Abstract
The increasing prevalence of antibiotic-resistant staphylococci has prompted the need for antibacterial controls other than antibiotics. In this study, a lytic bacteriophage (phage K) was assessed in vitro for its ability to inhibit emerging drug-resistant Staphylococcus aureus strains from hospitals and other species of Staphylococcus isolated from bovine infections. In in vitro inhibitory assays, phage K lysed a range of clinically isolated methicillin-resistant S. aureus (MRSA) strains, S. aureus with heterogeneous vancomycin resistance and vancomycin resistance, and teicoplanin-resistant strains. In these assays, 14 of the MRSA strains were initially only weakly sensitive to this phage. However, propagation of phage K on these less-sensitive strains resulted in all 14 being sensitive to the modified phages. The results enforce the principle that, while certain target bacteria may be relatively insensitive to lytic phage, this can be overcome by obtaining modified phage variants from passage of the phage through the insensitive strains. Model in situ hand wash studies using a phage-enriched wash solution resulted in a 100-fold reduction in staphylococcal numbers on human skin by comparison with numbers remaining after washing in phage-free solution. Infusion of the phage into a nonimmunogenic bismuth-based cream resulted in strong anti-Staphylococcus activity from the cream on plates and in broth.
The increasing prevalence of antibiotic resistance in clinical isolates of Staphylococcus aureus is a major problem, given that the bacterium causes a wide variety of human infections ranging from simple abscesses to fatal sepsis, as well as endocarditis, pneumonia, mastitis, phlebitis, meningitis, and toxinoses (for a review see reference 24). The rapid emergence of penicillin-resistant S. aureus in the 1950s led to the use of methicillin and related drugs for treatment of infections. In the 1960s, methicillin-resistant S. aureus (MRSA) strains emerged and have since become endemic in many hospital environments (14). In addition, these MRSA strains also frequently exhibit resistance to a variety of other common antibiotics (20). Indeed, over 95% of patients worldwide with S. aureus infections do not respond to first-line antibiotics, for example, ampicillin and penicillin (33). Recently, the SENTRY antimicrobial surveillance program reported that 36.8% of S. aureus isolates ribotyped belonged to multidrug-resistant, oxacillin-resistant S. aureus strains (7). In Ireland, Naylor et al. (23) found that MRSA was the commonest single organism cultured from patients with complex wound and graft infections after vascular surgery. In addition, the latest data from the European Antimicrobial Resistance Surveillance System showed an increase in MRSA from 39% in 1999 to 45% in 2002 in Ireland (37). Until recently, S. aureus has exhibited sensitivity to the glycopeptide antibiotics vancomycin and teicoplanin, and therefore these antibiotics represent one of the last lines of defense available against staphylococcal infection. However, the recent emergence of vancomycin-resistant S. aureus and also teicoplanin-resistant strains in hospital infections poses a major threat to this approach (13). As a result, investigations for new and alternative antimicrobials effective against S. aureus have become increasingly relevant.
Bacteriophages (phages) were investigated as far back as 1921 to eliminate bacteria including staphylococci in human infections (35). The majority of human phage therapy studies have been performed in Poland (29) and the former Soviet Union and have included challenges against Staphylococcus (for a review see reference 36). Although research on phage therapy diminished outside of the former Soviet Union with the advent of antibiotics, it has been revisited primarily as a result of the antibiotic resistance problem. This renewed interest is evident from the number of reviews published recently (2, 3, 5, 8, 9, 19, 22, 36). For S. aureus the potential of phage as an antibacterial therapeutic was shown by Matsuzaki and coworkers (21), who significantly reduced the mortality of mice previously injected with S. aureus by intraperitoneal injections of phage MR11 (21). Moreover, since the early 1990s, a variety of new companies that have placed major emphasis on bacteriophage research, with the aim of treating multidrug-resistant bacteria causing infections, have been established worldwide.
Phage K is a polyvalent phage with a broad host range, inhibiting both coagulase-positive and -negative staphylococci (32). It is a member of the family Myoviridae (1) and has been the subject of previous studies (15-17, 28-30). The origin of phage K is unclear. Both Rountree in 1949 (32) and Rippon in 1956 (31) state that phage K of Krueger and Northrop (18) is identical to phage Au2 described by Burnet and Lush in 1935 (4). Burnet and Lush also state that the phage used by Krueger and Northrop in 1930 (18) is Au2 and suggest that phage Au2 could be derived from the H strain of S. aureus of Gratia and De Namur described in 1922 (11) but noted that “this derivation is not positively known” (4). This study evaluates phage K for the reduction of pathogenic staphylococci, including a number of coagulase-positive and -negative staphylococci associated with bovine infections and antibiotic-resistant S. aureus associated with human infections.
