factory for the environmentally friendly and cost-effective production of pharmaceutically and biotechnologically relevant proteins is desirable. For this purpose, protease-deficient B. subtilis strains have been developed (57, 58), different secretion pathways (especially the Sec and Tat pathways) have been extensively studied (for reviews see references 48 and 51), and the response of B. subtilis cells to stresses caused by high-level protein secretion has been examined (1, 6, 23). Very recently, genome engineering was explored as a completely new approach to improve the B. subtilis cell factory. Six different regions were removed from the genome, including two prophages, three prophage-like regions, and the large pks gene cluster, which is involved in polyketide synthesis. The deletion of 7.7% of the genome affected neither the key physiological and developmental processes of B. subtilis nor its general capacity for protein production and secretion. Strikingly, however, one effect of the genome minimization was the significantly decreased production of LipA (56). The present studies focused on determining which genes of the deleted regions are required for LipA production. Our results show that high-level production of LipA depends on the presence of the skfA, skfB, skfC, and skfD gene cluster.
MATERIALS AND METHODS
Plasmids, bacterial strains, and media.
Table 1 lists the plasmids and bacterial strains used. B. subtilis was transformed as described by Kunst and Rapoport (34). Luria-Bertani (LB) medium contained Bacto tryptone (1%), Bacto yeast extract (0.5%), and NaCl (0.5%). Because B. subtilis cultures did not express LipA when grown on the regular S7 medium for pulse-chase labeling of B. subtilis proteins (50), S7-MAM (methionine assay medium) was used for this purpose. S7-MAM was basically prepared as S7 medium with the difference that the MAM amino acid mixture from Becton Dickinson, which consists of all amino acids except methionine, was used instead of the amino acid mixture normally used to supplement S7 medium. Proteins were labeled with Tran35S-label (ICN Biomedicals). Antibiotics were used in the following concentrations: chloramphenicol, 5 μg/ml; erythromycin, 2 μg/ml; hygromycin, 50 μg/ml; and kanamycin, 20 μg/ml.
TABLE 1.
Plasmids and strains
| Plasmid or strain | Relevant properties | Reference |
|---|---|---|
| Plasmids | ||
| pMA5 | pUB110 derivative; contains the HpaII promoter for the expression of cloned genes; ColE1; repB Kmr Apr | 10 |
| pLip2031 | pMA5 derivative; carries the B. subtilis lipA gene under control of the HpaII promoter; Kmr | 8 |
| pKTH10 | Vector containing the amyQ gene of B. amyloliquefaciens; Kmr | 38 |
| Strains | ||
| Escherichia coli | ||
| MC1061 | F−araD139 Δ(ara-leu)7696 Δ(lac)X74 galU galK hsdR2 mcrA mcrB1 rspL | 55 |
| B. subtilis | ||
| 168 | trpC2 | 33 |
| Δ1 | trpC2; Δprophage1 | 56 |
| Δpks | trpC2; pks::Cm; Cmr | 56 |
| TF8A | trpC2; ΔSPβ; Δskin; ΔPBSX | 56 |
| Δ5 | trpC2; ΔSPβ; Δskin; ΔPBSX; Δprophage 1; pks::Cm; Cmr | 56 |
| Δ6 | trpC2; ΔSPβ; Δskin; ΔPBSX; Δprophage1; pks::Cm; Δprophage 3; Cmr | 56 |
| 168 lipB::pMutin2 | trpC2; lipB::pMutin2; Emr | 32 |
| 168 ybcC::pMutin2 | trpC2; ybcC::pMutin2; Emr | 32 |
| 168 ybcD::pMutin2 | trpC2; ybcF::pMutin2; Emr | 32 |
| 168 ybcF::pMutin2 | trpC2; ybcF::pMutin2; Emr | 32 |
| 168 ybcH::pMutin1 | trpC2; ybcH::pMutin1; Emr | 32 |
| 168 ybcI::pMutin2 | trpC2; ybcI::pMutin2; Emr | 32 |
| 168 ybcL::pMutin2 | trpC2; ybcL::pMutin2; Emr | 32 |
| 168 ybcM::pMutin2 | trpC2; ybcM::pMutin2; Emr | 32 |
| 168 skfA::pMutin1 | trpC2; skfA::pMutin1; Emr | 32 |
| 168 skfB::pMutin2 | trpC2; skfB::pMutin2; Emr | 32 |
| 168 skfC::pMutin2 | trpC2; skfC::pMutin2; Emr | 32 |
| 168 skfD::pMutin1 | trpC2; skfD::pMutin1; Emr | 32 |
| 168 skfE::pMutin2 | trpC2; skfE::pMutin2; Emr | 32 |
| 168 skfF::pMutin2 | trpC2; skfF::pMutin2; Emr | 32 |
| 168 skfG::pMutin2 | trpC2; skfG::pMutin2; Emr | 32 |
| 168 skfH::pMutin2 | trpC2; skfH::pMutin2; Emr | 32 |
| TEB1030 | DB430 ΔlipA ΔlipB | 12 |
| TEB1030 skfA | DB430 ΔlipA ΔlipB; skfA::pMutin1; Emr | This study |
| TEB1030 skfB | DB430 ΔlipA ΔlipB; skfB::pMutin2; Emr | This study |
| TEB1030 skfC | DB430 ΔlipA ΔlipB; skfC::pMutin2; Emr | This study |
| TEB1030 skfD | DB430 ΔlipA ΔlipB; skfD::pMutin1; Emr | This study |
| Δ1 lipB | trpC2; Δprophage1; lipB::pMutin2; Emr | This study |
| WB800 | trpC2; nprE nprB aprE epr mpr bpf vpr wprA; Cmr Hygr | 57 |
| WB800 skfA | WB800; skfA::pMutin1; Cmr Hygr Emr | This study |
Transcript profiling.
Transcript profiling was performed with B. subtilis Panorama macroarrays from Sigma-Genosys. Duplicate cultures were grown in LB medium at 37°C until 1 h before and 3 h after the transition between exponential-phase and post-exponential-phase growth, after which total RNA from B. subtilis 168 and B. subtilis Δ6 was isolated with a High Pure RNA isolation kit (Roche Molecular Biochemicals) as described by Hamoen et al. (22). For simultaneous reverse transcriptase reactions on all mRNAs in the RNA sample, 4 μg of total RNA was added to 1 pmol of open reading frame-specific primers (Eurogentec). Reverse transcription was carried out as described by Westers et al. (56). Labeled cDNA was hybridized to B. subtilis Panorama macroarrays from Sigma-Genosys as described by the manufacturer. After hybridization and washing, Cyclone phosphorimager screens (Packard Instrument Company) were exposed for 2 or 3 days. The Cyclone readouts were analyzed with Array-Pro Analyzer 4.0 (Media Cybernetics). After background subtraction, duplicate spots were averaged and the signal was normalized against the total signal of all spots. The normalized array data were subjected to a statistical analysis using Cyber-T, a program based on a t test variant combined with a Bayesian statistical framework (43). Cyber-T is available for online use at the genomics website of the University of California at Irvine (http://visitor.ics.uci.edu/genex/cybert). The following parameters were used in Cyber-T: the minimum number of nonzero replicates was set to 2, a sliding window of 101 was used, and the recommended confidence value of 10 was chosen. Spots were associated with gene names by using the B. subtilis array information Excel spreadsheet provided by Sigma-Genosys.
