Abstract
Campylobacter coli is an infrequently studied but important food-borne pathogen with a wide natural distribution. We investigated its molecular epidemiology by use of amplified fragment length polymorphism (AFLP)-based genotyping and Penner serotyping. Serotype reference strains and 177 Danish isolates of diverse origin identified by routine phenotyping as C. coli were examined. Molecular tools identified some 12% of field isolates as Campylobacter jejuni, emphasizing the need for improved identification methods in routine laboratories. Cluster analysis of AFLP profiles of 174 confirmed C. coli isolates revealed a difference in the distribution of isolates from pig and poultry (chicken, duck, turkey, and ostrich) species and indicated the various poultry species, but not pigs, to be likely sources of human C. coli infection. A poor correlation was observed between serotyping and AFLP profiling, suggesting that the former method has limited value in epidemiological studies of this species.
Campylobacter spp. are the most frequently isolated bacteria in cases of human gastroenteritis in industrialized countries, with the vast majority of reported cases attributed to Campylobacter jejuni (90 to 95%) and Campylobacter coli (5 to 10%) (6, 27). Due to the apparently similar disease histories of the two species (7), the predominance of C. jejuni among cases, and the shortage of good biochemical markers for diagnostics (18), speciation is performed routinely in only a limited number of clinical laboratories. Consequently, most studies exploring the epidemiology of human Campylobacter infections have focused on C. jejuni or have treated C. jejuni-C. coli as one entity.
Clarification of the epidemiology of human campylobacteriosis has been hampered by the ubiquitous distribution of C. coli and C. jejuni in animals, foods, and the environment, as well as the mainly sporadic nature of human infections (6, 27). A recent case-case study has shown differences in exposures associated with human C. coli and C. jejuni infections, suggesting that epidemiological studies should be conducted at the species level to avoid masking and biasing epidemiological information (7). As C. coli is an economically important health burden causing a significant number of hospital bed days (31), the identification of sources and vehicles important for human C. coli infection would provide valuable information for improved public health protection.
Most case-control studies have identified handling or consumption of chicken or poultry as the major factor associated with human Campylobacter infections. However, some case-control studies have suggested that pork may also be an important source of human infection (9, 15, 30). In addition, a case-case study showed that patients infected by C. coli were more likely to have eaten products such as pāté or meat pie than were patients infected by C. jejuni (7). The primary reservoir for C. coli is pigs (ca. 95%), whereas C. coli constitutes only ca. 11% and 1 to 6% of the isolates from chicken and cattle, respectively (16, 17, 36). Thus, it is plausible that a significant proportion of human C. coli infections are related to the consumption of pork rather than chicken. However, in contrast with C. jejuni, only limited typing studies based on heat-stable (HS) (Penner) serotyping, ribotyping, and pulsed-field gel electrophoretyping have been performed on C. coli strains from human infections and diverse animal sources (12, 17, 28). Consequently, the major source(s) of human C. coli infections has not yet been determined, and relatively little information is available regarding the genetic diversity of the species, an essential prerequisite for effective interpretation of genotyping results (33).
Amplified fragment length polymorphism (AFLP) profiling is a technique which has proven useful for speciation and outbreak investigation of the related species of C. jejuni (5, 11, 20). Given the human health significance of C. coli and the paucity of epidemiological and population genetic information on this species, we applied AFLP profiling to (i) investigate the genetic diversity in C. coli, (ii) validate potential sources of human C. coli infections, and (iii) evaluate Penner serotyping as an epidemiological marker for C. coli. We mainly characterized Danish C. coli strains isolated from 1999 to 2001, representing as many sources and Penner serotypes per year as available.
MATERIALS AND METHODS
Isolates.
In Denmark, nationwide surveillance programs for Campylobacter are conducted by the Danish Institute for Food and Veterinary Research (DFVF) and Statens Serum Institut. From this surveillance and individual research projects, animal, food, and clinical isolates previously identified as C. coli by routine diagnostic methods at DFVF (2) were obtained.
Isolates (n = 177) came from all available sources (see Table 1 for details). In general, one or two isolates from all available C. coli serotypes were selected for each source and year (primarily from 1999 to 2001). Four isolates reacting with antisera originally raised against C. jejuni (23) were included.
