Abstract
Deep-sea mussels of the genus Bathymodiolus (Bivalvia: Mytilidae) harbor symbiotic bacteria in their gills and are among the dominant invertebrate species at cold seeps and hydrothermal vents. An undescribed Bathymodiolus species was collected at a depth of 3,150 m in a newly discovered cold seep area on the southeast Atlantic margin, close to the Zaire channel. Transmission electron microscopy, comparative 16S rRNA analysis, and fluorescence in situ hybridization indicated that this Bathymodiolus sp. lives in a dual symbiosis with sulfide- and methane-oxidizing bacteria. A distinct distribution pattern of the symbiotic bacteria in the gill epithelium was observed, with the thiotrophic symbiont dominating the apical region and the methanotrophic symbiont more abundant in the basal region of the bacteriocytes. No variations in this distribution pattern or in the relative abundances of the two symbionts were observed in mussels collected from three different mussel beds with methane concentrations ranging from 0.7 to 33.7 μM. The 16S rRNA sequence of the methanotrophic symbiont is most closely related to those of known methanotrophic symbionts from other bathymodiolid mussels. Surprisingly, the thiotrophic Bathymodiolus sp. 16S rRNA sequence does not fall into the monophyletic group of sequences from thiotrophic symbionts of all other Bathymodiolus hosts. While these mussel species all come from vents, this study describes the first thiotrophic sequence from a seep mussel and shows that it is most closely related (99% sequence identity) to an environmental clone sequence obtained from a hydrothermal plume near Japan.
Symbiotic associations with thiotrophic (sulfur-oxidizing) and methanotrophic (methane-oxidizing) bacteria occur in a wide array of animal species that live in reducing environments with high sulfide and methane concentrations, such as hydrothermal vents, whale skeletons, sunken wood, and cold seeps (4, 6, 11, 13, 15, 40). Cold seeps occur worldwide on both active and passive margins (39), and these ecosystems harbor a high proportion of invertebrates associated with symbiotic bacteria (34), including highly specialized annelids (Siboglinidae), as well as bivalve clams (Thyasiridae, Vesicomyidae, Lucinidae) and bathymodiolid mussels (Mytilidae).
Mussels of the genus Bathymodiolus are found worldwide in vents and seeps at depths from 400 to 3,600 m (41). The bacterial symbionts occur in specialized cells of the gill called bacteriocytes (13, 15) and have been characterized in about 10 of the 22 known species by using transmission electron microscopy (TEM), stable isotopes, enzymology, and molecular analyses (5, 7, 10, 14, 16, 30, 31, 37). Some species, like Bathymodiolus thermophilus from east Pacific vents, harbor only thiotrophic bacteria, while others, like Bathymodiolus childressii from the Gulf of Mexico, have only methanotrophic symbionts (6). A dual symbiosis, in which a single host harbors both thiotrophic and methanotrophic bacteria, has been described for four species, two from cold seeps in the Gulf of Mexico (Bathymodiolus brooksii and Bathymodiolus heckerae) (5, 16) and two from vents along the Mid-Atlantic Ridge (Bathymodiolus azoricus and Bathymodiolus puteoserpentis) (10, 14). The following data have been presented as evidence for the presence of dual symbionts in these species. Two distinct morphotypes have been shown to cooccur within the bacteriocytes by using TEM (5, 10, 14, 16). Enzyme assays and immunohistochemistry analyses have confirmed the presence of enzymes used by thiotrophic and methylotrophic bacteria (5, 14, 16, 30). Stable isotope analyses of gill tissues (5, 7, 16, 37) and lipid biomarkers (31) indicated that methanotrophy and thiotrophy are sources of nutrition, with filter feeding a possible further source, as a functional gut is still present in most bathymodiolids (29). Phylogenetic evidence for dual symbiosis so far only exists for B. puteoserpentis from the Mid-Atlantic Ridge (10). By using comparative 16S rRNA sequence analysis and fluorescence in situ hybridization (FISH), two distinct γ-proteobacterial phylotypes were shown to coexist within the host bacteriocytes, and the thiotrophic and methanotrophic symbionts were most closely related to the symbionts of mussels harboring only a single symbiont phylotype (10).
While symbiotic invertebrates obtained from vent and seep sites in the north Atlantic have been described, symbioses of vent or seep invertebrates from the south Atlantic have not been described previously. The discovery of a large active pockmark area (depth, 3,150 m) on the Gabon margin (southeast Atlantic) (28) provided the opportunity to study a possible new Bathymodiolus species (R. von Cosel, personal communication). Mussels up to 175 mm long form dense beds and dominate the macrofaunal community at this site (3). In this study, the morphology of symbiotic bacteria in this Bathymodiolus sp. was investigated by using TEM, the identities and phylogenetic relationships of the bacteria were determined by comparative 16S rRNA analysis, and the distribution of the bacteria was characterized by FISH. Measurement of methane and sulfide concentrations within mussel beds prior to specimen collection provided an ecological basis for comparing different habitats.
MATERIALS AND METHODS
Sampling and storage.