Proteomics.
B. subtilis strains were grown at 37°C under vigorous agitation in LB medium. After 1 h of post-exponential-phase growth, cells were separated from the growth medium by centrifugation. The secreted proteins in the growth medium were collected for two-dimensional (2D) PAGE as previously described (2). Protein identification by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry was performed as described earlier (2, 30).
Western blotting and immunodetection.
To detect LipA or AmyQ, B. subtilis cells were separated from the growth medium by centrifugation (2 min at 6,400 × g, followed by 3 min at 13,000 × g at room temperature). Proteins in the growth medium were concentrated upon precipitation with 5% trichloroacetic acid (TCA). Samples for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were prepared as described previously (50). After separation by SDS-PAGE, proteins were transferred to a Protran nitrocellulose transfer membrane (Schleicher and Schuell) as described by Kyhse-Andersen (35). LipA and AmyQ were visualized with specific antibodies and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G conjugates (Biosource International).
Pulse-chase protein labeling, immunoprecipitation, SDS-PAGE, and autoradiography.
Pulse-chase labeling of B. subtilis cells grown in S7-MAM, immunoprecipitation, SDS-PAGE, and autoradiography were performed essentially as described previously (50). Immunoprecipitations were performed with specific antibodies against LipA.
Enzyme activity assays.
To determine lipase activity, the colorimetric assay described by Lesuisse et al. (36) was applied with some modifications. In short, a semiautomated analysis was performed, using a MultiPROBEIIex robotic liquid handling system (Packard), in which 180 μl of a reaction buffer (0.1 M potassium phosphate buffer [pH 8.0], 0.1% gum arabic, 0.36% Triton X-100) was supplemented with 10 μl of the substrate 4-nitrophenyl caprylate (10 mM in methanol). The reaction was started by the addition of 10 μl of culture supernatant. Lipase activity was determined by measuring the increase in absorbance at 405 nm per min of incubation at room temperature, per unit of optical density at 600 nm (OD600) of the culture at the time of sampling. Experiments were performed with growth medium fractions of four different pLip2031 transformants per tested strain. The lipase activity in each growth medium fraction was determined in triplicate. Suspect lipase activity measurements (i.e., lipase activity values that seemed to differ significantly from the other three obtained values) were assessed for significance by performing Dixon's Q-test for outliers.
Spent LB media of B. subtilis 168, 168 skfA::pMutin1, 168 skfB::pMutin2, 168 skfC::pMutin2, and 168 skfD::pMutin1, which were used to monitor the possible degradation of purified LipA, were obtained from overnight cultures. For this purpose, cells were separated from the growth medium by centrifugation (2 min at 6,400 × g, followed by 3 min at 13,000 × g at room temperature). Purified LipA, produced in B. subtilis 168, was added to the medium up to a final concentration of 0.0011 mg/ml. As a control, LipA was added to fresh LB medium. Lipase activity measurements were performed immediately after addition of purified LipA to spent media, after 1 h of incubation in spent media, and upon overnight incubation in spent media (37°C).
α-Amylase activity was tested with a halo assay. Cells were grown overnight and subsequently separated from the growth medium by centrifugation (5 min at 13,000 × g at room temperature). Durapore membrane filters (Millipore) were placed on LB agar plates containing 1% starch (Merck), and then the medium fractions, were spotted. The amounts of medium spotted on the filters were corrected for the OD600 of each culture. After overnight incubation at 37°C, the plates were analyzed for starch degradation after staining with iodine vapor. Diameters of the resulting halos were measured.
Proteolytic activity was assayed with the resorufin-labeled casein substrate from Roche Molecular Biochemicals. Cells were grown overnight and subsequently separated from the growth medium by centrifugation (2 min at 5,000 × g followed by 2 min at 16,000 × g at room temperature). Next, 100 μl of supernatant was incubated for 1 h at 37°C in the presence of 0.4% resorufin-labeled casein in 200 mM Tris (pH 7.8)-2 mM CaCl2 buffer. Undigested substrate was removed by precipitation with 5% TCA and subsequent centrifugation. Finally, the release of resorufin-labeled peptides in the supernatant fractions was measured spectrophotometrically at 574 nm after addition of the assay buffer (500 mM Tris, pH 8.8).
RESULTS
Secretion of LipA.
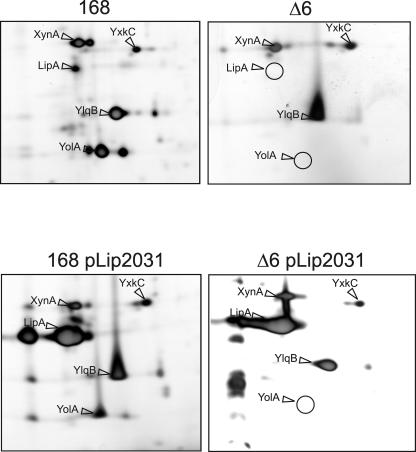
Previously, the effects of deleting six large genomic regions (i.e., the prophages PBSX and SPβ, the prophage-like regions skin, prophage 1, and prophage 3, and the pks operon) on the composition of the extracellular proteome of B. subtilis were studied by 2D gel electrophoresis and mass spectrometry (56). A remarkable observation was that the secreted lipase LipA was absent from the growth medium of the sixfold-deletion strain B. subtilis Δ6 (Fig. 1, top), despite the fact that the lipA gene itself was not deleted. In contrast, the secretory protein of unknown function YolA was absent from the medium of B. subtilis Δ6, because the corresponding gene that is located in the SPβ region had been removed (Fig. 1) (56). In these earlier studies it was not verified whether the Δ6 strain was still able to produce and secrete LipA when the lipA gene was expressed from a plasmid. In the present work, this question was addressed by a 2D gel electrophoretic analysis with post-exponential-phase growing cells containing plasmid pLip2031 for high-level lipA expression. As shown in Fig. 1 (bottom), B. subtilis Δ6 pLip2031 was still able to secrete LipA into the growth medium. Furthermore, the composition of the total extracellular proteome remained unaffected compared to the parental strain 168 transformed with pLip2031 (Fig. 1 [bottom] and data not shown). Because the 2D gel analysis, as performed in the present studies, is only semiquantitative, the amounts of LipA secreted by B. subtilis Δ6 and the parental strain were compared by using a LipA activity assay and Western blotting. The data shown in Fig. 2 demonstrate that the extracellular levels of LipA produced by B. subtilis Δ6 are ∼70% lower than those of B. subtilis 168. Notably, in independent experiments performed on different days, this level of reduction varied between ∼70 and ∼50%. These observations indicate that the capacity of the Δ6 strain for LipA production is significantly reduced due to the absence of one or more genes that are present in the parental strain 168. Because the results of the lipase activity assays correlate reasonably well with those of the Western blotting analyses (Fig. 2, compare left and right panels), only the results of the lipase activity assays are shown below. Importantly, the presence or absence of an intact lipB gene did not detectably affect the extracellular lipase activity when LipA was overproduced with the help of pLip2031 (data not shown). Therefore, indirect effects of LipB production on the extracellular levels of overproduced LipA, as assayed in the present studies, can be ruled out.