TABLE 1.
Species identification by AFLP and multiplex PCR of isolates from diverse sources first submitted for study as C. coli
| Source | No. of isolates (%)
|
||
|---|---|---|---|
| Total | C. coli | C. jejuni | |
| Human | 42 | 33 (78.6) | 9 (21.4) |
| Pig | 57 | 57 (100.0) | 0 (0.0) |
| Chicken | 32 | 29 (90.6) | 3 (9.4) |
| Turkey | 19 | 17 (89.5) | 2 (10.5) |
| Ostrich | 8 | 7 (87.5) | 1 (12.5) |
| Duck | 4 | 2 (50.0) | 2 (50.0) |
| Cattle | 4 | 3 (60.0) | 1 (20.0) |
| Sheep | 3 | 2 (66.6) | 1 (33.3) |
| Food or poultry | 6 | 4 (66.6) | 2 (33.3) |
| Dog | 2 | 1 (50.0) | 1 (50.0) |
Penner serotype reference strains (Culture Collection, University of Göteborg, Göteborg, Sweden) for C. coli (six strains isolated from humans, four strains isolated from pigs, one strain isolated from turkeys, two strains isolated from sheep, one strains isolated from marmosets, and five strains of unknown origin) and C. jejuni (35 strains from humans, 11 strains of unknown origin, and 1 strain from a goat) were included in the study.
Penner serotyping.
Serotyping of HS antigens by passive hemagglutination was carried out according to the Penner serotyping scheme (23) as previously described (17). Hippurate hydrolysis-negative strains were tested against the antisera for the 19 C. coli Penner reference strains.
AFLP fingerprinting.
In a total volume of 20 μl, ca. 625 ng of genomic DNA was simultaneously digested with 1 U of MfeI and 1 U of BspDI in NEB4 buffer (New England Biolabs) for 1 h at 37°C. Ligation was performed directly in the restriction digestion by adding 1 U of T4 DNA ligase, 2 μl of 10× T4 DNA ligase buffer (USB Corporation, Cleveland, Ohio), 2 μM FC adaptor, and 20 μM RC adapter complementary to the MfeI and BspDI restriction sites, respectively. Adaptor sequences and preparation were as described by Kokotovic and On (10). The final volume was adjusted to 40 μl, and ligation was performed at 37°C for 3 h. The digestion-ligation was subsequently diluted by the addition of 960 μl of Milli-Q H2O. PCR was performed as previously described (10) with the following exceptions. Five microliters of the diluted digestion-ligation mixture was used per reaction mixture; the PCR primer sequences were MfeI-F, synonymous with BGL2F-0 (10); 5′ GAG AGC TCT TGG AAT TG 3′; 6-carboxylfluorescein labeled at the 5′ end; and BspDI, (5′ GTG TAC TCT AGT CCG AT 3′) (DNA Technology, Århus, Denmark). The number of cycles was reduced from 30 to 25. AFLP fragments were detected with an ABI 377 automated sequencing machine (Amersham Biosciences AB, Uppsala, Sweden) as previously described (26) and processed with GeneScan, version 3.1 (Applied Biosystems, Foster City, Calif.) and BioNumerics, version 2.5 (Applied Maths, Kortrijk, Belgium).
Validation of AFLP protocol.
The robustness of the AFLP method was evaluated by performing the protocol with two separate modifications: (i) the amount of DNA added to digestion-ligation 1× (625 ng) versus 2× (1,250 ng) (eight analyses) and (ii) digestion of genomic DNA (625 ng) with 1× (1 U) as well as 5× (5 U) of the two restriction enzymes (nine analyses). Repeatability was tested by four separate duplicate analyses: (i) different DNA extractions from the same strains (16 analyses), (ii) AFLP-PCR analysis of different digestion-ligation templates from the same strains (14 analyses), (iii) different AFLP-PCR analyses performed on the same strain DNA sample at different times (16 analyses), and (iv) independent samples on random gels (33 analyses). Samples from each individual test were run on the same AFLP gel if not otherwise stated.
Data analysis and optimization.