Bathymodiolus sp. individuals were collected with the remotely operated vehicle VICTOR 6000 during the Biozaire 2 cruise (2001; IFREMER; sponsored by Total; chief scientist, Myriam Sibuet) to the Congo-Angola-Gabon margin, close to the Zaire channel (equatorial east Atlantic). Specimens were collected at a depth of 3,150 m from three mussel beds (mussel beds M1, M2 and M3) in a pockmark area called Régab (05°52.8134′S, 009°37.9419′E). Mussel beds M2 and M3 were 30 m from each other, while M1 was about 130 m from both M2 and M3. Specimens were collected at the center of mussel beds M1 to M3 and at the periphery of mussel bed M3. All specimens were prepared in the following manner. One gill of each mussel was fixed for transmission electron microscopy (see below). The other gill was divided into two parts, one of which was immediately frozen in liquid nitrogen and the other of which was fixed for FISH (see below).
Methane and sulfide measurements.
Seawater samples for determination of methane and sulfide concentrations were collected a few minutes before mussel samples were collected from the center of each of the three mussel beds (mussel beds M1, M2, and M3) and at the periphery of mussel bed M3. At each of these four collection sites, two replicate samples were taken at the bottom of the mussel bed with a syringe inserted into the bed and positioned 2 to 3 cm above the seafloor, and two replicate samples were taken just above the top of the mussel bed, where the mussels were surrounded by seawater. Methane concentrations were measured by gas chromatography by using headspace injection (33), and sulfide concentrations were determined photometrically by the method described by Fonselius (19).
Transmission electron microscopy.
Gill pieces from eight individuals were fixed in 3% glutaraldehyde in 0.4 M NaCl buffered with 0.1 M cacodylate (pH 7.4) for 2 h and were postfixed in 1% osmium tetroxide for 1 h in the same buffer. Fixed gills were dehydrated in a graded ethanol series and embedded in Araldite. Semithin sections were stained with toluidine blue. Ultrathin sections were contrasted with uranyl acetate and lead citrate and examined with an HITACHI H-7500 transmission electron microscope.
DNA extraction.
DNA was extracted individually from gill tissues of three mussels, one from each site, by the method described by Zhou et al. (44) by using proteinase K for cell digestion and a standard chloroform-isoamyl alcohol extraction procedure. DNA was precipitated in isopropanol, washed with ethanol, resuspended in sterile-filtered water, and stored in aliquots at −20°C.
16S rRNA PCR amplification.
Bacterial 16S rRNA was amplified from gill tissue DNA by using the universal bacterial primers 8F and 1492R (26). The reaction mixture contained 50 pmol of each primer, 5 μg of bovine serum albumin, 2.5 μmol of each deoxynucleoside triphosphate, 1× ExTaq buffer, and 1 U of Taq polymerase (TaKaRa, Otsu, Japan), and the volume was adjusted with sterile water to 50 μl. An initial denaturation step (96°C for 5 min) was followed by 25 cycles of 94°C for 1 min, 45°C for 1 min, and 72°C for 3 min and a final elongation step at 72°C for 5 min. PCR bias was minimized by using only 25 amplification cycles (32) and pooling four separate PCRs for each mussel. Amplified DNA was purified with a QIAquick PCR purification kit (QIAGEN, Hilden, Germany).
Cloning and sequencing.
PCR products of the correct size (∼1,500 bp) were cloned with a TOPO-TA kit (Invitrogen, Carlsbad, Calif.). A 16S rRNA clone library was constructed for each of the three mussels. The insert size of white Escherichia coli colonies was controlled after lysis of the cells in cracking buffer (0.1 M NaOH, 10 mM EDTA, 1% sodium dodecyl sulfate [SDS], 10% glycerol) and analysis of the supernatant by gel electrophoresis. Positive clones were grown overnight in 1.5 ml of Luria-Bertani medium, and plasmids were prepared from the pelleted cells with a QIAprep Miniprep kit (QIAGEN). For each of the three individuals, 51 to 59 clones were sequenced partially (∼500 bp) in a variable region of the 16S rRNA (E. coli positions 518 to ∼1,000). After alignment with BioEdit (22), manual correction, elimination of three chimeras by using ChimeraCheck (8), and visual examination of the alignments, seven representative clones were fully sequenced in both directions (Table 1). Sequencing reactions were performed by using ABI BigDye and an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, Calif.).
TABLE 1.
Numbers of partial sequences (from nucleotide 518 to nucleotide ∼1,000 based on E. coli numbering) and nearly full sequences (∼1,500 nucleotides) obtained from cloned PCR products amplified from gill DNA of three Bathymodiolus sp. specimens
| Specimen source | No. of partial sequences
|
No. of full sequences
|
|||||
|---|---|---|---|---|---|---|---|
| Thiotrophs | Methanotrophs | Chimeras | Total | Thiotrophs | Methanotrophs | Total | |
| M1 | 40 | 19 | 0 | 59 | 1 | 2 | 3 |
| M2 | 57 | 0 | 1 | 58 | 2 | 0 | 2 |
| M3 | 39 | 10 | 2 | 51 | 1 | 1 | 2 |
| Total | 136 | 29 | 3 | 168 | 4 | 3 | 7 |
Phylogenetic reconstruction.