FIG. 1.
Extracellular proteomes of B. subtilis 168 and Δ6. (Top) Cells of B. subtilis 168 or B. subtilis Δ6 were grown in LB medium, and extracellular proteins were harvested 1 h after entry of the cells into the post-exponential-growth phase. The extracellular proteins were separated by 2D PAGE and stained with silver nitrate. Protein spots identified by MALDI-TOF mass spectrometry are indicated. Positions on the gel of spots missing from the extracellular proteome of B. subtilis Δ6 are indicated by circles. (Bottom) B. subtilis 168 and Δ6, both containing pLip2031 for high-level production of LipA, were grown in LB medium. Secreted proteins were analyzed as described for the top panel.
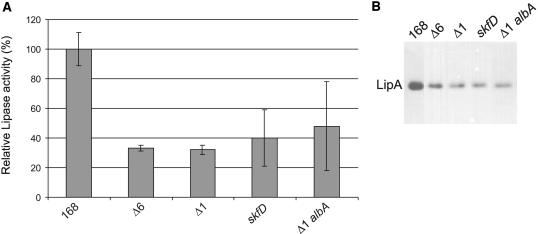
FIG. 2.
Identification of prophage 1-specific genes involved in LipA production. (A) LipA activities in the growth media of B. subtilis 168, Δ6, Δ1, 168 skfD, and Δ1 albA containing pLip2031 for the overproduction of LipA. Cells were grown overnight in LB medium at 37°C and separated from the growth medium by centrifugation. Growth medium fractions were used for lipase activity determinations as described in Materials and Methods. Numbers are averages for four independent cultures. LipA activity was calculated as the increase in the absorbance at 405 nm · min−1 per OD unit and is presented as a percentage of the lipase activity in the growth medium of the parental strain 168 (100%). Error bars indicate the standard deviations of the biological repeats. (B) LipA production and secretion. Cells from B. subtilis 168, Δ6, Δ1, 168 skfD, and Δ1 albA containing pLip2031 for the overproduction of LipA were grown overnight in LB medium at 37°C and separated from the growth medium by centrifugation. Proteins in the growth medium were concentrated by precipitation with TCA, and samples for SDS-PAGE and Western blotting were prepared. LipA was visualized by using LipA-specific antibodies.
Four prophage 1 genes are required for high-level LipA production.
As a first approach to determine which genes within the six deleted regions are required for high-level LipA production, the Δ5, TF8a, Δpks, and Δ1 strains (Table 1), which lack different prophages, prophage-like regions, and/or the pks operon, were transformed with plasmid pLip2031. As shown by Western blotting and activity assays, the production of LipA by the TF8a and Δpks strains was not reduced (data not shown). In contrast, LipA production by the Δ5 (data not shown) and Δ1 strains was significantly decreased. Since the Δ5 strain contains the combined mutations of the TF8a, Δpks, and Δ1 strains, these findings indicated that one or more genes of prophage 1 are involved in LipA production by B. subtilis (Fig. 2).
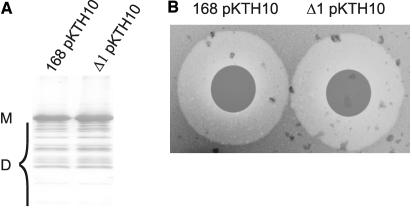
To verify whether genes in the prophage 1 region affect the high-level production of secretory proteins other than LipA, the production of the α-amylase AmyQ of Bacillus amyloliquefaciens by the B. subtilis Δ1 strain was studied by Western blotting and a plate activity assay. As shown in Fig. 3, the prophage 1 region is not involved in the secretion and extracellular accumulation of active AmyQ. Together with the proteomics data described above, our observations show that one or more genes in the prophage 1 region are specifically involved in LipA production.
FIG. 3.
Secretion of AmyQ by B. subtilis cells lacking prophage 1. (A) AmyQ production and secretion. B. subtilis 168 and Δ1 were transformed with pKTH10, directing overproduction of AmyQ. Cells were grown overnight in LB medium at 37°C and separated from the growth medium by centrifugation. Proteins in the growth medium were concentrated by precipitation with TCA, and samples for SDS-PAGE and Western blotting were prepared. AmyQ was visualized with specific antibodies. Note that several degradation products (D) of AmyQ are detectable. M, mature AmyQ. (B) AmyQ plate assay. B. subtilis 168 and Δ1 containing pKTH10 for AmyQ overproduction were grown overnight in LB medium at 37°C. Filters were placed on starch-containing LB agar plates, and then fractions of the growth medium were collected and spotted on the filters. The size of halos observed upon iodine staining of the plates is indicative of the secretion of active AmyQ.
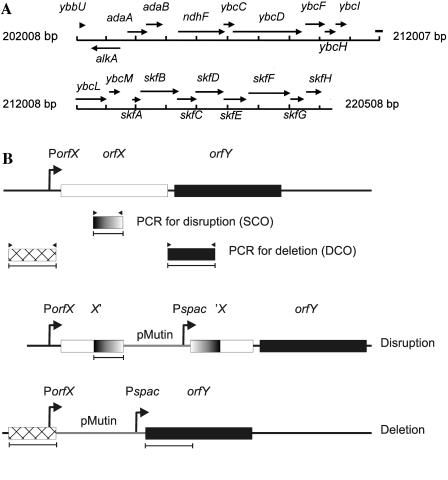
Prophage 1 contains 20 genes, some of which have a known function (Fig. 4A). For example, the alkA, adaA, and adaB genes encode proteins involved in DNA restriction/modification and repair, while ndhF encodes subunit 5 of an NADH dehydrogenase. In contrast, the function of the 15 prophage 1 genes downstream of ndhF was unknown at the time we started our investigations. Therefore, we addressed the question of which of these 15 genes might affect LipA production. To address this question, mutants constructed in the context of the B. subtilis Systematic Gene Function Analysis Programme (Table 1) (32) were transformed with pLip2031. Notably, the mutations in these genes involved either disruptions or deletions created with the pMutin system that is based on a chromosomal integration plasmid (Fig. 4B). The advantage of this system is that genes located downstream of a disrupted or deleted gene are transcribed from the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter present on the integrated pMutin plasmid. As shown by Western blotting and activity assays, the four genes originally annotated as ybcO, ybcP-Q, ybcS, and ybcT affect LipA production to similar extents. These genes were recently renamed skfA, skfB, skfC, and skfD, respectively, because they are required for the synthesis of the sporulation killing factor SkfA (20). Only the results obtained for the skfD (ybcT) mutant are documented in Fig. 2. In contrast, the 11 remaining prophage 1 genes downstream of ndhF are not involved in LipA production (Table 2; only the results for mutations in the region downstream of ybcL are listed). This conclusion is supported by experiments in which mutant strains, created with the pMutin system, were grown in the presence of IPTG to promote the transcription of genes located downstream of integrated pMutin plasmids. Notably, the results in Table 2 represent a single set of data that has been obtained with cultures (with and without IPTG) that were grown in parallel. In this particular set of data, the LipA activities in the growth media of the skfC mutant cells were relatively high, while the LipA activities in the growth media of the skfH mutant cells were relatively low. In other experiments, which were not included in Table 2 because they were performed on different dates, the LipA levels produced by skfC mutant cells were comparable to those produced by skfA, skfB, or skfD mutant cells, whereas the levels of LipA produced by skfH mutant cells were comparable to those obtained for the parental strain 168.