MATERIALS AND METHODS
Bacterial strains and growth media.
Phage K was purchased from the American Type Culture Collection (ATCC 19685-B1). Staphylococcal strains are listed in Table 1. Strains with the prefix DPC are held in the Dairy Products Research Centre culture collection. Mu3, Mu50, ST3550, ST2573, and 8325 were purchased from the Public Health Laboratory Service (London, United Kingdom). Human MRSA strains were isolated from hospital staff, outpatients, and inpatients from Irish hospitals over a 3-year period and are held at the Cork Institute of Technology (Table 1). Strains were grown at 37°C in brain heart infusion (BHI) broth (Oxoid, Basingstoke, Hampshire, United Kingdom). Solid media contained 1.0% (wt/vol) bacteriological agar (Oxoid). All strains were stocked in BHI containing 40% glycerol and stored at −80°C.
TABLE 1.
Phage K sensitivity and details of bacterial strains
| Host | Strain | Strain detailsi | Methicillin sensitivityh | Phage sensitivityf | EOPg | Phage sensitivity after modification | EOP after phage modification |
|---|---|---|---|---|---|---|---|
| S. aureus | 8325 | Type straina | S | + | NC | ||
| S. aureus | St3550 | Teicoplanin resistanta | S | + | 0.087 | ||
| S. aureus | St2573 | Teicoplanin resistanta | R | + | 0.11 | ||
| S. aureus | Mu50 | VRSAa | R | + | NC | ||
| S. aureus | Mu3 | hVRSAa | R | + | NC | ||
| S. aureus | M249318 | Human MRSAb | R | − | + | 6.75 × 10−5 | |
| S. aureus | W64352 | Human MRSAb | R | − | + | 2.3 × 10−1 | |
| S. aureus | W65216 | Human MRSAb | R | − | + | 2.8 × 10−4 | |
| S. aureus | M231003 | Human MRSAb | R | + | 1.03 × 10−1 | ||
| S. aureus | M249180 | Human MRSAb | R | − | + | 1.09 × 10−3 | |
| S. aureus | MS811 | Human MRSAb | R | − | + | 2.26 × 10−3 | |
| S. aureus | DPC5646 | Human MRSAb | R | + | 0.77 | ||
| S. aureus | DPC5645 | Human MRSAb | R | + | 0.45 | ||
| S. aureus | DPC5647 | Human MRSAb | R | + | 8.46 × 10−7 | ||
| S. aureus | M249954 | Human MRSAc | R | + | 1.12 × 10−1 | ||
| S. aureus | M250594 | Human MRSAc | R | + | 3.23 × 10−1 | ||
| S. aureus | M254959 | Human MRSAc | R | − | + | 7.33 × 10−5 | |
| S. aureus | M255039 | Human MRSAc | R | − | + | 1 | |
| S. aureus | M255409 | Human MRSAc | R | − | + | 7.6 × 10−2 | |
| S. aureus | M253472 | Human MRSAc | R | + | 1 | ||
| S. aureus | M249739 | Human MRSAc | R | + | 1.57 × 10−1 | ||
| S. aureus | M249892 | Human MRSAc | R | + | 4.10 × 10−1 | ||
| S. aureus | M252776 | Human MRSAc | R | + | 1 | ||
| S. aureus | M251760 | Human MRSAc | R | + | 1.32 × 10−1 | ||
| S. aureus | W71683 | Human MRSAc | R | + | 5.89 × 10−2 | ||
| S. aureus | M253206 | Human MRSAc | R | + | 8.57 × 10−2 | ||
| S. aureus | W73365 | Human MRSAc | R | − | + | 3.4 × 10−1 | |
| S. aureus | M253470 | Human MRSAc | R | + | 6.93 × 10−1 | ||
| S. aureus | M249025 | Human MRSAc | R | + | 1.41 × 10−1 | ||
| S. aureus | M249138 | Human MRSAc | R | + | 1 | ||
| S. aureus | M249807 | Human MRSAc | R | + | 1.48 × 10−1 | ||
| S. aureus | M250108 | Human MRSAc | R | + | 7.3 × 10−1 | ||
| S. aureus | M249671 | Human MRSAc | R | − | + | 1.46 × 10−4 | |
| S. aureus | W69939 | Human MRSAc | R | − | + | 1.177 × 10−3 | |
| S. aureus | M253164 | Human MRSAc | R | − | + | 2.65 × 10−2 | |
| S. aureus | M249678 | Human MRSAc | R | − | + | 1.75 × 10−4 | |
| S. aureus | M251955 | Human MRSAc | R | − | + | 2.1 × 10−1 | |
| S. aureus | M250564 | Human MRSAc | R | + | 5.1 × 10−3 | ||
| S. aureus | MM77438 | Human MRSAc | R | + | 2.76 × 10−2 | ||
| S. aureus | MM257671 | Human MRSAc | R | + | 5.46 × 10−3 | ||
| S. aureus | MM234150 | Human MRSAc | R | + | 6.2 × 10−1 | ||
| S. aureus | DPC5245 | Bovined | S | + | 1 | ||
| S. aureus | DPC5246 | Bovined | S | + | 1 | ||
| S. aureus | DPC5247 | Bovined | S | + | 1 | ||
| S. aureus | DPC5971 | Bovined | S | + | 0.21 | ||
| S. epidermidis | DPC6010e | Bovined | S | + | 0.46 | ||
| S. saprophyticus | DPC6011e | Bovined | S | + | 0.025 | ||
| S. chromogenes | DPC6012e | Bovined | S | + | 0.16 | ||
| S. capilis | DPC6013e | Bovined | S | + | NC | ||
| S. hominis | DPC6014e | Bovined | S | + | 2.1 × 10−8 | ||
| S. haemolyticus | DPC6015e | Bovined | S | + | NC | ||
| S. caprae | DPC6016e | Bovined | S | + | 0.022 | ||
| S. hyicus | DPC6017e | Bovined | S | + | 0.087 |
Public Health Laboratory Service.