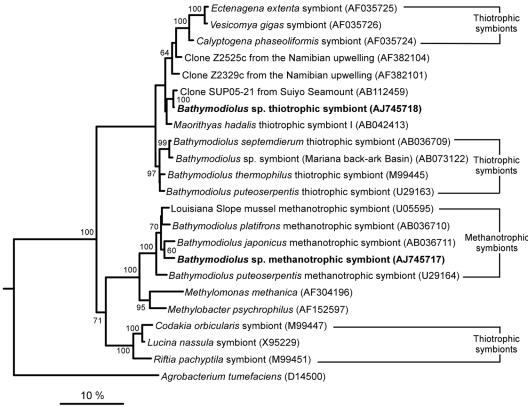
Sequences were compared with the database by using BLAST (1), and highly similar sequences were included in the analysis. Sequences were aligned with ARB (25) and were manually corrected. Preliminary analyses of the seven full sequences were performed by distance and parsimony methods by using the PHYLIP package (12) to select an appropriate data set (choice and number of sequences and positions). For the selected data set 36 sequences were used, but only 23 of these sequences are shown in Fig. 2 (the accession numbers of the sequences not shown in Fig. 2 are AF035728, AF035723, AF035727, AF035721, AB044744, X72767, AY029915, L34955, U77481, U62131, L40809, AF069959, and U11021). The 36 sequences were analyzed by the maximum-likelihood method with TREEFINDER (23) by using a general time reversible model and an eight-category discrete approximation of a Γ distribution (estimated α, 0.209) to account for among-site evolutionary rate heterogeneity. Maximum-likelihood bootstrap values were obtained from 1,000 replicates analyzed by the same method.
FIG. 2.
Phylogenetic relationships, based on maximum-likelihood analyses of 16S rRNA sequences, of the thiotrophic and methanotrophic endosymbionts of Bathymodiolus sp. (boldface type) in the γ-Proteobacteria (1,266 sites analyzed; L = −8,744). Two α-proteobacterial species were used as an outgroup (only A. tumefaciens is shown). Bootstrap percentages were obtained by using 1,000 maximum-likelihood replicates, and values greater than 60% are indicated at the nodes. Scale bar = 10% estimated base substitutions.
FISH.
Two mussels from each collection site (center of mussel beds M1, M2, and M3 and periphery of mussel bed M3) were fixed for FISH in 2% formaldehyde in sterile seawater at 4°C for 2 h. After two washes in 1× phosphate-buffered saline (10 mM sodium phosphate, 130 mM NaCl), samples were stored at −20°C in 0.5× phosphate-buffered saline-50% ethanol (1:1). Fixed gill fragments were dehydrated in an increasing ethanol series and xylene before they were embedded in paraffin. Transverse sections (thickness, 4 μm) were cut with an RM 2165 microtome (Leica, Wetzlar, Germany) and collected on Superfrost slides (Fisher, Pittsburgh, Pa.) coated with 3-aminopropyltriethyloxysilane. Paraffin was removed from the sections with xylene (three 10-min treatments), and the sections were rehydrated in a decreasing ethanol series, permeabilized for 10 min in 0.2 M HCl, rinsed for 10 min in 20 mM Tris buffer (Tris-HCl, pH 8), permeabilized again for 5 min at 37°C in Tris buffer containing 0.5 μg of proteinase K ml−1, and rinsed for 10 min in 20 mM Tris buffer. After air drying, the sections were circled with a Pap Pen (G. Kisker, Mühlhausen, Germany). Hybridization was performed in preheated chambers containing tissue wetted with hybridization buffer (0.9 M NaCl, 0.02 M Tris-HCl, 0.01% SDS, 30% formamide). The circled sections were covered with a mixture containing 100 ng of probe in 30 μl of hybridization buffer and incubated for 3 h at 46°C. The slides were washed in a buffer (0.1 M NaCl, 0.02 M Tris-HCl, 0.0001% SDS, 5 mM EDTA) at 48°C for 15 min, rinsed with MilliQ water, air dried, and covered with VectaShield (Vector, Burlingame, Calif.) and a coverslip. A total of 43 gill sections obtained from eight mussels collected from the four different sites (Table 2) were visually examined for differences in relative symbiont abundance by using an LSM 510 confocal microscope (Zeiss, Jena, Germany).
TABLE 2.
Methane and sulfide concentrations in the water collected at the bottom and top of three mussel beds and numbers of individuals and gill filament sections examined at each sampling site by fluorescence in situ hybridization
| Mussel bed | Concn (μM) ofa:
|
No. of specimens | Total no. of sections | |||
|---|---|---|---|---|---|---|
| CH4 (bottom) | CH4 (top) | H2S (bottom) | H2S (top) | |||
| M1 (center) | 0.7 | 1.6 | <1 | <1 | 2 | 10 |
| M2 (center) | 33.7 | 23.2 | <1 | <1 | 2 | 5 |
| M3 (center) | 23.7 | 12.5 | <1 | <1 | 2 | 4 |
| M3 (periphery) | 11.8 | 6.3 | <1 | <1 | 2 | 24 |
The detection limit for methane was 50 nM, and the detection limit for sulfide was 1 μM.