FIG. 4.
Schematic representations of the prophage 1 region and the pMutin mutagenesis system. (A) Genes in the B. subtilis prophage 1 region. (B) Disruption or deletion of chromosomal genes of B. subtilis with pMutin plasmids as described by Vagner et al. (49). To disrupt a certain gene (orfX), an internal fragment of orfX is amplified by PCR and cloned into pMutin. Transformation of B. subtilis with the pMutin plasmid containing the cloned fragment of orfX will result in the chromosomal integration of this plasmid by single-crossover recombination (SCO) into orfX. As a consequence of this integration, orfX is disrupted and the genes downstream of orfX (depicted as orfY) are placed under the control of the IPTG-inducible Pspac promoter of the integrated pMutin plasmid. To delete orfX, the flanking regions of orfX is amplified by PCR and cloned into pMutin in the same orientation but opposite order relative to that on the chromosome. Transformation of B. subtilis with the resulting pMutin plasmid results in the chromosomal integration of this plasmid by double-crossover recombination (DCO). Consequently, orfX is replaced by the integrated pMutin plasmid, while the genes downstream of orfX (depicted as orfY) are placed under the control of the IPTG-inducible Pspac promoter. PorfX, promoter of orfX; X′, 3′-truncated orfX; ′X, 5′-truncated orfX.
TABLE 2.
Involvement of prophage 1 genes in LipA production by B. subtilisa
| Strain | IPTG | Lipase activity (103) |
|---|---|---|
| 168 pLip2031 | − | 6.8 ± 0.8 |
| + | 7.3 ± 0.9 | |
| 168 ybcM::pMutin2 pLip2031 | − | 9.1 ± 0.4 |
| + | 5.6 ± 2.7 | |
| 168 skfA::pMutin1 pLip2031 | − | 1.3 ± 0.1 |
| + | 1.3 ± 0.1 | |
| 168 skfB::pMutin2 pLip2031 | − | 1.4 ± 0.2 |
| + | 1.5 ± 0.3 | |
| 168 skfC::pMutin2 pLip2031 | − | 3.2 ± 0.9 |
| + | 2.7 ± 0.5 | |
| 168 skfD::pMutin1 pLip2031 | − | 1.8 ± 0.6 |
| + | 1.6 ± 0.8 | |
| 168 skfE::pMutin2 pLip2031 | − | 7.3 ± 0.9 |
| + | 8.2 ± 2.4 | |
| 168 skfF::pMutin2 pLip2031 | − | 6.5 ± 0.3 |
| + | 6.8 ± 1.4 | |
| 168 skfG::pMutin2 pLip2031 | − | 6.0 ± 1.5 |
| + | 5.9 ± 0.6 | |
| 168 skfH::pMutin2 pLip2031 | − | 4.1 ± 0.4 |
| + | 2.9 ± 1.6 |
To determine which prophage 1 genes are involved in LipA production, cells of different B. subtilis mutant strains, transformed with plasmid pLip2031, were grown overnight in the absence (−) or presence (+) of 1 mM IPTG. Cultures were grown in parallel. Cells and growth media were separated by centrifugation, and 10 μl of culture supernatant was used for lipase activity determinations as described in Materials and Methods. LipA activities are indicated as the increase in the absorbance at 405 nm · min−1 per OD unit.
Earlier studies by Zheng and coworkers and by Zuber on the synthesis of the bacteriocin subtilosin (Sbo) by B. subtilis 168 revealed that one of the proteins involved in Sbo synthesis, known as AlbA (for “antilisterial bacteriocin production”), displays significant amino acid sequence similarity to SkfB (60, 61). To verify whether AlbA contributes to LipA production, an albA single mutant and an albA prophage 1 double mutant were transformed with pLip2031. Next, the production of LipA by the resulting strains was assessed. The absence of an intact albA gene resulted in neither a reduction of LipA production by the single albA mutant (data not shown) nor a further reduction of LipA production by the albA prophage 1 double mutant (Fig. 2).
The possibility that the reduced amount of LipA in the growth medium of the Δ6 strain is due to a regulatory effect on the transcription of the lipA gene was verified by using the DNA macroarray technique. For this purpose, total RNA was isolated from cultures of B. subtilis Δ6 and the 168 strain that had been grown until 1 h before or 3 h after the transition between exponential-phase and post-exponential-phase growth. Although the expression levels of lipA were relatively low, the results suggested that the transcription of lipA was not influenced by the combined deletions of the Δ6 strain. Since the five different independently obtained data sets showed some fluctuation in the absolute lipA expression levels, a statistical verification of the lack of effect of the prophage 1 deletion on lipA expression was difficult. Therefore, a transcriptional lipA-lacZ fusion (kindly provided by T. Eggert) was used to monitor lipA promoter activity during growth, in both B. subtilis Δ1 and the parental strain (data not shown). In this case, the results again suggest that the absence of prophage 1 does not affect lipA expression, but due to the low expression levels, it is difficult to evaluate the statistical significance of the data. Taken together, these findings suggest that skfABCD mutants have a posttranscriptional LipA production defect.
To test whether SkfA, SkfB, SkfC, and/or SkfD is important for the processing of LipA precursors by signal peptidase, the processing kinetics was studied by pulse-chase labeling experiments using cells grown on S7-MAM. As a control, the parental strain transformed with pLip2031 was used. The results showed that processing of pre-LipA and the secretion of mature LipA into the medium were not affected in the different skf mutants (data not shown). Thus, the skfABCD genes are dispensable for LipA processing and secretion, at least when B. subtilis is grown in S7-MAM. However, under these growth conditions, the LipA production levels in the skfA, skfB, skfC, and skfD mutant strains containing pLip2031 were indistinguishable from those of the parental strain 168 containing pLip2031. Thus, it seems that the involvement of the skfABCD gene products in LipA production is growth medium dependent.