Cork University Hospital, Cork, Ireland.
Waterford Regional Hospital, Waterford, Ireland.
Dairy Products Centre, Fermoy, County Cork, Ireland.
Coagulase negative.
Spot assay results. +, phage sensitive; −, not phage sensitive.
EOP, efficiency of plating; NC, not countable (plaques are too small to count, but confluent lysis occurs at >107 PFU/ml.
Sensitivity to methicillin at 5 μg/ml. S, sensitive; R, resistant.
VRSA, vancomycin-resistant S. aureus; hVRSA, heterogeneous vancomycin-resistant S. aureus.
Phage propagation.
Phage K was routinely propagated on S. aureus DPC5246 in BHI broth. Concentrated phage K preparations were obtained by CsCl density gradient centrifugation following polyethylene glycol (molecular weight, 8,000) precipitation of phage lysates of BHI cultures. Phage propagation protocols were used as described previously (25). Phage preparations were dialyzed in 10 mM sodium phosphate buffer, pH 7, and filter sterilized prior to use (0.45-μm-pore-size filter). Propagation of phage K on staphylococci which exhibited reduced phage sensitivity was achieved by incubating 100 μl of phage K (approximately 108 PFU/ml) with 20 ml of BHI broth containing a 1% inoculum from an overnight culture of the required host strain. Samples were incubated at 37°C overnight. Samples were then centrifuged, the supernatant was filter sterilized, and phage plaque assays were repeated. Modified phages were named according to the propagating strain.
Electron microscopy.
Phage stocks were prepared from CsCl density gradients to achieve titers in excess of 109 PFU/ml. Each sample was stained negatively with 1% uranyl acetate, and electron micrographs were taken at various magnifications with a JEM EX 1200 electron microscope.
Phage plaque assays.
Phage plaque assays and phage sensitivity tests were performed as described previously (27). Briefly, 50 μl of the appropriate overnight culture, 20 μl of 1 M CaCl2, and 1 ml of the appropriate phage dilution were added to 5 ml of BHI overlay (0.7% agar). The contents were mixed and poured onto BHI plates and incubated at 37°C for 18 h.
Phage host range and bacterial challenge.
Phage K was assessed for its ability to form a clearing on a lawn of each of the staphylococcal strains. The lawn was prepared by adding 50 μl of overnight culture (grown from a 1% inoculum with shaking at 37°C) to a molten 3-ml agar (0.7%) overlay based on BHI medium (Oxoid), which was poured over the BHI plate. After the overlay had solidified, a 10-μl aliquot of phage was spotted onto the surface. Plates were dried and incubated at 37°C for 18 h. Clearing indicated phage sensitivity. Results were confirmed by the plaque assay technique (above). Phage challenge experiments were performed in BHI broth with shaking at 100 rpm at 37°C. Generally, overnight cultures were pregrown in BHI broth and inoculated into BHI broth such that the initial titer was approximately 105 CFU/ml. Phage K was added at a multiplicity of infection (MOI) of 1 after the culture had reached approximately 107 CFU/ml. Samples were then removed and plated in triplicate at regular intervals (the lower limit of detection was 10 CFU/ml). Plates were incubated overnight at 37°C. Plate counts were recorded in triplicate, and standard deviations were determined. Phage titer changes over the course of the challenge were monitored by plaque assay simultaneously.