A specific probe was designed for each of the two 16S rRNA phylotypes found in Bathymodiolus sp. by using the PROBE_DESIGN function of ARB, and the specificity was checked by using BLAST. The probe specific for the methanotrophic symbiont, BangM-138 (5′-ACCAGGTTGTCCCCCACTAA-3′), was labeled with Cy3 (Thermo, Waltham, Mass.), and the probe specific for the thiotrophic symbiont, BangT-642 (5′-CCTATACTCTAGCTTGCCAG-3′), was labeled with Cy5 (Biomers.net, Ulm, Germany). Gills of the dual-symbiont mussel B. azoricus were used to test the specificity, as the thiotrophic and methanotrophic symbionts of this mussel have a 1-bp mismatch in the 16S rRNA region targeted by the probes designed for this study. Both the BangM-138 probe and the BangT-642 probe were highly specific with 30% formamide, as no signal was observed in B. azoricus gill sections, while a strong signal was visible in Bathymodiolus sp. gill sections. The general Bacteria probe EUB338 (2) was used as a positive control, and the NON338 (42) probe was used as a negative control (both were labeled with Cy3).
Nucleotide sequence accession numbers.
The EMBL accession numbers for the two sequences described in this paper are AJ745717 (methanotrophic symbiont) and AJ745718 (thiotrophic symbiont).
RESULTS
Methane and sulfide concentrations in the mussel beds.
The methane concentrations measured at the bottom of mussel beds M2 and M3 close to the seafloor were in the same range (33.7 and 23.7 μM, respectively) and decreased at the top of the beds by 30% at mussel bed M2 and by 47% at mussel bed M3 (Table 2). At the bottom of the M1 mussel bed, the methane concentrations were 33- to 48-fold lower (0.7 μM) than they were at the bottom of the two other beds, and the concentration increased slightly at the top of mussel bed M1 to 1.6 μM. At the periphery of the M3 mussel bed, the methane concentrations both at the bottom and at the top of the bed were 50% lower than the concentration at the center of this mussel bed (Table 2). The sulfide concentrations were below the detection limit (1 μM) at all sampling sites. No obvious differences in mussel biomass were observed for mussel beds M1, M2, and M3.
Transmission electron microscopy.
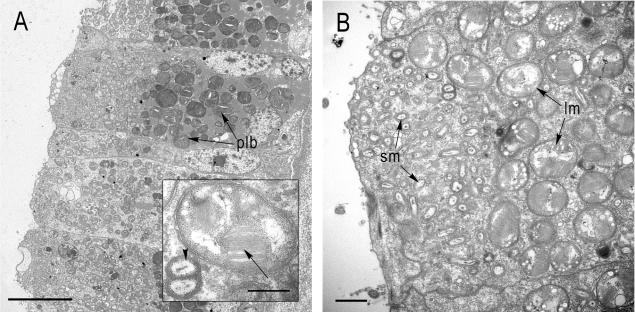
Bacteria were abundant in the apical half of the gill bacteriocytes, and numerous phagolysosome-like bodies occurred in the basal region of the cells (Fig. 1A). Two distinct bacterial morphotypes coexisted within each bacteriocyte (Fig. 1B). The smaller morphotype (0.42 ± 0.07 by 0.33 ± 0.06 μm) was a rod-shaped or coccoid bacterium, and the larger morphotype (1.23 ± 0.16 by 1.07 ± 0.10 μm) was coccoid with stacked membranes in its cytoplasm. In almost all bacteriocytes examined, the smaller morphotype was most abundant in the apical region of the cell (Fig. 1B), while the larger morphotype occurred more basally.
FIG. 1.
Transmission electron micrographs of Bathymodiolus sp. gill sections with endosymbiotic bacteria. (A) Transverse section showing an overview of a bacteriocyte (plb, phagolysosome-like bodies). Scale bar = 10 μm. (Inset) Large morphotype with stacked internal membranes (arrow) and a dividing stage of the small morphotype (arrowhead). Scale bar = 0.5 μm. (B) Apical part of a bacteriocyte showing the distinct distribution of the two morphotypes. The smaller morphotype (sm) occupies the apical part of the bacteriocyte toward the mantle fluid, while the larger morphotype (lm) is located more basally. Scale bar = 1 μm.
16S rRNA phylogeny of the symbionts.
Comparative 16S rRNA sequence analysis of 165 clones from three Bathymodiolus sp. individuals revealed two distinct bacterial phylotypes (Table 1). The sequence variation within each phylotype was very low (0 to 0.2%). Phylogenetic trees constructed from a variety of data sets and by using a variety of methods displayed topologies almost identical to that of the maximum-likelihood tree shown in Fig. 2. Both phylotypes clustered in the γ-Proteobacteria subdivision. One phylotype belongs to a monophyletic group that includes all known sequences of methanotrophic symbionts associated with bathymodiolid hosts (from the Gulf of Mexico, the Mid-Atlantic Ridge, and Japan). Its closest relative (97.8% identity) is the methanotrophic symbiont of Bathymodiolus japonicus from vents in the Okinawa Trough, near Japan (21).