AFLP fragments (48- to 487-bp size range) in digitized strain profiles were identified by automatic band scoring (7% of minimum peak height relative to maximum value) with subsequent manual editing. Numerical analyses were performed using band alignment parameters of 0.01 (optimization) and 0.04 and 0.2 (start and finish band tolerances, respectively). Ten positions representing bands or band complexes with unsatisfying reproducibility were identified and excluded from profile comparisons. Interstrain relationships were evaluated by calculating pattern similarities with the Dice coefficient and subsequent clustering by the unweighted pair-group method using arithmetic averages (UPGMA) method. All data handling was performed using BioNumerics software, version 2.5 (Applied Maths).
Species validation of atypical isolates.
Campylobacter isolates displaying AFLP profiles atypical for C. coli were subjected to a multiplex PCR for identification of C. jejuni and C. coli (34) under conditions previously described (21). Hippurate hydrolysis was retested by a standardized and previously validated procedure (19). Additional species confirmation of certain strains was performed by the use of a C. coli-specific PCR assay targeting a putative aspartokinase gene, as validated previously (21).
RESULTS
Speciation.
An initial cluster analysis of AFLP patterns from 177 field isolates previously identified as C. coli and 66 Penner serotype reference strains for C. coli and C. jejuni revealed two distinct clusters of isolates. Isolates within each cluster shared a minimum of 35.2% similarity (S) to each other, whereas the two groups clustered together at the 13.3% S level (data not shown). The first cluster comprised 155 field isolates (including 1 isolate assigned to serotype HS 42, originally prepared from a C. jejuni strain) and all 19 C. coli Penner reference strains. The second cluster contained 22 field isolates and all 47 C. jejuni Penner reference strains. C. jejuni- and C. coli-specific multiplex PCR analyses (see Materials and Methods) confirmed that all 22 isolates in cluster two were C. jejuni. Retesting the 22 strains for hippurate hydrolysis activity confirmed that all but 1 strain were hippurate positive, as expected for C. jejuni. Thus, 21% of the human isolates and approximately 10% of the poultry isolates originally identified as C. coli were identified as C. jejuni by AFLP and multiplex PCR analysis (Table 1). In contrast, the outlier strains in the C. coli AFLP cluster analysis (phenons F to H; see below) all yielded the appropriate size amplicon when tested by the C. coli-specific PCR assay.
General features of the AFLP analysis for C. coli isolates.
The mean pairwise similarity among 33 representative duplicates obtained independently throughout the study was 99.56% (standard deviation, 0.86), and the observed range of similarity was 96.35 to 100%.
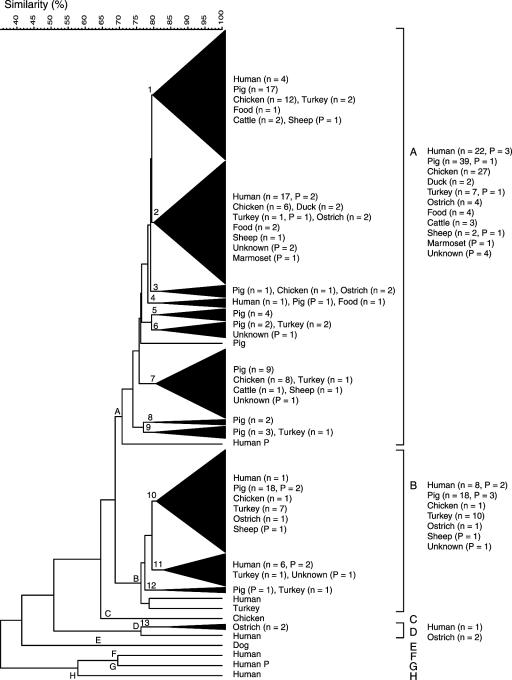
Cluster analysis of all confirmed 174 C. coli isolates identified eight phenons (A to H) at the 70.8% S level with distinct AFLP patterns (Fig. 1). Some 95% of all isolates studied were distributed between phenons A and B, which comprised 121 and 45 isolates, respectively; these phenons clustered together at the 68.8% S level. Phenon D contained one human isolate and two ostrich isolates (of seven studied); phenons C, E, F, G, and H each comprised single strains of human (three isolates), chicken, and canine origin. The number of assigned bands in each AFLP pattern ranged from approximately 33 to 65, with the number of bands per pattern in phenon A approximately 43 and those in phenon B approximately 35. AFLP patterns with more than 50 bands were only observed in phenons F and G.