Skf proteins protect LipA from proteolytic degradation.
Previous studies showed that the degradation of secretory proteins by extracellular proteases of B. subtilis is significantly reduced when cells are grown in minimal media (44). As a first approach to investigating a possible involvement of the skfABCD gene products in the extracellular degradation of LipA, purified LipA was added to spent LB media of different skf mutant strains. After incubation for various times at 37°C, lipase activity assays were performed. However, the levels of activity of the purified LipA added to spent media derived from cultures of skf mutant cells were comparable to the activity of purified LipA added to spent medium of a culture of the parental strain 168 (data not shown). In fact, no degradation of purified LipA was detectable upon overnight incubation in spent media. These observations show that the skfABCD gene products do not affect the stability or activity of correctly folded LipA in the growth medium.
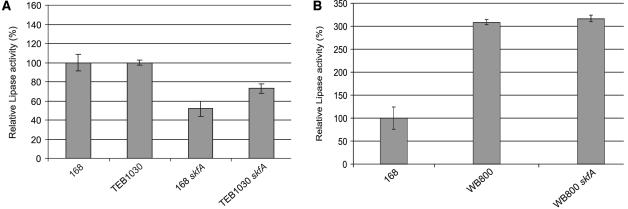
To test whether the extracellular proteolytic activity of B. subtilis Δ1 strain transformed with pLip2031 differed significantly from that of parental strain 168 transformed with pLip2031, the degradation of resorufin-labeled casein by (LB) growth medium fractions from overnight cultures of both strains was assayed. These experiments showed that the extracellular proteolytic activities of B. subtilis Δ1 and 168 did not differ significantly (data not shown). This suggested that the LipA produced by the Δ1 strain could be more prone to proteolytic degradation during its synthesis or secretion than the LipA produced by the parental strain 168. To verify this idea, the skfA, skfB, skfC, or skfD mutation was introduced into the protease mutant strain TEB1030 (containing pLip2031) by transformation with chromosomal DNA from the respective single-mutant strains. B. subtilis TEB1030 lacks the cytoplasmic protease IspA and the extracellular proteases AprE, NprE, and Bpf. In addition, TEB1030 lacks the lipB gene. Interestingly, the presence of skfA, skfB, skfC, and skfD mutations in TEB1030 resulted in a less severe decrease in LipA production than the presence of these mutations in strain 168. Specifically, skfABCD mutations resulted in a reduction of LipA activity of about 25% in the TEB1030 strain, and a reduction of about 50% in the 168 strain (Fig. 5A,; only the results for skfA mutant cells grown in LB medium are shown). These observations indicated that the skf gene products can protect LipA from proteolytic degradation.
FIG. 5.
LipA production in protease-deficient strains. To determine the influence of different proteases of B. subtilis on the extracellular accumulation of LipA, protease-deficient strains were transformed with pLip2031 for high-level expression of lipA. Cells were grown overnight in LB medium at 37°C and separated from the growth medium by centrifugation. Growth medium fractions were used for lipase activity determinations as described in Materials and Methods. Numbers are averages for four independent cultures. LipA activity was calculated as the increase in the absorbance at 405 nm · min−1 per OD unit and presented as the percentage of the lipase activity in the growth medium of the parental strain 168 (100%). Error bars indicate the standard deviations of the biological repeats. (A) Relative LipA activities in the growth media of B. subtilis 168, TEB1030, 168 skfA, and TEB1030 skfA. (B) Relative LipA activities in the growth media of B. subtilis 168, WB800, and WB800 skfA.
An intriguing question that remained to be answered was to what extent the reduced LipA production of the B. subtilis skfABCD mutant strains could be suppressed by the absence of additional proteases. To address this question, the skfA mutation was introduced into B. subtilis WB800. This strain lacks the extracellular proteases NprE, NprB, AprE, Epr, Mpr, Bpf, and Vpr, as well as the cell wall-associated protease WprA. Strikingly, the levels of LipA production in B. subtilis WB800 (containing pLip2031) were increased threefold compared to levels in the parental strain 168 (containing pLip2031), irrespective of the presence or absence of a skfA mutation (Fig. 5B). These findings show that the LipA overproduced in B. subtilis is prone to proteolytic degradation and that this degradation is substantially reduced by the absence of eight proteases from strain WB800. Moreover, the present studies show for the first time that the skfABCD gene products protect LipA against extracytoplasmic proteolysis.
DISCUSSION
Previously we have shown that the deletion of six large regions from the B. subtilis chromosome results in a severe reduction in the production of the extracellular lipase LipA (56). This was remarkable because the lipA gene is not located in the deleted regions. The results of our present studies show that four genes from prophage 1, which is one of the previously deleted genomic regions, affect the production of LipA most likely at a posttranscriptional level. Remarkably, these four prophage 1 genes are skfA, skfB, skfC, and skfD, which have been implicated in the production of the bacteriocin SkfA (20, 37). Specifically, our unprecedented observations show that the presence of skfABCD is required to protect LipA against degradation by extracytoplasmic proteases of B. subtilis.
SkfA is a peptide of 55 amino acids and may have an N-terminal leader peptide. It contains a double-glycine motif, which is a typical cleavage site in so-called type AII lantibiotics. After their modification, these lantibiotics are normally processed and transported by dual-function ABC transporters, which cleave the leader sequence immediately behind the double-glycine motif (21). A first involvement of the skfA, skfB, and skfC genes in bacteriocin production was demonstrated in B. subtilis SO113, which protects rice against infection by Xanthomonas oryzae (37). More recently, the skfABCDEFGH operon of B. subtilis 168 was implicated in sporulation. In fact, the name skf stands for “sporulation killing factor,” because the SkfA produced by sporulating cells was shown to act as a killing factor that blocks the sporulation of nonsporulating sister cells and causes their lysis (20). While the entire skfABCDEFGH operon is required for this sibling killing effect, only the skfABCD genes are involved in LipA production. Thus, it seems that the SkfE to SkfH proteins have a specific role in sibling killing, whereas the killing factor SkfA and the SkfB to SkfD proteins have dual functions in sibling killing and LipA production. Taken together, the observations of González-Pastor et al. (20) concerning SkfA production by B. subtilis and our present findings imply that the bacteriocin SkfA is involved in LipA production. Accordingly, SkfB to SkfD could be required for SkfA synthesis and export from the cytoplasm. This view is supported by the fact that SkfB shows amino acid sequence similarity with the AlbA protein, which is involved in the production of the antimicrobial peptide subtilosin (Sbo) of B. subtilis 168. AlbA is a member of the MoaA/NifB/PqqE protein family and is thought to be critical for modification of the presubtilosin peptide (60). Furthermore, sequence analyses indicate that SkfD is an integral membrane protein that belongs to the CAAX family of N-terminal proteases. These proteases have been implicated in bacteriocin production by Lactobacillus plantarum (39), and, presumably, they confer immunity to bacteriocins (9). Although SkfC shows no similarity with other known proteins, its predicted membrane attachment via one transmembrane segment makes it conceivable that this protein is involved in SkfA production or immunity. The precise role of SkfE to SkfH in SkfA production and sibling killing is not known, but it has been suggested that some of these components are involved in SkfA transport. If so, this raises the question of why the skfEFGH genes are not involved in LipA production.