The second Bathymodiolus sp. phylotype belongs to a large clade that includes thiotrophic symbionts associated with bivalves belonging to three families (Vesicomyidae, Thyasiridae, and Mytilidae), as well as three environmental sequences, clones ZA2525c and ZA2329c from an upwelling zone off the coast of Namibia and clone SUP05 from the Suiyo Seamount hydrothermal plume, south of Japan (36). The second Bathymodiolus sp. phylotype is most closely related to the SUP05 clone (99.3% identity). Among symbiotic bacteria, its sequence is more similar to the sequence of symbiont I from the thyasirid Maorithyas hadalis (97.8%) (20) than to any of the four previously published sequences for thiotrophic symbionts of bathymodiolid mussels (<97.4%). As determined by all three treeing methods, this sequence never fell within the well-supported monophyletic group of sequences from thiotrophic mussel symbionts (maximum-likelihood bootstrap value, 97.0). Instead, it formed a weakly supported monophyletic group with clone SUP05, M. hadalis symbiont I, and the symbionts of vesicomyids (maximum-likelihood bootstrap value, <60). Therefore, except for its closest relative, clone SUP05, the exact phylogenetic position of the second Bathymodiolus sp. phylotype remains unclear.
FISH.
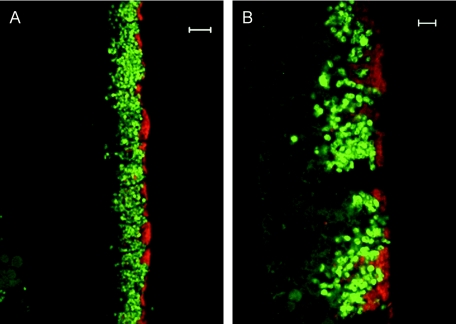
Hybridization with gill tissue sections by using specific probes confirmed the coexistence of the two symbionts in the gill bacteriocytes (Fig. 3). The probe specific for the methanotroph-related sequence (BangM-138) hybridized to large coccoid bacteria assumed to be the large morphotype observed by TEM. The probe specific for the thiotroph-related sequence (BangT-642) hybridized to bacteria similar in size and shape to the smaller TEM morphotype.
FIG. 3.
Fluorescence in situ hybridization images of Bathymodiolus sp. symbionts in gill filaments. The thiotrophic symbionts (red) occupy the apical region of the bacteriocytes, while the methanotrophic symbionts (green) are located more basally. (A) Scale bar = 10 μm. (B) Scale bar = 5 μm.
A nonquantitative, visual comparison of gill sections from two individuals collected from each of the mussel beds (center of mussel beds M1, M2, and M3 and periphery of mussel bed M3) revealed a distinct distribution of the two symbiont phylotypes in most sections (34 of 43 sections examined) (Table 1). The methanotroph-related symbionts occurred in the more basal region of the bacteriocytes, while the thiotroph-related symbionts consistently occupied the apical end of the bacteriocytes exposed to the environment (Fig. 3B). The most apical part of the bacteriocytes (width, 1 to 3 μm) was often almost exclusively occupied by the thiotroph-related symbionts. No obvious differences in the distribution and relative amounts of the thiotroph- and methanotroph-related symbionts were observed among gill sections of Bathymodiolus sp. collected from mussel beds M1 to M3.
DISCUSSION
The Bathymodiolus sp. mussels studied here from the Gabon margin harbor two morphologically distinct bacteria that resemble the methanotrophic (5, 14, 16) and thiotrophic (13, 17) symbionts of other Bathymodiolus species. Comparative 16S rRNA sequence analysis and FISH confirmed that Bathymodiolus sp. lives in a dual symbiosis with bacteria related to methane- and sulfide-oxidizing symbionts.
As expected, the Bathymodiolus sp. methanotroph 16S rRNA sequence falls within the monophyletic clade that includes all known methanotrophic symbionts of bathymodiolid mussels (9, 10, 21). In contrast, the phylogenetic position of the Bathymodiolus sp. thiotroph 16S rRNA sequence is surprising, because the sequence does not fall in the monophyletic group of thiotrophic symbionts from all other Bathymodiolus hosts (9, 10, 21). This monophyletic group consists exclusively of thiotrophic symbionts from hydrothermal vent mussels (from the Mid-Atlantic Ridge, the East Pacific Rise, and the western Pacific), while the thiotroph sequence obtained in this study is the first thiotroph sequence from a cold-seep mussel. A further unexpected result is the close relationship of the Bathymodiolus sp. thiotrophic symbiont to the environmental clone sequence SUP05 (36) and the placement of two environmental clone sequences obtained from surface waters off the Namibian coast (accession no. AF382104 and AF382101) (Fig. 2) in the Bathymodiolus-Vesicomya/Calyptogena clade. In all previous analyses this clade consisted exclusively of clam and mussel symbionts (9, 10, 20, 21).