FIG. 1.
Dendrogram derived from numerical comparison of AFLP profiles of 174 C. coli isolates of human and animal origin, by use of Dice similarity and UPGMA clustering coefficients. Phenons (A to H) are defined at the 70.8% S level, and subphenons (including two or more isolates) (1 to 13, triangulated) at the 79.6% S level. Source and number of field isolates (n), and Penner reference strains (P) in phenons and subphenons are indicated to the right.
At the 79.6% S level, 13 subphenons that were statistically significant by use of jackknife group separation analysis and that comprised characteristic AFLP profiles were defined. The strain compositions of these subphenons were biologically and epidemiologically relevant (discussed below).
AFLP cluster analysis: correlation with source of isolation.
Forty-five (75%) of the 60 isolates from various poultry sources (chickens, ducks, turkeys, and ostriches) appeared in phenon A, with 12 (20%) in phenon B and the remaining 3 (5%) isolates in two other phenons. Most (27 of 29; 93%) of the chicken isolates appeared in phenon A, with only 1 (3%) isolate occurring in phenons B and C (Fig. 1). In contrast, the 18 turkey isolates were almost evenly distributed between phenons A (44%) and B (56%) (8 and 10 isolates, respectively). Furthermore, 40 (65%) of the 61 pig isolates were observed in phenon A, with the remaining 21 (35%) isolates delineated to phenon B. The 39 human isolates were widely distributed and occurred among six phenons, with 64% (25 of 39) and 26% (10 of 39) of the isolates assigned to phenons A and B, respectively, and 10% (4 of 39) among phenons D, F, G, and H (Fig. 1).
Subphenons defined at the 79.6% S level also displayed a skewed distribution of isolates with respect to source. Many of the human isolates fell into two distinct subphenons (one subphenon in each of phenons A and B), and thus were nonrandomly distributed (Fig. 1). These two subphenons (2 and 11) contained no pig isolates, and subphenons including pig isolates contained no or very few human isolates. In contrast, all five subphenons including human isolates included isolates associated with poultry. Among the poultry isolates, 4 of 29 chicken isolates, 2 of 2 duck isolates, 3 of 17 turkey isolates, 3 of 7 ostrich isolates, and 2 of 4 food isolates were most closely related to a human isolate. Only 2 of 57 pig isolates were more similar to human isolates than isolates from any other source. As many as 50% (16 of 33) of the human isolates were most closely related to other human isolates.
Thirty-two isolates were distributed among 11 genotypically identical groups (Fig. 2), i.e., isolates with AFLP profiles that shared or exceeded the minimum 96.3% similarity cutoff level for strain identity, as determined by the reproducibility analysis described above. Six groups included isolates from only one source (i.e., humans, pigs, or turkeys), whereas no group consisted of only chicken isolates. Four groups of isolates were from diverse sources; two of these groups contained a human isolate and a chicken isolate or chicken, duck, and turkey isolates, whereas the two remaining groups contained pig and cattle isolates or sheep and chicken isolates. One human isolate grouped with an isolate of unknown source: both were Penner serotype reference strains.
FIG. 2.
AFLP profiles of the 11 genotypically identical groups of C. coli isolates of human and animal origin. Strain relationships represented in the dendrogram were determined by use of Dice similarity and UPGMA clustering coefficients. Phenons A and B correspond to Fig. 1. Strains: CCUG, Culture Collection University of Gothenburg; SVS, Danish Institute for Food and Veterinary Research, DFVF; TICO, Statens Serum Institut and DFVF.
AFLP analysis and Penner serotype.