Determination of the frequency of emergence of BIMs insensitive to phage K.
The frequency of bacteriophage-insensitive mutant (BIM) development was determined as described previously (26). The plaque assay technique was performed with an overnight culture of strain DPC5246 of known titer and phage at an MOI of 1. Plates were incubated overnight at 37°C. Any resulting colonies encountered were counted, and the BIM frequency (number of surviving colonies divided by the original bacterial titer) was determined. All experiments were performed in triplicate.
Antibiotic susceptibility testing.
The methicillin resistance phenotype of the staphylococcal strains was determined by the use of antibiotic susceptibility disks obtained from Oxoid. BHI plates were overlaid with each staphylococcal strain after overnight growth. Antibiotic disks were dispensed onto each plate and, after overnight incubation at 37°C, each plate was scored for antibiotic sensitivity by the Kirby-Bauer plate method (12).
Hand wash studies.
Model hand wash experiments were performed by dipping three human fingers in a suspension of S. aureus DPC5246 which had been diluted from an overnight culture to approximately 105 CFU/ml in quarter-strength Ringer's (Oxoid) solution for 10 s and allowing the bacterium to dry on the skin for 15 min. Finger A was then washed in Ringer's solution containing 108 PFU of phage K/ml for 10 s and allowed to dry. Finger B was washed in phage-free Ringer's solution in an identical fashion. Finger C was not washed. Each of the three fingers was then rinsed in Ringer's solution and thoroughly scraped with a plastic rod for 10 s to recover the material. Staphylococcal plate counts were performed in triplicate immediately after recovery of the material from the skin on each of the three washings.
Phage cream inhibiting staphylococci.
A commercial, oil-based cream containing bismuth subnitrate (Cross Vetpharm Group Ltd., Dublin, Ireland) (34) was combined with phage K at 108 PFU/g. Phage cream was placed in the center of a BHI plate, which was then overlaid with the test staphylococcal strain and incubated overnight at 37°C. The phage cream was also tested in a broth culture of S. aureus. Briefly, 1 g of phage cream was added to 10 ml of broth that contained 106 CFU of DPC5246/ml. A control tube was set up under the same conditions but without the phage cream. Samples and controls were incubated at 37°C, and bacterial-recovery counts were performed by plating 100 μl of dilutions onto BHI agar plates after 4 h.
RESULTS
Phage K exhibits the morphology of the Myoviridae.
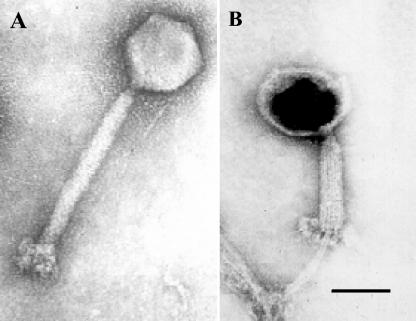
In a previous study we have shown that phage K is a member of a new taxonomic group within the family Myoviridae based on molecular characterization of the similarity between phage genomes (25). The morphology of phage K supports this grouping in that electron microscopy exhibits characteristics of the family Myoviridae. Electron micrographs show that phage K has an isometric head with contractile tail (Fig. 1). Also, the basal tuft of phage K is evident; Fig. 1B clearly shows knob-like appendages extending from the baseplate. In this electron micrograph (Fig. 1B) the tail is contracted, the DNA has been ejected (head is black), and the protruding core of the tail is evident.
FIG. 1.
Electron micrograph images of phage K negatively stained with 1% uranyl acetate. (A) Phage K with a contractile tail. (B) Phage K with tail contracted and empty phage head. Scale bar, 100 nm.
Phage K inhibits recently emerged drug-resistant bacteria.
Phage K does not require the addition of CaCl2 to BHI broth in order to infect, since there was no difference in plaque-forming ability when CaCl2 was omitted from the plaque assays. In addition, increasing the concentration of CaCl2 (0.1, 0.5, 1, 5, 10, and 20 mM) had no effect on plaque-forming ability (data not shown). To test the host range and potency of phage K, bacterial challenge experiments were performed. Details of the bacterial strains are shown in Table 1. These include an S. aureus type strain, 36 human MRSA strains, 4 glycopeptide-resistant strains, 4 distinct clinical isolates from bovine mastitis (10, 38), and 8 coagulase-negative non-S. aureus species of Staphylococcus. The MRSA strains have previously been shown by motif-dependent PCR to be distinct (M. Daly, personal communication; 6). Of the 53 strains, 39 were successfully lysed by phage K, as indicated by phage spot test and confirmed by plaque assay (Table 1). Plaque sizes generally ranged from 1 to 1.5 mm in diameter. Fourteen of the strains from the MRSA group were relatively insensitive to phage K in the initial challenge (Table 1). Plaque formation did not occur with any of these when phage K was used, although there was inhibition in the lawn of bacterial growth, typically at phage concentrations of 108, 107, and 106 PFU/ml, as determined by using the plaque assay technique. This inhibition of growth at the lower dilutions occurred with all the apparently insensitive MRSA strains. When phage K was incubated with these strains in broth, modified phage K variants, which were capable of forming clear plaques on their respective hosts, could be obtained for all of the 14 insensitive strains (Table 1).