One explanation for the lack of monophyly for vent and seep mussel thiotrophs is that the symbioses may have evolved independently of each other. It is intriguing that in a recent study of vent mussels from the Mid-Atlantic Ridge acquisition of the thiotrophic symbiont from the environment was suggested (43). Uptake of a free-living bacterial species from the environment is one way in which these associations could have developed independently during convergent evolution. However, environmental symbiont transmission is not necessarily a good indication of convergent evolution as some symbioses in which environmental transmission occurs have clearly been established through cospeciation, like the symbiosis between luminescent Vibrio bacteria and squid (27).
An alternative explanation for the lack of monophyly for the thiotrophic symbionts of Bathymodiolus sp. and the thiotrophic symbionts of vent mussels is that there is indeed a sister group relationship between them, but sequences that could allow resolution of this relationship are not yet known. In this case a single acquisition event would have occurred, with consequent speciation leading to separation of vent and seep mussels, followed by diversification in the two environments. Clearly, sequences from additional mussel species and related free-living bacteria are needed to fully resolve the evolutionary history of Bathymodiolus symbioses.
Ecology of the Bathymodiolus sp. symbiosis.
Sulfide and methane gradients over time and space are assumed to play a major role in determining the distribution, biomass, and productivity of symbiotic invertebrates at vents and seeps (18, 34, 35). However, little is known about how variations in these energy sources affect the nutrition of hosts living in dual symbioses with sulfide- and methane-oxidizing bacteria. Previous studies of Mid-Atlantic Ridge mussels with dual symbionts indicated that there is a nutritional response to fluid gradients, with an increase in the relative amounts of sulfide oxidizers and reliance on thiotrophy in mussels from sites with higher sulfide concentrations and, correspondingly, an increase in the relative amounts of methane oxidizers and reliance on methanotrophy in mussels exposed to higher methane concentrations (7, 37). In this study, no variations in the relative amounts of thiotrophic and methanotrophic symbionts were observed in Bathymodiolus sp., despite a nearly 50-fold difference in methane concentrations between sample sites. One explanation for this unexpected result is that even at the lowest concentration (0.7 μM), methane may not be limiting for the growth of the methanotrophic symbionts. Alternatively, the nonquantitive methods used here may not have been sufficient to recognize small differences in relative symbiont amounts. A further important consideration is that the snapshot quality of the one-time methane and sulfide measurements may not reflect the average concentrations over longer times, as some studies have indicated that seepage fluxes can vary greatly not only over space but also over time (24, 34, 38).
The site-independent distribution of the Bathymodiolus sp. symbionts within each bacteriocyte, with the thiotrophs occupying a more apical position than the methanotrophs, has not been described previously. The closer proximity of the thiotrophic symbionts to the apical edge of the bacteriocytes suggests that these bacteria are more dependent on exchange with the mantle fluids that contain seawater from the environment. Intriguingly, the sulfide concentrations in the seawater at the collection site were much lower than the methane concentrations. Thus, low sulfide concentrations could limit the distribution of the thiotrophs to the regions closest to the circulating mantle fluids, where sulfide is more readily available. Correspondingly, the methanotrophs are able to inhabit a more basal region of the bacteriocytes, because diffusive loss of methane through the bacteriocytes is compensated for by higher methane concentrations. While FISH has not been used previously to obtain a general overview of symbiont distribution, ultrastructural analyses of other Bathymodiolus species have revealed a more regular distribution of methanotrophs and thiotrophs throughout the bacteriocyte (10, 14, 16, 43).
This study shows the importance of examining physicochemical parameters, such as sulfide and methane concentrations, at spatial and temporal scales relevant to the organisms living in cold seeps. Time series measurements of gradients of these parameters and correlation with symbiont distribution, relative amounts, and biomass are needed to obtain a better understanding of the influence of the environment on the relationships established in nature between hosts and their symbiotic bacteria.
Acknowledgments
We thank the pilots and crew of the N/O L'Atalante and the ROV Victor 6000 for efficient cooperation during the Biozaire 2 cruise (2001; IFREMER; chief scientist, Myriam Sibuet). We thank Karine Olu, Alexis Fifis, and Ann Andersen for the maps of macrofaunal communities, for on-board dissections, and for sample preparation, and R. Von Cosel is acknowledged for providing unpublished taxonomic results. The technical assistance of B. Rivière and D. Saint-Hilaire with microscopy and sectioning is gratefully acknowledged. We also thank Claudia Bergin, Anna Blazejak, and Silke Wetzel for great technical assistance with molecular methods.
We thank the oil and gas company Total for sponsoring the Biozaire project. S.D. is a student in the International Max Planck Research School of Marine Microbiology Ph.D. program, and his grant is cofunded by IFREMER and MPI. This work was supported by the Max Planck Society, IFREMER, University Pierre-et-Marie Curie, and CNRS (UMR 7621).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen, A., S. Hourdez, B. Marie, D. Jollivet, F. H. Lallier, and M. Sibuet. Escarpia southwardae sp. nov., a new species of vestimentiferan tubeworm (Annelida, Siboglinidae) from West African cold seeps. Can. J. Zool. 82:980-999.
