Abstract
A quantitative method based on a real-time PCR assay to enumerate Listeria monocytogenes in biofilms was developed. The specificity for L. monocytogenes of primers targeting the listeriolysin gene was demonstrated using a SYBR Green I real-time PCR assay. The number of L. monocytogenes detected growing in biofilms was 6 × 102 CFU/cm2.
In the food industry, Listeria monocytogenes represents an important health risk. The consumption of food products contaminated with this gram-positive bacterium can cause listeriosis, a serious disease with a 30% mortality rate. Raw material can contain L. monocytogenes, but contamination of food products can also occur during processing. The rapid methods currently available for identifying L. monocytogenes are limited by a threshold of approximately 105 CFU/ml and consequently require enrichment procedures (22). Rapid quantification of L. monocytogenes is important for many reasons, such as in the Hazard Analysis and Critical Control Point method program to verify critical limits and monitor contamination levels and for quantitative risk assessment. L. monocytogenes, like several food-borne pathogens, can attach to the surfaces in contact with the products, leading to the formation of biofilms (12, 14). Adherent bacteria can acquire new physiological properties compared with planktonic cells conferring resistance to disinfectants or sanitizers, leading to hygiene problems, particularly for pathogens that can participate in the recontamination of products (7, 19). Rapid enumeration of L. monocytogenes in biofilms could thus be very useful in the identification of sources of food contamination. DNA-based methods, such as PCR, have been considerably developed to detect food-borne pathogens like L. monocytogenes (1, 6, 8, 21). Real-time PCR has previously been proposed to detect and quantify L. monocytogenes in food products like milk, cheese, and cabbage (10, 13, 18). However, to our knowledge, no study has been reported concerning real-time PCR for the detection and quantification of L. monocytogenes in biofilms.
The aim of this work was to develop a quantitative method to evaluate the population of L. monocytogenes in artificially made biofilms. The real-time PCR can be used to estimate the number of copies of a target gene in a sample and is reported to be more sensitive than conventional qualitative PCR. Real-time PCR is based on the detection and quantification of a fluorescent reporter, whose emission is directly proportional to the quantity of amplicons generated during the PCR. The fluorescent reporter used in this study was SYBR Green I, a nonspecific double-stranded DNA-binding dye. SYBR Green I has the advantage of not requiring the design of specific probes, and its binding is not affected by potential mutations of the target gene. The specificity and sensitivity of the primers used were determined, and then the effectiveness of four methods of extracting DNA from L. monocytogenes growing in biofilms was evaluated. Finally, the detection and quantification of L. monocytogenes in biofilms with the real-time PCR protocol developed in this paper were demonstrated.
Quantification of L. monocytogenes with the SYBR Green I real-time PCR assay.
A region of the L. monocytogenes listeriolysin O gene (hly) was used as a target for PCR amplification. The forward primer (5′-GGGAAATCTGTCTCAGGTGATGT-3′) and the reverse primer (5′-CGATGATTTGAACTTCATCTTTTGC-3′) (13) were used to amplify a 106-bp segment of the hly gene from bp 973 to 1078 (GenBank accession number AF253320). The primers were synthesized by Invitrogen (Cergy Pontoise, France).
The real-time PCR amplification reaction mixture (15 μl) contains a 4.5-μl DNA sample, 1.5 μl of each primer (2.5 μM), and 7.5 μl of QuantiTect SYBR Green PCR Master Mix (Qiagen, Courtaboeuf, France). Before amplification, the PCR mixture was heated to 95°C for 15 min followed by 45 cycles of 15 s at 95°C and then 1 min at 62°C. The final step consisted of a decrease of 0.5°C every 10 s (80 times). The amplification results were visualized and analyzed using the software iCycler provided with the thermocycler (iCycler iQ real-time PCR detection system; Bio-Rad Laboratories, Marnes-la-Coquette, France). Planktonic cultures used for the determination of the specificity of the real-time PCR assay were grown in conditions and medium suitable for each bacterium.