The 19 C. coli serotypes were widely distributed in the dendrogram. Fourteen serotypes, each represented by 4 to 20 isolates, were all distributed in more than one phenon (70.8% S level) (Fig. 1). Only five serotypes (HS 14, 28, 29, 39, and 61) represented by three or fewer field and reference isolates were restricted to phenon A, which contained 70% of C. coli isolates studied. Although most serotypes appeared almost randomly distributed in the cluster analysis, a pattern was observed for two serotypes. Serotype 46 (17 of 20) predominanted in subphenon 1 and serotype 48 (12 of 14) predominanted in subphenon 2 (Fig. 1). Among the 11 clusters of genotypically indistinguishable isolates, 7 were characterized by one serotype each (Fig. 2).
DISCUSSION
Species identification and genetic diversity of C. coli.
A prerequisite for performing epidemiological studies of infectious diseases is the ability to speciate accurately. In our study, close to 12% of the isolates previously characterized as C. coli by routine phenotypic testing were reidentified as C. jejuni. This may be due to difficulties in performing the hippurate test described by some workers (4), although our use of a recommended standard procedure (19) revealed only 1 atypical C. jejuni strain from those 22 initially misidentified. Thus, we found ca. 9 and 21% of C. coli isolates from chickens and humans, respectively, were in fact misidentified C. jejuni isolates, compared with 29% for poultry (35) and 24% for humans (32) in previous studies. It should be noted that C. coli may also be misidentified as C. jejuni (29). We caution epidemiologists performing studies at the species level to be aware of the problems with phenotypic identification of the two species, and we recommend the use of molecular identification tools, e.g., quality controlled PCR or hybridization-based tests (21, 32) for future studies.
AFLP profiling is established as a highly discriminatory genotyping method (24), and a correlation between AFLP analysis and results of multilocus sequence typing used for population genetic studies has also been noted for C. jejuni (25). Previous AFLP studies including a limited number of C. coli strains have suggested that the overall genetic diversity of C. coli is considerably smaller than in C. jejuni (3, 5, 20). However, population genetic studies applying multilocus enzyme electrophoresis showed equal diversity between the two species, despite more C. jejuni strains being studied (1, 14). Here, AFLP profiling of 174 C. coli and 69 C. jejuni isolates (including Penner serotype reference strains) clearly suggests the overall genetic diversity of the two species is the same, although a surprisingly high number (95%) of the C. coli isolates fell into two closely related phenons (A and B).
Correlation between AFLP profiling and Penner serotyping.
Serotyping methods were originally developed to facilitate epidemiological studies from a global perspective (22). Such methods are only useful if strains sharing a common set of antigens are also clonally related. The lack of concordance between major clusters defined in this study and heat-stable serotyping results support the observations of a more limited study of C. coli, where a lack of concordance between AFLP and serotyping methods different from those used in our study was also observed (8). In this respect, the results of the present investigation generally resemble those for a study of C. jejuni (26), although the latter study shows a few serotypes of this species share considerable genetic relatedness. The absence of any such correlation for C. coli shows that serotyping is a poor method for long-term epidemiological studies of this species, particularly in view of the cross-reactivity of C. coli sera with C. jejuni strains and vice versa (23), also observed here.
Sources of C. coli infections.
Case control studies of C. jejuni-C. coli frequently point to chicken or poultry as the most important source of Campylobacter infections (15). However, studies from Denmark, Sweden, Norway, The Netherlands, England, and Wales have also pointed to various cuts of pork as well as barbecued meat and sausages as sources of human infection (7, 15). A case-case study implicated food such as pāté, meat pies, and halal meat as more frequently associated with C. coli than C. jejuni infections (7). Since C. coli predominates in pigs (95%) (17, 27) but is uncommon (generally less than 10%) in other food animals such as chicken and cattle, it would be reasonable to presume that pork was the main source of human C. coli infections. However, we studied two to three times as many pig isolates as chicken and turkey isolates, and we found few examples of human isolates that shared only a moderate (i.e., subphenon level) genetic relationship with pig isolates. In fact, isolates from these two sources were largely inversely distributed among subphenons. Our observation that the majority of C. coli isolates from pigs are distinct from human isolates is corroborated by ribotyping data, where nine human isolates were different from 16 pig isolates (12). Thus, since many porcine C. jejuni isolates also appear distinct from human isolates by multilocus sequence typing analysis (13), pork may be an infrequent source of human campylobacteriosis.