- 4.Cavanaugh, C. M., S. L. Gardiner, M. L. Jones, H. W. Jannasch, and J. B. Waterbury. 1981. Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: possible chemoautotrophic symbionts. Science 213:340-342. [DOI] [PubMed] [Google Scholar]
- 5.Cavanaugh, C. M., P. R. Levering, J. S. Maki, R. Mitchell, and M. E. Lidstrom. 1987. Symbiosis of methylotrophic bacteria and deep-sea mussels. Nature 325:346-347. [Google Scholar]
- 6.Childress, J. J., C. R. Fisher, J. M. Brooks, M. C. Kennicutt II, R. Bidigare, and A. E. Anderson. 1986. A methanotrophic marine molluscan (Bivalvia, Mytilidae) symbiosis: mussels fueled by gas. Science 233:1306-1308. [DOI] [PubMed] [Google Scholar]
- 7.Colaço, A., F. Dehairs, D. Desbruyères, N. Le Bris, and P. M. Sarradin. 2002. δ13C signature of hydrothermal mussels is related with the end-member fluid concentration of H2S and CH4 at the Mid-Atlantic Ridge hydrothermal vent fields. Cah. Biol. Mar. 43:259-262. [Google Scholar]
- 8.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Distel, D. L., and C. M. Cavanaugh. 1994. Independent phylogenetic origins of methanotrophic and chemoautotrophic bacterial endosymbioses in marine bivalves. J. Bacteriol. 176:1932-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Distel, D. L., H. K. W. Lee, and C. M. Cavanaugh. 1995. Intracellular coexistence of methano- and thioautotrophic bacteria in a hydrothermal vent mussel. Proc. Natl. Acad. Sci. USA 92:9598-9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felbeck, H., J. J. Childress, and G. N. Somero. 1981. Calvin-Benson cycle and sulfide oxidation enzymes in animals from sulfide-rich habitats. Nature 293:291. [Google Scholar]
- 12.Felsenstein, J. 2002. PHYLIP (Phylogeny Inference Package), 3.6a3 ed. Department of Genome Sciences, University of Washington, Seattle.
- 13.Fiala-Médioni, A., C. Métivier, A. Herry, and M. Le Pennec. 1986. Ultrastructure of the gill filament of an hydrothermal vent mytilid Bathymodiolus sp. Mar. Biol. 92:65-72. [Google Scholar]
- 14.Fiala-Médioni, A., Z. P. McKiness, P. Dando, J. Boulegue, A. Mariotti, A. M. Alayse-Danet, J. J. Robinson, and C. M. Cavanaugh. 2002. Ultrastructural, biochemical and immunological characterisation of two populations of the mytilid mussel Bathymodiolus azoricus from the Mid Atlantic Ridge: evidence for a dual symbiosis. Mar. Biol. 141:1035-1043. [Google Scholar]
- 15.Fisher, C. R. 1990. Chemoautotrophic and methanotrophic symbioses in marine invertebrates. Rev. Aquat. Sci. 2:399-613. [Google Scholar]
- 16.Fisher, C. R., J. M. Brooks, J. S. Vodenichar, J. M. Zande, J. J. Childress, and R. A. Burke, Jr. 1993. The co-occurrence of methanotrophic and chemoautotrophic sulfur oxidizing bacterial symbionts in a deep-sea mussel. Mar. Ecol. 14:277-289. [Google Scholar]
- 17.Fisher, C. R., J. J. Childress, R. S. Oremland, and R. R. Bidigare. 1987. The importance of methane and thiosulphate in the metabolism of the bacterial symbionts of two deep-sea mussels. Mar. Biol. 96:59-71. [Google Scholar]
- 18.Fisher, C. R., J. J. Childress, A. J. Arp, J. M. Brooks, D. L. Distel, J. A. Favuzzi, H. Felbeck, R. Hessler, K. S. Johnson, M. C. Kennicutt, S. A. Macko, A. Newton, M. A. Powell, G. N. Somero, and T. Soto. 1988. Microhabitat variation in the hydrothermal vent mussel, Bathymodiolus thermophilus, at the Rose Garden vent on the Galapagos Rift. Deep-Sea Res. 35:1769-1791. [Google Scholar]
- 19.Fonselius, S. H. 1983. Determination of hydrogen sulfide, p. 73-80. In K. Grasshoff, M. Ehrhardt, and K. Kremling (ed.), Methods of seawater analysis, 2nd ed. Verlag Chemie, Kiel, Germany.
- 20.Fujiwara, Y., C. Kato, N. Masui, K. Fujikura, and S. Kojima. 2001. Dual symbiosis in the cold-seep thyasirid clam Maorithyas hadalis from the hadal zone in the Japan Trench, western Pacific. Mar. Ecol. Prog. Ser. 214:151-159. [Google Scholar]
- 21.Fujiwara, Y., K. Takai, K. Uematsu, S. Tsuchida, J. C. Hunt, and J. Hashimoto. 2000. Phylogenetic characterization of endosymbionts in three hydrothermal vent mussels: influence on host distribution. Mar. Ecol. Prog. Ser. 208:147-155. [Google Scholar]
- 22.Hall, T. 2001 1997. BioEdit. http://www.mbio.ncsu.edu/BioEdit/bioedit.html.
- 23.Jobb, G. 2003. TREEFINDER, version March 2003. [Online.] G. Jobb, Munich, Germany. www.treefinder.de.