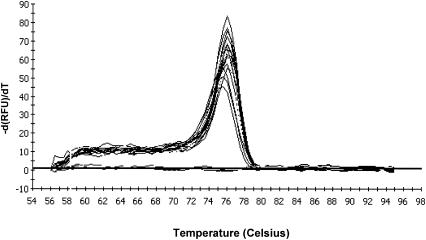
The specificity of the real-time PCR assay was evaluated by testing 22 strains belonging to 18 different species. The culture collection contained five strains of L. monocytogenes. Other strains were tested for their DNA sequence relatedness to L. monocytogenes or their occurrence in meat or fish products (Table 1). Bacterial DNA was extracted using the DNeasy tissue kit (Qiagen) as described below. In order to determine the primer specificity, the hly DNA fragment was amplified using DNA extracted from the 22 bacterial strains as a template (50 ng). Among all the bacteria tested, only L. monocytogenes strains produced a specific amplification reaction (data not shown). To determine the specificity of the real-time PCR assay, a melting-curve analysis was done (Fig. 1). The melting temperature of each product is defined as the temperature at which the corresponding peak maximum occurs. In this real-time PCR assay, the melt curves revealed peaks at a melting temperature of 76°C corresponding to the melting temperature of the specific amplified product. No other product has been detected, neither with melting curves nor after migration of the PCR products on an agarose gel. So there is no interference in the reading of the fluorescence, guaranteeing the specificity of the signal.
TABLE 1.
List of strains used in this study
| Species | Strain designationa |
|---|---|
| Listeria monocytogenes | 157 (serotype 1/2a) (ASEPT) |
| Listeria monocytogenes | 1421 (serotype 1/2a) (ASEPT) |
| Listeria monocytogenes | CIP 78.35 (serotype 3b) |
| Listeria monocytogenes | ATCC 35152 (serotype 1/2a) |
| Listeria monocytogenes | Scott A CIP 103575 (serotype 4a) |
| Listeria innocua | ATCC 33090 (serotype 6a) |
| Listeria welshimeri | ATCC 35897 (serotype 6b) |
| Listeria seeligeri | ATCC 35967 (serotype 1/2b) |
| Listeria ivanovii | ATCC 19119 (serotype 5) |
| Listeria grayi | ATCC 19120 |
| Carnobacterium divergens | ENITIAA V41 |
| Carnobacterium piscicola | ATCC 35586 |
| Lactobacillus sakei | ATCC 15521 |
| Brochothrix thermosphacta | ATCC 11509 |
| Photobacterium phosphoreum | ATCC 11040 |
| Vibrio fischeri | ATCC 14546 |
| Enterococcus faecalis | ATCC 19433 |
| Enterobacter cloacae | ATCC 23355 |
| Staphylococcus aureus | ATCC 25923 |
| Pseudomonas fluorescens | ATCC 13525 |
| Escherichia coli | ATCC 25922 |
| Bacillus subtilis | ATCC 9372 |
ASEPT, Association ASEPT, Laval, France; CIP, Collection de l'Institut Pasteur, Paris, France; ATCC, American Type Culture Collection, Manassas, Va. ENITIAA, Ecole Nationale d’Ingénieurs des Techniques des Industries Agricoles et Alimentaires, Nantes, France.
FIG. 1.
Melt curves corresponding to the amplification of L. monocytogenes DNA to 3.5 × 106 to approximately 35 copies of the genome.