In contrast to the results obtained with pig isolates, we were able to identify identical C. coli genotypes among human, chicken, turkey, and duck isolates. While chicken is considered the most important source of human campylobacteriosis, 93% of the chicken isolates in our study were restricted to phenon A, which harbored only 64% of the human isolates examined. These results suggest a significant proportion of human C. coli infections are not attributable to chicken. Indeed, it is noteworthy that a larger proportion of the C. coli isolates from turkeys, ostriches, and ducks were nearer neighbors to human isolates than were chicken isolates, which indicates that other poultry sources may be as important as chicken. Hopkins et al. (8) suggested that chicken and porcine C. coli strains represented host-specific populations, but their results were based upon a more limited AFLP analysis of 87 strains essentially restricted to these two sources (one cattle isolate was included). Our results do not support a clear division of strains with respect to host but do suggest that some strains have a wider host range than others. The marked paucity of chicken isolates (1 of 29) in phenon B may imply that most phenon B strains are unable (or poorly adapted) to colonize or persist in the chicken gut, whereas isolates in phenon A demonstrated no such host limitation and occurred among all food animal sources examined. The uneven distribution of chicken, turkey, and pig isolates among the subphenons in our analysis is also suggestive of differences in the host spectrum among C. coli strains that could prove useful in future risk assessment studies.
Some human strains yielded distinctive AFLP patterns (phenons F to H) (Fig. 1) not observed elsewhere. Stanley et al. (28) examined C. coli by serotyping, ribotyping, flaA typing, and macrorestriction profiling and found human isolates indistinguishable from those from sheep and cattle. The latter two sources, in addition to isolates from domestic pets, ducks, and ostriches, are poorly represented in our study and may account for the absence of any apparent reservoir for our human strains with unique AFLP profiles. Moreover, isolates from environmental sources were not available for study, and human infections could also have been acquired abroad. Since these are known risk factors for campylobacteriosis (6, 7), they could account for the absence of a matching profile among our (principally Danish) strain collection.
Our study indicates a continued need for improvements in routine identification procedures for C. coli and C. jejuni to ensure that the results of epidemiological studies of each species are accurate. Serotyping appears to be a poor epidemiological marker for C. coli. AFLP analysis identifies chickens, ducks, and turkeys as credible sources of human infection. In contrast, most porcine isolates appear genotypically distinct from human isolates, indicating that pigs are an infrequent source of campylobacteriosis. Additional C. coli isolates from cattle, sheep, and other sources should be AFLP typed to improve our understanding of the epidemiology of human C. coli infections.
Acknowledgments
We thank Birgitte Borck and Birthe Hald (DFVF), Niels L. Nielsen (Danish Veterinary and Food Administration), and the DANMAP project for isolates. We are indebted to Kenn Kristiansen, Penelope J. Jordan, Katja A. Kristensen, Sidsel Boelsen, and Sussie Kristoffersen for excellent technical assistance and to Branco Kokotovic for the BspDI PCR primer sequence and valuable discussions.
This work was supported by the Directorate for Food, Fisheries and Agri Business, grant no. FØSI00-SVS-4.
REFERENCES
- 1.Aeschbacher, M., and J.-C. Piffaretti. 1989. Population genetics of human and animal enteric Campylobacter strains. Infect. Immun. 57:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DANMAP 2002. 2003. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, foods and humans in Denmark. [Online.] http://www.dfvf.dk/Files/Filer/Zoonosecentret/Publikationer/Danmap/Danmap_2002.pdf.