In addition, two of the modified phages, namely, phage K.W64352 and phage K.W65216, were assayed for their ability to lyse or cross-react with all the other phage-insensitive strains in the study. Where phage K.W64352 was used, normal plaque formation was evident on the majority of strains but, notably, plaques were pinpoint on strains W65216, M249180, 254959, and M251955, although the plaque numbers (efficiencies of plating) were similar to those for strain DPC5246. These pinpoint plaques suggest that these strains may have phage resistance systems in addition to restriction modification (r/m), which permit only a relatively low burst size. Similarly, when phage K.W65216 was used, plaques were faint and pinpoint on strains W64352 (propagating host for phage K.W64352), M249138, M255409, and W69939. As with phage K.W64352, efficiency of plating on these strains was similar to that for strain DPC5246. This indicates that r/m is the principal cause of the phage insensitivity in the 14 isolates. It also indicates that there is common specificity among the r/m systems harbored by these strains.
Bacterial challenge experiments.
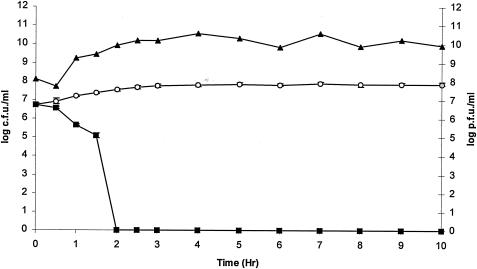
Challenge experiments were performed in BHI broth with MRSA strain DPC5645. This involved adding phage K to an exponential culture of DPC5645 at 37°C. The potency of phage K is illustrated in this challenge experiment (Fig. 2); within 30 min phage K started to reduce bacterial numbers, and within 2 h the phage had reduced the MRSA isolate (DPC5645) from 5.7 × 106 CFU/ml to an undetectable level. Plate counts confirmed that there were no viable bacteria remaining in the BHI broth 10 h after phage treatment, indicating that no BIMs had formed during the challenge (Fig. 2). Indeed, further plate counts 25 h after phage infection confirmed the absence of BIMs in this experiment.
FIG. 2.
Challenge of a MRSA isolate (DPC5645) with phage K at 37°C. Phage was added at an MOI of 1 when the culture reached approximately 107 CFU/ml. ▴, phage titer values. DPC5645 was undetectable in BHI broth 2 h after phage addition. ▪, DPC5645 plus phage K; ○, control DPC5645 without phage K. Values of CFU per milliliter are the means ± standard deviations of the means.
The absence of BIMs from this experiment agreed with the results of a plaque assay procedure to detect and enumerate BIMs, in that, at an MOI of 1, BIM formation on plates did not occur.
Inhibition of S. aureus on skin by phage wash.
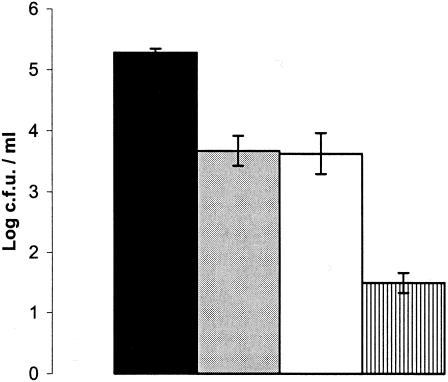
Phage K was assessed for its ability to reduce the numbers of MRSA on human skin with strain DPC5246 as a test strain. These human trials demonstrated that washing in phage-free Ringer's solution was associated with a slight reduction in the number of challenge organisms. When phage K was included in the wash at a titer of 1.4 × 108 PFU/ml, the number of staphylococci remaining was reduced a further 100-fold (Fig. 3). This experiment was performed 10 times, and results were consistent throughout.
FIG. 3.
Graph of phage K hand wash challenge against S. aureus DPC5246. Black bar, bacterial numbers in the original suspension; grey bar, unwashed control; white bar, washed control (washed with Ringer's solution containing no phage); hatched bar, titer after washing in Ringer's solution containing phage. Values shown are the means ± the standard deviations of the means.
Phage inhibition of S. aureus in a bismuth-based cream.