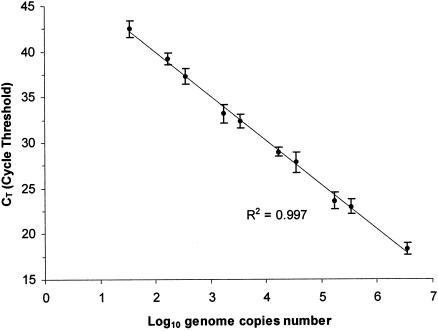
The sensitivity of DNA detection was determined by diluting L. monocytogenes Scott A DNA extracted with the DNeasy tissue kit. DNA was quantified at 260 nm with a UV spectrophotometer, UVIKON XS (Bio-Tek Instruments, Saint Quentin Yvelines, France). Positive amplification results were achieved from eight replicates with 10−8 to 10−13 g of L. monocytogenes DNA corresponding to 3.5 × 106 to approximately 35 copies of the genome, considering that the genome mass of L. monocytogenes is estimated to be 2.9 × 10−15 g (9). No amplification was observed in negative controls containing no DNA template. The relationship between cycle threshold (CT) values and the L. monocytogenes DNA quantities added to the PCR is illustrated in Fig. 2. The CT is defined as the PCR cycle at which the fluorescence signal of a sample rises above the determined baseline signal. The baseline is fixed by the user above the background, and the CT corresponds to the intersection point of each fluorescent curve with the baseline. The relationship was linear over 6 log cycles from 3.5 × 106 to 35 copies of the genome with a correlation coefficient of 0.9972. The standard deviations of the eight determinations were low (Fig. 2). In this assay, 35 copies of genomes (10−13 g of L. monocytogenes DNA) were detected. Previously, Hough et al. (13), using the same primers in a TaqMan real-time PCR protocol, reported the detection of nine genomes of L. monocytogenes. The difference could be explained by the techniques of fluorescence used. Such differences have been observed for the quantification of Staphylococcus aureus (11) with a slightly lower sensitivity with the SYBR Green I than with the TaqMan probe. This was probably due to an accumulation of dimers of oligonucleotide primers and nonspecific products to which SYBR Green I molecules bound.
FIG. 2.
Standard curve representing the detection of L. monocytogenes DNA (number of log genome copies). The means of eight determinations are shown with the corresponding standard deviations.
DNA extraction from L. monocytogenes growing in biofilms.
The procedure used for the preparation of clean stainless steel coupons (CSSC) has previously been described (16) and was slightly modified. Stainless steel coupons (AISI 304 L with two bright annealing finishes; diameter, 12 mm) were sonicated at room temperature in a vessel (Aerosec Industrie, Fécamp, France) for 10 min with a frequency of 28 kHz and a power of 300 W. Coupons were decontaminated in a peracetic acid bath (1%, vol/vol) for 30 min at room temperature, rinsed twice in distilled water for 5 min, and then washed using a 30-min immersion at 50°C with agitation in an 2% alkaline detergent RBS 35 solution (Traitement des surfaces; SARL, Frelinghien, France). Next, the coupons were rinsed with distilled water at 50°C for 30 min with agitation, followed by four further rinses in distilled water at room temperature, before they were dried in a laminar airflow hood and finally autoclaved (121°C, 15 min).
The biofilms were grown according to the procedure previously described (16) by inoculating the CSSC with 100 μl of an L. monocytogenes Scott A preculture (24 h at 30°C) in TSBYE (tryptone soy broth supplemented with yeast extract [6 g/liter]). The inoculated coupons were then incubated for 3 h at 30°C to allow cell adhesion, and then the nonadherent bacteria were removed by rinsing the coupons with 20 ml of peptone saline (tryptone, 1 g/liter; sodium chloride, 8.5 g/liter). To allow growth of adherent cells, 100 μl of sterile TSBYE was deposited on the inoculated coupons and then incubated at 30°C for 24 h. The nonadherent bacteria were removed by a rinse with 20 ml of peptone saline before sonication treatment or extraction directly on the coupons. Cells were detached from the stainless steel coupons by sonication for 4 min at a frequency of 28 kHz and 300 W of power in a flask containing 10 ml of peptone saline containing 0.5% (wt/vol) Tween 80. Epifluorescence microscopy with acridine orange as previously described (17) enabled us to verify that after sonication the cells were detached from the coupons. The viability of the cells was tested by submitting a cell suspension to sonication for different times. Cell enumeration was done before and after sonication; the maximum time for which there was no decrease in CFU number was chosen. Enumeration of L. monocytogenes from the biofilms growing on the coupons was performed by plating the detached cells on polymyxin-acriflavine-LiCl-ceftazidime-esculin-mannitol (PALCAM) agar, a selective medium for detection of L. monocytogenes or TSAYE (tryptone soy agar plus yeast extract [6 g/liter]), a nonselective medium.