- 3.de Boer, P., B. Duim, A. Rigter, J. van der Plas, W. F. Jacobs-Reitsma, and J. A. Wagenaar. 2000. Computer-assisted analysis and epidemiological value of genotyping methods for Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 38:1940-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denis, M., C. Soumet, K. Rivoal, G. Ermel, D. Blivet, G. Salvat, and P. Colin. 1999. Development of a m-PCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett. Appl. Microbiol. 29:406-410. [DOI] [PubMed] [Google Scholar]
- 5.Duim, B., P. A. Vandamme, A. Rigter, S. Laevens, J. R. Dijkstra, and J. A. Wagenaar. 2001. Differentiation of Campylobacter species by AFLP fingerprinting. Microbiology 147:2729-2737. [DOI] [PubMed] [Google Scholar]
- 6.Friedman, C. R., J. Neimann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 7.Gillespie, I. A., S. J. O'Brian, J. A. Frost, G. K. Adak, P. Horby, A. V. Swan, M. J. Painter, K. R. Neal, and Campylobacter Sentinel Surveillance Scheme Collaborators. 2002. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg. Infect. Dis. 8:937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopkins, K. L., M. Desai, J. A. Frost, J. Stanley, and J. M. J. Logan. 2004. Fluorescent amplified fragment length polymorphism genotyping of Campylobacter jejuni and Campylobacter coli strains and its relationship with host specificity, serotyping, and phage typing. J. Clin. Microbiol. 42:229-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapperud, G., E. Skjerve, N. H. Bean, S. M. Ostroff, and J. Lassen. 1992. Risk-factors for sporadic Campylobacter infections: results of a case-control study in southeastern Norway. J. Clin. Microbiol. 30:3117-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kokotovic, B., and S. L. W. On. 1999. High-resolution genomic fingerprinting of Campylobacter jejuni and Campylobacter coli by analysis of amplified fragment length polymorphisms. FEMS Microbiol. Lett. 173:77-84. [DOI] [PubMed] [Google Scholar]
- 11.Lindstedt, B. A., E. Heir, T. Vardund, K. K. Melby, and G. Kapperud. 2000. Comparative fingerprinting analysis of Campylobacter jejuni subsp. jejuni strains by amplified-fragment length polymorphism genotyping. J. Clin. Microbiol. 38:3379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manfreda, G., A. De Cesare, V. Bondioli, and A. Franchini. 2003. Ribotyping characterisation of Campylobacter isolates randomly collected from different sources in Italy. Diagn. Microbiol. Infect. Dis. 47:385-392. [DOI] [PubMed] [Google Scholar]
- 13.Manning, G., C. G. Dowson, M. C. Bagnall, I. H. Ahmed, M. West, and D. G. Newell. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370-6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meinersmann, R. J., C. M. Patton, G. M. Evins, I. K. Wachsmuth, and P. I. Fields. 2002. Genetic diversity and relationships of Campylobacter species and subspecies. Int. J. Syst. Evol. Microbiol. 52:1789-1797. [DOI] [PubMed] [Google Scholar]
- 15.Neimann, J., J. Engberg, K. Mølbak, and H. C. Wegener. 2003. A case-control study of risk factors for sporadic Campylobacter infections in Denmark. Epidemiol. Infect. 130:353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen, E. M. 2002. Occurrence and strain diversity of thermophilic campylobacters in cattle of different age groups in dairy herds. Lett. Appl. Microbiol. 35:85-89. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen, E. M., J. Engberg, and M. Madsen. 1997. Distribution of serotypes of Campylobacter jejuni and C. coli from Danish patients, poultry, cattle and swine. FEMS Immunol. Med. Microbiol. 19:47-56. [DOI] [PubMed] [Google Scholar]
- 18.On, S. L. W. 1996. Identification methods for campylobacters, helicobacters, and related organisms. Clin. Microbiol. Rev. 9:405-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.On, S. L. W., and B. Holmes. 1992. Assessment of enzyme detection tests useful in identification of campylobacteria. J. Clin. Microbiol. 30:746-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.On, S. L. W., and C. S. Harrington. 2000. Identification of taxonomic and epidemiological relationships among Campylobacter species by numerical analysis of AFLP profiles. FEMS Microbiol. Lett. 193:161-169. [DOI] [PubMed] [Google Scholar]
- 21.On, S. L. W., and P. J. Jordan. 2003. Evaluation of 11 PCR assays for species-level Identification of Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 41:330-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patton, C. M., and I. K. Wachsmuth. 1992. Typing schemes: Are current methods useful?, p. 110-128. In I. Nachamkin, M. J. Blaser, and L. S. Tompkins (ed.), Campylobacter jejuni: current status and future trends. ASM Press, Washington, D.C.