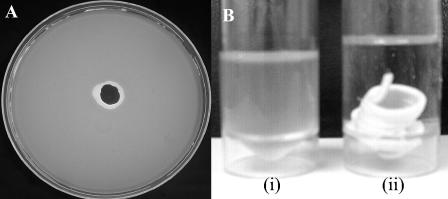
Phage K also exhibited inhibition of the indicator strain DPC5246 in a bismuth-based cream. The phage cream was placed in the center of an overlaid plate with the indicator strain DPC5246. A zone of inhibition is seen surrounding the phage cream (Fig. 4A), which shows that the cream containing phage has killed the surrounding bacteria by bacteriolysis. Importantly, the phage cream was also shown to kill the indicator strain when placed in broth cultures of the challenge organism. Following a 4-h incubation at 37°C, the control sample contained 8 × 107 CFU of DPC5246/ml (Fig. 4B, i) whereas no viable bacteria were detected in the test sample (Fig. 4B, ii), showing complete kill by the phage cream within 4 h.
FIG. 4.
Activity of phage cream. (A) Activity of cream on an agar plate overlaid with the indicator strain DPC5246. (B) Activity of phage cream in BHI broth. (i) Control broth containing DPC5246 and no phage cream; (ii) broth containing DPC5246 and phage cream.
DISCUSSION
With the increased incidence of community-acquired and hospital-acquired drug-resistant staphylococci, the need for new approaches to combat this versatile pathogen is paramount. Phage K is a polyvalent or broad-host-range antistaphylococcal phage. Based on morphology, phage K has previously been assigned to the family Myoviridae, order Claudovirales (1). In this study we demonstrate that phage K inhibits nine different species of Staphylococcus, namely, S. aureus, S. epidermidis, S. saprophyticus, S. chromogenes, S. capitis, S. hominis, S. haemolyticus, S. caprae, and S. hyicus. It is inhibitory to a wide range of distinct S. aureus strains from different hospital sources which were isolated over the last 3 years and also veterinary sources; we feel that these are representative of the problematic strains presently associated with infections in Ireland. Of particular interest is the inhibitory effect on recently emerged methicillin-resistant strains (obtained from hospital staff, outpatients, and inpatients). These studies show that phage K could be modified to hit less-sensitive strains, especially the MRSA strains, with better efficiency simply by passing the phage through the target strain, which ordinarily would not allow plaque formation. Modified derivatives of phage K still produced clear plaques on phage K-sensitive strains, and, in addition, individual modified variants were capable of inhibiting the other strains, which were previously only weakly sensitive to phage K. These data suggest the presence of restriction modification activity in these MRSA isolates and also that there is a large degree of common specificity among the r/m systems harbored by these strains. The modified phages generated in this study could be combined with phage K in a cocktail to increase the host range of the phage preparation.
The bacterial challenge with phage K indicated that no bacteria remained 2 h after phage addition in cases where up to 107 CFU/ml were used. Moreover, no BIMs had emerged 25 h after phage infection. The plate assay confirmed that BIM formation frequency was zero with this phage. Hence we suggest that BIM formation should not compromise the use of phage K to prevent or treat infections. Our laboratory has recently made similar observations with respect to BIM formation in Escherichia coli O157:H7, which again should not compromise the application of phage to combat this pathogen (26).
The mode of application of phage is an area, which warrants study for possible application in the hospital environment. Therefore we designed preliminary experiments exhibiting the potential of phage K for use in a hand wash or cream. Antibacterial activity from the phage cream is evident, and indeed this cream exhibits antibacterial activity for several days at room temperature.
In the context of the hand wash experiment, and in particular the interpretation of the bacterial counts after treatment with phage, it is noteworthy that the killing of bacteria by phage could continue during the washing, recovery, and plating process. Nevertheless, the washing and plating (which involved significant dilution in Ringers solution) were done immediately, and it is likely that the killing rate during this process would not be significantly different from that in a situation where the material was left on the skin. It is noteworthy that washing in Ringers solution in the absence of phage or any other antibacterial agent reduced the titer of staphylococci on skin only slightly. It is also noteworthy that, while the presence of phage reduced the staphylococcal titer on skin, it did not eliminate the bacterium completely. Nevertheless it is apparent from the results that washing hands in the presence of phage K has the potential to significantly reduce the numbers of problematic S. aureus strains that are resident on human hands. As mentioned earlier, additional modified phages could also be incorporated into the hand wash to improve the host range. If applied, this approach would be very likely to have the effect of reducing transmission of staphylococci from hands to patients in a hospital environment.