- 24.Levin, L. A., W. Ziebis, G. F. Mendoza, V. A. Growney, M. D. Tryon, K. M. Brown, C. Mahn, J. M. Gieskes, and A. E. Rathburn. 2003. Spatial heterogeneity of macrofauna at northern California methane seeps: influence of sulfide concentration and fluid flow. Mar. Ecol. Prog. Ser. 265:123-139. [Google Scholar]
- 25.Ludwig, W. O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W.Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic. Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannash. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 27.Nishiguchi, M. K., E. G. Ruby, M. J. McFall-Ngai. 1998. Competitive dominance among strains of luminous bacteria provides an unusual form of evidence for parallel evolution in sepiolid squid-Vibrio symbioses. Appl. Environ. Microbiol. 64:3209-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ondréas, H., K. Olu, Y. Fouquet, J. L. Charlou. A. Gay, B. Dennielou, J. P. Donval, A. Fifis, T. Nadalig, P. Cochonat, E. Cauquil, and M. Sibuet. Integrated “in situ” study of a deep giant pockmark on the Gabon margin during the ZAIANGO and BIOZAIRE projects. Submitted for publication.
- 29.Page, H., A. Fiala-Médioni, C. Fisher, and J. Childress. 1991. Experimental evidence for filter-feeding by the hydrothermal vent mussel, Bathymodiolus thermophilus. Deep-Sea Res. Part I 38:1455-1461. [Google Scholar]
- 30.Pimenov, N. V., M. G. Kalyuzhnaya, V. N. Khmelenina, L. L. Mityushina, and Y. A. Trotsenko. 2002. Utilization of methane and carbon dioxide by symbiotrophic bacteria in gills of Mytilidae (Bathymodiolus) from the Rainbow and Logatchev hydrothermal fields on the Mid-Atlantic Ridge. Microbiology 71:587-594. [PubMed] [Google Scholar]
- 31.Pond, D. W., M. V. Bell, D. R. Dixon, A. E. Fallick, M. Segonzac, and J. R. Sargent. 1998. Stable-carbon-isotope composition of fatty acids in hydrothermal vent mussels containing methanotrophic and thiotrophic bacterial endosymbionts. Appl. Environ. Microbiol. 64:370-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu, X., L. Wu, H. Huang, P. McDonel, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Evaluation of PCR-generated chimaeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 67:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarradin, P. M., and J. C. Caprais. 1996. Analysis of dissolved gases by headspace sampling gas chromatography with column and detector switching. Preliminary results. Anal. Commun. 33:371-373. [Google Scholar]
- 34.Sibuet, M., and K. Olu. 1998. Biogeography, biodiversity and fluid dependence of deep sea cold seep communities at active and passive margins. Deep-Sea Res. Part II 45:517-567. [Google Scholar]
- 35.Sibuet, M., and K. Olu-Le Roy. 2002. Cold seep communities on continental margins: structure and quantitative distribution relative to geological and fluid venting patterns, p. 235-251. In G. Weffer, D. Billett, D. Hebbeln, B. B. Jörgensen, and T. J. Van Weering (ed.). Ocean margin system. Springer Verlag, Berlin Germany.
- 36.Sunamura, M., Y. Higashi, C. Miyako, J. Ishibashi, and A. Maruyama. 2004. Two Bacteria phylotypes are predominant in the Suiyo Seamount hydrothermal plume. Appl. Environ. Microbiol. 70:1190-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trask, J. L., and C. L. Van Dover. 1999. Site-specific and ontogenetic variations in nutrition of mussels (Bathymodiolus sp.) from the Lucky Strike hydrothermal vent field, Mid-Atlantic Ridge. Limnol. Oceanogr. 44:334-343. [Google Scholar]
- 38.Tryon, M. D., K. M. Brown, and M. E. Torres. 2002. Fluid and chemical flux in and out of sediment hosting methane hydrate deposits on Hydrate Ridge, OR, II: hydrological processes. Earth Planet. Sci. Lett. 201:541-557. [Google Scholar]
- 39.Tunnicliffe, V., S. K. Juniper, and M. Sibuet. 2003. Reducing environments of the deep-sea floor, p. 81-110. In P. A. Tyler (ed.), Ecosystems of the world: the deep-sea. Elsevier Press, Amsterdam, The Netherlands.
- 40.Van Dover, C. L. 2000. The ecology of deep-sea hydrothermal vents. Princeton University Press, Princeton, N.J.
- 41.Von Cosel, R. 2002. A new species of bathymodioline mussel (Mollusca, Bivalvia, Mytilidae) from Mauritania (West Africa), with comments on the genus Bathymodiolus Kenk & Wilson, 1985. Zoosystema 24:259-271. [Google Scholar]
- 42.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ-hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 43.Won, Y. J., S. J. Hallam, D. O'Mullan, I. L. Pan, K. R. Buck, and R. C. Vrijenhoek. 2003. Environmental acquisition of thiotrophic endosymbionts by deep-sea mussels of the genus Bathymodiolus. Appl. Environ. Microbiol. 69:6785-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou, J., M. A. Bruns, and M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]