The efficiencies of four different methods of DNA extraction from L. monocytogenes cells growing in a biofilm were compared. The extractions were tested either on cells adherent to the coupons or on cells detached from the coupons before extraction. The four methods used were (i) the DNeasy tissue kit with the manufacturer's instructions for gram-positive bacteria in a final volume of 200 μl of elution buffer, (ii) the previously described method with potassium acetate (23), (iii) the phenol chloroform method (3), and (iv) the boiling method as previously described (4). In the last three methods, the DNA was resuspended in a final volume of 190 μl of distilled water and 10 μl of DNase-free RNase (1 mg/ml; Sigma-Aldrich, Saint Quentin Fallavier, France). For the adherent cells, the lysis was performed directly on the coupons. Consequently, the preliminary centrifugation steps were not necessary and excluded from the protocols. For the DNeasy tissue kit, the potassium acetate, and the phenol chloroform methods, the first step of extraction of adherent cells consisted of adding the lysis buffer on the coupons covered by cells. The coupons were incubated at 37°C. Then the lysis suspension was removed from the coupons and placed in a 1.5-ml tube, and the extractions were performed as described for the protocols above. For the extraction of adherent cells by the boiling method, sterile water was added on the surface of the coupons. The coupons were incubated at 100°C and then chilled on ice. The lysis suspension was placed into a 1.5-ml tube, and the extraction was performed as described for the protocol above. In regard to cells detached from the coupons by sonication (nonadherent cells), DNA extractions were performed as described above for the protocols. The effectiveness of the four DNA extraction methods was determined by comparing the CT (defined as the number of cycles after which fluorescence is significantly different from the background) obtained in a real-time PCR assay with 4.5 μl of purified DNA as a template. A Student t test was used to compare the CT of adherent and nonadherent bacteria (Table 2). At the 95% confidence level, the DNA extractions performed using adherent cells gave significantly different results from those with nonadherent cells. Thus, the CT values obtained with adherent cells were lower than those obtained with nonadherent cells. An analysis of variance with Fisher's least significant difference procedure among the four DNA extraction methods with adherent cells (Table 1) showed a statistical difference between the method using phenol chloroform and that using potassium acetate. The DNeasy tissue kit, which gave a low CT for adherent cells, was chosen for its ease of use and its reproducibility in the experiments designed to quantify L. monocytogenes in biofilms.
TABLE 2.
Comparison of the real-time PCR results for DNA extracted from biofilms by four methods
| DNA extraction method |
CT ± SDa
|
P valueb | |
|---|---|---|---|
| Adherent cellsc | Nonadherent cells | ||
| Phenol-chloroform (3) | 19.27(A) ± 0.91 | 25.47 ± 1.70 | 0.001 |
| DNeasy tissue kit (Qiagen) | 20.94(AB) ± 0.81 | 23.53 ± 1.29 | 0.014 |
| Boiling (4) | 21.71(AB) ± 1.46 | 28.75 ± 2.87 | 0.004 |
| Potassium acetate (23) | 22.70(B) ± 2.58 | 31.28 ± 1.77 | 0.001 |
Values are means of four determinations.
Results of a Student t test for comparison of the means between DNA extraction methods for adherent and nonadherent biofilms.
Means with no common letters are statistically different at the 0.05 significance level according to Fisher's least significant difference procedure.
Quantification of L. monocytogenes growing in biofilms.
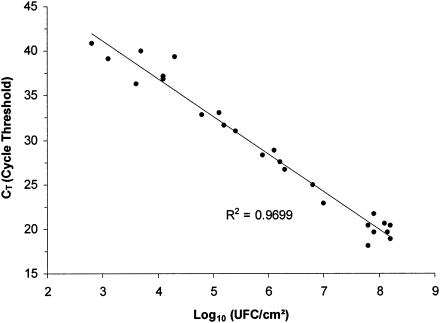
A set of 25 CSSC were inoculated with L. monocytogenes concentrations from 1 to 108 CFU/cm2 and then incubated at 30°C for 24 h. A standard curve was determined using DNA extracted from adherent cells with different L. monocytogenes population levels after 24 h of growth (Fig. 3). This standard curve allowed validation of the real-time PCR assay on 24-h biofilms by showing whether there was a linear relationship between real-time PCR results and enumerations. DNA extraction and enumeration of L. monocytogenes on TSAYE were performed on two different coupons inoculated and incubated in similar conditions. The hly gene fragment of L. monocytogenes DNA extracted from each biofilm was quantified by real-time PCR. The correlation between the CT and the L. monocytogenes enumeration is shown in Fig. 3. A linear regression analysis gave a correlation coefficient of 0.9699. This result revealed a linear relationship between the quantification of L. monocytogenes growing in a biofilm by the real-time PCR protocol and enumeration on an agar plate. A growth of the biofilm population of 1 logarithm corresponded to four cycles of real-time PCR. The sensitivity limit of detection of L. monocytogenes biofilm cells was estimated at 6 × 102 CFU/cm2. However, in practice considering the dispersal of the points, below a concentration of 104 cells per cm2, the detection was more difficult. Considering that only 4.5 μl of the 200 μl extracted was used for the PCR, this assay allowed approximately 14 CFU of L. monocytogenes to be detected per reaction. With the same primers, Hough et al. detected 1.4 × 102 CFU of L. monocytogenes per reaction in 25 g of cabbage. The detection limit of the real-time PCR assay in dairy products was approximately 6 to 60 CFU of L. monocytogenes per reaction (18). The 16S rRNA gene method developed to detect Enterococcus faecalis in water biofilms showed a detection limit of 6 CFU per reaction (20).