- 23.Penner, J. L., J. N. Hennessy, and R. V. Congi. 1983. Serotyping of Campylobacter jejuni and Campylobacter coli on the basis of thermostable antigens. Eur. J. Clin. Microbiol. 2:378-383. [DOI] [PubMed] [Google Scholar]
- 24.Savelkoul, P. H. M., H. J. M. Aarts, J. de Haas, L. Dijkshoorn, B. Duim, M. Otsen, J. L. W. Rademaker, L. Schouls, and J. A. Lenstra. 1999. Amplified-fragment length polymorphism analysis: the state of an art. J. Clin. Microbiol. 37:3083-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schouls, L. M., S. Reulen, B. Duim, J. A. Wagenaar, R. J. L. Willems, K. E. Dingle, F. M. Colles, and J. D. A. Van Embden. 2003. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J. Clin. Microbiol. 41:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siemer, B. L., C. S. Harrington, E. M. Nielsen, B. Borck, N. L. Nielsen, J. Engberg, and S. L. W. On. 2004. Genetic relatedness among Campylobacter jejuni serotyped isolates of diverse origin as determined by numerical analysis of amplified fragment length polymorphism (AFLP) profiles. J. Appl. Microbiol. 96:795-802. [DOI] [PubMed] [Google Scholar]
- 27.Skirrow, M. B. 1994. Diseases due to Campylobacter, Helicobacter and related bacteria. J. Comp. Pathol. 111:113-149. [DOI] [PubMed] [Google Scholar]
- 28.Stanley, J., D. Linton, K. Sutherland, C. Jones, and R. J. Owen. 1995. High-resolution genotyping of Campylobacter coli identifies clones of epidemiologic and evolutionary significance. J. Infect. Dis. 172:1130-1134. [DOI] [PubMed] [Google Scholar]
- 29.Steinhauserova, I., J. Ceskova, K. Fojtikova, and I. Obrovska. 2001. Identification of thermophilic Campylobacter spp. by phenotypic and molecular methods. J. Appl. Microbiol. 90:470-475. [DOI] [PubMed] [Google Scholar]
- 30.Studahl, A., and Y. Andersson. 2000. Risk factors for indigenous Campylobacter infection: a Swedish case-control study. Epidemiol. Infect. 125:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tam, C. C., S. J. O'Brien, G. K. Adak, S. M. Meakins, and J. A. Frost. 2003. Campylobacter coli—an important foodborne pathogen. J. Infect. 47:28-32. [DOI] [PubMed] [Google Scholar]
- 32.Totten, P. A., C. M. Patton, F. C. Tenover, T. J. Barrett, W. E. Stamm, A. G. Steigerwalt, J. Y. Lin, K. K. Holmes, and D. J. Brenner. 1987. Prevalence and characterization of hippurate-negative Campylobacter jejuni in King County, Washington. J. Clin. Microbiol. 25:1747-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Belkum, A., M. Struelens, A. de Visser, H. Verbrugh, and M. Tibayrenc. 2001. Role of genomic typing in taxonomy, evolutionary genetics, and microbial epidemiology. Clin. Microbiol. Rev. 14:547-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandamme, P., L. J. Van Doorn, S. T. Al Rashid, W. G. Quint, V., J. Van der Plas, V. L. Chan, and S.-L. W. On. 1997. Campylobacter hyoilei Alderton et al. 1995 and Campylobacter coli Veron and Chatelain 1973 are subjective synonyms. Int. J. Syst. Bacteriol. 47:1055-1060. [DOI] [PubMed] [Google Scholar]
- 35.Wainø, M., D. D. Bang, M. Lund, S. Nordentoft, J. S. Andersen, K. Pedersen, and M. Madsen. 2003. Identification of campylobacteria isolated from Danish broilers by phenotypic tests and species-specific PCR assays. J. Appl. Microbiol. 95:649-655. [DOI] [PubMed] [Google Scholar]
- 36.Wedderkopp, A., E. Rattenborg, and M. Madsen. 2000. National surveillance of Campylobacter in broilers at slaughter in Denmark in 1998. Avian Dis. 44:993-999. [PubMed] [Google Scholar]