Our present observations show that LipA is sensitive to degradation by extracytoplasmic proteases of B. subtilis. This is most clearly seen when the genes for eight of these proteases are mutated. Under these conditions, mutations in skfABCD no longer affect LipA production, indicating that the SkfABCD proteins are required to protect LipA against degradation. This protection must occur after LipA export from the cytoplasm, since the proteases that are responsible for LipA degradation are located either in the cell wall (i.e., WprA), or in the growth medium. Notably, the results of our experiments in which purified LipA was incubated in spent medium of B. subtilis 168 show that the activity and stability of correctly folded LipA are not affected by extracellular proteases. These observations imply that LipA degradation occurs after membrane translocation but prior to its folding into a stable and active conformation. As SkfA appears to be the only exported protein of the four SkfABCD gene products, it seems most likely that SkfA is the factor that protects LipA against degradation.
Studies by Jongbloed et al. (30) demonstrated that LipA is translocated via the Sec machinery, which can transport proteins only in an unfolded state (48, 54). This implies that LipA has to fold posttranslocationally at the membrane-cell wall interface. Consequently, at least two possible mechanisms for SkfA-mediated LipA protection are conceivable. Firstly, SkfA could act as an extracytoplasmic folding catalyst of LipA. If this is the case, SkfA would interact transiently with LipA, because no SkfA was detectable upon nano-LC (liquid chromatography)-ESI (electrospray ionization)-QTOF (quadrupole orthogonal time-of-flight) mass spectrometry of trypsin- or LysC-digested samples of purified LipA. However, it cannot be ruled out that SkfA was lost during the purification procedure or that SkfA is a modulator of the activity of an as-yet-unknown folding catalyst of LipA. Secondly, SkfA could act like a protease inhibitor. Although mutation of skfABCD does not affect the overall proteolytic activity in the growth medium, SkfA may specifically inhibit proteases at the membrane-cell wall interface, like HtrA, HtrB, or WprA. In this respect, it is noteworthy that the branched cyclic dodecylpeptide antibiotic bacitracin is known as a protease inhibitor that can even be used for the affinity purification of certain proteases (59). This bacteriocin, produced by Bacillus licheniformis and some strains of B. subtilis but not B. subtilis 168 (4, 25), inhibits the biosynthesis of peptidoglycan by binding to the lipid carrier undecaprenyl pyrophosphate, which is required for the translocation of cell envelope building blocks (46, 47). By analogy to bacitracin, SkfA of B. subtilis might be both a bacteriocin and a protease inhibitor. Though not conclusive, the observation that cells lacking skfABCD can still overproduce LipA (compared to wild-type levels of LipA production) can be regarded as an argument in favor of the idea that SkfA interferes with the activity of one or more proteases.
In conclusion, the present study shows that genes involved in the production of the sibling killing factor SkfA are required for optimal production of extracellular LipA. Whether SkfA acts as a folding catalyst or a protease inhibitor remains to be determined. This will involve in vitro LipA unfolding-refolding experiments in which the effects of the presence or absence of SkfA and proteases of B. subtilis are monitored. Studies of this type will aid our understanding of the mechanisms involved in bacterial sibling killing. Moreover, they will provide valuable leads to further improve B. subtilis as a cell factory, certainly for the production of lipases.
Acknowledgments
We thank Sierd Bron and other members of the Groningen Bacillus Secretion Groups and the ExporteRRs consortium for valuable discussions. Furthermore, we thank Junichi Sekiguchi for providing the lipB::pMutin2 strain, Ronald van Merkerk for establishing the semiautomated activity assay for LipA, Aäron Beekman for support in the macroarray analyses, and Dörte Becher and Patty Mulder for mass spectrometric analyses with purified LipA.
Funding for the project of which this work is a part was provided by the CEU projects BIO4-CT98-0250, QLK3-CT-1999-00413, QLK3-CT-1999-00917, LSH-503468, and LSH-05257.
REFERENCES
- 1.Antelmann, H., E. Darmon, D. Noone, J. W. Veening, H. Westers, S. Bron, O. P. Kuipers, K. M. Devine, M. Hecker, and J. M. van Dijl. 2003. The extracellular proteome of Bacillus subtilis under secretion stress conditions. Mol. Microbiol. 49:143-156. [DOI] [PubMed] [Google Scholar]
- 2.Antelmann, H., H. Tjalsma, B. Voigt, S. Ohlmeier, S. Bron, J. M. van Dijl, and M. Hecker. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11:1484-1502. [DOI] [PubMed] [Google Scholar]
- 3.Arpigny, J. L., and K. E. Jaeger. 1999. Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343:177-183. [PMC free article] [PubMed] [Google Scholar]
- 4.Azevedo, E. C., E. M. Rios, K. Fukushima, and G. M. Campostakaki. 1993. Bacitracin production by a new strain of Bacillus subtilis. Extraction, purification, and characterization. Appl. Biochem. Biotechnol. 42:1-9. [DOI] [PubMed] [Google Scholar]
- 5.Chihara-Siomi, M., K. Yoshikawa, N. Oshima-Hirayama, K. Yamamoto, Y. Sogabe, T. Nakatani, T. Nishioka, and J. Oda. 1992. Purification, molecular cloning, and expression of lipase from Pseudomonas aeruginosa. Arch. Biochem. Biophys. 296:505-513. [DOI] [PubMed] [Google Scholar]
- 6.Darmon, E., D. Noone, A. Masson, S. Bron, O. P. Kuipers, K. M. Devine, and J. M. Van Dijl. 2002. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J. Bacteriol. 184:5661-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dartois, V., A. Baulard, K. Schanck, and C. Colson. 1992. Cloning, nucleotide sequence and expression in Escherichia coli of a lipase gene from Bacillus subtilis 168. Biochim. Biophys. Acta 1131:253-260. [DOI] [PubMed] [Google Scholar]