FIG. 3.
Standard curve representing CT values obtained with real-time PCR for the different quantities of L. monocytogenes in the biofilms.
In conclusion, to our knowledge, the method developed in this paper is the first real-time PCR approach reported to quantify L. monocytogenes in an artificial biofilm. The real-time PCR assay with SYBR Green I enabled 35 genome copies of planktonic cultured L. monocytogenes to be detected. The DNA extraction method specially designed for biofilms does not require a detachment step for the microorganisms, like sonication, scraping, vortexing, or shaking with beads. When L. monocytogenes bacteria are cultured in a biofilm, 6 × 102 CFU/cm2 can be detected. Other methods have been successfully used for the quantification of bacteria growing in biofilms, such as in situ hybridization (15), microtiter plate assay (5), or confocal scanning laser microscopy (2). Real-time PCR offers the advantages of being quite fast and easy to use.
The technique described in this paper is a first laboratory step to develop a method to study L. monocytogenes biofilms in the food industry where they can constitute a contamination source for food products. To apply this technique to surfaces, further experiments will be necessary, such as testing DNA extraction directly on biofilms sampled from the industry. In our research, real-time PCR will help us to study the behavior of L. monocytogenes in a biofilm in the presence of other bacteria showing inhibitory activity against this pathogen.
Acknowledgments
We thank M. Amgar for welcoming M.G. to the ASEPT company. We also thank P. Courcoux for his support in statistical analyses and are very grateful to C. Cherbut and M. Champ (UFDNH INRA) for allowing access to the real-time PCR thermocycler.
REFERENCES
- 1.Almeida, P. F., and R. C. C. Almeida. 2000. A PCR protocol using inl gene as a target for specific detection of Listeria monocytogenes. Food Control 11:97-101. [Google Scholar]
- 2.Chae, M. S., and H. Schraft. 2000. Comparative evaluation of adhesion and biofilm formation of different Listeria monocytogenes strains. Int. J. Food Microbiol. 62:103-111. [DOI] [PubMed] [Google Scholar]
- 3.de los Reyes-Gavilan, C. G., G. K. Y. Limsowtin, P. Tailliez, L. Sechaud, and J. Accolas. 1992. A Lactobacillus helveticus-specific DNA probe detects restriction fragment length polymorphisms in this species. Appl. Environ. Microbiol. 58:3429-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Medici, D., L. Croci, E. Delibato, S. Di Pasquale, E. Filetici, and L. Toti. 2003. Evaluation of DNA extraction methods for use in combination with SYBR Green I real-time PCR to detect Salmonella enterica serotype Enteritidis in poultry. Appl. Environ. Microbiol. 69:3456-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Djordjevic, D., M. Wiedmann, and L. A. McLandsborough. 2002. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 68:2950-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ericsson, H., and P. Stalhandske. 1997. PCR detection of Listeria monocytogenes in ‘gravad’ rainbow trout. Int. J. Food Microbiol. 35:281-285. [DOI] [PubMed] [Google Scholar]
- 7.Frank, J. F., and R. Koffi. 1990. Surface-adherent growth of Listeria monocytogenes is associated with increased resistance to surfactant sanitizers and heat. J. Food Prot. 53:550-554. [DOI] [PubMed] [Google Scholar]