With regard to the inclusion of phage in the cream, its antibacterial effect is self-evident from the in vitro experiment described. This suggests that such phage creams could find applications in the treatment of local skin infections. Nevertheless, the results on the host range of unmodified phage K described in Table 1 suggest that creams should also contain modified phage to improve their antibacterial activity.
In this study we demonstrate that the exclusively lytic phage K (25) has particular applications in the prevention and/or treatment of infections caused by antibiotic-resistant staphylococci. In this respect, we have shown its ability to kill a broad range of newly isolated pathogenic staphylococci, including both human and veterinary strains. Moreover, the study details some preliminary findings which show the potential of delivering the phage in an antistaphylococcal cream or hand wash.
Acknowledgments
We thank Nina Chanishvili from the Eliava Institute of Bacteriophage, Microbiology and Virology, Tbilisi, Georgia, for electron microscopy of phages.
This research was funded by the Irish Government under the FIRM program as part of National Development Plan 2000-2006, by EU structural funds, and by the Science Foundation of Ireland. Sarah O'Flaherty is in receipt of a Teagasc Walsh Fellowship.
REFERENCES
- 1.Ackermann, H. W., and M. S. DuBow. 1987. Viruses of prokaryotes, vol. 1. CRC Press, Boca Raton, Fla.
- 2.Alisky, J., K. Iczkowski, A. Rapoport, and N. Troitsky. 1998. Bacteriophages show promise as antimicrobial agents. J. Infect. 36:5-15. [DOI] [PubMed] [Google Scholar]
- 3.Breithaupt, H. 1999. The new antibiotics. Nat. Biotechnol. 17:1165-1169. [DOI] [PubMed] [Google Scholar]
- 4.Burnet, F. M., and D. Lush. 1935. The staphylococcal bacteriophages. J. Pathol. Bacteriol. 40:455-469. [Google Scholar]
- 5.Carlton, R. M. 1999. Phage therapy: past history and future prospects. Arch. Immunol. Ther. Exp. 47:267-274. [PubMed] [Google Scholar]
- 6.Cotter, L., M. Daly, P. Greer, B. Cryan, and S. Fanning. 1998. Motif-dependent DNA analysis of a methicillin-resistant Staphylococcus aureus collection. Br. J. Biomed. Sci. 55:99-106. [PubMed] [Google Scholar]
- 7.Deshpande, L. M., T. R. Fritsche, and R. N. Jones. 2004. Molecular epidemiology of selected multidrug-resistant bacteria: a global report from the SENTRY antimicrobial surveillance program. Diagn. Microbiol. Infect. Dis. 49:231-236. [DOI] [PubMed] [Google Scholar]
- 8.Duckworth, D. H., and P. A. Gulig. 2002. Bacteriophages: potential treatment for bacterial infections. BioDrugs 16:57-62. [DOI] [PubMed] [Google Scholar]
- 9.Fischetti, V. A. 2001. Phage antibacterials make a comeback. Nat. Biotechnol. 19:734-735. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald, J. R., W. J. Meaney, P. J. Hartigan, C. J. Smyth, and V. Kapur. 1997. Fine-structure molecular epidemiological analysis of Staphylococcus aureus recovered from cows. Epidemiol. Infect. 119:261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gratia, A., and M. De Namur. 1922. Individualite des principes lytiques staphylococciques de provenances differentes. C. R. Soc. Biol. 87:364-366. [Google Scholar]
- 12.Harley, J. P. 2004. The effects of chemical agents on bacteria. II. Antimicrobial agents (Kirby-Bauer method). McGraw-Hill, New York, N.Y.