- 8.Dartois, V., J. Y. Coppee, C. Colson, and A. Baulard. 1994. Genetic analysis and overexpression of lipolytic activity in Bacillus subtilis. Appl. Environ. Microbiol. 60:1670-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diep, D. B., L. S. Havarstein, and I. F. Nes. 1996. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178:4472-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dröge, M. J., R. Bos, and W. J. Quax. 2001. Paralogous gene analysis reveals a highly enantioselective 1,2-O-isopropylideneglycerol caprylate esterase of Bacillus subtilis. Eur. J. Biochem. 268:3332-3338. [DOI] [PubMed] [Google Scholar]
- 11.Duong, F., C. Soscia, A. Lazdunski, and M. Murgier. 1994. The Pseudomonas fluorescens lipase has a C-terminal secretion signal and is secreted by a three-component bacterial ABC-exporter system. Mol. Microbiol. 11:1117-1126. [DOI] [PubMed] [Google Scholar]
- 12.Eggert, T., U. Brockmeier, M. J. Dröge, W. J. Quax, and K. E. Jaeger. 2003. Extracellular lipases from Bacillus subtilis: regulation of gene expression and enzyme activity by amino acid supply and external pH. FEMS Microbiol. Lett. 225:319-324. [DOI] [PubMed] [Google Scholar]
- 13.Eggert, T., G. Pencreac'h, I. Douchet, R. Verger, and K. E. Jaeger. 2000. A novel extracellular esterase from Bacillus subtilis and its conversion to a monoacylglycerol hydrolase. Eur. J. Biochem. 267:6459-6469. [DOI] [PubMed] [Google Scholar]
- 14.Eggert, T., G. van Pouderoyen, B. W. Dijkstra, and K. E. Jaeger. 2001. Lipolytic enzymes LipA and LipB from Bacillus subtilis differ in regulation of gene expression, biochemical properties, and three-dimensional structure. FEBS Lett. 502:89-92. [DOI] [PubMed] [Google Scholar]
- 15.Ferrari, E., A. S. Jarnagin, and B. F. Schmidt. 1993. Commercial production of extracellular enzymes, p. 917-937. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 16.Filloux, A., G. Michel, and M. Bally. 1998. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol. Rev. 22:177-198. [DOI] [PubMed] [Google Scholar]
- 17.Frenken, L. G., J. W. Bos, C. Visser, W. Muller, J. Tommassen, and C. T. Verrips. 1993. An accessory gene, lipB, required for the production of active Pseudomonas glumae lipase. Mol. Microbiol. 9:579-589. [DOI] [PubMed] [Google Scholar]
- 18.Gerritse, G., R. Ure, F. Bizoullier, and W. J. Quax. 1998. The phenotype enhancement method identifies the Xcp outer membrane secretion machinery from Pseudomonas alcaligenes as a bottleneck for lipase production. J. Biotechnol. 64:23-38. [DOI] [PubMed] [Google Scholar]
- 19.Godtfredsen, A. E. 1990. Microbial lipases, p. 255-274. In W. M. Fogarty and K. T. Kelly (ed.), Microbial enzymes and biotechnology. Elsevier Applied Science, London, United Kingdom.
- 20.González-Pastor, J. E., E. C. Hobbs, and R. Losick. 2003. Cannibalism by sporulating bacteria. Science 301:510-513. [DOI] [PubMed] [Google Scholar]
- 21.Guder, A., I. Wiedemann, and H. G. Sahl. 2000. Posttranslationally modified bacteriocins—the lantibiotics. Biopolymers 55:62-73. [DOI] [PubMed] [Google Scholar]
- 22.Hamoen, L. W., W. K. Smits, A. de Jong, S. Holsappel, and O. P. Kuipers. 2002. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 30:5517-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyyryläinen, H. K., A. Bolhuis, E. Darmon, L. Muukkonen, P. Koski, M. Vitikainen, M. Sarvas, Z. Prágai, S. Bron, J. M. van Dijl, and V. P. Kontinen. 2001. A novel two-component regulatory system of Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41:1159-1172. [DOI] [PubMed] [Google Scholar]
- 24.Ihara, F., I. Okamoto, K. Akao, T. Nihira, and Y. Yamada. 1995. Lipase modulator protein (LimL) of Pseudomonas sp. strain 109. J. Bacteriol. 177:1254-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishihara, H., M. Takoh, R. Nishibayashi, and A. Sato. 2002. Distribution and variation of bacitracin synthetase gene sequences in laboratory stock strains of Bacillus licheniformis. Curr. Microbiol. 45:18-23. [DOI] [PubMed] [Google Scholar]
- 26.Jaeger, K. E., B. W. Dijkstra, and M. T. Reetz. 1999. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu. Rev. Microbiol. 53:315-351. [DOI] [PubMed] [Google Scholar]
- 27.Jaeger, K. E., and T. Eggert. 2002. Lipases for biotechnology. Curr. Opin. Biotechnol. 13:390-397. [DOI] [PubMed] [Google Scholar]
- 28.Jaeger, K. E., S. Ransac, B. W. Dijkstra, C. Colson, M. van Heuvel, and O. Misset. 1994. Bacterial lipases. FEMS Microbiol. Rev. 15:29-63. [DOI] [PubMed] [Google Scholar]
- 29.Jarnagin, A. S., and E. Ferrari. 1992. Extracellular enzymes: gene regulation and structure function relationship studies, p. 191-219. In R. Doi and M. McGloughlin (ed.), Biology of bacilli: applications to industry. Butterworth-Heinemann, Boston, Mass. [PubMed]
- 30.Jongbloed, J. D., H. Antelmann, M. Hecker, R. Nijland, S. Bron, U. Airaksinen, F. Pries, W. J. Quax, J. M. van Dijl, and P. G. Braun. 2002. Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis. J. Biol. Chem. 277:44068-44078. [DOI] [PubMed] [Google Scholar]
- 31.Kawasaki, K., H. Kondo, M. Suzuki, S. Ohgiya, and S. Tsuda. 2002. Alternate conformations observed in catalytic serine of Bacillus subtilis lipase determined at 1.3 A resolution. Acta Crystallogr. D 58:1168-1174. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. M. L. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA. 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Dusterhoft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S. Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Henaut, H. Hilbert, S. Holsappel, S. Hosono, M. F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. KlaerrBlanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauer, V. Lazarevic, S. M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauel, C. Medigue, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. OReilly, K. Ogawa, A. Ogiwara, B. Oudega, S. H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, G. Rapoport, M. Rey, S. Reynolds, M. Rieger, C. Rivolta, E. Rocha, B. Roche, M. Rose, Y. Sadaie, T. Sato, E. Scanlan, S. Schleich, R. Schroeter, F. Scoffone, J. Sekiguchi, A. Sekowska, S. J. Seror, P. Serror, B. S. Shin, B. Soldo, A. Sorokin, E. Tacconi, T. Takagi, H. Takahashi, K. Takemaru, M. Takeuchi, A. Tamakoshi, T. Tanaka, P. Terpstra, A. Tognoni, V. Tosato, S. Uchiyama, M. Vandenbol, F. Vannier, A. Vassarotti, A. Viari, R. Wambutt, E. Wedler, H. Wedler, T. Weitzenegger, P. Winters, A. Wipat, H. Yamamoto, K. Yamane, K. Yasumoto, K. Yata, K. Yoshida, H. F. Yoshikawa, E. Zumstein, H. Yoshikawa, and A. Danchin. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 34.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kyhse-Andersen, J. 1984. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods 10:203-209. [DOI] [PubMed] [Google Scholar]