- 8.Gilot, P., and J. Content. 2002. Specific identification of Listeria welshimeri and Listeria monocytogenes by PCR assays targeting a gene encoding a fibronectin-binding protein. J. Clin. Microbiol. 40:698-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 10.Hein, I., D. Klein, A. Lehner, A. Bubert, E. Brandl, and M. Wagner. 2001. Detection and quantification of the iap gene of Listeria monocytogenes and Listeria innocua by a new real-time quantitative PCR assay. Res. Microbiol. 152:37-46. [DOI] [PubMed] [Google Scholar]
- 11.Hein, I., A. Lehner, P. Rieck, K. Klein, E. Brandl, and M. Wagner. 2001. Comparison of different approaches to quantify Staphylococcus aureus cells by real-time quantitative PCR and application of this technique for examination of cheese. Appl. Environ. Microbiol. 67:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hood, S. K., and E. A. Zottola. 1997. Adherence to stainless steel by foodborne microorganisms during growth in model food systems. Int. J. Food Microbiol. 37:145-153. [DOI] [PubMed] [Google Scholar]
- 13.Hough, A. J., S. A. Harbison, M. G. Savill, L. D. Melton, and G. Fletcher. 2002. Rapid enumeration of Listeria monocytogenes in artificially contaminated cabbage using real-time polymerase chain reaction. J. Food Prot. 65:1329-1332. [DOI] [PubMed] [Google Scholar]
- 14.Kumar, C. G., and S. K. Anand. 1998. Significance of microbial biofilms in food industry: a review. Int. J. Food Microbiol. 42:9-27. [DOI] [PubMed] [Google Scholar]
- 15.La Cono, V., and C. Urzi. 2003. Fluorescent in situ hybridization applied on samples taken with adhesive tape strips. J. Microbiol. Methods 55:65-71. [DOI] [PubMed] [Google Scholar]
- 16.Leriche, V., D. Chassaing, and B. Carpentier. 1999. Behaviour of L. monocytogenes in an artificially made biofilm of a nisin-producing strain of Lactococcus lactis. Int. J. Food Microbiol. 51:169-182. [DOI] [PubMed] [Google Scholar]
- 17.Lunden, J., M. Miettinen, T. Autio, and H. Korkeala. 2000. Persistent Listeria monocytogenes strains show enhanced adherence to food contact surface after short contact times. J. Food Prot. 63:1204-1207. [DOI] [PubMed] [Google Scholar]
- 18.Nogva, H. K., K. Rudi, K. Naterstad, A. Holck, and D. Lillehaug. 2000. Application of 5′-nuclease PCR for quantitative detection of Listeria monocytogenes in pure cultures, water, skim milk, and unpasteurized whole milk. Appl. Environ. Microbiol. 66:4266-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng, J.-S., W.-C. Tsai, and C.-C. Chou. 2002. Inactivation and removal of Bacillus cereus by sanitizer and detergent. Int. J. Food Microbiol. 77:11-18. [DOI] [PubMed] [Google Scholar]
- 20.Santo Domingo, J. W., S. C. Siefring, and R. A. Haugland. 2003. Real-time PCR method to detect Enterococcus faecalis in water. Biotechnol. Lett. 25:261-265. [DOI] [PubMed] [Google Scholar]
- 21.Simon, M. C., D. I. Gray, and N. Cook. 1996. DNA extraction and PCR methods for the detection of Listeria monocytogenes in cold-smoked salmon. Appl. Environ. Microbiol. 62:822-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutherland, P. S., and R. J. Porritt. 1997. Listeria monocytogenes, p. 333-378. In A. D. Hocking, G. Arnold, I. Jenson, K. Newton, and P. Sutherland (ed.), Foodborne microorganisms of public health significance, 5th ed. AIFST (NSW Branch), Sydney, Australia.
- 23.Tudor, J. J., L. Marri, P. J. Piggot, and L. Daneo-Moore. 1990. Size of the Streptococcus mutans GS-5 chromosome as determined by pulsed-field gel electrophoresis. Infect. Immun. 58:838-840. [DOI] [PMC free article] [PubMed] [Google Scholar]