- 13.Hiramatsu, K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1:147-155. [DOI] [PubMed] [Google Scholar]
- 14.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9:486-493. [DOI] [PubMed] [Google Scholar]
- 15.Hotchin, J. E. 1954. The purification and electron microscopical examination of the structure of staphylococcal bacteriophage K. J. Gen. Microbiol. 10:250-260. [DOI] [PubMed] [Google Scholar]
- 16.Hotchin, J. E. 1951. Staphylococcus aureus and Staphylococcus K phage. J. Gen. Microbiol. 5:609-618. [DOI] [PubMed] [Google Scholar]
- 17.Hotchin, J. E., I. M. Dawson, and W. J. Elford. 1952. The use of empty bacterial membranes in the study of the adsorption of Staphylococcus K phage upon its host. Br. J. Exp. Pathol. 33:177-182. [Google Scholar]
- 18.Kreuger, A. P., and J. H. Northrop. 1930. The kinetics of the bacterium-bacteriophage reaction. J. Gen. Physiol. 14:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krylov, V. N. 2001. Phage therapy in terms of bacteriophage genetics: hopes, prospects, safety, limitations. Russ. J. Genet. 37:715-730. [PubMed] [Google Scholar]
- 20.Lowy, F. D. 2003. Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Investig. 111:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuzaki, S., M. Yasuda, H. Nishikawa, M. Kuroda, T. Ujihara, T. Shuin, Y. Shen, Z. Jin, S. Fujimoto, M. D. Nasimuzzaman, H. Wakiguchi, S. Sugihara, T. Sugiura, S. Koda, A. Muraoka, and S. Imai. 2003. Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phi MR11. J. Infect. Dis. 187:613-624. [DOI] [PubMed] [Google Scholar]
- 22.Merril, C. R., D. Scholl, and S. L. Adhya. 2003. The prospect for bacteriophage therapy in Western medicine. Nat. Rev. Drug Discov. 2:489-497. [DOI] [PubMed] [Google Scholar]
- 23.Naylor, A. R., P. D. Hayes, and S. Darke. 2001. A prospective audit of complex wound and graft infections in Great Britain and Ireland: the emergence of MRSA. Eur. J. Vasc. Endovasc. Surg. 21:289-294. [DOI] [PubMed] [Google Scholar]
- 24.Noble, W. C. 1998. Staphylococcal diseases, p. 231-256. In L. Collier, A. Balows, and M. Sussman (ed.), Topley and Wilsons microbiology and microbial infections, 9th ed., vol. 3. Oxford University Press, New York, N.Y. [Google Scholar]
- 25.O'Flaherty, S., A. Coffey, R. Edwards, W. Meaney, G. F. Fitzgerald, and R. P. Ross. 2004. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J. Bacteriol. 186:2862-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Flynn, G., R. P. Ross, G. F. Fitzgerald, and A. Coffey. 2004. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl. Environ. Microbiol. 70:3417-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Sullivan, D., D. P. Twomey, A. Coffey, C. Hill, G. F. Fitzgerald, and R. P. Ross. 2000. Novel type I restriction specificities through domain shuffling of HsdS subunits in Lactococcus lactis. Mol. Microbiol. 36:866-875. [DOI] [PubMed] [Google Scholar]
- 28.Rees, P. J., and B. A. Fry. 1981. The morphology of staphylococcal bacteriophage K and DNA metabolism in infected Staphylococcus aureus. J. Gen. Virol. 53:293-307. [DOI] [PubMed] [Google Scholar]
- 29.Rees, P. J., and B. A. Fry. 1981. The replication of bacteriophage K DNA in Staphylococcus aureus. J. Gen. Virol. 55:41-51. [DOI] [PubMed] [Google Scholar]
- 30.Rees, P. J., and B. A. Fry. 1983. Structure and properties of the rapidly sedimenting replicating complex of staphylococcal phage K DNA. J. Gen. Virol. 64:191-198. [DOI] [PubMed] [Google Scholar]
- 31.Rippon, J. E. 1956. The classification of bacteriophages lysing staphylococci. J. Hyg. 54:213-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rountree, P. M. 1949. The serological differentiation of staphylococcal bacteriophages. J. Gen. Microbiol. 3:164-173. [DOI] [PubMed] [Google Scholar]
- 33.Rubin, R. J., C. A. Harrington, A. Poon, K. Dietrich, J. A. Greene, and A. Moiduddin. 1999. The economic impact of Staphylococcus aureus infection in New York City hospitals. Emerg. Infect. Dis. 5:9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryan, M. P., J. Flynn, C. Hill, R. P. Ross, and W. J. Meaney. 1999. The natural food grade inhibitor, lacticin 3147, reduced the incidence of mastitis after experimental challenge with Streptococcus dysgalactiae in nonlactating dairy cows. J. Dairy Sci. 82:2108-2114. [DOI] [PubMed] [Google Scholar]
- 35.Sharp, R. 2001. Bacteriophages: biology and history. J. Chem. Technol. Biotechnol. 76:667-672. [Google Scholar]
- 36.Sulakveidze, A., Z. Alavidze, and J. G. Morris. 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tiemersma, E. W., S. L. A. M. Bronzwaer, O. Lyytikainen, J. E. Degener, P. Schrijnemakers, N. Bruinsma, J. Monen, W. Witte, H. Grundmann, and Eurpoean Antimicrobial Resistance Surveillance System Participants. 2004. Methicillin-resistant Staphylococcus aureus in Europe, 1999-2002. Emerg. Infect. Dis. 10:1627-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Twomey, D. P., A. I. Wheelock, J. Flynn, W. J. Meaney, C. Hill, and R. P. Ross. 2000. Protection against Staphylococcus aureus mastitis in dairy cows using a bismuth-based teat seal containing the bacteriocin, lacticin 3147. J. Dairy Sci. 83:1981-1988. [DOI] [PubMed] [Google Scholar]