- 36.Lesuisse, E., K. Schanck, and C. Colson. 1993. Purification and preliminary characterization of the extracellular lipase of Bacillus subtilis 168, an extremely basic pH-tolerant enzyme. Eur. J. Biochem. 216:155-160. [DOI] [PubMed] [Google Scholar]
- 37.Lin, D., L. J. Qu, H. Gu, and Z. Chen. 2001. A 3 center dot 1-kb genomic fragment of Bacillus subtilis encodes the protein inhibiting growth of Xanthomonas oryzae pv. oryzae. J. Appl. Microbiol. 91:1044-1050. [DOI] [PubMed] [Google Scholar]
- 38.Palva, I. 1982. Molecular cloning of alpha-amylase gene from Bacillus amyloliquefaciens and its expression in B. subtilis. Gene 19:81-87. [DOI] [PubMed] [Google Scholar]
- 39.Pei, J. M., and N. V. Grishin. 2001. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem. Sci. 26:275-277. [DOI] [PubMed] [Google Scholar]
- 40.Reetz, M. T., and K. E. Jaeger. 1998. Overexpression, immobilization and biotechnological application of Pseudomonas lipases. Chem. Phys. Lipids 93:3-14. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez, M., N. Prim, F. Randez-Gil, F. I. Pastor, and P. Diaz. 2002. Engineering of baker's yeasts, E. coli and Bacillus hosts for the production of Bacillus subtilis lipase A. Biotechnol. Bioeng. 78:339-345. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt-Dannert, C. 1999. Recombinant microbial lipases for biotechnological applications. Bioorg. Med. Chem. 7:2123-2130. [DOI] [PubMed] [Google Scholar]
- 43.Schreiber, J., J. Enderich, and M. Wegner. 1998. Structural requirements for DNA binding of GCM proteins. Nucleic Acids Res. 26:2337-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, H., S. Bron, J. Vanee, and G. Venema. 1987. Construction and use of signal sequence selection vectors in Escherichia coli and Bacillus subtilis. J. Bacteriol. 169:3321-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soberon-Chavez, G., and B. Palmeros. 1994. Pseudomonas lipases: molecular genetics and potential industrial applications. Crit Rev. Microbiol. 20:95-105. [DOI] [PubMed] [Google Scholar]
- 46.Stone, K. J., and J. L. Strominger. 1971. Mechanism of action of bacitracin—complexation with metal ion and C55-isoprenyl pyrophosphate. Proc. Natl. Acad. Sci. USA 68:3223-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Storm, D. R., and J. L. Strominger. 1973. Complex-formation between bacitracin peptides and isoprenyl pyrophosphates—specificity of lipid-peptide interactions. J. Biol. Chem. 248:3940-3945. [PubMed] [Google Scholar]
- 48.Tjalsma, H., A. Bolhuis, J. D. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 50.van Dijl, J. M., A. de Jong, H. Smith, S. Bron, and G. Venema. 1991. Non-functional expression of Escherichia coli signal peptidase I in Bacillus subtilis. J. Gen. Microbiol. 137:2073-2083. [DOI] [PubMed] [Google Scholar]
- 51.van Dijl, J. M., P. G. Braun, C. Robinson, W. J. Quax, H. Antelmann, M. Hecker, J. Muller, H. Tjalsma, S. Bron, and J. D. H. Jongbloed. 2002. Functional genomic analysis of the Bacillus subtilis Tat pathway for protein secretion. J. Biotechnol. 98:243-254. [DOI] [PubMed] [Google Scholar]
- 52.van Leen, R. W., J. G. Bakhuis, R. F. van Beckhoven, H. Burger, L. C. Dorssers, R. W. Hommes, P. J. Lemson, B. Noordam, N. L. Persoon, and G. Wagemaker. 1991. Production of human interleukin-3 using industrial microorganisms. Biotechnology (N.Y.) 9:47-52. [DOI] [PubMed] [Google Scholar]
- 53.van Pouderoyen, G., T. Eggert, K. E. Jaeger, and B. W. Dijkstra. 2001. The crystal structure of Bacillus subtilis lipase: a minimal alpha/beta hydrolase fold enzyme. J. Mol. Biol. 309:215-226. [DOI] [PubMed] [Google Scholar]
- 54.van Wely, K. H., J. Swaving, R. Freudl, and A. J. Driessen. 2001. Translocation of proteins across the cell envelope of Gram-positive bacteria. FEMS Microbiol. Rev. 25:437-454. [DOI] [PubMed] [Google Scholar]
- 55.Wertman, K. F., A. R. Wyman, and D. Botstein. 1986. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49:253-262. [DOI] [PubMed] [Google Scholar]
- 56.Westers, H., R. Dorenbos, J. M. van Dijl, J. Kabel, T. Flanagan, K. M. Devine, F. Jude, S. J. Séror, A. C. Beekman, E. Darmon, C. Eschevins, A. de Jong, S. Bron, O. P. Kuipers, A. M. Albertini, H. Antelmann, M. Hecker, N. Zamboni, U. Sauer, C. Bruand, D. S. Ehrlich, J. C. Alonso, M. Salas, and W. J. Quax. 2003. Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol. Biol. Evol. 20:2076-2090. [DOI] [PubMed] [Google Scholar]
- 57.Wu, S. C., J. C. Yeung, Y. Duan, R. Ye, S. J. Szarka, H. R. Habibi, and S. L. Wong. 2002. Functional production and characterization of a fibrin-specific single-chain antibody fragment from Bacillus subtilis: effects of molecular chaperones and a wall-bound protease on antibody fragment production. Appl. Environ. Microbiol. 68:3261-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, X. C., W. Lee, L. Tran, and S. L. Wong. 1991. Engineering a Bacillus subtilis expression-secretion system with a strain deficient in six extracellular proteases. J. Bacteriol. 173:4952-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaya, N. E., E. E. Vaughan, S. K. Shah, and D. Castignetti. 2002. Bacitracin: Substantiation and elimination of contaminating proteolytic activity and use as an affinity chromatography ligand to purify a siderophore-degrading enzyme. Curr. Microbiol. 44:71-74. [DOI] [PubMed] [Google Scholar]
- 60.Zheng, G. L., R. Hehn, and P. Zuber. 2000. Mutational analysis of the sbo-alb locus of Bacillus subtilis: identification of genes required for subtilosin production and immunity. J. Bacteriol. 182:3266-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuber, P. 2001. A peptide profile of the Bacillus subtilis genome. Peptides 22:1555-1577. [DOI] [PubMed] [Google Scholar]